Abstract
The Ku protein, a heterodimer of Ku70 and Ku80, binds to chromosomal replication origins maximally at G1-phase and plays an essential role in assembly of origin recognition complex. However, the mechanism regulating such a critical periodic activity of Ku remained unknown. Here, we establish human Ku70 as a novel target of cyclin B1-Cdk1, which phosphorylates it in a Cy-motif dependent manner. Interestingly, cyclin E1- and A2-Cdk2 also phosphorylate Ku70, and as a result, the protein remains in a phosphorylated state during S-M phases of cell cycle. Intriguingly, the phosphorylation of Ku70 by cyclin-Cdks abolishes the interaction of Ku protein with replication origin due to disruption of the dimer. Furthermore, Ku70 is dephosphorylated in G1-phase, when Ku interacts with replication origin maximally. Strikingly, the over-expression of Ku70 with non-phosphorylable Cdk targets enhances the episomal replication of Ors8 origin and induces rereplication in HeLa cells, substantiating a preventive role of Ku phosphorylation in premature and untimely licensing of replication origin. Therefore, periodic phosphorylation of Ku70 by cyclin-Cdks prevents the interaction of Ku with replication origin after initiation events in S-phase and the dephosphorylation at the end of mitosis facilitates its participation in pre-replication complex formation.
INTRODUCTION
The Ku protein, a heterodimer consisting of Ku70 and Ku80 subunits, is a multifunctional complex playing critical roles in important cellular processes such as non-homologous end joining (NHEJ), V(D)J recombination, apoptosis, telomere maintenance and DNA replication (1). The most well-studied function of Ku is its DNA-PKcs dependent role in NHEJ pathway, where it functions as a DNA binding protein complex holding broken DNA ends during the repair process of double-strand breaks (DSBs) (2,3). More recently, the involvement of Ku protein in DNA replication has been implicated by virtue of its purification through the binding to the monkey replication origin Ors8 (4,5) and its interaction with several other chromosomal replication origins in a sequence-specific and cell cycle dependent manner (6,7). It has been shown that after initial binding of initiation protein Orc2 to replication origin subsequent loading of Orc3, Orc4 and Orc6 requires the association of Ku-heterodimer with the origin demonstrating its critical role during the assembly of pre-replication complex (8). It is also established through co-immunoprecipitation studies that Ku interacts with other DNA replication proteins including DNA polymerases, PCNA, topoisomerase II, RFC and RPA (9). Thus, the involvement of Ku in initiation of DNA replication is well-established, but how the protein is regulated in a cell cycle dependent manner remains unclear.
Several studies identify Ku as a nuclear component (10–14) with dispersed appearance throughout the nucleus in interphase cell (14–16). In addition, a portion of Ku has been shown to be bound to chromatin DNA in interphase nuclei and involved in chromosome organization (15,17). Interestingly, some early studies show Ku to be dissociated from chromatin and localized on the periphery of condensed chromosomes at metaphase, suggesting its distinct functional involvement in mitosis (18–21). It is also reported that the localization of Ku80 is different from that of Ku70 at metaphase (20), though the functional implications of such an observation remain to be confirmed. Hence, further studies are essential to elucidate the distinctive role of Ku in mitosis related activities.
Previously, three substrates of an S-phase LdCyc1–CRK3 complex from Leishmania donovani, a protozoan parasite causing potentially fatal visceral leishmaniasis, have been identified at our laboratory (22–24). Interestingly, one of them contains the Ku70 related conserved domains, raising the possibility of human Ku70 to be a substrate of cell cycle kinases as well. In fact, cyclin A1- and A2-Cdk2 have been shown to interact with and phosphorylate Ku70. Among them, cyclin A1-Cdk2 dependent phosphorylation has been specifically found to play a crucial role in DSB repair (25). Ku70 has also been reported to be a substrate of cyclin E1-Cdk2 (25), but the significance of Ku70 phosphorylation by cyclin E1- and A2-Cdk2 remains unknown. Also, no information is available on the association of Ku70 with B-type mitotic cyclins.
It is known that as a mechanistic component of replication licensing system, the high cell cycle kinase activity during S, G2 and M phases prevents the pre-replication complex formation in eukaryotic cells, which can only be formed in early G1-phase starting from late mitosis concomitant with the destruction of cyclin B1 (26). So, it would be interesting to investigate the status of phosphorylation of Ku during cell cycle progression, which could contribute significantly in regulation of replication related and mitosis specific activities of the protein.
Therefore, a detailed characterization of Ku70 phosphorylation by cyclin B1-Cdk1 is carried out in the present studies. We establish that the phosphorylation of Ku70 by cell cycle kinases abrogates the interaction of Ku with replication origin sequence and inhibits its replication related function. In fact, prevention of Ku70 phosphorylation by cyclin-Cdks induces rereplication in cells. Our results strongly suggest that such phosphorylation prevents Ku from taking part in assembly of functional origin recognition complex prematurely. On degradation of cyclin B1 at the end of mitosis, Ku is dephosphorylated, and as a result, can participate in replication related activities.
MATERIALS AND METHODS
Cell culture, synchronization and transfection
Human cell lines HeLa and HEK293 were cultured in Dulbecco's modifiedEagle's medium (DMEM) containing 10% fetal bovine srum (FBS). To arrest at mitosis, the cells were incubated with nocodazole (100 ng/ml) for 14–16 h. To block at early S-phase, the cells were first incubated with 2 mM thymidine for 19 h, then for 10 h without thymidine, and finally, again in the presence of 2 mM thymidine for 17 h. Sf9 insect cells were grown in TNM-FH medium containing 10% FBS at 27°C. All the transfections were carried out with Lipofectamin 2000 (Invitrogen).
Protein expression and purification
Human Ku70 ORF was amplified from the cDNA clone (MHS1010-74186, Open Biosystem) and inserted into pET28a and pEBG (mammalian expression vector for GST-tagged protein). The protein induction in Escherichia coli BL21-DE3 strain was carried out overnight at 18°C with 1 mM IPTG. GST tagged cyclins (B1, E1 and A2), Cdk1, Cdk2, streptavidin binding peptide (SBP)-tagged Ku80 (MHS1011-76130, Open Biosystem) and also 6His-Ku70 were expressed in insect cells using Bac-to-Bac baculovirus expression system (Invitrogen).
The 6His-tagged proteins were purified using Ni-NTA agarose beads (Qiagen). The recombinant Ku dimer was purified also using the Ni-NTA agarose from Sf9 insect cells co-expressing 6His-Ku70 and SBP-Ku80 (Supplementary Figure S1). The active cyclin–Cdk complexes were purified over glutathione Sepharose beads (GE Healthcare) from the insect cells co-expressing appropriate GST-cyclin and Cdk partners.
