Abstract
The archaea-/eukaryote-specific 40S-ribosomal-subunit protein S31 is expressed as an ubiquitin fusion protein in eukaryotes and consists of a conserved body and a eukaryote-specific N-terminal extension. In yeast, S31 is a practically essential protein, which is required for cytoplasmic 20S pre-rRNA maturation. Here, we have studied the role of the N-terminal extension of the yeast S31 protein. We show that deletion of this extension partially impairs cell growth and 40S subunit biogenesis and confers hypersensitivity to aminoglycoside antibiotics. Moreover, the extension harbours a nuclear localization signal that promotes active nuclear import of S31, which associates with pre-ribosomal particles in the nucleus. In the absence of the extension, truncated S31 inefficiently assembles into pre-40S particles and two subpopulations of mature small subunits, one lacking and another one containing truncated S31, can be identified. Plasmid-driven overexpression of truncated S31 partially suppresses the growth and ribosome biogenesis defects but, conversely, slightly enhances the hypersensitivity to aminoglycosides. Altogether, these results indicate that the N-terminal extension facilitates the assembly of S31 into pre-40S particles and contributes to the optimal translational activity of mature 40S subunits but has only a minor role in cytoplasmic cleavage of 20S pre-rRNA at site D.
INTRODUCTION
Ribosomes are complex ribonucleoprotein particles that are responsible for protein synthesis. In all organisms, ribosomes are composed of two ribosomal subunits (r-subunits), the large one (LSU) being about twice the size of the small one (SSU) (1). Although the core of ribosomes is highly conserved in all kingdoms of life, eukaryotic ribosomes are larger and more complex than bacterial or archaeal ribosomes (1,2).
Structural analyses of the prokaryotic r-subunits have shown that many r-proteins possess long N- and C-terminal tails and/or extended internal loops (3,4). These extensions protrude from globular domains located on the r-subunit surface and often enter deep into the corresponding r-subunit structure forming close contacts with the rRNA (e.g. (5,6)). In addition to the conserved core, eukaryotic ribosomal subunits contain specific components that are mainly located on the periphery of the mature ribosome. These elements include archaea-/eukaryote- or exclusively eukaryote-specific r-proteins, additional internal loops and external tails within conserved r-proteins and unique rRNA sequences, referred to as expansion segments (e.g. (7)). Understanding the role of r-protein extensions in ribosome assembly and function, especially the eukaryote-specific ones, is a current challenge of the ribosome synthesis and translation fields that has clearly not been sufficiently addressed. As a plausible general role, it has been assumed that extensions, which are poorly ordered, highly flexible and of basic nature (8), stabilize the correct binding of r-proteins to the r-subunits since they increase the total interaction surface through their extensive contacts with several regions of rRNAs (3,9). During assembly, these contacts apparently facilitate the correct rRNA folding while concomitantly the extensions acquire their final three-dimensional conformation (e.g. (8–10)). Moreover, it has been proposed that r-protein extensions could also play direct roles during translation (e.g. (3,4,6,11,12)).
A handful of bacterial r-protein extensions have been studied in detail, among them, those of r-proteins S4 (uS4 according to the recently proposed r-protein nomenclature (13,14,15), S9 (uS9) (16), S12 (uS12) (17), S13 (uS13) (16), L4 (uL4) (18), L7/L12 (bL12) (19,20), L20 (bL20) (21) and L22 (uL22) (18,22) from Escherichia coli.
Extensions of eukaryotic r-proteins are currently under intensive investigation. Examples include the extensions of several yeast r-proteins from both r-subunits, such as the N-terminal region of S5 (uS7) (23,24), the C-terminal tail of S14 (uS14) (25), the N-terminal tail and internal loop of L3 (uL3) (26–30), the C-terminal extension and internal loop of L4 (uL4) (31–33), the N- and C-terminal extensions of L5 (uL18) (34–36), the internal loop of L10 (uL16) (37–39), the internal loop of L11 (uL5) (40,41), the C-terminal tail of L14 (eL14) (42), the N-terminal tail of L25 (uL23) (43), the N-terminal extension of L29 (eL29) (44) and the C-terminal extension of P0 (uL10) (45,46). These studies clearly indicate that many extensions provide binding sites for dedicated chaperones or harbour nuclear localization signals (NLSs); thus, promoting the soluble expression and active nuclear import of r-proteins (26,32,36,43,44,47–49). Moreover, extensions of distinct r-proteins are important for specific pre-rRNA processing reactions, such as the C-terminal tail of S14 (25) and the internal loop of L3 (30,50), which both are important for cleavage of 20S pre-rRNA at site D, and the internal loop of L4 (31), which influences cleavage of 27SB pre-rRNA at site C2 and exonucleolytic trimming of 7S pre-rRNA to mature 5.8S rRNA. Furthermore, extensions have been shown to facilitate early, late or cytoplasmic steps of ribosome assembly (30,32,33,39,40,42,46). For example, it has been reported that the internal loop of L10 might be involved in the efficient recruitment of the ribosome assembly factor Sdo1, which along with the GTPase Efl1, is required to release Tif6 from late cytoplasmic pre-60S r-particles (37,39). The eukaryote-specific C-terminal extension of L14 might be required for the correct interaction of a distinct set of assembly factors (Ebp2, Mak5, Nop16 and Rpf1) during early steps of the maturation of pre-60S r-particles (42). Finally, as described for bacterial r-proteins (i.e. (16,19,51)), the extensions of several eukaryotic r-proteins ensure the correct maturation of the functional sites of the ribosome, thus contributing to the efficiency and accuracy of translation; in this respect, the N-terminal tail of S5 has been suggested to participate in the recruitment of specific mRNAs and initiation factors to the ribosome (23,24), the C-terminal tail of human S15 (uS19) is involved in the formation of the 40S decoding site during elongation and termination of translation (52), the internal loops of L3, L10 and L11 have been proposed to modulate the interaction of tRNAs and translation elongation factors (eEFs) at the A- and P-sites (11,27–30) and the C-terminal extensions of the r-stalk P0, P1 and P2 proteins have been suggested to be important during the recruitment of eEFs (45,53).
We are interested in understanding the contribution of specific yeast r-proteins to ribosome biogenesis (54). We have previously studied yeast S31 (eS31) (55), which is an archaea-/eukaryote-specific SSU r-protein that, in eukaryotes, is synthesized as a linear ubiquitin-fusion precursor protein called Ubi3 (56). Our previous study revealed that the ubiquitin moiety of Ubi3 contributes to the efficient production of mature SSUs and, more importantly, that ubiquitin cleavage is not a prerequisite for S31 assembly but essential for ribosome function (55). S31 consists of a globular body and a eukaryote-specific N-terminal extension (Figure 1A and Supplementary Figure S1). In this study, we have examined the role of the eukaryote-specific N-terminal extension of S31 in ribosome biogenesis and function. Our data indicate that this extension harbours a NLS that facilitates the nuclear import and the assembly of S31 into pre-40S r-particles. More importantly, yeast strains expressing a truncated S31 variant lacking its N-terminal extension, either from the genomic locus or from a high-copy-number plasmid, are hypersensitive to aminoglycoside antibiotics, suggesting that the N-terminal part of S31 also contributes significantly to the accurate translational activity of mature SSUs.
Figure 1.
