Abstract
Poly(A)-binding protein (PABP) is a major component of the messenger RNA–protein complex. PABP is able to bind the poly(A) tail of mRNA, as well as translation initiation factor 4G and eukaryotic release factor 3a (eRF3a). PABP has been found to stimulate translation initiation and to inhibit nonsense-mediated mRNA decay. Using a reconstituted mammalian in vitro translation system, we show that PABP directly stimulates translation termination. PABP increases the efficiency of translation termination by recruitment of eRF3a and eRF1 to the ribosome. PABP's function in translation termination depends on its C-terminal domain and its interaction with the N-terminus of eRF3a. Interestingly, we discover that full-length eRF3a exerts a different mode of function compared to its truncated form eRF3c, which lacks the N-terminal domain. Pre-association of eRF3a, but not of eRF3c, with pre-termination complexes (preTCs) significantly increases the efficiency of peptidyl–tRNA hydrolysis by eRF1. This implicates new, additional interactions of full-length eRF3a with the ribosomal preTC. Based on our findings, we suggest that PABP enhances the productive binding of the eRF1–eRF3 complex to the ribosome, via interactions with the N-terminal domain of eRF3a which itself has an active role in translation termination.
INTRODUCTION
Poly(A)-binding protein (PABP) is one of the major mRNA-interacting proteins in eukaryotes. The protein is widespread and highly conserved among animals. Seven isoforms of PABP were identified in humans; the most abundant is the cytoplasmic isoform PABPC1 (1). The N-terminal domain of PABP contains four RNA recognition motifs (RRMs) (Figure 1A) each binding 12 adenines, while the whole protein covers 27 adenines (2,3). RRM1 and RRM2 are required for the specific recognition of poly(A) stretches, whereas RRM3 and RRM4 can associate with any RNA. RRM domains 1–4 bind the poly(A) tail from 3′ to 5′ (4). One of the main functions of the PABP is the protection of the poly(A) tail of cellular mRNAs from nuclease degradation (5). Besides, the protein can associate with other extensive poly(A) stretches like those occurring in certain 5′ untranslated regions (5′UTRs) that impact on translation initiation (6).
Figure 1.
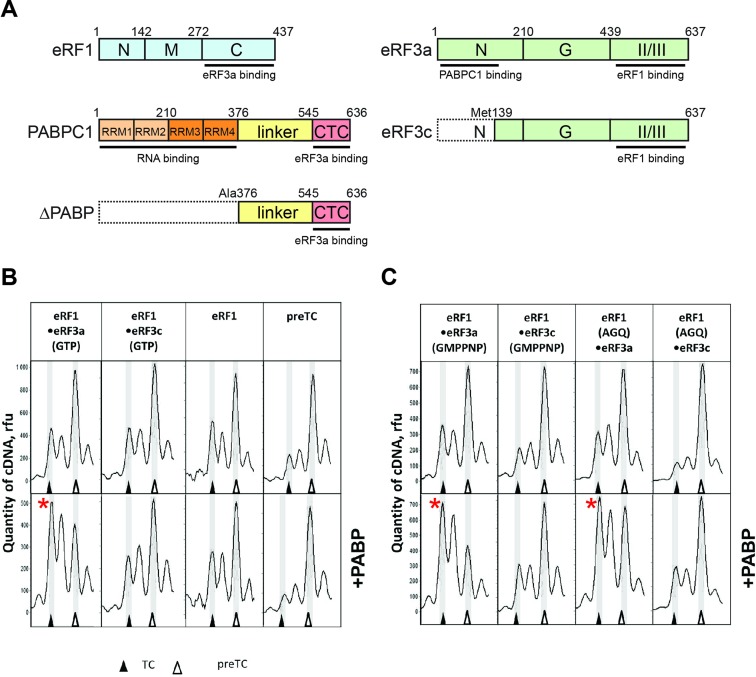
PABP increases the stop codon recognition by release factors. (A) A schematic representation of the release factors and PABP constructs used in this study. Domains involved in protein-protein interactions are indicated. Domains are color-coded and assigned functions and interaction partners are depicted at the corresponding position below the domain. Numbers above represent amino acid positions. (B and C) Toe-print analysis of termination complexes (TCs) formed by addition to the preTCs (B) of eRF1•eRF3a•GTP, eRF1•eRF3c•GTP, eRF1 and PABP; and of (C) eRF1•eRF3a•GMPPNP, eRF1•eRF3c•GMPPNP, eRF1(AGQ)•eRF3a•GTP, eRF1(AGQ)•eRF3с•GTP and PABP. Release factor complexes were associated before addition to the preTCs. Rfu—relative fluorescence unit. Positions of preTCs and TCs are labeled by white and black triangles respectively. Red stars mark the samples where stop codon recognition is enhanced.
RRM1 and RRM2 of PABP bind to the N domain of eukaryotic translation initiation factor (eIF) 4G (7). Thus, PABP's interactions with poly(A) and eIF4G together cause the formation of the 5′ cap-eIF4E-eIF4G-PABP–poly(A) complex where the 5′ and 3′ ends of mRNA approach each other to form the closed-loop structure (8). Proximity of the mRNA ends in the closed-loop structure is considered to facilitate the reinitiation of translation, since ribosomes are more easily engaged in the next round of initiation after termination (9). Also, the interaction between eIF4G and PABP is reported to increase the affinity of cap-binding factor eIF4E for the mRNA m7G cap (10,11). Thus, PABP can be regarded as a translation initiation-stimulating factor.
The C-terminal domain of PABP (CTC) is joined with the RNA-binding part of the protein by an unstructured proline-rich, ∼100 amino acid-long linker. The CTC binds proteins containing PAM2 motifs. Specifically, the PAM2 motif is found in the two main PABP regulators: polyadenylate-binding protein-interacting proteins (Paip) 1 and 2. Paip1 stimulates the activity of PABP in translation initiation (12). Paip1 comprises PAM1 and PAM2 motifs which interact with the N and C-terminal domains of PABP respectively. Moreover, Paip1 interacts with eIF3g and eIF4A (13) to form Paip1-eIF3-eIF4G and Paip1-PABP-eIF4G complexes, which increase the stimulatory effect of PABP in translation initiation. In contrast, Paip2 is a repressor of translation initiation (14). Paip2 also contains PAM1 and PAM2 motifs, but complex formation decreases PABP's affinity to the poly(A) tail and to eIF4G, leading to a disruption of the closed-loop structure (15).