Protein interaction assay
The interaction assays between cyclin B1 and Ku70 were carried out by incubating glutathione beads bound to 0.5 μg of GST or GST-cyclin B1 proteins expressed in insect cells with either bacterially expressed and purified 6His-Ku70 (10 μg) or Sf9 extract with overexpressed 6His-Ku70 or HEK293 whole cell extract or varying amounts of Ku dimer (6His-Ku70/SBP-Ku80) at 4°C for 1 h in a total volume of 60 μl of 50 mM Tris–HCl, pH 8.0 containing 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM ethylenediaminetetraaceticacid (EDTA), 2 mM DTT and protease inhibitors. For competition experiments, 150 μM of SM100 peptide (AMNKRLGSLV) containing the conserved 516KRLG putative Cy-motif of Ku70 (Supplementary Figure S2) or AM100 (ACRDDKDPVDSE) as control was added during the interaction assay. To show the interaction between Ku70 and cyclin B1 in cells, GST or GST-Ku70 was expressed in HeLa cells from pEBG or pEBG-Ku70 plasmid, respectively, and pulled down with glutathione beads. For all interaction assays, the pulled-down protein complexes were analyzed by immunoblotting with appropriate antibodies: anti-Ku70, anti-SBP and anti-GST (sc-9033, sc-101595 and sc-138, respectively; Santa Cruz Biotechnology); anti-cyclin B1 (05-373, Millipore).
Kinase assay
The kinase assay was routinely carried out at 30°C in 50 mM Tris–HCl, pH 8.0 containing 10 mM MgCl2, 50 μM ATP, 1 μg histone H1 or bacterially expressed Ku70 (wild-type and mutants) and 2 μCi of γ-32P[ATP] in a total volume of 15 μl for 15 min. The products were visualized by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by phosphorimager scanning using Typhoon (GE Healthcare).
Immunoprecipitation
The cells were crosslinked with 1% formaldehyde and extracted with 50 mM Tris–HCl, pH 7.5 containing 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 10% glycerol, 1 mM DTT, 1 mM PMSF, 50 mM Na-vanadate, 1 mM NaF and protease inhibitors by sonication at 4°C. The clarified extracts were subjected to immunoprecipitation with anti-Ku70 and anti-cyclin B1 antibodies and the precipitated complexes were analyzed by immunoblotting with antibodies against Ku70, Ku80 (sc-5280, Santa Cruz Biotechnology) and Cdk1 (0923, Millipore).
For determining the phosphorylation status of Ku70 in different phases of cell cycle, the protein extracts from the synchronized HeLa cells at various time points were subjected to immunoprecipitation with anti-phospho-threonine-proline antibody (ab9344, Abcam), followed by anti-Ku70 immunoblotting. The band intensities were quantified by ImageJ software (imagej.nih.gov/ij/).
DNA replication Assay
For episomal replication assay in mammalian cells, the 320 bp core fragment of human Ors8 origin (27) was cloned into bacterial plasmid pTZ57R/T (Fermentas). A control sequence of similar size from 2 kb downstream of the chromosomal origin was also cloned independently. Exponentially growing HeLa cells were co-transfected with the plasmids containing origin or control sequences, GST-tagged wild type or mutant Ku70 and GFP open reading frame (pEGFP-N1, for normalization of transfection efficiency) in a molar ratio of 5:5:1. Seventy two hours post-transfection, low-molecular-weight DNA was isolated from the cells by the modified Qiagen protocol (28). Half of the isolated plasmid sample was digested with DpnI. The DpnI digested as well as undigested DNA samples were used to transform E. coli (XLI-Blue) and the relative episomal replication efficiency of each plasmid was determined by counting the number of DpnI resistant colonies (27). BrdU incorporation assay was carried out according to the manufacturer (Novagen) protocol.
Electrophoretic mobility shift assay (EMSA)
To show the effect of phosphorylation on interaction with origin DNA, the purified Ku dimer and cyclin-Cdks were incubated for 15 min at 30°C with non-radioactive ATP. For dephosphorylation, Ku dimer was incubated with recombinant shrimp alkaline phosphatase (eBioscience) for 30 min at 37°C. EMSA was carried out by incubating the phosphorylated or the dephosphorylated Ku dimer with 100 fmol of 32P-labeled A3/4 oligo, a 36-bp mammalian replication origin sequence that is capable of supporting autonomous replication in vivo (4,29), or a non-specific NS oligo (30) in a final volume of 20 μl of 12 mM HEPES, pH 7.9 containing 5% glycerol, 60 mM KCl, 0.12 mM EDTA, 1 μg BSA, 1 μg poly dI-dC for 20 min at 20°C. The products were resolved on 6% polyacrylamide gel in 0.5× TBE and visualized by autoradiography. For super-shift, the Ku-specific antibody (ab80828, Abcam) was pre-incubated with Ku protein prior before adding to radioactive probe.
Chromatin immunoprecipitation (CHIP)
For ChIP experiments, performed essentially as described before (31), GST-Ku70 or GST-Ku70ΔK (Ku70T401/428/455A mutant) was expressed in HeLa cells and immunoprecipitated after crosslinking and chromatin shearing with anti-GST antibody. PCR were carried out using primer pair targeting the core of Ors8 origin (forward, 5′TTGCACTTCACAGAGCAGTCATG and reverse, 5′GACCCACAAAGGCAAAAGTACC). The intensities PCR product bands were quantified by ImageJ software (imagej.nih.gov/ij/).
Fluorescence microscopy
Exponentially growing HeLa cells, expressing GST, GST-Ku70 or GST-Ku70ΔK proteins, were fixed on coverslips with ice-cold methanol for 5 min at −20°C followed by blocking with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) and overnight incubation with anti-GST antibody at 4°C. The cells were then stained with AlexaFluor-488 conjugated secondary antibody and DAPI (for nuclei) and examined under a fluorescence microscope (AxioObserver.Z1, Zeiss). The nuclear size was determined by NIS AR 4.3 software (Nikon).
RESULTS
Ku70 interacts with cyclin B1 in vitro and in vivo
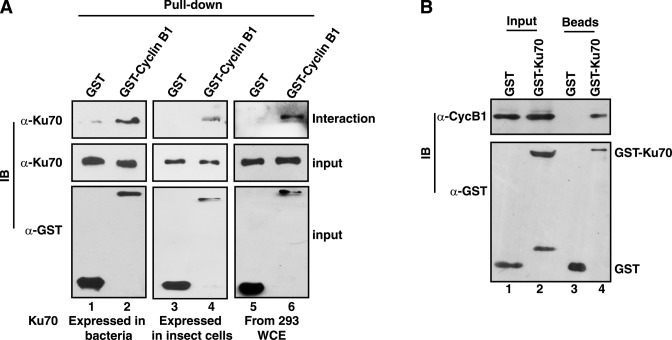
As human Ku70 protein contains six putative RxL type cyclin interacting Cy-motifs (Supplementary Figure S2), its ability to interact with cyclin B1 was tested by GST pull-down assay. Glutathione beads bound to GST or GST-cyclin B1 (expressed in insect cell system) were incubated with 6His-Ku70 protein expressed in bacteria or insect cells. As shown in Figure 1A, Ku70 from both sources interacted specifically with cyclin B1 (lanes 1–4). The assay was repeated with the extract of HEK293 cells and cyclin B1 was shown to interact also with Ku70 expressed endogenously (Figure 1A, lanes 5 and 6). To establish that such a stable interaction takes place in cells as well, GST-Ku70 was expressed in HeLa cells ectopically from pEBG-Ku70 plasmid, and expectedly, cyclin B1 could be detected specifically in the pulled-down complex with glutathione beads (Figure 1B). Therefore, the experimental result established that Ku70 interacts with cyclin B1, which is active in mitosis and forms mitotic kinase complex with Cdk1.