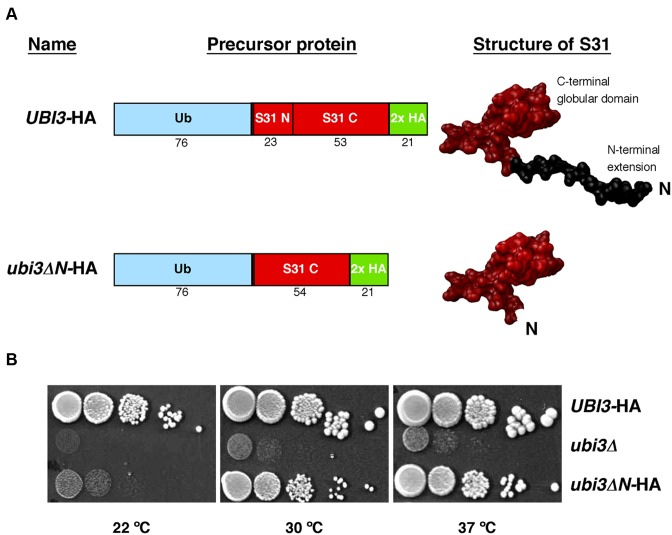
Growth phenotype of the ubi3ΔN mutant. (A) Schematic representation of the yeast ubiquitin-S31 fusion proteins present in the UBI3-HA and ubi3ΔN-HA strains. Ubi3 consists of a N-terminal ubiquitin moiety fused to the S31 r-protein. S31 consists of an N-terminal extension of 23 amino acids and a C-terminal globular domain that includes 53 amino acids. The N-terminal extension of S31 has been deleted in the ubi3ΔN mutant. The structure of the wild-type S31 protein is shown as found in the 40S r-subunit (PDB file 4V88, (7)). For the structure of the S31ΔN protein, the corresponding amino acids were removed from the wild-type S31 protein model. Both constructs contain a C-terminal 2x HA tag for western blot detection. (B) Growth defect of the ubi3ΔN mutant. Strains TLY56.D3 and TLY63.A3 harbour genomically integrated UBI3-HA or ubi3ΔN-HA alleles at the UBI3 locus, respectively. These two strains as well as the ubi3 null mutant (ubi3Δ) strain TLY12.1A were grown in liquid YPD medium and diluted to an OD600 of 0.05. Serial 5-fold dilutions were spotted onto YPD plates, which were incubated at 30 and 37°C for 3 days and at 22°C for 5 days.
MATERIALS AND METHODS
Strains and microbiological methods
All yeast strains used in this study are listed in Supplementary Table S1. Unless otherwise indicated, experiments were conducted in the W303 genetic background.
The UBI3-HA and ubi3Δ strains have been previously described (55). For the integration of the ubi3ΔN-HA allele at the genomic locus, a fusion PCR product containing the UBI3 promoter, the truncated ORF, followed by a double HA-tag, the kanMX4 marker module and a short UBI3 terminator flanking region was transformed into W303. Transformants were selected on YPD plates containing 200 μg/ml G418; selected candidates were chosen and the correctness of the integration verified by PCR. Some positive candidates were sporulated, tetrads dissected and the progeny examined. TLY63.A3 (MATa ubi3ΔN-HA::kanMX4) is a segregant of one of these candidates, which was used in further experiments. To ascertain that no additional mutations were introduced in the replaced sequence, the DNA construct was newly PCR amplified from genomic DNA of TLY63.A3 and sequenced. Strain SMY151 (rps13::kanMX4) is a haploid segregant of Y23999 (Euroscarf), which requires a plasmid-borne copy of RPS13 for cell viability.
Growth and handling of yeast and standard media were performed by established procedures (57). Strains were grown at selected temperatures either in rich YPD medium (1% yeast extract, 2% peptone and 2% glucose) or synthetic minimal medium (0.15% yeast nitrogen base, 0.5% ammonium sulphate) supplemented with the appropriate amino acids and bases as nutritional requirements, and containing either 2% glucose (SD) or 2% galactose (SGal) as carbon source. To prepare plates, 2% agar was added to the media before sterilization. Antibiotic-containing plates were prepared by adding the drugs from stock solutions into 2% YPD-agar medium before pouring the plates (58). Tetrad dissections were performed using a Singer MS micromanipulator. Escherichia coli strain DH5α was used for cloning and propagation of plasmids (59).
Plasmids and oligonucleotides
Plasmids used in this study are listed in Supplementary Table S2. For most of the plasmids constructed in this study, cloned DNA fragments were generated by PCR amplification. All constructs were verified by DNA sequencing. More information about the construction of the different plasmids and the used oligonucleotides is given in the Supplementary Data file or will be available upon request.
Sucrose gradient centrifugation
Cell extracts for polysome analyses were performed according to Foiani et al. (60), as previously described (61) using an ISCO UA-6 system equipped to continuously monitor A254. When needed, fractions of 0.5 ml were collected from the gradients; proteins were extracted from the different fractions as exactly described (62) and analysed by western blot analyses as described below.
Pulse-chase labeling of pre-rRNA
Pulse-chase labeling of pre-rRNA was performed as previously described (63), using 250 μCi of [methyl-3H] methionine (70 to 85 Ci/mmol; Amersham Biosciences) per 40 OD600 units of yeast cells. Cells were grown in liquid SD-Met medium to exponential growth phase at 30°C, pulse-labeled for 1 min, and chased for 2, 5, 15 min with SD medium containing an excess of cold methionine. Total RNA was extracted by the acid-phenol method (64). Radioactive incorporation was measured by scintillation counting and about 20,000 cpm per RNA sample were loaded and resolved on 1.2% agarose–6% formaldehyde gels. RNA was then transferred to a nylon membrane and visualized by fluorography (65).
Northern hybridization analyses
Steady-state levels of pre-rRNAs were assessed by northern blot analyses as described previously (65). In all experiments, RNA was extracted from samples corresponding to 10 OD600 units of exponentially grown cells. Equal amounts of total RNA (5 μg) were loaded on 1.2% agarose–6% formaldehyde or on 7% polyacrylamide–8M urea gels. Specific oligonucleotides, whose sequences are listed in Supplementary Table S3, were 5′-end labeled with [γ-32P] ATP and used as probes. Signal intensities were quantified using a FLA-5100 imaging system and Image Gauge (Fujifilm).
Preparation of ribosomal particles
Ribosomal particles were prepared from S30 cell extracts of the wild-type UBI3-HA or the mutant ubi3Δ-HA strain as previously described (58,66). Briefly, 200 ml of cells from each strain were grown in YPD to mid-log phase, washed, and concentrated in 500 μl of ice-cold buffer 1 (10 mM Tris–HCl, pH 7.4; 20 mM KCl; 12.5 mM MgCl2) containing a protease inhibitor mixture (Complete EDTA-free, Roche Applied Science). Cells were disrupted by vigorous shaking with glass beads in a Fast-prep-24 homogenizer (MP Biomedicals) at 4°C. S30 fractions were obtained by centrifuging the extract at 16 100 × g for 20 min at 4°C in an Eppendorf microcentrifuge. The S30 extracts were centrifuged in a TLA110 rotor in a Beckman-Coulter Optima™ Max centrifuge at 90 000 rpm for 60 min at 4°C. The pellets were resuspended in buffer 2 (20 mM Tris–HCl, pH 7.4; 100 mM MgCl2) and washed by centrifugation in a 20–40% discontinuous sucrose gradient prepared in buffer 2 containing 500 mM ammonium acetate at 90 000 rpm for 120 min at 4°C again in a TLA110 rotor. The pellets were resuspended in buffer 2 and immediately used or stored at −80°C. Ribosomal proteins were separated on NuPAGE 4–12% gradient polyacrylamide gels (Invitrogen), and different proteins were assayed by western blot using selected antibodies described in the next section.
Western blotting and antibodies
Western blot analysis was performed according to standard procedures using the following primary antibodies: mouse monoclonal anti-HA 16B12 (Covance), anti-GFP (Roche), anti-Pgk1 (Invitrogen) and anti-L3 (gift from J.R. Warner) (67); rabbit polyclonal anti-L1 (gift from F. Lacroute) (68), anti-L5 (gift from S.R. Valentini) (69), anti-P0 (gift from J.P.G. Ballesta) (70), anti-S0 (gift from L. Valasek) (71), anti-S2/L30 (gift from J.R. Warner) (67), anti-S3 (gift from M. Seedorf) (72), anti-S8 (gift from G. Dieci) (73), anti-S26 (gift from V. Panse) (74) and anti-Asc1 (gift from V.R. Gerbasi) (75). Secondary goat anti-mouse or anti-rabbit horseradish peroxidase-conjugated antibodies (Bio-Rad) were used. Proteins were detected using a chemiluminescence detection kit (Super-Signal West Pico, Pierce) and a ChemiDoc™ MP system (Bio-Rad).
