Abstract
Single-stranded DNA binding proteins (SSBs) play an important role in DNA processing events such as replication, recombination and repair. Human single-stranded DNA binding protein 1 (hSSB1/NABP2/OBFC2B) contains a single oligosaccharide/oligonucleotide binding (OB) domain followed by a charged C-terminus and is structurally homologous to the SSB from the hyperthermophilic crenarchaeote Sulfolobus solfataricus. Recent work has revealed that hSSB1 is critical to homologous recombination and numerous other important biological processes such as the regulation of telomeres, the maintenance of DNA replication forks and oxidative damage repair. Since the ability of hSSB1 to directly interact with single-stranded DNA (ssDNA) is paramount for all of these processes, understanding the molecular details of ssDNA recognition is essential. In this study, we have used solution-state nuclear magnetic resonance in combination with biophysical and functional experiments to structurally analyse ssDNA binding by hSSB1. We reveal that ssDNA recognition in solution is modulated by base-stacking of four key aromatic residues within the OB domain. This DNA binding mode differs significantly from the recently determined crystal structure of the SOSS1 complex containing hSSB1 and ssDNA. Our findings elucidate the detailed molecular mechanism in solution of ssDNA binding by hSSB1, a major player in the maintenance of genomic stability.
INTRODUCTION
Single-stranded DNA binding proteins (SSBs) are essential for almost all DNA processing events, most notably DNA repair (1). DNA damage and subsequent repair can result in the creation of single-stranded DNA (ssDNA), which is recognized and protected by SSBs. This family of SSBs contain a structurally conserved DNA binding domain termed the oligonucleotide/oligosacharide binding (OB) fold, which is comprised of five β-strands that associate in an anti-parallel fashion to create a β-barrel. However, both the number and DNA binding properties of OB domains vary among different SSBs. For example, human replication protein A (RPA), the most widely studied SSB in humans, contains multiple OB domains (only some of which bind ssDNA) in three different subunits (2–5). In contrast, bacterial and crenarchaeal SSBs have a ‘simple’ domain organization containing only one DNA binding OB domain followed by a divergent spacer region and a charged unstructured C-terminal tail that is known to interact with other important DNA processing proteins (6–9).
In recent years, two new SSBs, hSSB1 and hSSB2 have been discovered by data mining of the human genome using the protein sequence of the well characterized SSB from the crenarchaeote Sulfolobus solfataricus (SsoSSB) as template. In contrast to RPA, both of these proteins have a simple domain organization (10). One of these proteins, hSSB1, was demonstrated to be critical for homologous recombination (HR), which repairs of one of the most lethal types of DNA damage, double-stranded DNA breaks (DSBs). In this process, the OB domain of hSSB1 recognizes and protects ssDNA, while the C-terminal part of the protein becomes phosphorylated at T117 by the ataxia telangiectasia mutated kinase as part of a positive feedback loop in the response to DSB damage (10). Subsequent work demonstrated that hSSB1 is essential for the recruitment of the MRN (Mre11, Rad50 and NBS1) complex to DSBs and the efficient resection of DSBs (11,12). In addition to the hSSB1–MRN complex, hSSB1 was also shown to be part of the SOSS1 complex (consisting of hSSB1, INTS3 and C9orf80), which plays an important role in HR-mediated DNA repair (13–15) and stimulates DSB resection by human Exonuclease 1 (hExo1), a member of the Rad2 family of nucleases (16).
Besides its role in DNA repair, hSSB1 also regulates both the stability and the transcriptional activity of p53 (17) and binds and protects p21 from ubiquitin mediated degradation (18). More recently, the importance of hSSB1 in stabilizing and repairing DNA replication forks (19) and in the regulation of telomeres (20) was demonstrated. In the latter, it was revealed that the interaction of hSSB1 with single-stranded G-rich oligonucleotides (found in the G-overhangs of telomeres) is essential for its function in telomere maintenance. Overall, hSSB1 has been shown to play an essential role in a wide range of important biological processes where the ability of the protein to physically contact DNA via its OB domain is paramount. Moreover, we have demonstrated that hSSB1 acts very early in the DNA damage response by directly binding to ssDNA independent of any other molecule or component of either the SOSS1 or the MRN repair complexes (11,12,15). For this reason, determination of the molecular mechanism of ssDNA recognition by hSSB1 in isolation is of significant interest.
In a recent study, Ren at al. (21) solved the structure of the SOSS1 complex bound to ssDNA by X-ray crystallography. Binding to ssDNA was found to be exclusively mediated by the OB domain of hSSB1. The structure indicated that ssDNA recognition by hSSB1 within the SOSS1 complex is achieved mainly by base-stacking of two aromatic residues (W55 and F78). More recently, we found significant differences in the DNA binding mode between the ssDNA-bound structure of the structurally homologous SsoSSB (root mean square deviation (RMSD) 0.82Å) and the above mentioned crystal structure (22). For example, while base-stacking of two aromatic residues within hSSB1 is sufficient for ssDNA recognition in the crystal structure, SsoSSB utilizes three aromatic residues (all of which are conserved in hSSB1). Closer inspection of the hSSB1 crystal structure reveals a possible crystal packing effect caused by interaction of ssDNA with the complex subunit INTS3 (SOSSA), which may have distorted hSSB1 binding to the ssDNA and could have caused the differences in ssDNA binding observed between the SsoSSB and the hSSB1 complex structures. Another possible explanation for these differences is that the interaction of hSSB1 with INTS3 causes allosteric effects that induce structural changes in hSSB1 that can modulate its DNA binding mode.
In this study, we analysed the structural properties of ssDNA binding by hSSB1 in isolation using solution-state nuclear magnetic resonance (NMR) in combination with biophysical and functional experiments as well as in silico molecular modelling methods. We reveal that ssDNA recognition by hSSB1 in solution is modulated by base-stacking of four key aromatic residues (W55, Y74, F78 and Y85) and that the structural conformation of the ssDNA is conserved between hSSB1 and SsoSSB.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis
Both GST-tagged full length hSSB1 (1-221, hSSB11-221) and hSSB1 OB domain construct (1–123; hSSB11-123) were prepared by directional cloning into pGEX-6P using the restriction enzymes BamHI and EcoRI. All hSSB11-123 mutants used were synthesized by GeneArt (Regensburg, Germany). The full-length 3× FLAG hSSB1 mammalian expression construct has been described previously (23). Site-directed mutagenesis was used for the preparation of all point-mutants described in Figure 5, as well as siRNA resistant 3× FLAG hSSB1.