Figure 1.
Ku70 interacts with cyclin B1. (A) Glutathione Sepharose beads bound to GST or GST-cyclin B1 (expressed in insect cells) were incubated with bacterially expressed and purified Ku70 (lanes 1 and 2), insect cell extract containing over-expressed Ku70 (lanes 3 and 4) or HEK293 cell extract (lanes 5 and 6). The pulled-down complexes and the supernatant were analyzed by immunoblotting (IB) with antibodies against Ku70 (Interaction and Input) and GST (Input). (B) GST or GST-Ku70 were expressed in HeLa cells and pulled down from the cell-extract with glutathione beads. The presence or absence of cyclin B1 in the extract (lanes 1 and 2) and the pulled-down complexes (lanes 3 and 4) was tested by anti-cyclin B1 immunoblotting. The expression of GST and GST-Ku70 and their presence in the beads were also checked by anti-GST immunoblotting.
Cyclin B1 interacts with Ku70 but not with Ku80 in vitro and in vivo
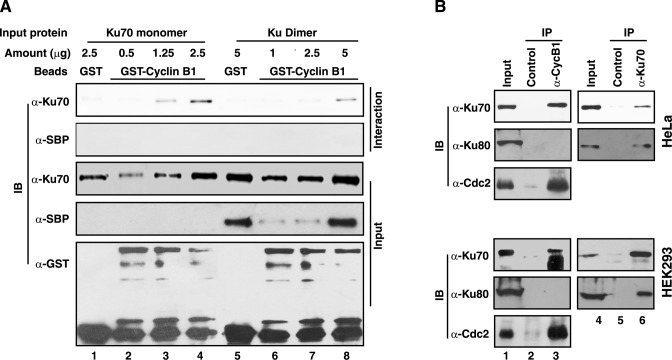
Ku70 is present in cell predominantly as a dimer with Ku80. However, some reports indicated the independent localization of Ku70 and Ku80 in mammalian cells during mitosis (21). Since Ku70 and cyclin B1 were shown to interact with each other in vivo and in vitro, it was tested by similar GST pull-down assay whether Ku80 could also bind to cyclin B1. The Ku70 monomer and the Ku dimer (6His-Ku70/SBP-Ku80), expressed in Sf9 insect cells and purified, were incubated with glutathione beads bound to GST or GST-cyclin B1 and it was found that 6His-Ku70 monomer and 6His-Ku70 subunit of the dimer interacted with cyclin B1 specifically (Figure 2A). However, SBP-Ku80 subunit of the dimer protein did not interact with cyclin B1, indicating that cyclin B1 interacts with Ku70 independently. To further clarify the observation in vivo, Ku70 and cyclin B1 were immunoprecipitated from nocodazole blocked HeLa and HEK293 cells. Expectedly, Ku80 could be detected only in Ku70 immunoprecipitates, but not in that of cyclin B1 in M-phase cells (Figure 2B). Similarly, Ku70 was detected in the immunoprecipitate of cyclin B1, confirming that the cyclin interacts with Ku70, and in the process, disrupts Ku70/Ku80 dimer.
Figure 2.
Cyclin B1 interacts with Ku70 in Ku-heterodimer in vitro and in vivo. (A) Varying quantities of 6His-Ku70 monomer (lanes 1–4) and 6His-Ku70/SBP-Ku80 dimer (lanes 5–8), purified from insect cell expression system, were incubated with glutathione agarose beads bound to GST or GST-cyclin B1 (expressed in insect cells). The pulled-down complexes were analyzed by anti-Ku70 and anti-SBP tag (for Ku80) immunoblotting (Interaction). The presence of Ku70 and Ku80, as appropriate, in the reaction mixtures was checked by indicated immunoblotting (Input panels). The presence of GST or GST-cyclin B1 was also tested by anti-GST immunoblotting. (B) Immunoprecipitation with antibodies against cyclin B1 and Ku70 were carried out from the extract of HeLa or HEK293 cells arrested at mitosis by nocodazole and the presence of Ku70, Ku80 or Cdc2 in the immunoprecipitates were determined by immunoblotting with appropriate antibodies as indicated.
Ku70 is phosphorylated by cyclin–Cdks in a Cy-motif dependent manner
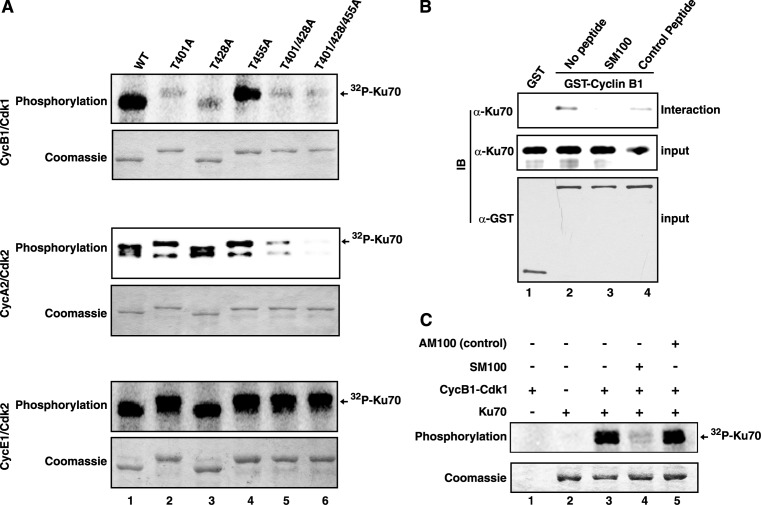
Since cyclin B1 was shown to interact with Ku70, which contains one canonical and several minimal Cdk phosphorylation sites (Supplementary Figure S2), it was explored whether Ku70 could be phosphorylated by cyclin B1–Cdk1. As shown in Figure 3A, Ku70 was phosphorylated when it was used as a substrate in an in vitro kinase assay with cyclin B1-Cdk1 (lane 1, top panel). As the canonical target sites along with nearby minimal sites are most likely to be phosphorylated by Cdks (32), the threonine residues at 401, 428 and 455 positions of Ku70 (Supplementary Figure S2) were mutated to alanine individually or in combination in order to identify the target phosphorylation sites. As observed, the phosphorylation level of Ku70-T401A and Ku70-T428A mutants, but not that of Ku70-T455A, by cyclin B1-Cdk1 was significantly reduced compared to that of the wild type protein (Figure 3A, lanes 1–4, top panel), indicating that Thr-401 and Thr-428 are the major phosphorylation target sites. However, some residual level of phosphate incorporation was visible when the single site mutants and Ku70-T401/428A double mutant were used (lanes 2, 3 and 5), which was further diminished with Ku70-T401/428/455A triple mutant as substrate in the kinase assay (lane 6, top panel). Therefore, the results described here strongly suggest that all the three sites in Ku70 were phosphorylated by cyclin B1–Cdk1, though in varying degree.