Similar to Paip1/2, the eukaryotic release factor 3 (eRF3) contains a PAM2 motif that recognizes the CTC domain of PABP (16). eRF3 is one of two factors required for translation termination (17,18). eRF3 comprises an unstructured N-terminal domain, a G domain which binds nucleotides, as well as II and III domains which interact with the second essential termination factor eRF1 (Figure 1A) (19). The PAM2 motif of human eRF3 is composed of two mini-domains, PAM2.1 and PAM2.2. These mini-domains are highly conserved among higher eukaryotes, while the remaining sequence of the N-terminal domain is highly variable (20). The PAM2.2 mini-domain of human eRF3 has a higher affinity for PABP when compared with the PAM2.1 mini-domain (21).
The II and III domains of eRF3 interact with the C-terminal domain of eRF1, leading to a conformational change in eRF1 (22). When eRF1 recognizes a stop codon (UAG, UAA, UGA) (23), which is the first step of translation termination, eRF3 hydrolyzes guanosine-5′-triphosphate (GTP). This results in a conformational rearrangement of eRF1 (18): The M domain of eRF1 enters the A-site of the large ribosomal subunit and reaches into the peptidyl-transferase center (PTC). The second step of translation termination, which is peptidyl–tRNA hydrolysis, is thereby triggered (24). Two human isoforms of eRF3 exist: eRF3a and eRF3b, encoded by different genes (GSPT1 and GSPT2) (25,26). The main isoform is most likely eRF3a which is ubiquitously expressed; in contrast, eRF3b expression is tissue-specific (27). eRF3a and eRF3b differ with respect to their N domains, but both proteins have conserved PAM2.1 and PAM2.2 motifs and thus are able to bind PABP.
PABP has been suggested to have a stimulatory effect on translation termination and was shown to interfere with nonsense-mediated mRNA decay (NMD) (28). Most likely, PABP exerts its function in a position-dependent manner (29,30). Normal stop codons are usually positioned in the last exon of the mRNA, followed by a relatively short 3′ untranslated region (3′ UTR). In contrast, in the case of a premature stop codon which elicits NMD a long 3′UTR and/or an exon junction complex (EJC) is present. This leads to a larger distance between the terminating ribosome and the PABP bound to the poly(A) tail. In the latter case, NMD factors can interact with the terminating ribosome and initiate the assembly of the mRNA decay-inducing complex. PABP was tethered to the mRNA such that it was positioned closer to the stop codon and upstream of an EJC (29,30). It was shown that this led to suppression of NMD (30,31). Evidence for a stimulatory effect of PABP on translation termination is indirect: It is based on in vivo experiments where it was found that stop codon readthrough is increased when PABP is knocked down (29).
The effect of the PABP analog in yeast (Pab1) on translation termination is controversial: On the basis of indirect data Pab1 is thought to decrease termination (32). However, overexpression of Pab1 in yeast strains is suggested to activate termination of translation (33).
The molecular mechanism of PABP's function in termination is enigmatic. Here, we characterize the impact of cytoplasmic human PABP (PABPC1, referred to as PABP here) on translation termination using an in vitro reconstituted mammalian translation system. We show that PABP directly stimulates stop codon recognition in vitro and that this function is independent of its RNA-binding activity. The termination stimulation effect is most likely caused by the optimal positioning of eRF3a on the ribosome, increasing the efficiency of eRF1 to recognize stop codons and to catalyze peptidyl–tRNA hydrolysis in the PTC.
MATERIALS AND METHODS
Ribosomal subunits and translation factors
The 40S and 60S ribosomal subunits, as well as eukaryotic translation factors eIF2, eIF3, eIF4F, eEF1H and eEF2, were purified from rabbit reticulocyte lysate as described (18). The human translation factors eIF1, eIF1A, eIF4A, eIF4B, eIF5B, eIF5, PABP, ΔPABP lacking RRM1—RRM4 motifs (first 375 amino acids residues), eRF1, eRF1(AGQ) and eRF3c lacking the N-terminal domain (138 amino acid residues including PAM2) were produced as recombinant proteins in Escherichia coli strain BL21 and subsequently purified via Ni-NTA agarose and ion-exchange chromatography (18).
Expression and purification of human eRF3a
Human full-length eRF3a (GSPT1) was cloned into the pFastBac-Htb vector (Life Technolgies) and expressed in insect cells Sf21 using the EMBacY baculovirus from the MultiBac expression system (34). Cells were lysed by sonication in 20 mM Tris–HCl pH 7.5, 100 mM KCl, 6 mM β-mercapthoethanol, 0.1% Tween-20 and 5% glycerol supplemented with protease inhibitors. eRF3a was purified by affinity chromatography using a HisTrap HP column (GE Healthcare) followed by anion-exchange chromatography using a MonoQ column (GE Healthcare). In the final size-exclusion chromatography step (Superdex-200 column, GE Healthcare) using 20 mM Tris–HCl pH 7.5, 100 mM KCl, 6 mM beta-mercapthoethanol, 0.1% Tween-20 and 5% glycerol, the protein elutes as a monomer.
In vitro transcription of mRNA
The mRNA was transcribed in vitro using T7 RNA polymerase. The MVHL-stop plasmid contains a T7 promoter, four CAA repeats, the β-globin 5′-UTR, open reading frame (encoding for the peptide MVHL), followed by the UAA stop codon with the various next base (U,A,G,C) and a 3′-UTR comprising the rest of the natural β-globin coding sequence (35). For run-off transcription the MVHL-stop plasmid was linearized by restriction digest with XhoI. The MVHC–polyA plasmid contains a T7 promoter, four CAA repeats, an MVHC open reading frame followed by an UAA stop codon, the complete human β-globin 3′-UTR and polyA tail (70 nucleotides). For run-off transcription mRNA plasmids were linearized with EcoRI.
Pre-termination complex assembly and termination analysis
Pre-termination complexes (preTCs) were assembled in vitro as described (36) and used in peptide release assays and conformational rearrangement analyses by toe-print assays (37). For peptide release assays aliquots containing 0.2 pmol of the preTCs were incubated at 37°C for 3 min with 0.6 pmol of eRF1/3 and 5 pmol of PABP. For conformational rearrangement analyses aliquots containing 0.2 pmol of the preTCs were incubated at 37°C for 15 min with 0.6 pmol of eRF1/3 and 5 pmol of PABP or 7 pmol of ΔPABP. In case of preTC assembly in the presence of PABP—18 pmol MVHC–polyA and/or 18 pmol MVHL mRNA were used and incubated with 80 pmol PABP at 37°C for 2 min before preTC formation. For toe-print assay of MVHL and MVHC–polyA mRNAs, 5′-FAM labeled toe-primers 1 (5′-GCATGTGCAGAGGACAGG-3′) and 2 (5′-GCAATGAAAATAAATTCC-3′) were used respectively.
preTC binding assay
Purified preTCs (160 μl) were incubated with 3.5 pmol eRF1(AGQ), 10 pmol eRF3a or eRF3c, 200 pmol PABP in buffer with 0.2 mM GTP and with 0.2 mM MgCl2 at 37°C for 10 min (37). Pre-incubation of PABP with eRF3 was performed to exclude a preliminary binding of eRF3 with eRF1, which can decrease the termination activity of eRFs (Figure 3). The reaction volume was 500 μl. Subsequently, TCs were incubated with 1% formaldehyde at 4°C for 1 h (38). Glycine was added up to 0.1 M to stop the cross-linking reaction. The TCs were purified in a 10–30% (w/w) linear sucrose density gradient (SDG) as described above. The gradients were fractionated into 14 equal fractions followed by precipitation in 10% trichloroacetic acid (TCA). The protein pellets were dried and analyzed by western blot.