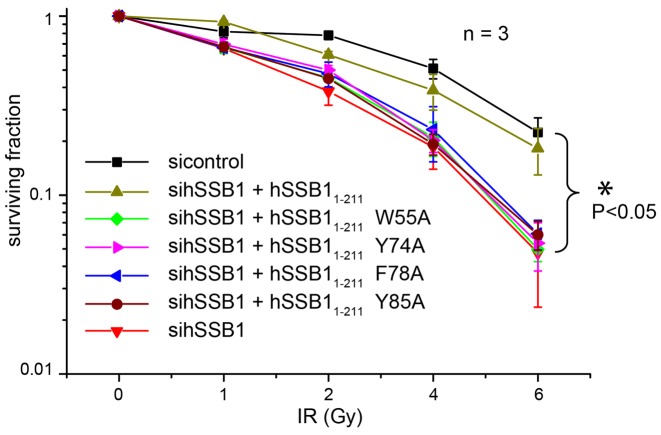
Figure 5.
Functional assay confirms the importance of the four key aromatics for ssDNA binding. Survival curves from a clonogenic assay of U2OS cells depleted for wild-type hSSB11-211. Non-depleting negative control (scramble), sihSSB11-211, siRNA-resistant flag-tagged hSSB11-211 (+hSSB11-211) and siRNA-resistant flag-tagged hSSB11-211 mutants (+hSSB11-211 W55A, + hSSB11-211 Y74A, + hSSB11-211 F78A and + hSSB11-211 Y85A), respectively, were transfected into cells. All points represent the mean ± standard error from three independent experiments. Note the significant difference between cell surviving fraction of control and all hSSB11-211 mutants (P < 0.05).
Recombinant protein expression
hSSB1 full-length and hSSB11-123 protein expression using the Escherichia coli Rosetta 2 (for BioLayer interferometry (BLI)) or E. coli BL21(DE3) (for NMR) strain was induced by addition of 0.2 mM IPTG at 25°C for 16 h. Cells were lysed by sonication in 10 mM MES, pH 6.0, 50 mM NaCl, 3 mM TCEP, 0.5 mM PMSF, 0.1% Triton X-100. Following centrifugation, the supernatant was subjected to GSH affinity chromatography followed by HRV-3C protease cleavage overnight at 4°C (leaving the 5-residue stretch GPLGS at the N-terminus of the OB domain). The solution was applied to a HiTrap HP Heparin (2 × 5 ml tandem, GE) column equilibrated with NMR buffer (10 mM MES, pH 6.0, 50 mM NaCl, 3 mM TCEP). A 500 ml linear gradient comprising 50–1000 mM NaCl was used to elute cationic proteins. Fractions corresponding to a distinct absorbance peak were analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, pooled, concentrated and loaded onto a Superdex-75 gel filtration column in NMR buffer or BLI buffer (10 mM Phosphate, pH 7.1, 50 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM DTT). 15N- and 15N13C-labelled hSSB1 protein was prepared using the procedure of (24) in a 5-l biofermenter and purified as described above. Protein concentrations were determined using the absorbance at 280 nm and the theoretical molar extinction coefficient for hSSB1.
Multi-angle laser light scattering (MALLS)
Size exclusion chromatography of hSSB11-123 coupled to multi-angle laser light scattering (MALLS) was carried out as described previously (25) in MALLS buffer (20 mM Tris, pH 7, 100 mM NaCl, 1 mM EDTA, 1 mM TCEP). Briefly, 250 μg hSSB1-123 was applied to a Superose 12 (10/300) analytical size exclusion column (GE healthcare) at 0.5 ml/min. MALLS was measured in tandem with size exclusion chromatography using a MiniDawn solid-state laser diode (Wyatt) measuring at three different angles (41.5°, 90° and 138.5°) at a wavelength of 690 nm. Monomeric BSA (66 kDa) was used as a reference to determine the molecular weight of the target protein.
BioLayer interferometry (BLI)
The BLI steady-state analysis (Figure 4) was carried out using a set of 8–10 appropriate protein concentrations. Proteins were bound to a 5′ biotinylated ssDNA oligonucleotide (5′-AAATTTTTT-3′) in triplicate, using the BLItz biosensor system (ForteBio). Streptavidin biosensors (ForteBio) were equilibrated in a buffer containing 20 mM HEPES and 100 mM NaCl (pH 7) for 24 h prior to use. For each individual binding curve, an initial baseline was performed (30 s), followed by the binding of the oligonucleotide to the biosensor until saturation (60 s). Two further baselines (30 s each) were carried out to transition to BLI buffer. Each construct (in BLI buffer) was allowed between 90 and 180 s to reach an equilibrium state, followed by a 60 s dissociation step. Average BLI equilibrium values were taken from the sensorgrams, plotted against the respective protein concentrations and fitted using the Hill equation (1:1 stoichiometry, steady-state model) in Origin 9.1.
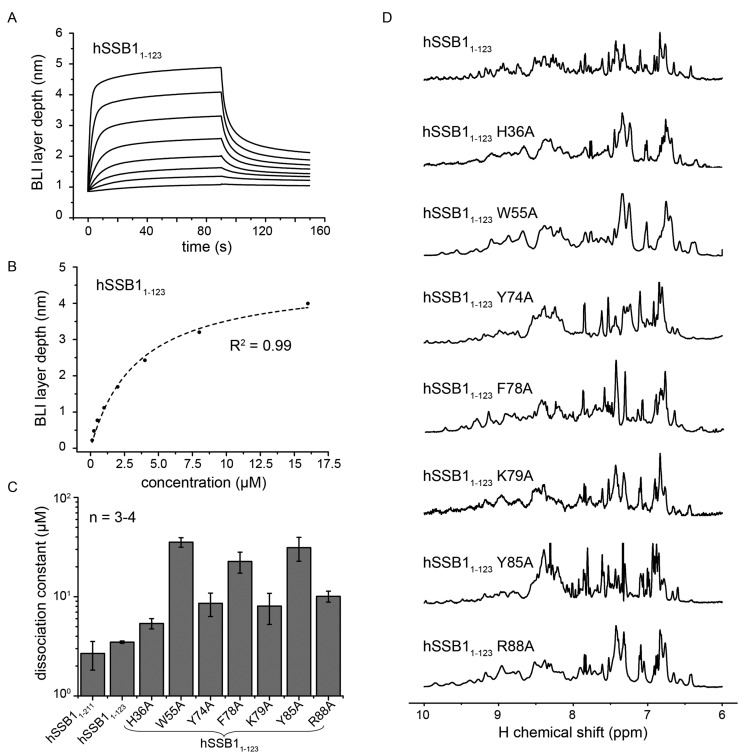
Figure 4.