Figure 3.
Ku70 is phosphorylated by cyclin-Cdks in a Cy-motif dependent manner. (A) The wild type and mutant (Thr to Ala, as indicated) Ku70 proteins were used in [γ-32P]ATP-based kinase assays with the cyclin-Cdks as marked. For each set of assays, the presence of equal amounts of Ku70 protein was ascertained by Coomassie blue staining of the reaction mixtures. The radioactive Ku70 bands due to incorporation of 32P are indicated. (B) GST or GST-cyclin B1 bound to glutathione sepharose beads were incubated with purified Ku70 protein either in absence of any peptide (lanes 1 and 2), or in presence of 150 μM competitor SM100 peptide (lane 3) or control AM100 peptide (lane 4). The pulled-down complexes were analyzed by immunoblotting with antibodies against Ku70 (interaction) and GST (input). The presence of equivalent amounts of Ku70 protein in interaction mixtures were checked by anti-Ku70 immunoblotting (input). (C) HsKu70 was used as substrate in [γ-32P]ATP based kinase assay with cyclin B1–Cdk1 in the absence or presence of 150 μM of the indicated peptides.
It was reported previously that Ku70 could also be a substrate of cyclin A1–Cdk2, cyclin A2–Cdk2 and cyclin E1–Cdk2 (25) and our results also confirmed the observations (Figure 3A, middle and bottom panels). Moreover, the kinase assays with mutant Ku70 established that the target phosphorylation sites of cyclin A2–Cdk2 are similar to that of cyclin B1–Cdk2 (Figure 3A, middle panel). However, cyclin E1–Cdk2 could phosphorylate additional target in Ku70, as the phosphorylation was not diminished even in case of the triple mutant (Figure 3A, bottom panel). The possible alternative target could be the other minimal site at 58TP. Further studies are essential to ascertain the target phosphorylation sites of cyclin E1–Cdk2 in Ku70.
It is well-known that RxL-type Cy-motif together with the phosphorylation target site (S/T)Px(K/R) constitute a bipartite substrate recognition sequence of Cdks (33). Interestingly, Ku70 contains six putative Cy-motifs (Supplementary Figure S2), among which 516KRLG is highly conserved and more similar to the canonical RRLFG containing Cy-motif first identified in p21 (34). Therefore, the effect of a synthetic peptide SM100 containing the 516KRLG sequence and a control peptide were tested in the interaction assay as well as in the kinase assay. Strikingly, SM100 completely abolished the interaction specifically between cyclin B1 and Ku70 (Figure 3B) and also inhibited phosphorylation of Ku70 by cyclin B1–Cdk1 kinase (Figure 3C) confirming the identity of a functional Cy-motif of Ku70 and its importance in the interaction and the phosphorylation events.
Ku70 remains phosphorylated during S, G2 and M phases and is dephosphorylated on exit from mitosis
In order to show that the Ku70 is phosphorylated by cyclin-Cdks in a periodic manner during cell cycle, the phosphorylation status of Thr residues of the protein in the Cdk target motif of Thr-Pro was checked in synchronously growing HeLa cells. To ensure that the phosphorylation status of Ku70 at the Thr residues of Cdk target sites was followed particularly, the presence of the protein specifically in the anti-phospho-threonine-proline immunoprecipitates from HeLa cell extract, but not in that with anti-phospho-serine antibody, was first confirmed (Supplementary Figure S3). The extent of synchronicity of the cells was ascertained by analysis in a flow cytometer and detecting differential expression pattern of cyclins E and B (Figure 4). S-phase was confirmed by the detection of cyclin E protein maximally at 2.5 h point, whereas M-phase was characterized by peak expression of cyclin B protein during 7.5 h point after the release from double thymidine arrest. Finally, Ku70 phosphorylated at the Thr residues of Thr-Pro motifs could be detected in significantly increasing amounts with maximum at 7.5 h point in the anti-phospho-threonine-proline immunoprecipitates from the cells progressing from S to M phase (Figure 4, lanes 2–5), and its level decreased drastically in the cells exiting M-phase, suggesting reduced phosphorylation of Ku70 during G1-phase (lane 6, 9.5 h point).
Figure 4.
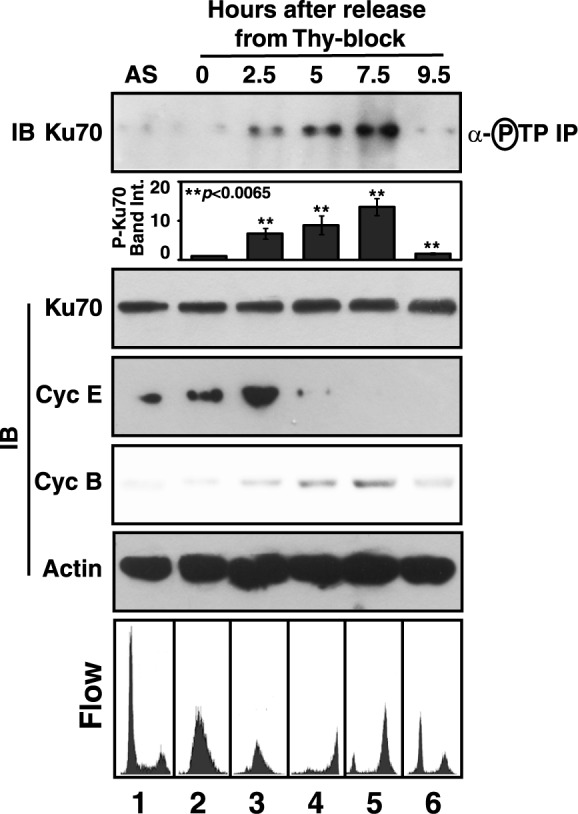
Differential phosphorylation of Ku70 during cell cycle. HeLa cells were collected at the indicated time intervals after release from the arrest induced by double thymidine treatment. The presence of Ku70 in the anti-phospho-threonine-proline immunoprecipitates from the cell extracts and the supernatants were determined by anti-Ku70 immunoblotting (IB). The mean fold-change (±S.D.) of the intensities of the phospho-Ku70 (P-Ku70) bands in the immunoblots (n = 3) in different time points with respect to that at 0 h point were represented as column plot. Synchronization of the cells was checked by anti-cyclin E and anti-cyclin B immunoblotting and flow cytometer analysis after propidium iodide staining. Equal loading of protein samples were ascertained by anti-actin immunoblotting of the supernatants. AS, asynchronous culture of cells.