Fluorescence microscopy
To address the sub-cellular assembly location of S31, the GAL::EMG1 strain (see Supplementary Table S1) was transformed with plasmids expressing GFP-tagged versions of either the wild-type S31 or the truncated S31ΔN protein. GFP-tagged constructs for other r-proteins were used as controls (Supplementary Table S2). Transformants were grown in selective SGal medium or subjected to in vivo depletion in selective SD medium for different times. When required, cultures were incubated for 20 min with Hoechst 33342 (1 mg/ml, final concentration) to visualize nuclear DNA.
To study the NLS of S31, the strain YKL500 (32), which expresses the red fluorescent protein yEmCherry fused to the nucleolar protein Nop58, was transformed with YCplac111-derived plasmids containing different NLS constructions fused to a 3xyEGFP tag (Supplementary Table S2).
To test pre-ribosomal particle export, the wild-type UBI3-HA strain as well as the ubi3Δ and ubi3ΔN-HA mutants were co-transformed with plasmid constructs (gift from J. Bassler), which co-express the nucleolar marker mRFP-Nop1 and either the L25-eGFP or the S3-eGFP reporter (Supplementary Table S2). Transformants were grown at either 30 or 22°C in selective SD medium.
In all cases, cells were inspected in an Olympus BX61 fluorescence microscope equipped with a digital camera. Images were analyzed using the Cell Sens software (Olympus) and processed with Adobe Photoshop.
Affinity purification of S31-yEGFP protein
yEGFP-tagged S31 and S31ΔN proteins were precipitated by following the one-step GFP-Trap_A procedure (Chromotek) with GFP-Trap_A beads, as described in (58). As a control for an early assembling 40S r-protein, a fully functional S13-yEGFP construct was used. A GFP-fused Pwp2 construct was also used as a control. Pre- and mature rRNAs were recovered from the beads by phenol-chloroform extraction and assayed by northern hybridization as indicated above.
Protein identification by LC–ESI-MS/MS analysis
Proteins from three independent preparations of r-particles from either wild-type (UBI3-HA) or ubi3ΔN-HA cells were digested overnight with trypsin (76). The digests were analysed by liquid chromatography–electrospray ionization-tandem mass spectrometry (LC–ESI-MS/MS) using a QExactive plus Orbitrap MS (Thermo Fisher Scientific) that was coupled to an Easy-nLC1000 (Proxeon Biosystems). Peptides were separated on a C18 analytical column (75 μm × 50 cm) at a 200 nl/min flow rate. The elution gradient was from buffer A (0.1% formic acid in water) to buffer B (0.1% formic acid in 99% acetonitrile) as follows: from 100% buffer A to 27% of buffer B for 240 min, from 27% to 90% of buffer B for 9 min and from 90% to 2% of buffer B for 33 min. The spray voltage was set to 2.1 kV, and the temperature of the heated capillary was 270°C. The MS scanned a mass range of 200–2000 Da. The top 15 most abundant peptides (ions), acquired every 80 ms during 240 min for each sample, were analyzed in data-dependent scan mode. The normalized collision energy was adjusted to 35%, and the dynamic exclusion was set to a repeat count of 1, repeat duration of 30 s, and ±2 m/z exclusion mass width.
The label-free quantitative analysis of peptides was performed by spectral counting analysis using the Proteome Discoverer software 1.4 (Thermo Fisher Scientific). Exported data were analyzed by the normalized spectral abundance factor (NSAF) method (77,78) to normalize run-to-run variations for all proteins independently (79). Then, the mean and the standard deviation were calculated for each detected protein; the Student's t-test (P < 0.05) was used to determine whether the difference in abundance of any protein in the r-particle preparations from either UBI3-HA or ubi3ΔN-HA was significant.
RESULTS
Truncation of the N-terminal extension of yeast S31 impairs cell growth
Yeast S31 is a quasi-essential small r-protein of 76 amino acids that is encoded by a single gene named UBI3 (56,80). S31 is found in archaea and eukaryotes (Supplementary Figure S1A) but not in eubacteria (81). Moreover, S31 seems to be functionally conserved as the human S31 protein (named S27A) fully complements a yeast ubi3Δ null mutant (82). Strikingly, in all eukaryotes, S31 as well as the r-protein L40 (eL40), are produced as C-terminal parts of ubiquitin-fused precursor proteins (56). Proteolytic removal of the ubiquitin moiety is a very rapid, likely co-translational, process (55,56), which must occur before the corresponding S31 and L40 r-proteins assemble into r-subunits, that is essential for ribosome function and cell growth ((55) and our unpublished results).
S31 consists of a globular domain, which is well conserved from archaea to eukaryotes, and a eukaryote-specific N-terminal extension (Figure 1A and Supplementary Figure S1B and C). Both, eukaryotic S31 and its archaeal orthologue are found in equivalent positions at the beak structure within the mature SSU and, in eukaryotes, the N-terminal tail of S31 extends toward the A-site (Supplementary Figure S1D and E).
To address the biological role of the N-terminal extension of yeast S31, we generated a genomic deletion allele (hereafter referred to as ubi3ΔN), which codes for an ubiquitin-fused S31 variant (hereafter referred to as S31ΔN) lacking the N-terminal fragment comprising amino acids K78 to K99, but keeping amino acid G77 to ensure proper cleavage of the ubiquitin moiety. The allele also codes for a C-terminal double HA tag to allow detection of the S31ΔN protein (Figure 1A and Supplementary Figure S2). As controls, we used a wild-type UBI3 allele also followed by a C-terminal double HA tag, which has been previously proven to support wild-type growth (55), and the ubi3Δ null allele, which has been described to display a severe slow-growth (sg) and cold-sensitive (cs) phenotype (55,56). Examination of the growth phenotype of the ubi3ΔN-HA strain on plates revealed that, compared to its wild-type counterpart, truncation of the N-terminal tail of S31 conferred a clear sg phenotype at 30 and 37°C that it is severely enhanced at 22°C. This growth defect is not as pronounced as that caused by the complete deletion of the UBI3 gene (Figure 1B). Doubling times of ∼1.6, 2.5 and 8.5 h for the UBI3-HA, ubi3ΔN-HA and ubi3Δ were obtained in liquid YPD medium at 30°C, respectively. At 22°C, the doubling time of the UBI3-HA strain was ∼2.3 h, whereas that of the ubi3ΔN-HA mutant was ∼6 h. We conclude that the N-terminal extension of S31 is required for optimal cell growth, especially at low temperatures.