Mutational analysis revealing critical ssDNA binding residues of hSSB1. (A) A representative BioLayer Interferometry (BLI) binding curve of wild-type hSSB11-123 (concentrations used were 125, 250, 500, 1000, 2000, 4000, 8000 and 16000 nM). (B) Graph showing steady state equilibrium values taken from A as a function of the protein concentration and fit to a 1:1 binding curve (Hill equation). (C) Summary of dissociation constants (± standard error) for wild-type hSSB11-211, wild-type hSSB1-123 and various hSSB1-123 alanine mutants, for binding to ssDNA, as measured by BLI. Three to four independent protein preparations of each mutant at different concentrations (ranging from 125–512 000 nM) have been utilized to calculate the dissociations constants. (D) 1H NMR spectra of all used mutants (recorded at 298 K) showing that each is correctly folded. The utilized protein concentrations were between 50 and 500 μM. Note the different resolutions of the recorded spectra due to different magnetic field strengths used (400, 600 or 800 MHz).
NMR spectroscopy and data processing
NMR experiments were carried out using 0.2–0.8 mM hSSB11-211 or hSSB1-123 in NMR buffer with 10% D2O and 1:1 complexes of hSSB1-123 with oligo(dT)6 ssDNA (purchased from Sigma Aldrich) at the same concentrations. Mutant hSSB1-123 proteins (Figure 4) were prepared at concentration between 50 and 500 μM in BLI buffer. Proton chemical shifts were referenced to 4,4-dimethyl-4-silapentanesulfonic acid at 0 ppm. 13C and 15N chemical shifts were referenced indirectly to the same signal. All NMR experiments were recorded at 298 K on Bruker 400, 600 or 800 MHz spectrometers (Bruker Avance III) equipped with 5-mm TCI cryoprobes. The spectra recorded included 1D, 2D 15N HSQC, 2D 13C HSQC (aliphatic and aromatic), 3D CBCA(CO)NH, 3D HNCACB, 3D HNCO, 3D HN(CA)CO, 3D CC(CO)NH TOCSY, 3D HCC(CO)NH TOCSY, 3D (H)CCH-TOCSY and 3D 15N NOESY. All data were processed using Topspin (Bruker Biospin) and assignments were made using Sparky (T. D. Goddard and D. G. Kneller, University of California at San Francisco). Calculation of weighted chemical shift changes (Figure 2C) was carried out as described in (26).
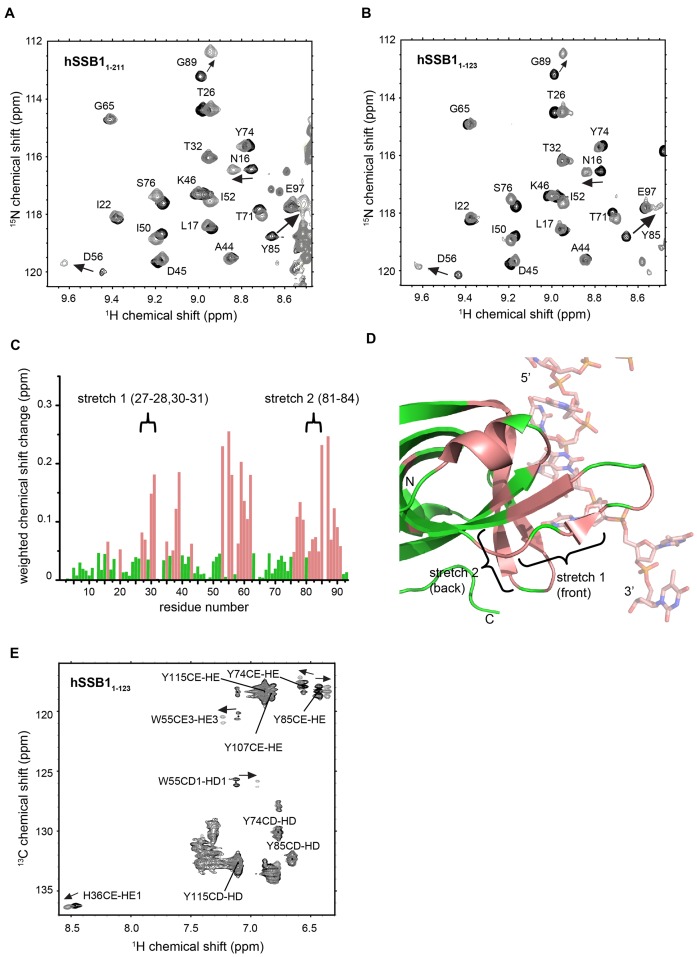
Figure 2.
NMR analysis of hSSB1 OB domain in complex with ssDNA (oligo(dT)6). Sections of 15N-HSQC spectrum of full-length hSSB1 (hSSB11-211) (A) and hSSB11-123 (B) in the absence (black) and presence (1:1 mixture, light grey) of oligo(dT)6, respectively. Assignments and directions of movement are indicated. (C) Weighted backbone chemical shift changes of HN and N (26) atoms for hSSB11-123 upon binding to ssDNA. Residues exhibiting changes larger than the average (solution binding residues) are coloured in salmon. Two stretches of residues (stretch 1 and 2) that exhibit larger than average chemical shift changes but are not involved in ssDNA binding in the published X-ray crystal structure of the SOSS1 complex (PDB ID: 4OWX) (21) are indicated. (D) Cartoon representation of the published crystal structure with solution binding residues coloured as in C and residue stretches 1 and 2 indicated. (E) Portion of 13C HSQC spectrum of hSSB1-123 in the absence (black) and presence (1:1 mixture, light grey) of oligo(dT)6, respectively.
HADDOCK modelling
The protein structure of hSSB1 (residue 5–111 containing the OB domain) was taken from the crystal structure of the SOSS1 complex (PDB ID: 4OWX) (21) and used as input for HADDOCK (27,28), together with a model of ssDNA (oligo(dT)6) constructed in silico using the structure of the ssDNA within the complex structure of SsoSSB as a template (22). hSSB1 protein residues 15–17, 26–40, 52–63 and 73–91 were defined as semi-flexible based on our NMR data (Figure 2) and all six thymines of the ssDNA were defined as semi-flexible and flexible. Eighty-three ambiguous interaction restraints for both the protein and the ssDNA were chosen based on our NMR data (Figure 2) and our mutant data (Figure 4) and fixed at 2 Å. Additional restraints to maintain base planarity between the four aromatic residues (W55, Y74, F78 and Y85) and ssDNA bases THY2, THY3 and THY5 were used in the calculations. The 10 conformers with the lowest value of total energy of the lowest-energy cluster were analysed and visualized using PYMOL (Schrödinger, NY). Protein resonance of all backbone residues have been deposited into the BMRB database (accession number 26 752), the structural coordinates of the hSSB1–ssDNA model were deposited into the Figshare data repository (DOI: 10.6084/m9.figshare.3422788) as the RCSB PDB database does not currently accept molecular models (29).