Phosphorylation of Ku by cyclin–Cdks inhibits its specific interaction with replication origin DNA
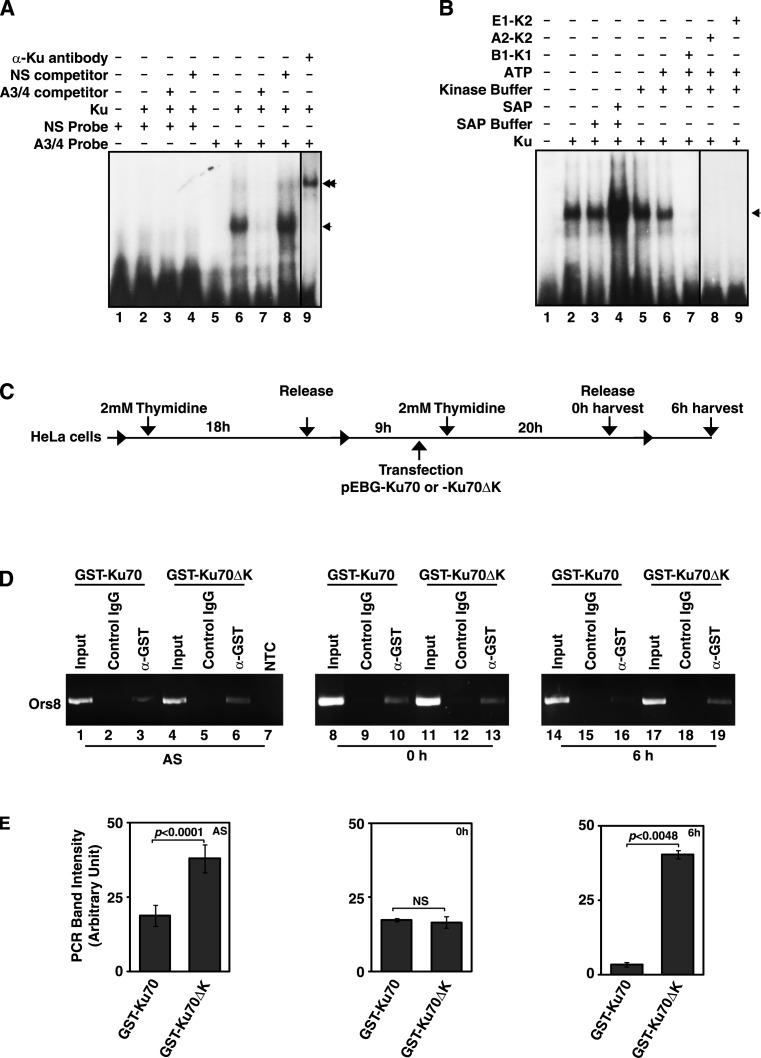
Intrigued by the observation of significant variation of phosphorylation of Ku70 at Cdk target sites during cell cycle, the influence of phosphorylation of the protein by cyclin–Cdks on its interaction with mammalian replication origin sequence was studied in detail. At first, the specificity of binding of Ku with A3/4 oligo carrying a functional mammalian replication origin was characterized by EMSA (Figure 5A). As shown in Figure 5A, Ku did not bind to a probe (NS) containing some random sequence (lanes 1–4), but interacted specifically with the A3/4 probe (lanes 5–8). The retarded radioactive band in EMSA with A3/4 and Ku was almost completely disappeared on competition with non-radioactive A3/4 probe (lane 7), but not with NS probe (lane 8), confirming the specificity of the interaction. The interaction was further confirmed by the super-shift of the radioactive band with the addition of antibody against Ku protein (lane 9). Next, the EMSA was repeated to study the effect of phosphorylation on the interaction of Ku with A3/4 probe. Since the purified recombinant Ku protein from the eukaryotic insect cell might carry endogenously incorporated phosphate moieties, the EMSA was done with the Ku protein treated with shrimp alkaline phosphatase. Interestingly, the intensity of the retarded band enhanced significantly (Figure 5B, lanes 1–4) upon treatment with the phosphatase. Furthermore, the retarded band was found to be almost completely disappeared in presence of the Ku protein that was phosphorylated by cyclin B1–Cdk1, cyclin A2–Cdk2 and cyclin E1–Cdk2 (Figure 5B, lanes 5–9), confirming the inhibitory effect of the phosphorylation of Ku by the kinases on its interaction of origin DNA.
Figure 5.
Phosphorylation of Ku abrogates its interaction with replication origin DNA. (A) To establish the specificity of origin DNA binding, the recombinant Ku (expressed and purified from Sf9 cells) was incubated with 100 fmol of 32P-labeled NS (non-specific oligo) probe (lanes 1–4) and A3/4 oligo probe (lanes 5–9) and the interaction was followed by the retarded bands (indicated by arrow-head) in EMSA. 50×molar concentrations of non-radioactive A3/4 oligo (lanes 3 and 7) and NS oligo (lanes 5 and 8) were used for competition. The interaction was further confirmed by super-shift of the retarded band (indicated by double arrow head) in presence of Ku specific antibody. (B) The effect of phosphorylation by the cyclin-Cdks and dephosphorylation of Ku on A3/4 probe binding was studied by EMSA. The addition of different components is indicated on top of the autoradiogram. (C) The workflow of transfection and synchronization of HeLa cells for the ChIP experiments. (D) PCR was carried out using the DNA samples obtained by ChIP from the extracts of asynchronously or synchronously growing (as shown in Figure 5C) HeLa cells expressing the indicated Ku70 proteins and the products were analyzed using agarose gel electrophoresis. NTC, no template control. (E) The average band intensities (±S.D., n = 3) of the Ors8 PCR products from the indicated ChIP (after subtracting the intensities of the band from the corresponding control IgG ChIP) were plotted as column graphs. NS, non-significant.
To extend the intriguing observation to cell, chromatin immunoprecipitation (ChIP) with antibody against GST tag was carried out at first from the asynchronously growing HeLa cells overexpressing GST-Ku70 or GST-Ku70ΔK (Ku70T401/428/455A mutant). The specific presence of Ors8 origin DNA in the precipitates was confirmed by PCR (Figure 5D, lanes 1–7 and Figure 5E). In order to determine the effect of the mutation on origin DNA binding activity of the protein in early S- and G2/M-phase, HeLa cells were transfected with plasmids expressing the wild type or the mutant Ku70 proteins and synchronized (Supplementary Figure S4) as per the experimental workflow shown in Figure 5C. In early S-phase (0 h cells after double thymidine block) the wild type Ku70 as well as Ku70ΔK were found to interact with Ors8 origin almost equally (Figure 5D, lanes 8–13 and Figure 5E). With the progress of cell cycle and DNA replication, the phosphorylation level of Ku70 was shown to be increased (Figure 4), and accordingly, the binding of wild-type Ku70 to the origin DNA was found to diminish significantly (Figure 5D, lanes 14–16 and Figure 5E) after S-phase (6 h cells, Supplementary Figure S4). Strikingly, the non-phosphorylable Ku70ΔK mutant was bound to the origin DNA even in G2/M-phase cells (Figure 5D, lanes 17–19 and Figure 5E), confirming that the reversible phosphorylation controls the origin DNA binding activity of Ku during cell cycle progression.