Truncation of the N-terminal extension of yeast S31 leads to a deficiency in 40S ribosomal subunits
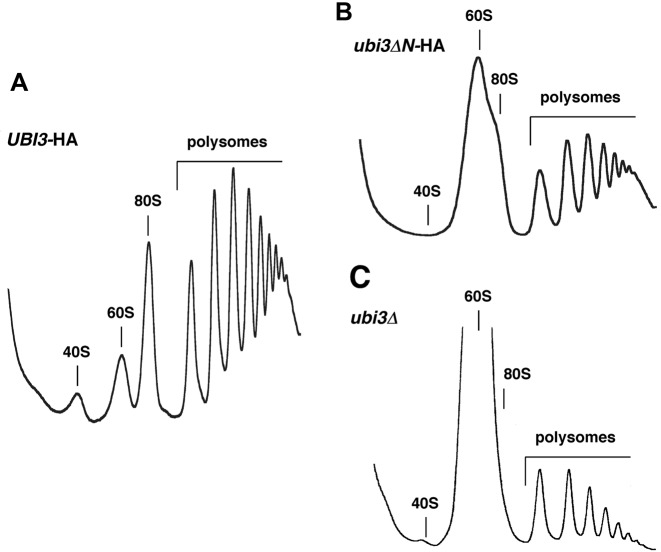
Cold sensitivity is a phenotype often associated with ribosome biogenesis defects (e.g. (83–85)). Consequently, we first examined whether the truncation of the N-terminal tail of S31 altered the polysome profiles obtained from cells grown at 22°C. As shown in Figure 2, the ubi3ΔN-HA mutant exhibits a clear imbalance in the amounts of free SSUs relative to LSUs and a reduction in the polysome content. A similar profile was obtained for the ubi3Δ null strain, although in this case, the severity of the imbalance was stronger (see also, (55)). In contrast, the polysome profile obtained for the UBI3-HA strain was identical to that of a wild-type strain (Figure 2A). We also performed polysome analysis of ubi3ΔN-HA grown at 30°C, and, in this case, similar but slightly less severe defects were observed (data not shown). Altogether, these results indicate that the ubi3ΔN-HA allele causes a clear deficit in SSU formation.
Figure 2.
The ubi3ΔN mutation leads to a deficit in 40S r-subunits. Strains TLY56.D3 (UBI3-HA) (A), TLY63.A3 (ubi3ΔN-HA) (B) and TLY12.1A (ubi3Δ) (C) were grown in YPD at 22°C to an OD600 of 0.8. Cell extracts were prepared and 10 A260 units of each extract were resolved in 7–50% sucrose gradients. The A254 was continuously measured. Sedimentation is from left to right. The peaks of free 40S and 60S r-subunits, 80S free couples/monosomes and polysomes are indicated.
The ubi3ΔN-HA mutation results in defective pre-rRNA processing
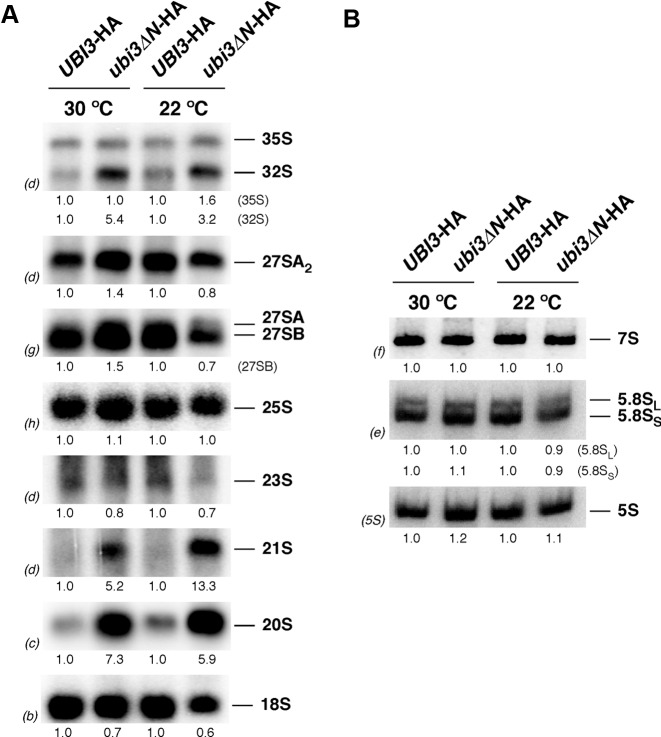
To determine whether the deficit in SSUs detected in the ubi3ΔN-HA mutant strain was the consequence of pre-rRNA processing defects (for a pre-rRNA processing scheme, see Supplementary Figure S3), we analyzed the steady-state levels of pre- and mature rRNA by northern blot hybridization. To this end, the ubi3ΔN-HA strain and its isogenic UBI3-HA counterpart were grown either at 30 or 22°C in liquid YPD medium until mid-log phase and harvested. Total RNA was isolated from each strain and analyzed. As shown in Figure 3A, a mild decrease in 18S rRNA relative to 25S rRNA was observed in the ubi3ΔN-HA strain, which is in agreement with the reduction in SSUs in this mutant. The 35S and especially 32S pre-rRNAs slightly accumulated in the ubi3ΔN-HA strain. More importantly, aberrant 21S (extending from site A1 to site A3) and 20S pre-rRNAs clearly accumulated in the ubi3ΔN-HA strain. All these features were more pronounced when ubi3ΔN-HA cells were grown at 22°C. Northern analysis of low-molecular-weight RNA species showed that the levels of the 7S pre-rRNAs and the mature 5.8S and 5S rRNAs were unaltered in the ubi3ΔN-HA strain (Figure 3B). These results are similar to those reported for the ubi3Δ null mutant (56,80), however, the diminution of the steady-state levels of mature 18S rRNA were clearly less pronounced in ubi3ΔN-HA cells (see also Supplementary Figure S8).
Figure 3.
The 20S pre-rRNA accumulates in the ubi3ΔN mutant. Strains TLY56.D3 (UBI3-HA) and TLY63.A3 (ubi3ΔN-HA) were grown in YPD medium at 30 or 22°C to an OD600 of 0.8. Total RNA was extracted from each strain. Equal amounts of RNA (5 μg) were subjected to northern hybridization. (A) Northern blot analysis of high-molecular-mass pre- and mature rRNAs. (B) Northern blot analysis of low-molecular-mass pre- and mature rRNAs. Signal intensities were measured by phosphorimager scanning; values (indicated below each panel) were normalized to those obtained for the wild-type control, arbitrarily set at 1.0. Probes, between parentheses, are described in Supplementary Figure S3A and Supplementary Table S3.
To better define the effects of the ubi3ΔN-HA mutation on pre-rRNA processing, we studied the kinetics of rRNA production in the ubi3ΔN-HA strain by pulse-chase labeling with [methyl-3H] methionine and compared it to that of UBI3-HA and ubi3Δ cells. In the UBI3-HA strain, the 35S pre-rRNA was rapidly converted into the 27S and 20S pre-rRNAs that were next processed to mature 25S and 18S rRNAs, respectively (Supplementary Figure S4). In contrast, in the ubi3Δ strain, processing of 35S pre-rRNA was mildly delayed; remarkably, conversion of 20S pre-rRNA to mature 18S rRNA was significantly delayed, although a considerable amount of 18S rRNA was still produced. Similarly to the deletion of UBI3 and consistent with the northern blot data, the ubi3ΔN-HA mutation also resulted in a delay in 20S pre-rRNA maturation (Supplementary Figure S4).
Altogether, these data reveal that the deficit in SSUs detected in ubi3ΔN-HA cells, as also observed in ubi3Δ cells, might be mainly attributed to a mild defect in 20S pre-rRNA processing as a consequence of the improper assembly of the S31ΔN protein. Moreover, pre-rRNA processing at site A2 seems to be slightly delayed as suggested by the accumulation of 32S pre-rRNA and the appearance of an aberrant 21S pre-rRNA.
Processing of the 20S pre-rRNA occurs in the cytoplasm (86), thus, the delay in 20S pre-rRNA processing of ubi3ΔN-HA cells may result from either improper export of pre-40S particles or reduced cleavage of cytoplasmic 20S pre-rRNA. It has been previously reported that export of 20S pre-rRNA is not impaired upon depletion of S31 (80). Likewise, we did not observe nuclear accumulation of the SSU reporter S2-eGFP in ubi3ΔN-HA cells (data not shown), therefore suggesting that, as for Ubi3-depleted cells, 20S pre-rRNA is exported from the nucleus but fails to be efficiently processed in the cytoplasm.