Cell culture, transfections and clonogenic survival assays
HeLa cells were obtained from the American Type Culture Collection and maintained in Roswell Park Memorial Institute medium supplemented with 10% foetal calf serum, and grown in a humidified atmosphere at 37°C and with 5% CO2. For the clonogenic survival assays, HeLa cells were transfected with 50 nM of Stealth siRNA (Thermo Fisher Scientific) targeting the hSSB1 transcript (siRNA sequence 5′-GCCCUUCCAGCAACCCUGUUAGUAA-3′) or with a negative control sequence (Stealth siRNA negative control, med GC), twice over 48 h using Lipofectamine RNAiMAX (Thermo Fisher Scientific). The following day, cells were transfected with plasmids encoding hSSB11-211 or W55A, F74A, F78A and Y85A siRNA-resistant 3× FLAG hSSB11-211 mutants or with an empty vector, using Lipofectamine 2000 (Thermo Fisher Scientific). After 6 h, cells were then seeded into wells of a 6-well plate at a density of 400 cells per well. Twenty-four hours post-seeding, cells were exposed to 1, 2, 4 or 6 Gy of ionizing radiation (IR) using a Gammacell 40 Exactor caesium-source irradiator, or left untreated. Following 10 days of culture, cells were fixed and stained with 4% methylene blue in methanol and colonies manually counted. Assays were performed three times and results displayed as the average relative colony count ± standard error. Statistical analysis was performed using Student's t-test with a P-value of <0.05 considered significant.
Immunoblotting
Whole cell lysates were prepared by suspension of HeLa cells in radioimmunoprecipitation buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% sodium dodecyl sulphate, 0.5% sodium deoxycholate, 1% Triton X100) containing protease inhibitors (cOmplete, EDTA free; Roche), followed by sonication (Vibra-Cell, 3 mm probe; Sonics and Materials). A total of 20 μg of lysate was then separated by electrophoresis (4–20% Bolt Bis–Tris Plus gel; Thermo Fisher Scientific) and transferred to nitrocellulose. Blots were blocked with fish gelatin, before probing with primary antibodies. These were subsequently detected with IRDye 680RD or 800CW-conjugated donkey secondary antibodies (Li-Cor) and visualized using the Odyssey imaging system (Li-Cor). Sheep antiserum against hSSB1 has been described previously (10). Antibodies against FLAG and actin were purchased from Sigma-Aldrich and BD Bioscience, respectively.
RESULTS
Four key aromatic residues in hSSB1 mediate ssDNA binding in solution
We have recently reported backbone chemical shift assignments of a hSSB1 construct containing the OB domain (hSSB11-123) in solution and have mapped the ssDNA binding interface (30). To first confirm that indeed the OB domain but not the carboxyl-tail (for OB domain sequence information see Figure 1) mediates ssDNA binding we recorded HSQC spectra of full-length hSSB1 (hSSB11-211) in the presence (grey) and absence (black) of oligodeoxithymidine ssDNA (oligo(dT)6) (Figure 2A) and compared these with the previously recorded spectra of hSSB11-123 (30) (Figure 2B). No significant difference between the two spectra was observed indicating that the flexible carboxyl-tail of hSSB1 is not involved in ssDNA binding. Calculation of weighted chemical shift changes (26) for hSSB1-123 upon binding to ssDNA revealed residues that undergo substantial changes in backbone structure and are thus highly likely to be involved in ssDNA binding (Figure 2C). We have mapped these residues onto the existing X-ray crystal structure of hSSB1 (PDB ID 4OWX) (21) (Figure 2D, coloured in salmon). Surprisingly, we were able to identify two stretches of hSSB1 residues (as indicated in Figure 2C and D) that exhibit large backbone chemical shift changes but are not involved in ssDNA binding in the crystal lattice of the published structure.
Figure 1.
hSSB1 OB domain sequence information. Sequence alignment of the OB domains of hSSB1, hSSB2, SsoSSB and RPA70B. Boxed residues and residues in bold indicate aromatics that intercalate with ssDNA and residues involved in hydrogen bonding with ssDNA, respectively, whereas grey areas indicate high sequence conservation. Note that the hSSB1 construct used in this study (hSSB11-123) comprises of the OB domain (sequence shown in this Figure, residue 1–93) as well as parts of the flexible carboxyl-tail region (residues 94–123).
The interaction between the closely related (sequence similarity of ∼55% and RMSD of 0.82 Å over all atoms of the OB domain) SsoSSB (Figure 1) and ssDNA is strongly mediated by base-stacking of three aromatic residues (W56, W75 and F79) that are all conserved in hSSB1. In order to test for the presence of NOEs between the homologous aromatics in hSSB11-123 (W55, Y74 and F78) and ssDNA we initially recorded 3D filtered aromatic NOESY experiments at different temperatures in analogy to our SsoSSB study (22). However, attempts to increase the temperature above 298 K resulted in substantial protein degradation (as expected for a human protein), whereas at 298 K and below, some of the signals were experiencing intermediate exchange, preventing the observation of any intermolecular NOEs. Despite the absence of any intermolecular NOEs further NMR experiments (carried out at 298 K) enabled us to partly assign an aromatic 13C-HSQC in the presence and absence of ssDNA (Figure 2E). As expected, we observed significant chemical shift changes of the side chain protons of W55 and Y74, whereas F78 could not be unambiguously assigned. In good agreement with our 15N HSQC data (Figure 2B and C), side chain protons of a fourth aromatic residue (Y85) also exhibited large chemical shift changes, indicating that this residue plays a major role in ssDNA recognition. Overall, the magnitude of the observed chemical shifts changes of all four aromatics in both 15N HSQC and 13C HSQC experiments is comparable with changes seen for the three conserved aromatic residues in the SSoSSB protein (W56, W75 and F79) upon ssDNA binding (22), indicating a major involvement of these residues in the recognition of ssDNA in solution.
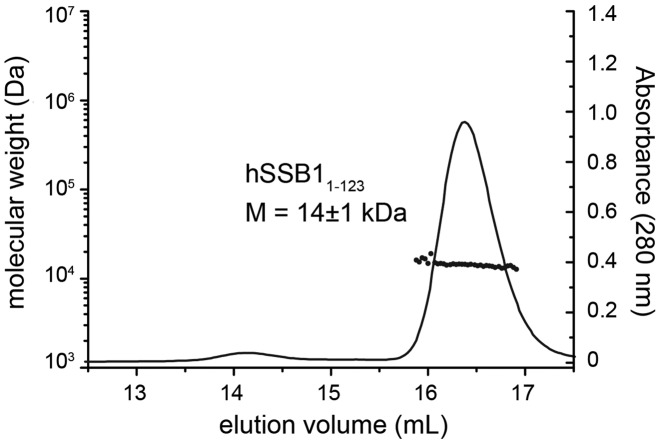
Close inspection of the protein sequence of hSSB1 reveals that the protein has two cysteine residues within the OB domain (C41 and C81; Figure 1) and one just outside (C99) that may facilitate the formation of higher order oligomers dependent on the presence or absence of any reducing agents. To confirm that hSSB11-123 is monomeric under the conditions used in our NMR and BLI experiments, we employed tandem size exclusion chromatography and MALLS (Figure 3). The observed MALLS peak of hSSB11-123 corresponded to the theoretical size of a single molecule in solution (Figure 3).