Figure 3.
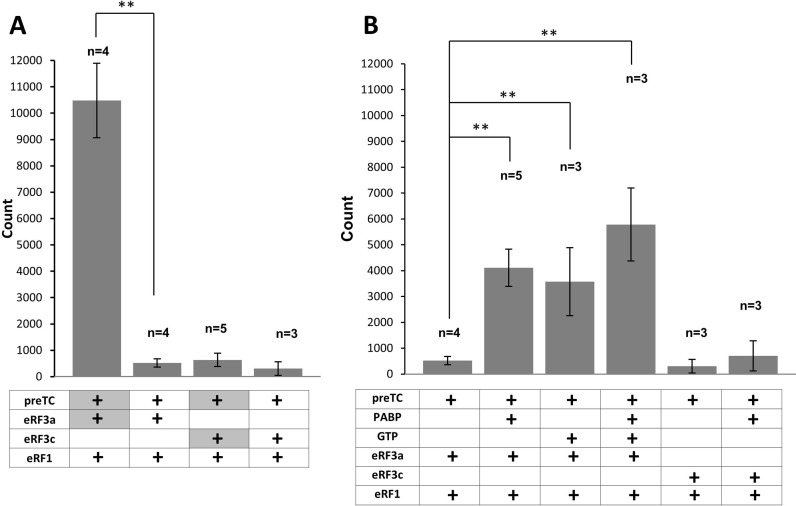
PABP increases the efficiency of peptidyl–tRNA hydrolysis in the presence of release factors. (А) Hydrolysis of peptidyl–tRNA induced by the addition of eRF1 and eRF3 or their preformed complexes. GTP was added first to the reaction. (В) Hydrolysis of peptidyl–tRNA induced by the eRF1•eRF3a or eRF1•eRF3с complexes in the presence/absence of PABP. In experiments 1, 2, 5 and 6, GTP was added to preTCs before the eRFs. In most cases proteins added to the preTCs were pre-associated. The only exceptions are the experiments in panel 3A (lines 1 and 3), where eRF3a/c were pre-incubated with the preTC first (highlighted in gray). n corresponds the number of measurement repeats. The stars (**) mark a significant difference from the respective control P < 0,01.
RESULTS
PABP stimulates stop codon recognition activity of release factors
PABP and eRF3a were shown to interact directly (16). Therefore, we decided to determine how PABP affects the activity of eRF3a and eRF1 in translation termination. In the presence of PABP, both release factors were added to preTCs which were assembled in vitro from individual components on the MVHL mRNA and purified by SDG centrifugation. We then performed toe-print analyses of the ribosomal complexes in order to assess stop codon recognition and termination complex (TC) formation. In Supplementary Figure S1, we show examples of raw data of the toe-print analyses, obtained by capillary electrophoresis of cDNA products generated with fluorescently labeled primers. During stop codon recognition of eRF1, the ribosome protects additional nucleotides on the mRNA, which can be detected in toe-printing assays as a two-nucleotide shift of the ribosomal complex (18,39,40).
Our preTCs contain a UAA stop codon in the ribosomal A-site. Addition of release factors leads to the appearance of a peak, corresponding to the TC. For our experiments, we applied limiting concentrations of release factors (0.6 pmol of eRF1 and eRF3a/c) such that the 2-nt shift of the ribosomal complex was rather inefficient. This allowed us to detect any enhancement of release factor activity in the presence of PABP.
Addition of 5 pmol PABP, release factors and GTP to the preTCs significantly increases the 2-nt shift, i.e. PABP stimulates stop codon recognition (Figure 1B). This stimulatory effect is specific, as it is observed only after the addition of full-length eRF3 (eRF3a) to the reaction: the N-terminally truncated version, eRF3c, which is unable to bind PABP does not show any stimulation of stop codon recognition. Moreover, the stimulatory effect of PABP is not observed in the absence of release factors, or in the presence of eRF1 only (Figure 1B). Addition of PABP to the preTCs in the presence of higher amounts of eRFs (5 pmol of each, corresponding to a 25-fold molar excess of eRFs over preTCs) also stimulates stop codon recognition, but the effect is rather weak (Supplementary Figure S1B).
Our model MVHL mRNA contains an uracil nucleotide (U) in the +4 position after the stop codon. Recently, it was shown that eRF1 binding to a stop codon in the decoding site leads to a conformational change in the 18S rRNA which pulls the +4 nt into the decoding site (39,40). This compaction due to mRNA U-turn motif formation is the basis of stop codon recognition by eRF1. It pulls downstream mRNA further into the mRNA channel and thus provides an explanation for the toe-printing peak shift, which is observed upon TC formation (Figure 1B) (39,40). We tested the effect of the stop codon context on stimulation of stop codon recognition by PABP: we generated three additional model mRNAs with the various nucleotides (adenine, guanine and cytosine) in the fourth position. We assembled preTCs on these mRNAs and used them for toe-printing assays in the presence and absence of PABP (Supplementary Figure S2). We found that in all possible contexts of the UAA stop codon, PABP equally stimulates stop codon recognition.
Notably, the same stimulatory effect of PABP on stop codon recognition is observed when a non-hydrolysable analog of GTP (GMPPNP) is added to the reaction (Figure 1C). Similarly, PABP stimulates stop codon recognition in the presence of eRF3a and an AGQ mutant of eRF1 (eRF1(AGQ)), which is unable to hydrolyze peptidyl–tRNA (Figure 1С). We conclude that PABP activates TC formation independently of GTP or peptidyl–tRNA hydrolysis. This suggests that the stimulatory effect of PABP on translation termination occurs when the release factors bind to the ribosome and recognize the stop codon.