Ku70ΔK mutant induces re-replication
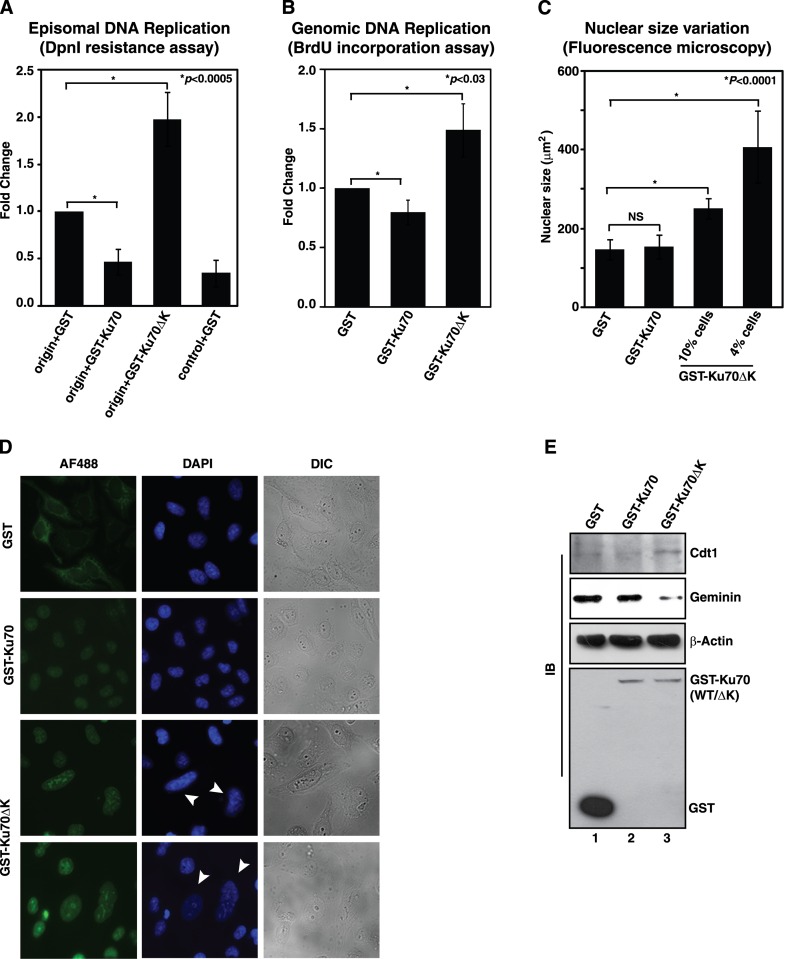
Since origin DNA binding activity of Ku70 can be modulated through reversible phosphorylation by cell cycle kinases, it would be interesting to study the effect of the post-translational modification on replication related activities of the protein. Hence, the effect of Ku70 and its mutants on DNA replication were first studied in HeLa cells using episomal plasmid based DpnI resistance assay. Interestingly, the overexpression of wild type Ku70 inhibited the replication in HeLa cells, but that of Ku70ΔK mutant enhanced the episomal replication of Ors8 origin by 2-fold (Figure 6A), suggesting an inhibitory role of phosphorylation of Ku70 by cell cycle kinases in DNA replication. Similar significant stimulatory role (1.5-fold) of the mutant Ku70 was also observed on genomic DNA replication of the cell as depicted by BrdU incorporation assay (Figure 6B). Since such enhancement of DNA replication could be due to the induced rereplication by the non-phosphorylable variant of Ku70, the nuclear sizes of the cells overexpressing GST tagged Ku70 or its mutant were determined microscopically. As observed, there was no significant difference in sizes of the nuclei in cells expressing either GST or GST-Ku70 proteins (Figure 6C). Remarkably, the nuclear sizes were found to be increased by 1.7-fold in 10% of the cells expressing GST-Ku70ΔK protein and 2.8-fold in 4% of such cells (Figure 6C), suggesting significant increase in DNA content in these cells. The representative images of the observed giant nuclei specifically in the cells expressing GST-Ku70ΔK protein were shown in Figure 6D, which were considered to be hallmarks of the cells undergoing rereplication (35,36). Moreover, increased amount of pre-replication complex component Cdt1 and reduced expression of Cdt1-sequestering factor Geminin were observed in the cells overexpressing the mutant Ku70 compared to that overexpressing the wild type protein (Figure 6E). Since Geminin depletion or overexpression of Cdt1 was shown to be associated with rereplication (37), the results shown in Figure 6E further ascertained the rereplication driven by Ku70ΔK protein.
Figure 6.
Overexpression of Ku70 with altered Cdk phosphorylation sites drives rereplication in HeLa cells. Exponentially growing HeLa cells were transfected with plasmids expressing GST, GST-Ku70 or GST-Ku70ΔK as indicated. (A) The replication of plasmids containing the HsOrs8 origin sequence or the control sequence was followed. The results were plotted as mean fold-change (±S.D., n = 4) with respect to colony numbers obtained in origin + GST experiments. (B) Genomic DNA replication was followed by BrdU incorporation assay. The results from three independent experiments were plotted as mean fold-change (±S.D.) in BrdU incorporation with respect to that in the cells expressing GST protein only. (C) Area of the DAPI stained nuclei were determined in the cells overexpressing the indicated proteins that were identified by anti-GST Alexa Flour 488 staining. In cells overexpressing GST-Ku70ΔK, the percentage of Alexa Flour 488 positive cells showing increased nuclear size were indicated. NS, non-significant change. (D) Representative fluorescence microscopy images showing giant nuclei in the cells overexpressing GST-Ku70ΔK (indicated by arrowheads). (E) Immunoblot analyses of the cell extracts with antibodies against Cdt1 (sc-28262), geminin (sc-74496), β-actin and GST proteins.
DISCUSSION
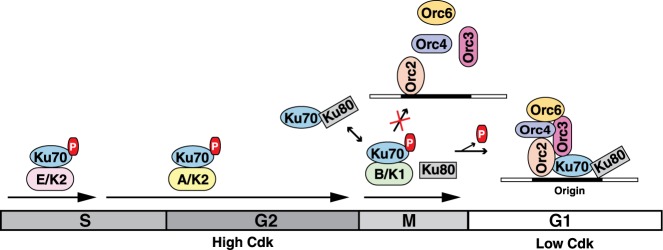
The critical role of Ku protein in formation of pre-replication complex (pre-RC) in mammalian cell is well established. However, the mechanism of cell cycle dependent periodic regulation of Ku protein activity related to replication initiation has not been addressed before. In this context, prior identification of a Ku70 related protein as a substrate of the S-phase cell cycle kinase from L. donovani in our laboratory (24) and the presence of putative Cy-motifs and Cdk target sites, along with the report of uncharacterized phosphorylation events of human Ku70 by S-phase Cdks (25), have prompted us to explore the contribution of the post-translational modification by cell cycle kinases on the regulation of replication initiation related function of Ku. In the present studies, we have shown that cyclin B1–Cdk1, along with cyclin E1–Cdk2 and cyclin A2–Cdk2, phosphorylates Ku70 subunit of the Ku dimer inhibiting its interaction with replication origin. Our results confirm that the phosphorylation of Ku by cell cycle kinases during high kinase phases prevents its interaction with the origin of replication. The phosphorylation of Ku during S, G2 and M phases is extremely critical to avoid untimely replication initiation during cell cycle progression as we have observed that the overexpression of non-phosphorylable mutant of Ku70 subunit induces rereplication in HeLa cells. Due to the low kinase activity at the end of mitosis, the non-phosphorylated state of Ku allows its interaction with origin DNA facilitating the role in pre-replication formation in G1-phase (Figure 7). Therefore, cell cycle kinases regulate the periodic origin association activities of Ku protein like other players of replication licensing system including Cdc6, cdt1 and MCMs (36).