Figure 3.
MALLS data of hSSB11-123 protein. Size-exclusion chromatography traces of hSSB1-123 in MALLS buffer. The corresponding molecular weight is indicated. Note that hSSB1-123 exist solely as monomer.
Mutational analysis confirms ssDNA binding interface
To confirm the involvement of these four aromatic residues and to further define the ssDNA binding interface, we made a series of hSSB11-123 alanine mutants based on our HSQC data (Figure 2) and the sequence alignment between the OB domains of hSSB1 and SsoSSB (Figure 1). Dissociation constants of the binding between full-length hSSB1-211, hSSB11-123 as well as mutant hSSB11-123 proteins and ssDNA (oligo(dT)6) were calculated using a steady-state analysis (1:1 stoichiometry) from BLI data (Figure 4A–C and Supplementary Figure S1). We confirmed that all constructs were correctly folded by 1H NMR spectroscopy (Figure 4D). In good agreement with our NMR data (Figure 2A and B), no significant difference of the ssDNA binding affinity between full-length hSSB11-211 and hSSB11-123 could be observed, providing further evidence that the flexible carboxyl-tail of hSSB1 is not involved in ssDNA recognition. All hSSB11-123 mutants revealed decreased binding affinities compared to wild-type hSSB11-123 underscoring the importance of these residues for DNA binding (Figure 4C and Supplementary Table S1). Notably, replacing W55, F78 or Y85 with alanines resulted in very large increase in the dissociation constants (∼6.5-10 times that of hSSB11-123) further confirming that these aromatic residues play a major role in the recognition of ssDNA.
Functional data confirm the importance of aromatic residues for ssDNA recognition
To further corroborate our biophysical findings in a functional environment, we carried out a clonogenic survival assay using HeLa cells depleted of endogenous full-length hSSB1 and transiently expressing siRNA resistant full-length 3× FLAG tagged wild-type hSSB11-211 or W55A, Y74A, F78A and Y85A mutants (Figure 5 and Supplementary Figure S2). As can be seen from Figure 5, expression of wild-type hSSB11-211 was able to rescue depletion of endogenous hSSB1 following induction of DNA damage by IR. IR is routinely used to introduce DSBs in living cells (31). In contrast to wild-type hSSB11-211, mutation of any of the four aromatic residues led to significantly decreased cell survival compared to the control. Taken together, these data provide further strong evidence that ssDNA binding by hSSB1 in solution is mediated by four key aromatic residues.
A structural model of a hSSB1–ssDNA complex reveals molecular details of ssDNA recognition in solution
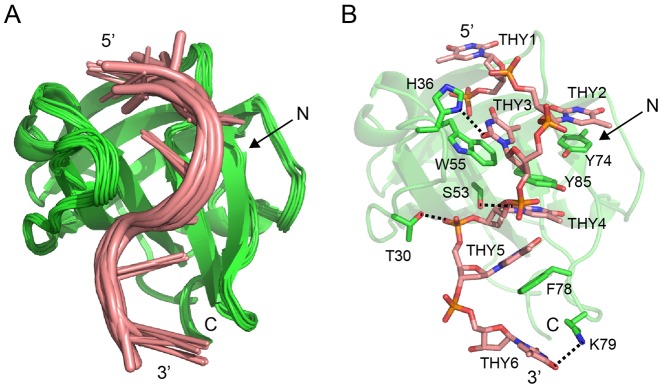
Notably, a full structural calculation of hSSB1 alone or of an ssDNA-bound complex was not possible due to the low quality of our 3D NOESY experiments at 298 K and the inability to change the temperature significantly (see also above). However, our 13C and 15N HSQC data in combination with data from our mutational and functional assays as well as the existing crystal structure of hSSB1 enabled us to calculate a structural model of an hSSB1–ssDNA complex (Figure 6). Figure 6A depicts the 10 best structures calculated from a total of 1000 HADDOCK structures displaying an RMSD of 0.24 Å. Overall, the interaction of hSSB1 with ssDNA occurs via the OB domain and is predominantly mediated by base-stacking of the four aromatic residues W55, Y74, F78 and Y85 (boxed in Figure 1), consistent with the large chemical shift changes of these residues observed in the 15N and 13C HSQCs (Figure 2). Hydrogen bonds observed in at least 50% of the family of 10 energy-best structures (Table 1 and Figure 6B) were identified between side chain protons of T30, H36, S53 and K79 (bold in Figure 1) and base as well as backbone protons of THY3,THY4,THY5 and THY6. Notably, THY1 is disordered in the structural model and does not form any contacts with the protein.
Figure 6.
The hSSB1–ssDNA complex solution model. (A) Overlay of family of 10 hSSB1–ssDNA HADDOCK complex structures with the lowest total energy in cartoon representation. (B) Cartoon (hSSB1) and stick (ssDNA) representation of the energy-lowest complex structure. The four aromatic residues (W55, Y74, F78 and Y85) that intercalate with the ssDNA, all residues that form hydrogen bonds (black, dashed line) as well as all DNA bases are indicated.
Table 1. Hydrogen bonds.
| H-atom pair | D0istance (Å) | Angle (º)a | Number (of 10) |
|---|---|---|---|
| T30.HG1–THY5.O1P | 2.3 ± 0.0 | 26 ± 4 | 9 |
| H36.HD1–THY3.O2 | 2.1 ± 0.0 | 12 ± 1 | 7 |
| S53.HG–THY4.O2 | 2.2 ± 0.0 | 28 ± 1 | 5 |
| K79.HZ1–THY6.O4 | 2.2 ± 0.0 | 23 ± 2 | 9 |
aAngle between the line from the atom connected to the donor and the donor and the line from the atom connected to the donor and the acceptor.
DISCUSSION
Mechanism of DNA and protein binding of hSSB1 in contrast to human RPA
Our data-driven hSSB1–ssDNA structural model provides insight into the molecular details of ssDNA recognition by the single OB domain of hSSB1. In the context of DNA binding, the main difference between RPA, the other important SSB in humans and hSSB1 is the presence of additional OB domains within RPA.