PABP stimulates stop codon recognition as a cis- and trans-acting factor
We demonstrate in Figure 1 that PABP activates stop codon recognition on the model mRNA lacking a poly(A) tail (MVHL). However, within cells, most molecules of PABP are bound to the poly(A) tail of mRNAs. Therefore, we decided to investigate whether the poly(A) binding of PABP affects its stimulation of translation termination. Therefore, we used MVHC–polyA mRNA which contains the same leader sequence as MVHL mRNA (but a different coding sequence (MVHC)), as well as an UAA stop codon followed by the β-globin 3′UTR and by the poly(A) tail. Different 3′UTRs were used to distinguish cDNA products produced by toe-printing primers 1 and 2 in the mixture of mRNAs (see below).
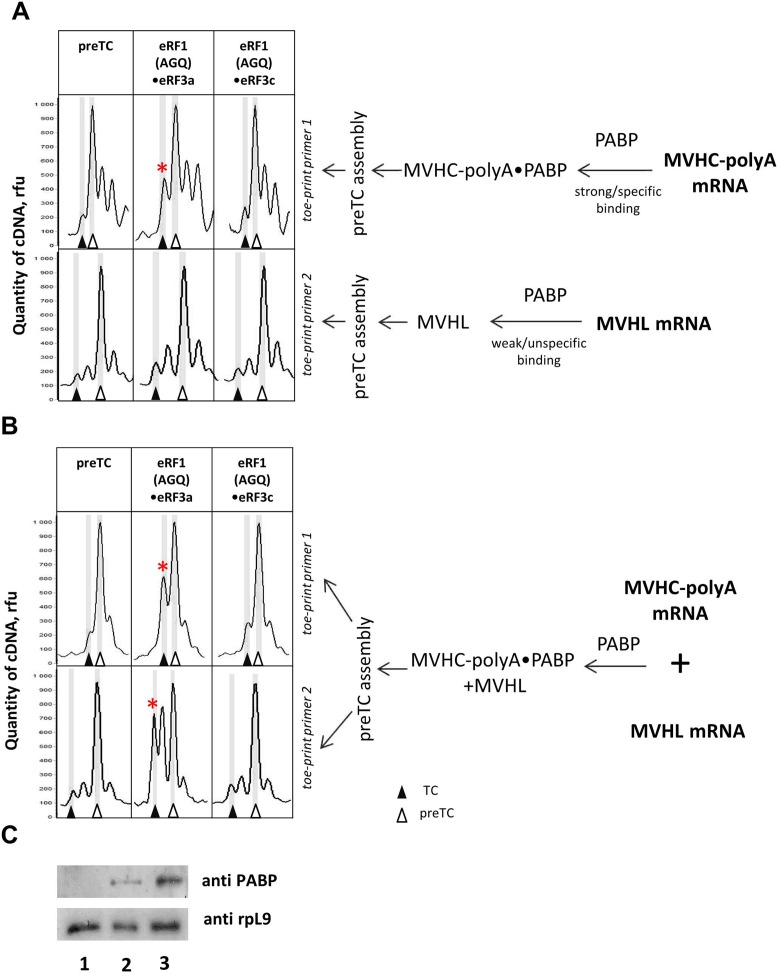
The MVHC–polyA mRNA was first incubated with PABP and then used for preTC assembly (Figure 2A). Subsequently, the preTCs were purified by SDG centrifugation, subjected to termination reactions and tested in toe-printing assays. We speculate that the different 3′UTR sequence in the MVHC–polyA mRNA leads to a different mobility of the corresponding cDNAs, and TC formation is detected as +1 nt toe-print shift for the MVHC–polyA construct (Figure 2 A, B and Supplementary Figure S3). The addition of release factors to preTCs, assembled on MVHC–polyA mRNA and incubated with PABP, results in a more efficient stop codon recognition of the eRF1•eRF3a complex compared to the eRF1•eRF3c complex (Figure 2A). In the absence of PABP, the activities of eRF3a and eRF3c in stop codon recognition are very low and almost equal using the MVHC–polyA construct and limiting amounts of eRFs (Supplementary Figure S3). As a control, we assembled preTCs on MVHL mRNA in the presence of PABP and found almost no differences in stop codon recognition in the presence of eRF3a versus eRF3c (Figure 2A). Moreover, the efficiency of stop codon recognition was similar, irrespective of incubation of MVHL mRNA with PABP (Supplementary Figure S3 and Figure 2A). Western blot analysis of the SDG-purified preTCs shows that PABP binds only very weakly to preTCs assembled on MVHL mRNA and more efficiently binds to the PreTCs assembled on MVHC–poly(A) mRNA (Figure 2C, compare lanes 2 and 3). Thus, the amount of PABP unspecifically bound to MVHL mRNA is apparently not sufficient to detectably activate translation termination (Figure 2A). Taken together, these experiments show that PABP remained bound to the MVHC–poly(A) mRNA during SDG (Figure 2C) and stimulated stop codon recognition in cis by interaction with the N-terminal domain of eRF3a.
Figure 2.
PABP increases the stop codon recognition by release factors in cis and trans. Toe-print analysis of TCs formed by addition to the preTCs of eRF1(AGQ)•eRF3a•GTP or eRF1(AGQ)•eRF3с•GTP. (A) MVHC–polyA or MVHL mRNAs were incubated with PABP separately and used for reconstitution of preTCs. (B) The mixture of MVHC–polyA and MVHL mRNAs was incubated with PABP and used for reconstitution of preTCs. Release factor complexes were associated before addition to preTCs. Rfu—relative fluorescence unit. Positions of preTC and TC are labeled by white and black triangles respectively. (C) Western blot analysis of fractions after SDG of preTCs assembled on the mixture of MVHL and MVHC-polyA (lane 1), MVHL (lane 2) or MVHC–polyA (lane 3) mRNAs in the absence (lane 1) or presence (lanes 2 and 3) of PABP. Stars mark the samples where stop codon recognition is enhanced.
To test whether PABP can also act in trans, MVHC–poly(A) mRNA was mixed with equal amounts of MVHL mRNA and incubated with PABP. After preTC assembly and SDG purification, translation termination reactions were performed followed by toe-printing assays (Figure 2B). The toe-printing results show that stop codon recognition is enhanced for both preTCs. Thus, PABP bound to the poly(A) tail of MVHC–polyA mRNA is able to stimulate termination on the own mRNA (i.e. in cis) and on preTCs with MVHL mRNA (e.g. in trans) (Figure 2B). The observed stimulation is specific because it depends on the presence the N-terminal domain of eRF3a and accordingly is not observed for eRF3c.