Figure 7.
Modulation of replication initiation related function of Ku through phosphorylation by cyclin-Cdks. Ku70 is phosphorylated by cyclin–Cdks during high Cdk phases—S, G2 and M. The phosphorylation of Ku70 by cyclin B1-Cdk1 disrupts its association with Ku80 subunit, abrogating the interaction of Ku with replication origin. With the removal of cyclin B1 at the end of mitosis, Ku70 is dephosphorylated and as a result the functional Ku heterodimer can be formed for the assembly of origin recognition complex at replication origin in early G1-phase.
Ku70 contains one well-conserved canonical Cdk phosphorylation site (401TPRR) with two nearby minimal sites (428TP and 455TP). In addition, it also contains another minimal site (58TP) on the N-terminal region. Our study using site directed mutagenesis has confirmed that Thr residues at 401 and 428 positions are the major target phosphorylation sites of cyclin B1–Cdk1. On the other hand, cyclin A2–Cdk2 phosphorylates Thr residues at 401, 428 and 455 positions apparently with equal specificity. However, cyclin E1-Cdk2 also phosphorylates some other target residue—probably Thr-58, which could have additional functional implication. Again, like other Cdk targets, the phosphorylation of Ku70 depends on its interaction with cyclin B1 through the RxL type Cy-motif located at amino acid positions 516–519. It is established that the region spanning amino acids 378–482 of Ku70 is involved in heterodimerization with Ku80 (16). Interestingly, the identified phosphorylation sites of cyclin B1-Cdk2 are present in the region of Ku70 that is responsible for dimerization with Ku80. Also, the identified functional RxL type Cy-motif of Ku70 is located very close to the dimerization domain on the C-terminal. This explains our findings of the disruption Ku heterodimerization due to the interaction of Ku70 with cyclin B1-Cdk1 and the incorporation of phosphates. Our study has established by co-immunoprecipitation that in mitotic cells, there are two pools of Ku70 - one is in complex with cyclin B1 and another with Ku80. This supports the observation in several previous studies indicating independent regulation of Ku70 and Ku80 in mitosis (20,21).
Licensing in eukaryotes starts in late mitosis/early G1-phase through sequential assembly of a multiprotein complex—pre-RC (38). Cdk activity is then required for the onset of S-phase (39,40). After firing, origins are kept in an unlicensed state until the completion of mitosis in the cell cycle. High Cdk activity inhibits pre-RC assembly and licensing during S, G2 and M phases in eukaryotes (41,42). Ku as an origin binding protein, associates maximally with chromosomal replication origins in G1-phase. The binding of Ku to chromosomal origins decreases as cells enter S-phase. Ku acts in the initiation of DNA replication and dissociates after origin firing (6). Though C-terminal of Ku70 shows similarity with the SAP domain that is involved in DNA interaction (43,44), binding of Ku to origin of DNA replication requires heterodimerization (9). Our results confirm that the increased phosphorylation of Ku70 by Cdks from S-phase onwards till M-phase is responsible for its non-interaction with origin DNA during these phases. This is further supported by the observed binding of non-phosphorylable variant of Ku to origin DNA even in G2/M phase cells. Therefore, the prevention of the interaction of the Ku with origin DNA in S, G2 and M phases is also critical for replication licensing mechanism. The conclusion is supported by the observed rereplication in the cells overexpressing the non-phosphorylable variant of Ku70.
Taken together, the results described here provide an insight how the replication related function of Ku can be periodically regulated in cells. Due to the formation of the complexes of Ku70 with cyclin–Cdks and its phosphorylation, the Ku dimer is disrupted. This results in the decrease of the amount of functional Ku dimer available for binding to replication origins during the high Cdk activity phases, thus preventing premature assembly of origin recognition complex till the end of mitosis (Figure 7). After the degradation of cyclin B1, Ku70 can be converted to dephosphorylated state and is able to form heterodimer with ku80 for taking part in origin licensing during G1-phase.
Supplementary Material
Acknowledgments
We thank Dr Susanta Debnath for his technical help during the course of the work.
Footnotes
Present address: Prabal Chakraborty, Towa Optics (India) Pvt. Ltd., 30 Gold Park, Kolkata 700107, India.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Atomic Energy (DAE), Govt. of India (to S.M. and P.C.). Funding for open access charge: Intramural institutional funding [RS1210].
Conflict of interest statement. None declared.
REFERENCES
- 1.Downs J.A., Jackson S.P. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell. Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 2.Weterings E., van Gent D.C. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst.) 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Lees-Miller S.P., Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz M.T., Matheos D., Price G.B., Zannis-Hadjopoulos M. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Mol. Biol. Cell. 1999;10:567–580. doi: 10.1091/mbc.10.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiz M.T., Pearson C.E., Nielsen T., Price G.B., Zannis-Hadjopoulos M. Cofractionation of HeLa cell replication proteins with ors-binding activity. J. Cell Biochem. 1995;58:221–236. doi: 10.1002/jcb.240580211. [DOI] [PubMed] [Google Scholar]
- 6.Novac O., Matheos D., Araujo F.D., Price G.B., Zannis-Hadjopoulos M. In vivo association of Ku with mammalian origins of DNA replication. Mol. Biol. Cell. 2001;12:3386–3401. doi: 10.1091/mbc.12.11.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sibani S., Price G.B., Zannis-Hadjopoulos M. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/- cells. J. Cell Sci. 2005;118:3247–3261. doi: 10.1242/jcs.02427. [DOI] [PubMed] [Google Scholar]
- 8.Sibani S., Price G.B., Zannis-Hadjopoulos M. Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry. 2005;44:7885–7896. doi: 10.1021/bi047327n. [DOI] [PubMed] [Google Scholar]
- 9.Matheos D., Ruiz M.T., Price G.B., Zannis-Hadjopoulos M. Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication. Biochim. Biophys. Acta. 2002;1578:59–72. doi: 10.1016/s0167-4781(02)00497-9. [DOI] [PubMed] [Google Scholar]
- 10.Reeves W.H. Antibodies to the p70/p80 (Ku) antigens in systemic lupus erythematosus. Rheum. Dis. Clin. North Am. 1992;18:391–414. [PubMed] [Google Scholar]