In contrast to hSSB1, human RPA is trimeric (RPA70, RPA32 and RPA14) and possesses four ssDNA binding OB domains within two of the three subunits (RPA70 and RPA32). To date, four different DNA binding configurations have been described. The first configuration is facilitated by the second and third RPA70 OB domains (denoted DBD A and B) binding in a linear arrangement to ssDNA with low-affinity, occluding a region of ∼8 nucleotides (nt) (32). The second configuration (12–23 nt mode) represents DBD A, B and C (the first RPA70 OB domain) binding to ssDNA (33,34). The additional contribution of the DBD D OB-fold (the single RPA32 OB domain) then allows RPA to bind ssDNA with high-affinity, where either ∼23–27 nt (33) or ∼30 nt are occluded (35,36). Major structural rearrangements are linked to transitions between these discrete states, and associated with that is a difference in the ability to contact other proteins (in particular through DBD A and B domains) (35).
hSSB1, on the other hand, recognizes ssDNA solely through its single OB domain. While our recent data demonstrated that hSSB1 is able to recognize ssDNA at DNA damage sites independently of any other molecules, the protein is also part of two well-characterized multi-protein complexes that are essential for DNA DSB repair (SOSS1 and MRN complex) (11–13,21). Whereas INTS3 in the SOSS1 complex contacts the OB domain opposite to the DNA binding site, protein binding in the MRN complex is via the flexible carboxyl tail of hSSB1. It is possible that the interaction with either INTS3 or MRN modulates the DNA binding mode of hSSB1. Further, although hSSB1 exists as a monomer in both complexes, the existence of hSSB1 homo-dimers and tetramers have recently been described as a consequence of hSSB1 oxidation in the response to oxidative DNA damage (23,37). In this context, it was found that oxidized hSSB1 binds with increased affinity to DNA containing 8-oxoguanines that form by oxidation with reactive oxygen species. This is in contrast to RPA, which exhibits varying DNA binding affinities depending on the number and structural arrangement of its individual OB domains. Further biophysical and structural studies are required to explore the possibility that oxidation-induced oligomerization of hSSB1 results in a change in ssDNA binding modality.
Binding affinity and specificity of the hSSB1–ssDNA interaction
In this study, we have used BLI to measure dissociation constants for the interaction of hSSB1 with ssDNA of ∼3.5 μM, which is slightly weaker than obtained by ITC (1.5 μM) in an earlier study (10). However, in the latter study substantially longer oligomers (30mers) were utilized under slightly different experimental conditions. In contrast, binding of the closely related SSB from SsoSSB to ssDNA is significantly tighter (dissociation constant of ∼180 nM) (22). Although it has been shown that an increasing number of hydrogen bonds between proteins is often correlated with stronger binding affinity (38), this concept remains controversial as the hydrogen bonding process continuously competes with bulk water (39). However, given the high structural similarity between hSSB1 and SsoSSB (RMSD over OB fold is 0.82Å), the large difference in the dissociation constant is likely due to the different number of hydrogen bonds present (4 in hSSB1 versus 7 in SsoSSB).
Although SSBs from the OB domain family are generally classified as non-specific DNA binders, it was previously found that the identity of the DNA bases also plays an important role for hSSB1 binding capacity (10,20); the larger adenine base is not as thermodynamically favourable for binding which is thought to be due to steric hindrance and/or inefficient base-stacking with the aromatic side chains (40–42). This is consistent with other SSB proteins from both prokaryotes and eukaryotes, which generally bind with highest affinity to poly-thymine and poly-cytidine and with lower affinity to poly-adenines. Our hSSB1–ssDNA structural model displays base-stacking of four key aromatic residues as opposed to two in the E. coli SSB–ssDNA (43) complex and three in the SsoSSB–ssDNA complex (22), indicating that the dependency of the binding affinity on the identity of the DNA base is strongest in the human protein.
The solution structure of hSSB1–ssDNA differs significantly from the crystal structure
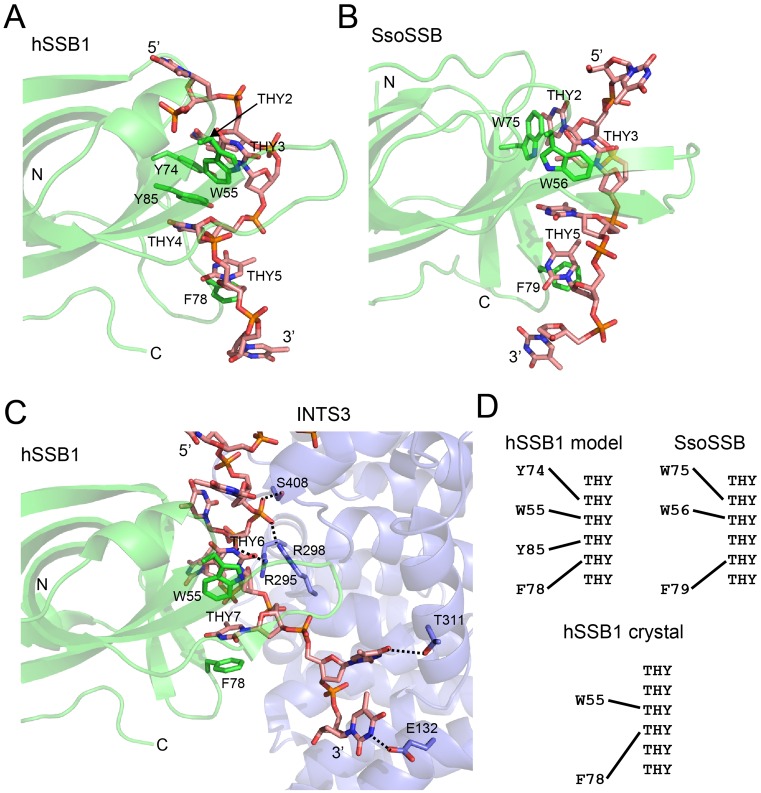
Comparison of ssDNA binding by hSSB1 in solution versus the crystal (21) reveals three important differences (Figure 7):
Figure 7.
DNA binding mode of hSSB1 in solution versus crystal and comparison with SsoSSB. Cartoon and stick representation of complex structures of hSSB1 (solution model) (A), Sulfolobus solfataricus SSB (SsoSSB, PDB ID: 2MNA) (B) and hSSB1 (X-ray crystal structure, PDB ID: 4OWX) (C), respectively, bound to ssDNA. The structure in A has been rotated by 90° counter clockwise about the vertical axis when looking from above relative to Figure 6; the structures in B and C are shown in the same orientation as in A. All protein aromatic residues that intercalate with the ssDNA are indicated. Note that panel C additionally depicts the symmetry related molecule INTS3 (SOSSA) as part of the crystal structure of the entire SOSS1 complex (PDB ID: 4OWX) as well as hydrogen bonds and electrostatic interactions between the backbone and bases of the ssDNA and INTS3. (D) Schematic showing DNA binding mode of hSSB1–ssDNA solution complex (top left), SsoSSB–ssDNA (top right) and hSSB1–ssDNA crystal structure (bottom), respectively.