PABP increases peptidyl–tRNA hydrolysis by the release factors
To test whether PABP also affects the peptidyl–tRNA hydrolysis reaction we assembled preTCs on the MVHL mRNA using S35-labeled initiator-tRNA. The efficiency of peptidyl–tRNA hydrolysis was determined by quantification of the radioactive MVHL peptide released from the ribosomal complexes. We observed that the efficiency of peptide release depends on the order of addition of release factors to the preTCs. Incubation of eRF3a with the preTCs and GTP, followed by addition of eRF1, causes effective termination. In contrast, addition of the pre-associated complex of eRF1 and eRF3a to the preTCs decreases termination efficiency by a factor of ∼20 (Figure 3A compare lanes 1 and 2). In the same experiment using eRF3c, the efficiency of termination does not depend on the order of factor addition and the peptide release measured after pre-incubation with eRF3c with preTCs, followed by eRF1 addition was virtually identical to the addition of the pre-associated eRF1•eRF3c complex or eRF1•eRF3a complex to preTCs (Figure 3A compare lanes 2,3,4). Thus, the observed high efficiency of peptide release requires the N-terminal part of eRF3a.
Pre-association of PABP with the eRF1•eRF3a complex stimulates peptidyl–tRNA hydrolysis by approximately 8-fold (Figure 3B, lanes 1 and 2). In contrast, incubation of PABP with the eRF1•eRF3c complex does not significantly affect the efficiency of peptide release (Figure 3B, lanes 5 and 6). We found that the optimal condition for translation termination is the addition of eRF1 after the pre-incubation of eRF3a with the preTCs. Under these conditions, PABP does not change the efficiency of peptide release (Supplementary Figure S4B). However, when we use limiting concentrations of eRFs (three times lower) and the same order for the addition of release factors, the level of peptide release diminishes significantly. Under these sub-optimal conditions, PABP exerts a stimulating effect on termination and increases the efficiency of peptidyl–tRNA hydrolysis ∼2-fold (Supplementary Figure S4B). In summary, PABP has a stimulatory effect when the termination efficiency is reduced, for instance due to limiting eRF concentrations or due to the formation of eRF1•eRF3a complexes which seem to be less active in termination.
Notably, pre-association of GTP with eRF1•eRF3a increases the efficiency of peptide release (Figure 3B, lane 3). Formation of eRF1•eRF3a•GTP•PABP complexes also results in a pronounced, 11-fold stimulatory effect on translation termination indicating a moderate additional stimulatory effect compared to eRF1•eRF3a•GTP complexes (7-fold) or eRF1•eRF3a•PABP complexes (8-fold) (Figure 3B, compare lane 4 to lanes 2 and 3).
The C-terminal domain of PABP is essential for improved efficiency of stop codon recognition
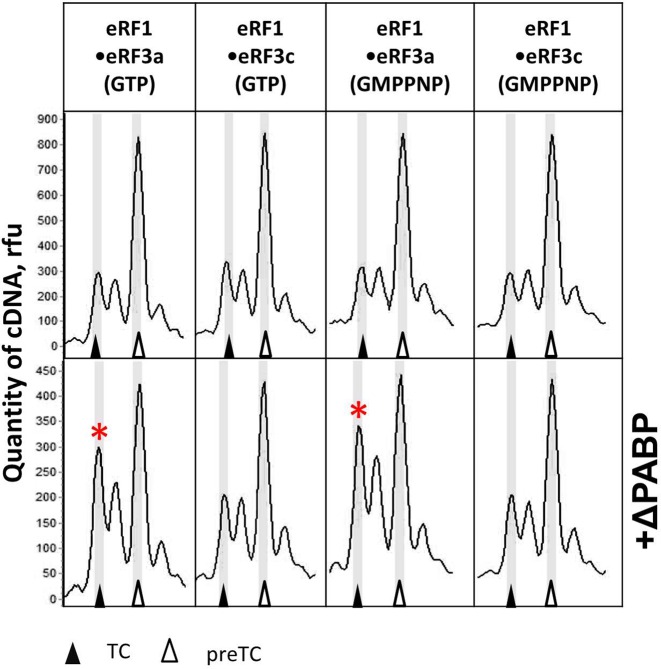
We generated an N-terminally truncated PABP protein (ΔPABP) lacking the four RRMs which are necessary to bind mRNA (Figure 1A). The deletion of the RNA-binding motifs of PABP allows assessing whether the RRMs are required for stop codon recognition. ΔPABP comprises the unstructured proline-rich linker and the CTC domain which is able to interact with PAM2 motif of eRF3a (41). To determine an effect of ΔPABP on stop codon recognition, we performed toe-printing assays with reconstituted preTCs on MVHL mRNA. We observed that ΔPABP also promoted stop codon recognition upon addition to eRF1 and eRF3a, although less efficiently than the full-length PABP, requiring high ΔPABP concentrations to achieve a visible effect (compare Figures 1B and 4). The stimulatory effect on stop codon binding can be reproduced in the presence of GMPPNP, which was added instead of GTP (Figure 4). These experiments indicate that the C-terminal part of PABP is required for stimulation of stop codon recognition.
Figure 4.
The C-terminal part of PABP (ΔPABP) stimulates stop codon recognition by release factors. Тое-print analysis of TCs formed in the presence of eRF1•eRF3a•GTP, eRF1•eRF3c•GTP, eRF1•eRF3a•GMPPNP, eRF1•eRF3c•GMPPNP in the absence (above) and presence (below) of ΔPABP. Release factor complexes were associated before addition to the preTCs. Rfu—relative fluorescence unit. Positions of preTCs and TCs are labeled by white and black triangles respectively. Red stars mark the samples where stop codon recognition is enhanced.