Two mature 40S ribosomal subunit subpopulations are present in ubi3ΔN-HA cells
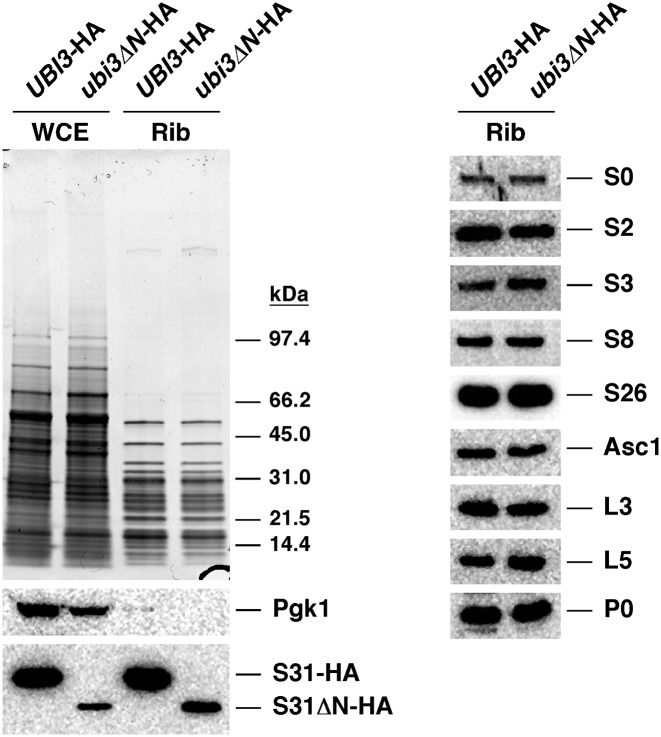
We next investigated whether the truncated S31ΔN r-protein was efficiently assembled into SSUs. To this end, two sets of experiments were performed. First, we isolated r-particles from UBI3-HA and ubi3ΔN-HA cells by a fractionation protocol. These preparations are composed mainly of mature ribosomes, although they also contain pre-ribosomal particles, as previously described (70). After the r-particle isolation, proteins were separated by SDS-PAGE and analyzed by Coomassie staining and western blotting. As shown in Figure 4, Coomassie staining showed no apparent differences in the protein composition of the r-particles from the two strains. However, reduced levels of S31ΔN-HA relative to a subset of other r-proteins from both r-subunits were detected by western blotting. A similar result was found when levels of S31ΔN-HA were inspected in whole cell extracts (Figure 4), indicating that the reduction was not the consequence of a specific loss of the S31ΔN-HA protein during the purification of the r-particles. MS analyses confirmed the reduction in the levels of S31ΔN-HA (Supplementary Figure S5), ruling out the possibility that the HA epitope could have been differentially recognized by the anti-HA antibody in the S31-HA versus the S31ΔN-HA protein. Altogether, these results indicate that truncation of the N-terminal extension of S31 does not impede the incorporation of the S31ΔN-HA protein into r-particles. Since S31ΔN is present in sub-stoichiometric amounts relative to the other tested SSU r-proteins (S0, S2, S3, S8, S26 and Asc1) within r-particle preparations from the ubi3ΔN-HA cells, it can also be deduced that two major subpopulations of ribosomes are present in ubi3ΔN-HA cells, one lacking and another one containing the truncated S31ΔN-HA r-protein.
Figure 4.
Two subpopulations of mature 40S r-subunits, one lacking and another one containing truncated S31ΔN, are present in the ubi3ΔN mutant. Strains TLY56.D3 (UBI3-HA) and TLY63.A3 (ubi3ΔN-HA) were grown in YPD medium at 30°C to an OD600 of 0.8. Whole cell extracts (WCE) were prepared from each strain and used for purification of ribosomes (Rib). Proteins (100 μg) were resolved by SDS-PAGE and analysed by Coomassie staining or western blotting. Specific antibodies against the HA-tag and the r-proteins S0, S2, S3, S8, S26, Asc1, L3, L5 and P0 were used. Anti-Pgk1 was also used as a control.
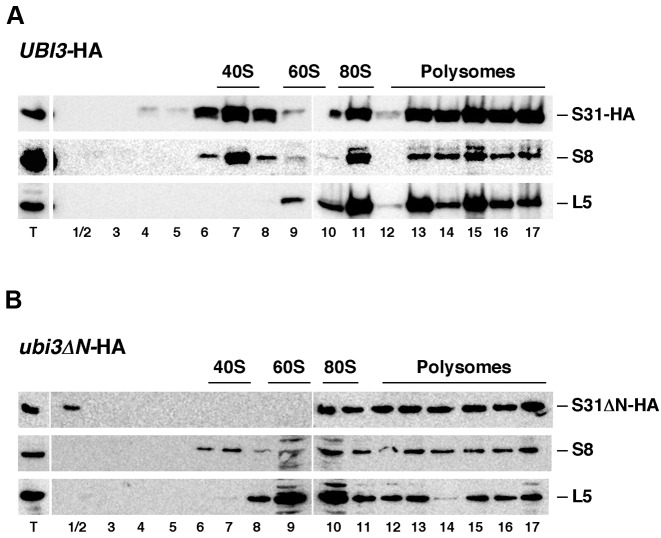
Second, we prepared cell extracts from wild-type (UBI3-HA) and ubi3ΔN-HA cells grown at 30°C under polysome-preserving conditions and analyzed them by sucrose gradient centrifugation and fractionation. Then, the fractions were analyzed by SDS-PAGE and western blotting using anti-HA antibodies to detect S31 and, as controls, anti-S8 and anti-L5 antibodies to monitor the sedimentation behavior of SSUs and LSUs, respectively (Figure 5). In UBI3-HA cells, as expected for SSU r-proteins, S31 and S8 were peaking in the 40S, 80S/monosome and polysomal fractions (Figure 5A). In ubi3ΔN-HA cells, S31ΔN and S8 were found mainly in 80S/monosome and the polysomal fractions (Figure 5B). Weaker signals were detected in fractionations of extracts from this strain, most likely due to its deficit in SSUs. However, S8 but not S31ΔN was identified in the 40S peak. These results indicated that the removal of the N-terminal extension of S31 does not impede the truncated S31ΔN protein to assemble into SSUs and suggest that SSUs either containing or lacking S31ΔN are present in ubi3ΔN-HA cells. Notably, the ones lacking S31ΔN are apparently less efficiently recruited into polysomes.
Figure 5.
S31ΔN protein is incorporated into translating polyribosomes. Strains TLY56.D3 (UBI3-HA) (A) and TLY63.A3 (ubi3ΔN-HA) (B) were grown in YPD medium at 30°C to mid-log phase. Cell extracts were prepared and 10 A260 units of each extract were resolved in 7–50% sucrose gradients and fractionated. Proteins were extracted from each fraction and equal volumes analyzed by western blotting using anti-HA, anti-S8 and anti-L5 antibodies. The position of free 40S and 60S r-subunits, 80S vacant ribosomes/monosomes and polysomes, obtained from the recorded A254 profiles, are shown. T stands for aliquots of the applied total cell extracts.