Firstly, as mentioned previously, the number aromatic residues that intercalate with the ssDNA is different; in addition to W55 and F78 our solution model also revealed intercalation of residues Y74 and Y85 with ssDNA (Figure 7A and C). Interestingly, in contrast to our BLI experiments, electrophoretic mobility shift assays (EMSAs) using Y74A and Y85A mutant proteins and ssDNA did not reveal any change in binding affinity (21). However, these discrepancies are likely due to differences between the two techniques. For example, whereas BLI has been shown to be able to detect very small differences in interactions with affinity constants in the nM or subnanomolar range (44,45), dissociation of protein–DNA complexes as well as diffusion of both free ssDNA and ssDNA–protein complexes within the gel matrix can make it challenging to accurately measure complex formation with small association constants or small differences thereof in EMSA experiments (46,47). Notably, whereas Y74 was found not to contact the ssDNA at all in the crystal, the terminal hydroxyl group of Y85 was shown to form a hydrogen bond with the ssDNA (21). However, our NMR data (Figure 2) revealed major structural changes in both backbone and sidechain of Y85, consistent with ssDNA base-stacking of this residue in solution.
The second important difference between our hSSB1–ssDNA solution model and the crystal structure is the spacing between all four aromatics with respect to the ssDNA (Figure 7D). Whereas both W55 and Y78 base-stack with two adjacent ssDNA bases in the crystal, a one-base gap exists (the base which stacks with Y85) in the corresponding ssDNA sequence in solution.
Finally, the structural conformation of the ssDNA in our hSSB1 complex structure resembles the one found in the closely related SsoSSB structure (Figure 7B), but is substantially different to the hSSB1 crystal structure.
Importantly, in the crystal structure a symmetry-related INTS3 (SOSSAN) molecule interacts with the DNA supported by a large network of hydrogen bonds and electrostatic interactions between five residues (E132, R295, R298, T311, S408) and backbone as well as side-chain base atoms of the ssDNA (Figure 7C) (21). These interactions may have distorted hSSB1 binding to the DNA and may have caused the unusual conformation of the ssDNA in the crystal lattice. We also cannot rule out the possibility that the direct interaction of INTS3 with hSSB1 (at the opposite site to the ssDNA) has caused the described differences in the DNA binding mode of the crystal structure.
In conclusion, the defining feature of the hSSB1–ssDNA complex solution structure is the base-stacking of four aromatic residues (W55, Y74, F78, F85), three of which (W55, Y74 and F78) are conserved in the closely related SsoSSB, with four ssDNA bases. Our structural analysis has also revealed that significant differences exist between ssDNA recognition by hSSB1 in solution compared to the crystal environment. The data presented here is important in understanding the molecular mechanism of the interaction between hSSB1 and ssDNA, especially since blocking ssDNA binding by hSSB1 in tumour cells may be of significant interest for the development of novel cancer therapeutics.
Supplementary Material
Acknowledgments
We would like to thank Dr Ann Kwan from the University of Sydney and Dr Allan Torres from the University of Western Sydney for expert advice and maintenance of NMR spectrometers.
Footnotes
Present address: Dr Roland Gamsjaeger/Dr Liza Cubeddu, School of Science and Health, Western Sydney University, Penrith, NSW 2751, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NHMRC project grant [1066550]; UWS Women's Research Fellowship (to L.C.); Cancer Council Queensland Scholarship (to N.W.A.); NHMRC Early Career Fellowship [1091589 to M.N.A.]. Funding for open access charge: School HDR funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.Richard D.J., Bolderson E., Khanna K.K. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit. Rev. Biochem. Mol. Biol. 2009;44:98–116. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- 2.Chen R., Wold M.S. Replication protein A: single-stranded DNA's first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays. 2014;36:1156–1161. doi: 10.1002/bies.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanning E., Klimovich V., Nager A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iftode C., Daniely Y., Borowiec J.A. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 5.Oakley G.G., Patrick S.M. Replication protein A: directing traffic at the intersection of replication and repair. Front. Biosci. (Landmark Ed) 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadman C.J., McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlov A.G., Cox M.M., Lohman T.M. Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 2010;285:17246–17252. doi: 10.1074/jbc.M110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryzhikov M., Korolev S. Structural studies of SSB interaction with RecO. Methods Mol. Biol. 2012;922:123–131. doi: 10.1007/978-1-62703-032-8_7. [DOI] [PubMed] [Google Scholar]
- 9.Wadsworth R.I., White M.F. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 2001;29:914–920. doi: 10.1093/nar/29.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richard D.J., Bolderson E., Cubeddu L., Wadsworth R.I., Savage K., Sharma G.G., Nicolette M.L., Tsvetanov S., McIlwraith M.J., Pandita R.K., et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 11.Richard D.J., Cubeddu L., Urquhart A.J., Bain A., Bolderson E., Menon D., White M.F., Khanna K.K. hSSB1 interacts directly with the MRN complex stimulating its recruitment to DNA double-strand breaks and its endo-nuclease activity. Nucleic Acids Res. 2011;39:3643–3651. doi: 10.1093/nar/gkq1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richard D.J., Savage K., Bolderson E., Cubeddu L., So S., Ghita M., Chen D.J., White M.F., Richard K., Prise K.M., et al. hSSB1 rapidly binds at the sites of DNA double-strand breaks and is required for the efficient recruitment of the MRN complex. Nucleic Acids Res. 2011;39:1692–1702. doi: 10.1093/nar/gkq1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J., Gong Z., Ghosal G., Chen J. SOSS complexes participate in the maintenance of genomic stability. Mol. Cell. 2009;35:384–393. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Bolderson E., Kumar R., Muniandy P.A., Xue Y., Richard D.J., Seidman M., Pandita T.K., Khanna K.K., Wang W. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J. Biol. Chem. 2009;284:23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skaar J.R., Richard D.J., Saraf A., Toschi A., Bolderson E., Florens L., Washburn M.P., Khanna K.K., Pagano M. INTS3 controls the hSSB1-mediated DNA damage response. J. Cell Biol. 2009;187:25–32. doi: 10.1083/jcb.200907026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran P.T., Erdeniz N., Symington L.S., Liskay R.M. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Xu S., Wu Y., Chen Q., Cao J., Hu K., Tang J., Sang Y., Lai F., Wang L., Zhang R., et al. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res. 2013;23:423–435. doi: 10.1038/cr.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S., Feng Z., Zhang M., Wu Y., Sang Y., Xu H., Lv X., Hu K., Cao J., Zhang R., et al. hSSB1 binds and protects p21 from ubiquitin-mediated degradation and positively correlates with p21 in human hepatocellular carcinomas. Oncogene. 2011;30:2219–2229. doi: 10.1038/onc.2010.596. [DOI] [PubMed] [Google Scholar]