PABP promotes binding of eRF3a to preTCs
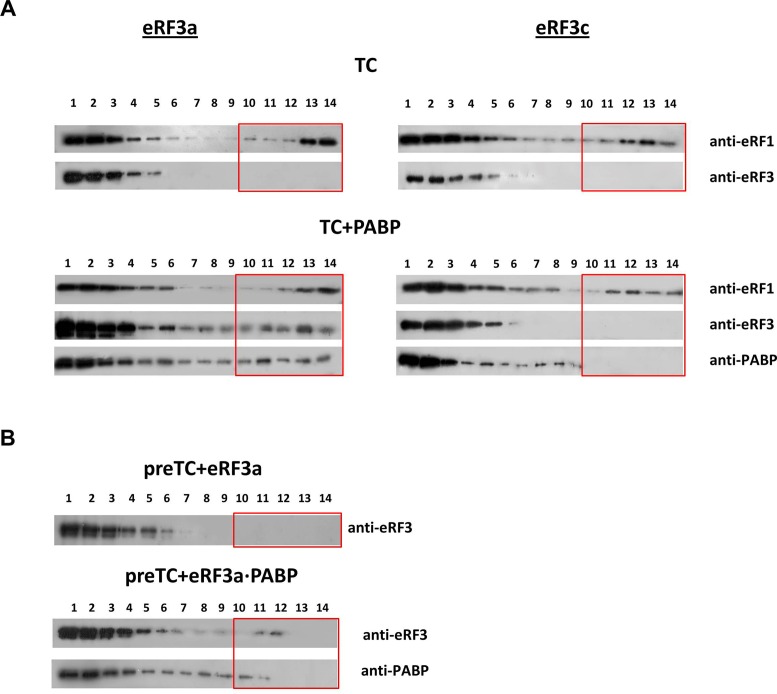
In order to study how PABP affects the interaction of release factors with ribosomes and improves stop codon recognition we tested the ability of this protein to bind preTCs in the presence or absence of eRFs (Figure 5). PreTCs were incubated with PABP, eRF1(AGQ), eRF3a or eRF3c, and GTP. Subsequently, the complexes were centrifuged into an SDG. After fractionation, the proteins were detected by western blotting. The eRF1(AGQ) mutant was used to stabilize TCs, since this mutant is able to bind the stop codon, but is inactive in peptide release. Therefore, it can stabilize the TCs.
Figure 5.
eRF3a and PABP bind preTCs cooperatively. (A) Western blot analysis of TC fractions after SDG of preTCs incubated with GTP, eRF1(AGQ) (upper panel) and eRF3a (left) or eRF3c (right) in presence of PABP (lower panel). Antibodies raised against eRF1, eRF3 and PABP were used for detection. (B) Western blot analysis of preTCs incubated with eRF3a + GTP (above) and eRF3a•PABP + GTP (below) using antibodies against eRF3 and PABP. The fractions of the SDG are indicated above the Western blots; fraction 1 corresponds to the top of the gradient, 14 to the bottom. Boxes indicate the fractions that contain ribosomal complexes.
PreTCs migrate in fractions 10–14 (Supplementary Figure S5A). To exclude aggregation of proteins in solution, we determined their distribution in the SDG in the absence of ribosomes (Supplementary Figure S5B). All proteins (eRF1, eRF3a, eRF3c and PABP and their complexes) are detected only in the top fractions of the gradient (fractions 1–5). PABP can interact with preTCs. However, the complex is likely low affinity and dissociates during SDG. Accordingly, PABP is found not only in fraction 1–3 (Supplementary Figure S5B), but also in fractions 4–9 in the presence of preTCs (Supplementary Figure S5C).
In the absence of PABP, both eRF3a and eRF3c do not form stable complexes with the preTCs, irrespective of the presence (Figure 5A) or absence of eRF1 (Figure 5B). The presence of PABP in the binding reactions stabilizes the binding of eRF3a to preTCs (Figure 5). In contrast, eRF3c is still not found in the preTC fractions despite the addition of PABP. This was expected because PABP does not interact with either eRF3c or eRF1. It is important to note that the interaction of eRF1(AGQ) with the preTCs is independent from the addition of PABP and from the eRF3 variant (full-length versus truncated).
We conclude that PABP stabilizes the binding of eRF3a to ribosomal complexes. This finding is in agreement with the stimulating effect of PABP on stop codon recognition and peptidyl–tRNA hydrolysis induced by the release factors (Figures 1 and 4).
DISCUSSION
PABP is an important player in eukaryotic translation and its control (42). The role of PABP in stimulation of translation initiation is well-established (10). However, the functions of PABP in translation are not limited to regulation of initiation. Several studies suggest that PABP interferes with NMD and that PABP deletion increases read-through of stop codons, thereby indirectly providing evidence for a role of PABP in stimulation of termination (29–31). Here we show, that PABP directly stimulates translation termination in vitro, confirming PABP's role as a regulatory factor in translation termination.
Using a reconstituted in vitro translation system we show that PABP stimulates the stop codon recognition activity of eRF1 in the presence of full-length eRF3a (Figure 1). Apparently, this PABP activity is independent of GTP hydrolysis by eRF3a. It can be observed in the presence of a non-hydrolysable GTP analog (GMPPNP) and independent of peptidyl–tRNA hydrolysis, which is inhibited by GMPPNP and by the eRF1(AGQ) mutant (Figure 1B). Moreover, it is independent of the stop codon context (Supplementary Figure S2). In addition, PABP increases the efficiency of peptidyl–tRNA hydrolysis under non-optimal termination conditions, e.g. the formation of eRF1•eRF3а complexes (Figure 3B) or in the presence of limiting amounts of release factors (Supplementary Figure S4B). We show that PABP does not interact with eRF1 directly, but exerts its effect via eRF3a (Figure 1B). Taken together, our experiments show that PABP increases the efficiency of stop codon recognition leading to enhanced formation of TCs. PABP interacts with the N-terminus of eRF3a and the GTPase activity of eRF3a is not required for stimulation. Therefore, we suggest that the observed increase in efficiency of peptide release is a consequence of more efficient TC formation. Importantly, the observed stimulatory effect does not depend on the RNA-binding motifs of PABP (Figure 4). The C-terminal part of PABP comprising the proline-rich linker and the CTC, is sufficient to elicit increased TC formation in toe-print assays. However, it should be noted that RRM motifs of PABP can participate in the stimulation of termination by binding to the poly(A) tail and thereby positioning PABP closer to the preTCs. Indeed, we demonstrated that the cis-action of PABP in the presence of a mRNA with a poly(A) tail requires significantly less PABP and thus is more efficient compared to the trans-reaction (compare Figures 1 and 2).
We observe that PABP stabilizes the binding of eRF3a to the ribosome (Figure 5). eRF3a is not associated with the TCs after sucrose gradient centrifugation, most likely it dissociates from the complexes in the SDG. In the presence of PABP, eRF3a can be detected in the ribosome-containing fractions indicating that the binding of eRF3a to ribosomes is stabilized by PABP. Previous studies showed that PABP specifically binds ribosomes in vivo (43–45). For yeast PABP (Pab1), a specific interaction was shown with the 60S subunit via the C-terminal part of Pab1 comprising the linker region and CTC domain (43). Moreover, it has been suggested that Pab1 interacts with the ribosomal protein rpL39, which is located near the exit of the ribosomal tunnel (44). It should be noted that the linker sequences of the yeast and human PABP are very different, therefore this interaction might not be conserved from yeast to human. Furthermore, rpL39 is located very distant from the eRF3-binding site on the ribosome (46). We assume that the relevant ribosomal binding site for human PABP is not rpL39, but close to the ribosomal A site.