The N-terminal extension of S31 harbours a functional nuclear localization signal
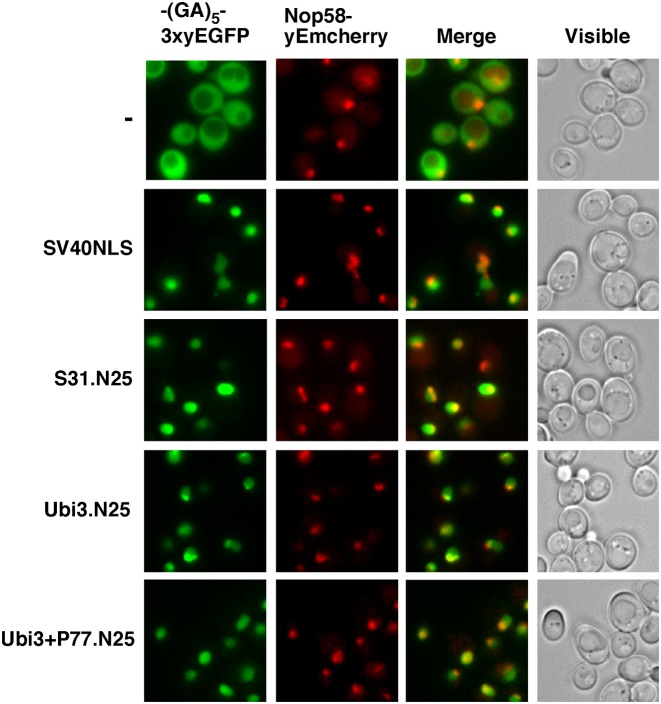
Our initial examination of the S31 N-terminal primary sequence with the cNLS mapper program (87) predicted a bipartite NLS with a significant score (55). To experimentally demonstrate whether this sequence would be able to target S31 to the nucleus, we transformed a wild-type strain expressing the nucleolar marker protein Nop58-yEmCherry from its genomic locus with plasmids expressing from the ADH1 promoter specific fragments of the N-terminal tail of S31 fused in frame, via a (GA)5 decapeptide linker, with a C-terminal triple yeast-enhanced green fluorescent protein (3xyEGFP). One of these fragments consisted of the first 11 amino acids of S31 (from G77 to T87; hereafter S31.N11), while another one comprised the first 25 amino acids (from G77 to A101; hereafter S31.N25). As a positive control for nuclear targeting, we used the NLS of the SV40 large T-antigen fused similarly to 3xyEGFP and, as a negative, primarily cytoplasmic control, a construct expressing the (GA)5-3xyEGFP alone (Figure 6). While the S31.N11-(GA)5-3xyEGFP construct displayed a mainly cytoplasmic localization (data not shown), the S31.N25-(GA)5-3xyEGFP construct was efficiently directed to the nucle(ol)us (Figure 6).
Figure 6.
The N-terminal tail of S31 contains a functional NLS sequence. Strain YKL500, which expresses a yEmCherry-tagged Nop58, was transformed with plasmids, expressing from the ADH1 promoter, a (GA)5-3xyEGFP reporter alone (-), the first 25 amino acids of S31 fused in frame with the (GA)5-3xyEGFP reporter (S31.N25), a wild-type ubiquitin moiety plus the first 25 amino acids of S31 (Ubi3.N25) or a ubiquitin moiety variant that carries a supernumerary proline at the junction between ubiquitin and S31 followed by the first 25 amino acids of S31 (Ubi3+P77.N25). Note that this extra proline blocks the proteolytic maturation of Ubi3 (55). As a positive control for these analyses, we used a plasmid harbouring the NLS sequence of the SV40 large T-antigen fused to the (GA)5-3xyEGFP reporter also expressed from the ADH1 promoter (SV40NLS). Cells were grown in SD-Leu medium at 30°C and the localization of the GFP fusions was inspected by fluorescence microscopy. Nucleoli were revealed by the nucleolar marker protein Nop58-yEmCherry. Cells were identified under brightfield illumination (Visible). The nuclear localization of the reporter protein indicates the ability of the N-terminal region of S31 to work as a NLS sequence. Approximately 200 cells were examined for each reporter, and practically all cells gave the same results as the selected cells shown in the pictures.
Cleavage of Ubi3 into ubiquitin and S31 may occur co-translationally (55,56); thus, we studied whether the presence of the ubiquitin moiety interfered with the nuclear import signal present within the first 25 amino acids of S31. To this end, a new fragment consisting of the complete ubiquitin sequence followed by the first 25 amino acids of S31 (hereafter Ubi3.N25) were fused in frame to the (GA)5-3xyEGFP tag. Likewise, we generated an Ubi3.N25 construct harbouring a supernumerary proline at the junction between ubiquitin and S31 (hereafter Ubi3+P77.N25); we have previously shown that the +P77 insertion impairs almost completely cleavage of the Ubi3 precursor (55). As also shown in Figure 6, in both cases, independently of the efficiency of ubiquitin removal, the GFP-tagged S31.N25 fragment could be efficiently targeted to the nucle(ol)us.
Altogether, these data indicated that the first 25 amino acids of the N-terminal extension of S31 code for a bona fide NLS, which is responsible for the nuclear import of S31 independently of the release of ubiquitin from the Ubi3 precursor by proteolytic cleavage.
S31 assembles in the nucle(ol)us
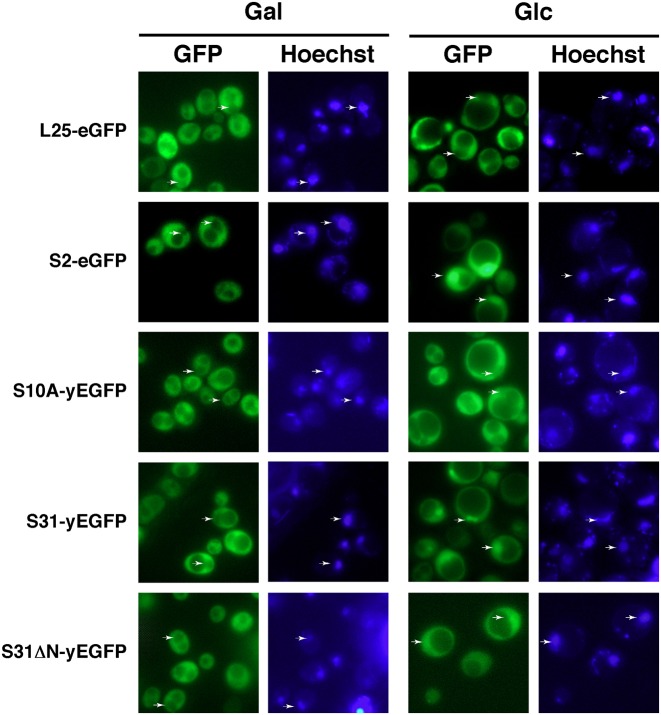
The above results suggested that S31 assembles in the nucleus. To further explore in which cellular compartment occurs the assembly of S31, we made use of plasmids expressing functional C-terminal fusions of a single yEGFP monomer to either Ubi3 or Ubi3ΔN precursors. These precursors were efficiently processed to ubiquitin and S31 or S31ΔN r-proteins, respectively, and these r-proteins were incorporated into translating ribosomes (Supplementary Figure S6). We monitored the localization of the GFP-fused constructs in a conditional GAL::EMG1 strain by fluorescent microscopy. Emg1/Nep1 is an essential trans-acting factor required for early events of SSU biogenesis (88). It has been well reported that depletion of many assembly factors involved in SSU biogenesis leads to nuclear retention of pre-40S r-particles (e.g. (89–93)). As shown in Figure 7, the SSU reporter S2-eGFP, as expected for a r-protein that assembles in the nucleus (93), is primarily located in the cytoplasm of GAL::EMG1 cells under permissive culture conditions, as revealed by fluorescence microscopy; when shifted to the non-permissive culture condition, S2-eGFP accumulated inside the nucleus. In contrast, yEGFP-tagged S10A r-protein, which has been suggested to predominantly assemble late in the cytoplasm (94), did not change its cytoplasmic localization upon the shift (Figure 7). Interestingly, S31-yEGFP localized similarly as S2-eGFP (Figure 7), thus, it clearly accumulated in the nucleus only following a shift to non-permissive culture conditions. These results were quite specific since nuclear export of pre-60S r-particles, followed by a L25-eGFP reporter (95), was not impaired in the GAL::EMG1 strain upon the shift to glucose medium. We conclude that S31 already assembles in the nucle(ol)us into pre-40S r-particles.
Figure 7.