- 19.Bolderson E., Petermann E., Croft L., Suraweera A., Pandita R.K., Pandita T.K., Helleday T., Khanna K.K., Richard D.J. Human single-stranded DNA binding protein 1 (hSSB1/NABP2) is required for the stability and repair of stalled replication forks. Nucleic Acids Res. 2014;42:6326–6336. doi: 10.1093/nar/gku276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandita R.K., Chow T.T., Udayakumar D., Bain A.L., Cubeddu L., Hunt C.R., Shi W., Horikoshi N., Zhao Y., Wright W.E., et al. Single-strand DNA-binding protein SSB1 facilitates TERT recruitment to telomeres and maintains telomere G-overhangs. Cancer Res. 2015;75:858–869. doi: 10.1158/0008-5472.CAN-14-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren W., Chen H., Sun Q., Tang X., Lim S.C., Huang J., Song H. Structural basis of SOSS1 complex assembly and recognition of ssDNA. Cell Rep. 2014;6:982–991. doi: 10.1016/j.celrep.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Gamsjaeger R., Kariawasam R., Gimenez A.X., Touma C., McIlwain E., Bernardo R.E., Shepherd N.E., Ataide S.F., Dong Q., Richard D.J., et al. The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus. Biochem. J. 2015;465:337–346. doi: 10.1042/BJ20141140. [DOI] [PubMed] [Google Scholar]
- 23.Paquet N., Adams M.N., Leong V., Ashton N.W., Touma C., Gamsjaeger R., Cubeddu L., Beard S., Burgess J.T., Bolderson E., et al. hSSB1 (NABP2/ OBFC2B) is required for the repair of 8-oxo-guanine by the hOGG1-mediated base excision repair pathway. Nucleic Acids Res. 2015;43:8817–8829. doi: 10.1093/nar/gkv790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai M., Huang Y., Sakaguchi K., Clore G.M., Gronenborn A.M., Craigie R. An efficient and cost-effective isotope labeling protocol for proteins expressed in Escherichia coli. J. Biomol. NMR. 1998;11:97–102. doi: 10.1023/a:1008222131470. [DOI] [PubMed] [Google Scholar]
- 25.Cubeddu L., Joseph S., Richard D.J., Matthews J.M. Contribution of DEAF1 structural domains to the interaction with the breast cancer oncogene LMO4. PLoS One. 2012;7:e39218. doi: 10.1371/journal.pone.0039218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayed A., Mulder F.A., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 27.de Vries S.J., van Dijk A.D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A.M. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 28.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 29.Berman H.M., Burley S.K., Chiu W., Sali A., Adzhubei A., Bourne P.E., Bryant S.H., Dunbrack R.L., Jr, Fidelis K., Frank J., et al. Outcome of a workshop on archiving structural models of biological macromolecules. Structure. 2006;14:1211–1217. doi: 10.1016/j.str.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Kariawasam R., Touma C., Cubeddu L., Gamsjaeger R. Backbone H, C and N resonance assignments of the OB domain of the single stranded DNA-binding protein hSSB1 (NABP2/OBFC2B) and chemical shift mapping of the DNA-binding interface. Biomol. NMR Assign. 2016 doi: 10.1007/s12104-016-9687-6. doi:10.1007/s12104-016-9687-6. [DOI] [PubMed] [Google Scholar]
- 31.Vignard J., Mirey G., Salles B. Ionizing-radiation induced DNA double-strand breaks: a direct and indirect lighting up. Radiother. Oncol. 2013;108:362–369. doi: 10.1016/j.radonc.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Bochkarev A., Pfuetzner R.A., Edwards A.M., Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 33.Bastin-Shanower S.A., Brill S.J. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai L., Roginskaya M., Qu Y., Yang Z., Xu Y., Zou Y. Structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry. 2007;46:8226–8233. doi: 10.1021/bi7004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan J., Pavletich N.P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salas T.R., Petruseva I., Lavrik O., Saintome C. Evidence for direct contact between the RPA3 subunit of the human replication protein A and single-stranded DNA. Nucleic Acids Res. 2009;37:38–46. doi: 10.1093/nar/gkn895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paquet N., Adams M.N., Ashton N.W., Touma C., Gamsjaeger R., Cubeddu L., Leong V., Beard S., Bolderson E., Botting C.H., et al. hSSB1 (NABP2/OBFC2B) is regulated by oxidative stress. Sci. Rep. 2016;6:27446. doi: 10.1038/srep27446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klebe G., Bohm H.J. Energetic and entropic factors determining binding affinity in protein-ligand complexes. J. Recept. Signal Transduct. Res. 1997;17:459–473. doi: 10.3109/10799899709036621. [DOI] [PubMed] [Google Scholar]
- 39.Chen D., Oezguen N., Urvil P., Ferguson C., Dann S.M., Savidge T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2:e1501240. doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrari M.E., Lohman T.M. Apparent heat capacity change accompanying a nonspecific protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry. 1994;33:12896–12910. doi: 10.1021/bi00209a022. [DOI] [PubMed] [Google Scholar]
- 41.Kozlov A.G., Lohman T.M. Calorimetric studies of E. coli SSB protein-single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. J. Mol. Biol. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 42.Kozlov A.G., Lohman T.M. Adenine base unstacking dominates the observed enthalpy and heat capacity changes for the Escherichia coli SSB tetramer binding to single-stranded oligoadenylates. Biochemistry. 1999;38:7388–7397. doi: 10.1021/bi990309z. [DOI] [PubMed] [Google Scholar]
- 43.Raghunathan S., Kozlov A.G., Lohman T.M., Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 44.Abdiche Y., Malashock D., Pinkerton A., Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 45.Concepcion J., Witte K., Wartchow C., Choo S., Yao D., Persson H., Wei J., Li P., Heidecker B., Ma W., et al. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb.Chem. High Throughput Screen. 2009;12:791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- 46.Flores J.K., Kariawasam R., Gimenez A.X., Helder S., Cubeddu L., Gamsjaeger R., Ataide S.F. Biophysical characterisation and quantification of nucleic acid-protein interactions: EMSA, MST and SPR. Curr. Protein Pept. Sci. 2015;16:727–734. doi: 10.2174/1389203716666150505230806. [DOI] [PubMed] [Google Scholar]
- 47.Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.