- 11.Reeves W.H., Satoh M., Wang J., Chou C.H., Ajmani A.K. Systemic lupus erythematosus. Antibodies to DNA, DNA-binding proteins, and histones. Rheum. Dis. Clin. North Am. 1994;20:1–28. [PubMed] [Google Scholar]
- 12.Yaneva M., Busch H. A 10S particle released from deoxyribonuclease-sensitive regions of HeLa cell nuclei contains the 86-kilodalton-70-kilodalton protein complex. Biochemistry. 1986;25:5057–5063. doi: 10.1021/bi00366a013. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Chou C.H., Blankson J., Satoh M., Knuth M.W., Eisenberg R.A., Pisetsky D.S., Reeves W.H. Murine monoclonal antibodies specific for conserved and non-conserved antigenic determinants of the human and murine Ku autoantigens. Mol. Biol. Rep. 1993;18:15–28. doi: 10.1007/BF01006891. [DOI] [PubMed] [Google Scholar]
- 14.Yaneva M., Jhiang S. Expression of the Ku protein during cell proliferation. Biochim. Biophys. Acta. 1991;1090:181–187. doi: 10.1016/0167-4781(91)90099-8. [DOI] [PubMed] [Google Scholar]
- 15.Mimori T., Hardin J.A., Steitz J.A. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J. Biol. Chem. 1986;261:2274–2278. [PubMed] [Google Scholar]
- 16.Koike M., Miyasaka T., Mimori T., Shiomi T. Subcellular localization and protein-protein interaction regions of Ku proteins. Biochem. Biophys. Res. Commun. 1998;252:679–685. doi: 10.1006/bbrc.1998.9368. [DOI] [PubMed] [Google Scholar]
- 17.Yaneva M., Ochs R., McRorie D.K., Zweig S., Busch H. Purification of an 86-70 kDa nuclear DNA-associated protein complex. Biochim. Biophys. Acta. 1985;841:22–29. doi: 10.1016/0304-4165(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 18.Reeves W.H. Antinuclear antibodies as probes to explore the structural organization of the genome. J. Rheumatol. Suppl. 1987;14(Suppl. 13):97–105. [PubMed] [Google Scholar]
- 19.Li L.L., Yeh N.H. Cell cycle-dependent migration of the DNA-binding protein Ku80 into nucleoli. Exp. Cell Res. 1992;199:262–268. doi: 10.1016/0014-4827(92)90433-9. [DOI] [PubMed] [Google Scholar]
- 20.Higashiura M., Shimizu Y., Tanimoto M., Morita T., Yagura T. Immunolocalization of Ku-proteins (p80/p70): iocalization of p70 to nucleoli and periphery of both interphase nuclei and metaphase chromosomes. Exp. Cell Res. 1992;201:444–451. doi: 10.1016/0014-4827(92)90293-h. [DOI] [PubMed] [Google Scholar]
- 21.Koike M., Ikuta T., Miyasaka T., Shiomi T. Ku80 can translocate to the nucleus independent of the translocation of Ku70 using its own nuclear localization signal. Oncogene. 1999;18:7495–7505. doi: 10.1038/sj.onc.1203247. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee S., Banerjee R., Das R., Duttagupta S., Saha P. Isolation, characterization and expression of a cyclin from Leishmania donovani. FEMS Microbiol. Lett. 2003;226:285–289. doi: 10.1016/S0378-1097(03)00606-2. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee S., Sen A., Das P., Saha P. Leishmania donovani cyclin 1 (LdCyc1) forms a complex with cell cycle kinase subunit CRK3 (LdCRK3) and is possibly involved in S-phase-related activities. FEMS Microbiol. Lett. 2006;256:75–82. doi: 10.1111/j.1574-6968.2006.00105.x. [DOI] [PubMed] [Google Scholar]
- 24.Maity A.K., Goswami A., Saha P. Identification of substrates of an S-phase cell cycle kinase from Leishmania donovani. FEBS Lett. 2011;585:2635–2639. doi: 10.1016/j.febslet.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 25.Ji P., Baumer N., Yin T., Diederichs S., Zhang F., Beger C., Welte K., Fulda S., Berdel W.E., Serve H., et al. DNA damage response involves modulation of Ku70 and Rb functions by cyclin A1 in leukemia cells. Int. J. Cancer. 2007;121:706–713. doi: 10.1002/ijc.22634. [DOI] [PubMed] [Google Scholar]
- 26.Mendez J., Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callejo M., Sibani S., Di Paola D., Price G.G., Zannis-Hadjopoulos M. Identification and functional analysis of a human homologue of the monkey replication origin ors8. J. Cell Biochem. 2006;99:1606–1615. doi: 10.1002/jcb.20868. [DOI] [PubMed] [Google Scholar]
- 28.Wasia Rizwani S.P.C. In: Chromatin Protocols. Chellappan SP, editor. Vol. 523. Springer; 2009. pp. 203–216. [Google Scholar]
- 29.Price G.B., Allarakhia M., Cossons N., Nielsen T., Diaz-Perez M., Friedlander P., Tao L., Zannis-Hadjopoulos M. Identification of a cis-element that determines autonomous DNA replication in eukaryotic cells. J. Biol. Chem. 2003;278:19649–19659. doi: 10.1074/jbc.M207002200. [DOI] [PubMed] [Google Scholar]
- 30.Schild-Poulter C., Matheos D., Novac O., Cui B., Giffin W., Ruiz M.T., Price G.B., Zannis-Hadjopoulos M., Hache R.J. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA Cell Biol. 2003;22:65–78. doi: 10.1089/104454903321515887. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee M., Datta M., Majumder P., Mukhopadhyay D., Bhattacharyya N.P. Transcription regulation of caspase-1 by R393 of HIPPI and its molecular partner HIP-1. Nucleic Acids Res. 2010;38:878–892. doi: 10.1093/nar/gkp1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moses A.M., Heriche J.K., Durbin R. Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol. 2007;8:R23. doi: 10.1186/gb-2007-8-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeda D.Y., Wohlschlegel J.A., Dutta A. A bipartite substrate recognition motif for cyclin-dependent kinases. J. Biol. Chem. 2001;276:1993–1997. doi: 10.1074/jbc.M005719200. [DOI] [PubMed] [Google Scholar]
- 34.Chen J., Saha P., Kornbluth S., Dynlacht B.D., Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol. Cell. Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorn E.S., Chastain P.D. 2nd, Hall J.R., Cook J.G. Analysis of re-replication from deregulated origin licensing by DNA fiber spreading. Nucleic Acids Res. 2009;37:60–69. doi: 10.1093/nar/gkn912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machida Y.J., Hamlin J.L., Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 37.Saxena S., Dutta A. Geminin-Cdt1 balance is critical for genetic stability. Mutat. Res. 2005;569:111–121. doi: 10.1016/j.mrfmmm.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 38.Wyrick J.J., Aparicio J.G., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 39.Jallepalli P.V., Kelly T.J. Cyclin-dependent kinase and initiation at eukaryotic origins: a replication switch? Curr. Opin. Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- 40.Krude T., Jackman M., Pines J., Laskey R.A. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 41.Dahmann C., Diffley J.F., Nasmyth K.A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 42.Petersen B.O., Wagener C., Marinoni F., Kramer E.R., Melixetian M., Lazzerini Denchi E., Gieffers C., Matteucci C., Peters J.M., Helin K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 44.Aravind L., Koonin E.V. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.