Fundamental differences may exist concerning the role of human PABP and yeast Pab1 in translation termination. However, the exact role of Pab1 in termination is enigmatic: in dual luciferase assays increased stop codon read-through is observed in the presence of Pab1. This is likely due to interaction of Pab1 and eRF3 leading to decreased termination efficiency (32). Based on this, it was suggested that Pab1 is required for maintaining a basic level of read-through in yeast. In contrast, an earlier study showed that overexpression of Pab1 in yeast strains with a mutant eRF3 causes an anti-suppression effect indicating that the interaction of eRF3 and Pab1 stimulates translation termination (33). Importantly, yeast eRF3 interacts via the N and M domains with the linker and C domain of Pab1 (32). This indicates that the underlying protein–protein interactions and molecular mechanisms differ between yeast and human.
Interestingly, we observe that the N domain of eRF3a significantly enhances translation termination, even in the absence of PABP. Pre-association of full-length eRF3a with the preTCs dramatically increases the efficiency of translation termination compared to the N-terminally truncated variant (eRF3c) (Figure 3). The latter was used in most previous termination studies (see below). Pre-association of eRF1•eRF3a complexes in solution abolishes the stimulating effect of the eRF3a N domain on termination (Figure 3A). However, addition of PABP or GTP to these complexes partly resumes the activity of full-length eRF3a (Figure 3B). It should be noted that the N-terminal parts of eRF3a and eRF3b vary widely between different species but the PABP-binding motif (PAM2) is conserved (21).
Previous in vitro characterizations of release factor activity and all available structures of TCs used truncated eRF3c which is unable to bind PABP, but is active in termination. Until now, the activity of eRF3c was considered to be identical to the one of full-length eRF3a and eRF3b, and thus the N-terminal part of eRF3 was assumed to be dispensable for termination due to its sequence variability (47). This hypothesis is based on a thermodynamic study of eRF1, eRF3a and nucleotide interactions that showed no effect of the N domain of eRF3a on the eRF1–eRF3 association constant in the absence of the ribosome (21). Importantly, the authors noted a change in entropy and enthalpy of the eRF1–eRF3 interaction depending on the presence of the N domain of eRF3. This was interpreted as an enthalpy–entropy compensation, meaning that interactions of eRF1 with either eRF3c or eRF3a use different pathways, but reach the same final state. Notably, that this study was performed in the absence of the ribosome and the association constant of the full-length eRF3a with the ribosome was not determined. Our data indicates that the N domain of eRF3a plays an important role in peptidyl–tRNA hydrolysis and that it is essential for the stimulation of termination by PABP.
We observe that the association of eRF1 with eRF3a in solution prevents efficient binding of the release factors to the preTCs (Figure 3A). One possible explanation could be an unspecific interaction of eRF1 with the N domain of eRF3a. Based on structural data, the PAM2 motif is considered unstructured (41). eRF1–eRF3a interactions may lead to an unfavorable conformation for ribosome binding.
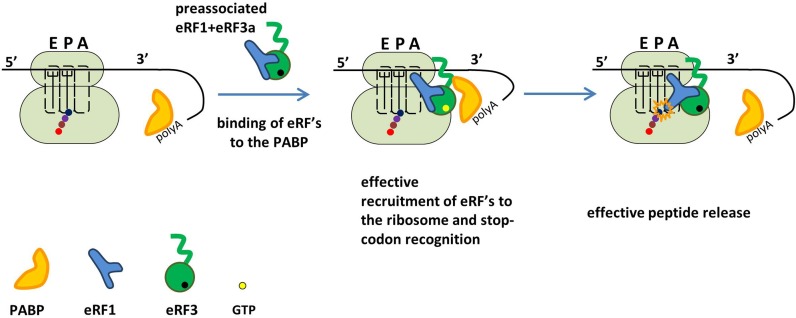
Our data, showing a role of the N domain of eRF3a in translation termination, changes the current concept of termination in higher eukaryotes. Based on our findings, we propose a model for the interaction of release factors with the preTCs and the impact of PABP (Figure 6). We suggest that PABP can specifically bind to the N domain of eRF3a and thereby recruits eRF3a to the preTCs. Possibly, PABP promotes the correct orientation of eRF3a and eRF1 on the ribosome, allowing efficient stop codon recognition. To exert this effect, PABP needs to interact with the ribosome near the factor binding site. Otherwise, PABP could not stimulate termination and would reduce the pool of free eRF3a which would interfere with termination. Efficient peptidyl–tRNA hydrolysis in the presence of PABP may be a consequence of the increased stop codon recognition.
Figure 6.
Model for PABP-stimulated translation termination. In the presence of PABP, eRF3a binds efficiently to preTCs and optimally positions eRF1 in the ribosomal complex for stop codon recognition. Accordingly, the termination efficiency is high as indicated by more efficient peptide release. When PABP is pre-bound to the mRNA, it recruits eRF1/3a to the preTCs, thus increasing the local eRF1/3a concentration and stabilizing the eRF binding to the ribosome.
In vivo, a ribosome encountering a normal stop-codon is assumed to be in close vicinity to the poly(A) tail which is bound by PABP (Figure 6). Here, we show that in vitro PABP triggers eRF3a binding to ribosomes via its C-terminal domain and thus helps to recruit eRF3a and eRF1. We demonstrate that through the interaction of PABP with the N-terminal domain of eRF3a, the stop codon recognition and peptidyl–tRNA hydrolysis is stimulated under limiting concentrations of eRFs which is likely to be the case in vivo. Moreover in vivo, PABP is suggested to compete with NMD factor UPF1 for eRF binding thus interfering with NMD. In cases, where the 3′UTR is very long, or when an EJC is bound in the 3′UTR, UPF1 may outcompete PABP for preTC binding and trigger mRNA decay (29–31).
In summary, we show here that the N-terminus of eRF3a is important for translation termination in the presence and absence of PABP. The C-terminal part of PABP is sufficient for stimulation of stop codon recognition. PABP enhances eRF3a ribosome binding, stop codon recognition and peptidyl–tRNA hydrolysis. The increased efficiency of TC formation is most likely achieved by optimal positioning of eRF3a on the ribosome.