S31 assembles in the nucle(ol)us. Localization of S31-yEGFP and S31ΔN-yEGFP upon depletion of the 40S r-subunit assembly factor Emg1. A GAL::EMG1 strain was transformed with centromeric plasmids expressing L25-eGFP, S2-eGFP, S10A-yEGFP, Ubi3-yEGFP and Ubi3ΔN-yEGFP from their respective cognate promoters. Transformants were grown in SGal-Leu medium (Gal) or shifted overnight to SD-Leu medium (Glc) to fully deplete Emg1. Hoechst was used to stain chromatin DNA. The GFP and Hoechst staining signals were simultaneously inspected by fluorescence microscopy. Arrows point to nuclear fluorescence. Approximately 200 cells were examined for each reporter, and practically all cells gave the same results as the selected cells shown in the pictures.
To study the contribution of the N-terminal extension of S31 in assembly, we transformed the GAL::EMG1 strain with a plasmid expressing a GFP-tagged Ubi3ΔN construct. As expected, S31ΔN-yEGFP was predominantly located in the cytoplasm of GAL::EMG1 cells in galactose medium. Strikingly, when GAL::EMG1 cells were shifted from galactose to glucose medium, S31ΔN-yEGFP was initially localized in the cytoplasm (ca. first 6–9 h in glucose medium; data not shown), but then, at later times after the transfer to glucose medium, distributed evenly between the nucleus and the cytoplasm (Figure 7). These results strongly suggest that the S31ΔN-yEGFP construct, which lacks the above-described NLS and has a molecular mass of ca. 37 kDa, may passively diffuse into the nucleus where it could assemble over the time course of the shift into pre-40S particles.
Altogether, these findings indicate that the NLS sequence of S31 is required for normal import of the protein from the cytoplasm to the nucleus where it assembles into pre-40S r-particles; however, it cannot be totally excluded that some cytoplasmic assembly, especially of the S31ΔN protein, might occur (but see Discussion).
Overexpression of N-terminally truncated S31 suppresses almost completely the ribosome biogenesis defects of the ubi3ΔN-HA strain
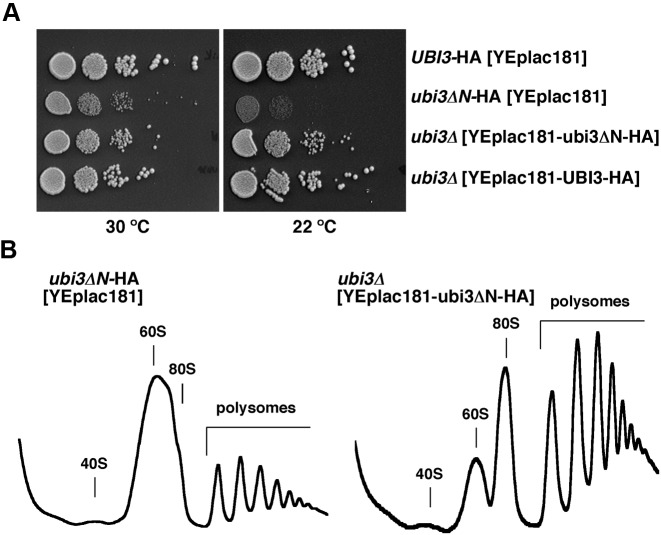
Since the amount of S31ΔN is limited relative to other r-proteins in the ubi3ΔN-HA strain (see Figure 4), we addressed whether multiple copies of the ubi3ΔN-HA allele could alleviate the growth and ribosome biogenesis defects linked to this allele. To this end, we cloned either the wild-type UBI3-HA or the mutant ubi3ΔN-HA allele on a high-copy-number plasmid and transformed the sg ubi3Δ and ubi3ΔN-HA mutants. As shown in Figure 8A and Supplementary Figure S7, increased dosage of ubi3ΔN-HA partially suppressed the sg and cs phenotypes of both the ubi3Δ and the ubi3ΔN-HA mutants. The suppression was clearly dosage-dependent as the ubi3ΔN-HA allele on a low-copy-number plasmid was also able to ameliorate the growth defects of the ubi3Δ strain, albeit slightly less efficiently than on a high-copy-number plasmid (see Supplementary Figure S7, upper panel at 22°C). Western blot analysis showed a good correlation between the ubi3ΔN-HA allele dosage and the total amount of S31ΔN-HA protein in whole cell extracts of cells grown at 30°C (Supplementary Figure S8A) or 22°C (data not shown). To determine whether the suppression of the growth defects was due to restoration of the SSU deficiency, we analyzed polysomes of cells grown at 22°C. As shown in Figure 8B, supplying ubi3ΔN-HA on a multicopy plasmid yielded almost wild-type polysomes profiles, as evidenced by an increase in the peaks of 80S and polysomes, although a slight imbalance in the free SSU peak relative to that of LSU was still observed. In agreement with this, overexpression of Ubi3ΔN-HA also almost completely rescued the rRNA maturation defects of the ubi3Δ mutant (Supplementary Figure S8B and C), as observed by the almost complete restoration of the amounts of the 35S and 20S pre-rRNAs and the mature 18S rRNA to wild-type levels.
Figure 8.
Multiple copies of the ubi3ΔN-HA allele partially alleviate the growth and ribosome biogenesis defects linked to a sole genomic copy of this allele. (A) Growth test of mutants harbouring single or multiple copies of the ubi3ΔN-HA allele. Strains TLY56.D3 and TLY63.A3, which harbour genomically integrated UBI3-HA and ubi3ΔN-HA constructs at the UBI3 locus, respectively, were transformed with the empty high-copy-number plasmid YEplac181. The ubi3 null mutant (ubi3Δ) strain TLY12.1A was transformed with the high-copy-number plasmids YEplac181-UBI3-HA or YEplac181-ubi3ΔN-HA. Transformants were grown in liquid SD-Leu medium and diluted to an OD600 of 0.05. Serial 5-fold dilutions were spotted onto SD-Leu plates, which were incubated at either at 30°C or 22°C for 3 and 5 days, respectively. (B) Overexpression of the S31ΔN protein leads to nearly wild-type polysome profiles. The indicated strains were grown in SD-Leu medium at 22°C to mid-log phase. Cell extracts were prepared and 10 A260 units of each cell extracts were resolved in 7–50% sucrose gradients. The A254 was continuously measured. Sedimentation is from left to right. The peaks of free 40S and 60S r-subunits, 80S vacant ribosomes/monosomes and polysomes are indicated.
We conclude that the amount of S31ΔN-HA r-protein is limiting when genomically expressed, thus leading to an impaired growth and a SSU deficiency. These defects can be efficiently suppressed by increasing the dosage of the ubi3ΔN-HA allele. These results also indicated that truncation of the N-terminal extension of S31 by itself has only a minor impact on SSU biogenesis and 20S pre-rRNA processing.