Supplementary Material
Acknowledgments
We are grateful to Nahum Sonnenberg for plasmid pET3b-PABPC1, to Ludmila Frolova for providing plasmids encoding release factors and to Tatyana Pestova and Christopher Hellen who provided us with plasmids encoding initiation factors. Sequencing of plasmids, coding truncated PABP and cDNA fragment analyses were performed by the center of the collective use ‘Genome’ of EIMB RAS. We would like to thank Karine Huard, Frederic Garzoni and Etienne Raimondeau for technical assistance and discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The investigation of the activity of PABP in translation termination: RFBR [15-04-08174]; Program of fundamental research for state academies for 2013-2020 years (Subprogram No. 58 Molecular genetics, mechanisms of realization of genetic information, bioengineering); European Research Council Starting grant [ComplexNMD, 281331 to C.S.]. The study of the effect of the eRF3a N domain on the efficiency of translation termination: Russian Science Foundation [14-14-00487]. Funding for open access charge: European Research Council Starting grant [ComplexNMD, 281331].
Conflict of interest statement. None declared.
REFERENCES
- 1.Eliseeva I.A., Lyabin D.N., Ovchinnikov L.P. Poly(A)-binding proteins: structure, domain organization, and activity regulation. Biochemistry (Mosc.) 2013;78:1377–1391. doi: 10.1134/S0006297913130014. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn U., Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 3.Baer B.W., Kornberg R.D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deo R.C., Bonanno J.B., Sonenberg N., Burley S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein P., Peltz S.W., Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J., Bag J. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J. Biol. Chem. 1998;273:34535–34542. doi: 10.1074/jbc.273.51.34535. [DOI] [PubMed] [Google Scholar]
- 7.Imataka H., Gradi A., Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 9.Rajkowitsch L., Vilela C., Berthelot K., Ramirez C.V., McCarthy J.E. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 2004;335:71–85. doi: 10.1016/j.jmb.2003.10.049. [DOI] [PubMed] [Google Scholar]
- 10.Kahvejian A., Svitkin Y.V., Sukarieh R., M'Boutchou M.N., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haghighat A., Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 12.Craig A.W., Haghighat A., Yu A.T., Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 13.Martineau Y., Derry M.C., Wang X., Yanagiya A., Berlanga J.J., Shyu A.B., Imataka H., Gehring K., Sonenberg N. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol. Cell. Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khaleghpour K., Svitkin Y.V., Craig A.W., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 15.Karim M.M., Svitkin Y.V., Kahvejian A., De Crescenzo G., Costa-Mattioli M., Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino S., Imai M., Kobayashi T., Uchida N., Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 17.Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 19.Kong C., Ito K., Walsh M.A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 20.Atkinson G.C., Baldauf S.L., Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol. Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kononenko A.V., Mitkevich V.A., Atkinson G.C., Tenson T., Dubovaya V.I., Frolova L.Y., Makarov A.A., Hauryliuk V. GTP-dependent structural rearrangement of the eRF1:eRF3 complex and eRF3 sequence motifs essential for PABP binding. Nucleic Acids Res. 2010;38:548–558. doi: 10.1093/nar/gkp908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Z., Saito K., Pisarev A.V., Wada M., Pisareva V.P., Pestova T.V., Gajda M., Round A., Kong C., Lim M., et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seit-Nebi A., Frolova L., Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D.E., Berninghausen O., Green R., Becker T., Beckmann R. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014;8:59–65. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakobsen C.G., Segaard T.M., Jean-Jean O., Frolova L., Justesen J. [Identification of a novel termination release factor eRF3b expressing the eRF3 activity in vitro and in vivo] Mol. Biol. (Mosk) 2001;35:672–681. [PubMed] [Google Scholar]
- 26.Hoshino S., Imai M., Mizutani M., Kikuchi Y., Hanaoka F., Ui M., Katada T. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 1998;273:22254–22259. doi: 10.1074/jbc.273.35.22254. [DOI] [PubMed] [Google Scholar]
- 27.Chauvin C., Salhi S., Le Goff C., Viranaicken W., Diop D., Jean-Jean O. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell. Biol. 2005;25:5801–5811. doi: 10.1128/MCB.25.14.5801-5811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behm-Ansmant I., Gatfield D., Rehwinkel J., Hilgers V., Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov P.V., Gehring N.H., Kunz J.B., Hentze M.W., Kulozik A.E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eberle A.B., Stalder L., Mathys H., Orozco R.Z., Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh G., Rebbapragada I., Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roque S., Cerciat M., Gaugue I., Mora L., Floch A.G., de Zamaroczy M., Heurgue-Hamard V., Kervestin S. Interaction between the poly(A)-binding protein Pab1 and the eukaryotic release factor eRF3 regulates translation termination but not mRNA decay in Saccharomyces cerevisiae. RNA. 2015;21:124–134. doi: 10.1261/rna.047282.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosson B., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trowitzsch S., Bieniossek C., Nie Y., Garzoni F., Berger I. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 2010;172:45–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Alkalaeva E., Eliseev B., Ambrogelly A., Vlasov P., Kondrashov F.A., Gundllapalli S., Frolova L., Soll D., Kisselev L. Translation termination in pyrrolysine-utilizing archaea. FEBS Lett. 2009;583:3455–3460. doi: 10.1016/j.febslet.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kryuchkova P., Grishin A., Eliseev B., Karyagina A., Frolova L., Alkalaeva E. Two-step model of stop codon recognition by eukaryotic release factor eRF1. Nucleic Acids Res. 2013;41:4573–4586. doi: 10.1093/nar/gkt113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Susorov D., Mikhailova T., Ivanov A., Sokolova E., Alkalaeva E. Stabilization of eukaryotic ribosomal termination complexes by deacylated tRNA. Nucleic Acids Res. 2015;43:3332–3343. doi: 10.1093/nar/gkv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valasek L., Szamecz B., Hinnebusch A.G., Nielsen K.H. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- 39.Brown A., Shao S., Murray J., Hegde R.S., Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matheisl S., Berninghausen O., Becker T., Beckmann R. Structure of a human translation termination complex. Nucleic Acids Res. 2015;43:8615–8626. doi: 10.1093/nar/gkv909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozlov G., Gehring K. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One. 2010;5:e10169. doi: 10.1371/journal.pone.0010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proweller A., Butler J.S. Ribosomal association of poly(A)-binding protein in poly(A)-deficient Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:10859–10865. doi: 10.1074/jbc.271.18.10859. [DOI] [PubMed] [Google Scholar]
- 44.Sachs A.B., Davis R.W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 45.Kuyumcu-Martinez N.M., Joachims M., Lloyd R.E. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 2002;76:2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 47.Kushnirov V.V., Ter-Avanesyan M.D., Telckov M.V., Surguchov A.P., Smirnov V.N., Inge-Vechtomov S.G. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene. 1988;66:45–54. doi: 10.1016/0378-1119(88)90223-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.