Truncation of the N-terminal extension of yeast S31 affects translation
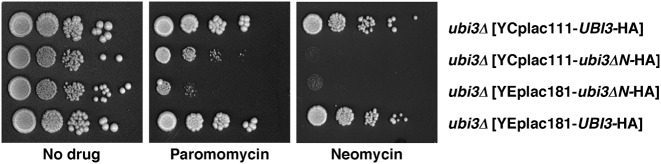
To study whether the N-terminal extension of S31 has a role in translation, we tested the sensitivity of a ubi3::HIS3MX6 strain containing centromeric or multicopy plasmids expressing either wild-type Ubi3-HA or Ubi3ΔN-HA to several protein synthesis inhibitors. We did not include the strain TLY63.A3, which harbours a genomically integrated ubi3ΔN-HA allele, since it also carries the G418-resistant kanMX6 marker that interferes with this antibiotic sensitivity assay. As shown in Figure 9 and Supplementary Figure S9, the full deletion of UBI3 and the expression of the Ubi3ΔN-HA protein, either from a low-copy (YCplac111) or high-copy number (YEplac181) plasmid, conferred hypersensitivity to the aminoglycosides paromomycin and neomycin B. Strikingly, the cells containing YEplac181-ubi3ΔN-HA were slightly more sensitive to paramomycin than those containing YCplac111-ubi3ΔN-HA. However, the ubi3ΔN-HA mutation did not alter the sensitivity to anisomycin or cycloheximide when supplied from either plasmid (Supplementary Figure S9). At the molecular level, aminoglycosides interact with bacterial 16S or eukaryotic 18S rRNA at the upper part of helix H44 (A-site) near the SSU decoding center (12,96) and induce translational misreading (97,98) by perturbing the conformation of the A-site and altering the stringency of correct decoding of aminoacyl-tRNAs (99). Sensitivity to aminoglycosides has been previously reported for mutants defective in SSU biogenesis factors (e.g. (63,100–102)), in r-proteins or 18S rRNA (e.g. (103–107)) and in translation factors and ribosome-associated chaperones (e.g. (63,108,109)). Importantly, increased sensitivity to aminoglycosides has often been linked to reduced translational accuracy (e.g. (103–105,110)). Given that the N-terminal tail of S31 extends toward the A-site (7) and that the ubi3Δ [YEplac181-ubi3ΔN-HA] strain is hypersensitive to aminoglycosides while practically not showing any ribosome biogenesis defects, our results strongly suggest that the N-terminal extension of S31 likely plays a role in ribosome function, most specifically in maintaining the fidelity of translation. In agreement with this, when we analysed the kinetics of S31 protein and ribosome production in cells containing YEplac181-ubi3ΔN-HA after a treatment with a lethal concentration of neomycin, we observed that neither the biogenesis of 40S nor 60S r-subunit was significantly impaired by the drug during the time course of the experiment (Supplementary Figure S10).
Figure 9.
The ubi3ΔN allele confers hypersensitivity to aminoglycoside antibiotics. Haploid ubi3::HIS3MX6 cells (ubi3Δ) containing the YCplac111-UBI3-HA, YCplac111-ubi3ΔN-HA, YEplac181-UBI3-HA or YEplac181-ubi3ΔN-HA plasmids were grown in SD-Leu medium and spotted in 5-fold serial dilution steps onto YPD plates without antibiotics or containing 1 mg/ml paramomycin or 5 mg/ml neomycin. Plates were incubated at 30°C for 3–5 days.
DISCUSSION
In eukaryotes, most r-proteins have evolved large extensions, mostly highly positively charged and intrinsically disordered, that extend far away from their respective globular cores into distant rRNA domains. While few eukaryote-specific extensions have been studied in some detail (see Introduction), the precise role of these extensions in ribosome assembly and function remain largely unknown. Here we have analyzed the phenotypic consequences of deleting the eukaryote-specific N-terminal tail from r-protein S31 on cell growth, ribosome production, pre-rRNA processing and translation. In all eukaryotes, S31 is expressed as the C-terminal part of an ubiquitin-fused Ubi3 precursor protein (111) that requires precise and rapid, most likely co-translational, proteolytic maturation for proper assembly into SSUs (55,56).
In yeast, S31 is a practically essential protein whose complete deletion causes a severe sg phenotype and a strong 40S r-subunit shortage due to delayed pre-rRNA cleavage at site A2 and cytoplasmic maturation of 20S pre-rRNA (55,56,80). The expression of S31 lacking the ubiquitin moiety from its genomic locus under the control of the cognate promoter (ubi3Δub allele) also conferred a pronounced sg phenotype at any tested temperature, a 40S r-subunit deficit and a delayed conversion of cytoplasmic 20S pre-rRNA to mature 18S rRNA; however, these defects were overall not as severe as those observed for the ubi3Δ null mutant (55). In this study, we show that the deletion of the N-terminal domain of S31 (ubi3ΔN mutant) also results in a mild growth defect that is enhanced at low temperatures, a deficit in 40S r-subunits and a delay in cleavages at sites A2 and D. These phenotypes are again similar, but much less pronounced, to those reported for the isogenic ubi3Δ and ubi3Δub strains (Supplementary Figures S4 and S8B-C, and data not shown). In this sense, it is remarkable that the ubiquitin moiety apparently contributes more to cell viability and ribosome synthesis than the N-terminal extension of S31. Ubiquitin has been proposed to act as a chaperone that facilitates the correct folding and hence the efficient synthesis of S31 (55,56). In agreement with this observation, we have previously shown that the levels of S31 were significantly reduced in an ubi3Δub strain. This reduction similarly affected other 40S r-proteins (for further details, see (55)). When Ubi3ΔN was examined, no precursor band could be detected, thus, the Ub-S31ΔN fusion protein was as rapidly and efficiently cleaved as the wild-type Ubi3 precursor. In this study, we have observed a large reduction, both in total cell extracts and in r-particles, in the levels of the S31ΔN protein compared to those of the wild-type S31 protein; however, levels of other r-proteins from either r-subunit remained similar in wild-type and ubi3ΔN cells. Semi-quantitative MS confirmed that the relative protein levels of ribosome-associated S31ΔN were notably reduced in the ubi3ΔN strain compared to those of S31 in the wild-type strain and, as above, the levels of other r-proteins were not significantly altered when compared to each other in ubi3ΔN versus wild-type cells. A plausible explanation for this result is that stable SSUs can be assembled in the presence and in the absence of S31ΔN in the ubi3ΔN strain. The existence of stable S31-deficient SSUs is not surprising since they are regularly produced in the ubi3Δ strain. Similar results have been described for rpl1bΔ cells, which lack the RPL1B paralogue that codes for yeast r-protein L1 (uL1) (112). In this strain, it was reported that LSUs could be assembled with or without L1; further, polysome profile analyses showed that L1-deficient r-subunits were more abundant in the free 60S r-subunit peak than in the polysomal fractions, suggesting that these LSUs were less competent for subunit joining and hence less efficiently engaged in translation (112). Similarly, our polysome fractionation results revealed that the S31ΔN/S8 ratio was reduced in ubi3ΔN cells compared to wild-type cells only in the free 40S peak but not in the polysome fractions; thus, suggesting that, as observed for r-subunits lacking L1, S31-deficient r-subunits were selected against integration into polysomes.
Here, we have also addressed the course of assembly of S31, which has been generally assumed to occur at a relatively late stage during ribosome biogenesis. We have identified with the cNLS mapper program (87) a potential bipartite NLS within the N-terminal extension of yeast S31, which is conserved among other eukaryotes (113) (see also Supplementary Figure S1). Our data show that this sequence is sufficient to actively target a triple GFP reporter to the yeast nucleus. In agreement with the presence of an active NLS within the N-terminal part of S31, our study also shows that a functional GFP-tagged S31 protein notably accumulates in the nucleus upon the depletion of the 90S assembly factor Emg1, which leads to the nuclear retention of pre-40S r-particles. The truncation of the N-terminal tail of S31 impedes an active accumulation of GFP-tagged S31ΔN reporter in the nucleus; however, this fusion protein was present in both the cytoplasm and the nucleoplasm, most likely as the result of passive diffusion across nuclear pore complexes during the long time course of the depletion experiment. These results, thus, reflect that S31 is carried to the nucleus by an active transport process, which is dependent on its N-terminal eukaryote-specific NLS and must be mediated by a distinct importin. As shown for several other importins (114), this factor would be also likely responsible for protecting newly synthesized S31 from its putative tendency to aggregate by shielding the highly basic patch of its N-terminal extension. In yeast, the non-essential ß-karyopherin Kap123/Yrb4 has been shown to be the main nuclear importer of r-proteins (115,116), although other ß-karyopherins, such as Kap104, Kap108/Sxm1 or Kap121/Pse1, have a redundant role in r-protein import (116,117). Most likely due to this redundancy, we have not yet been able to reliably identify the specific importin of S31. In vitro, it has been shown that recombinant S31 and Kap123 are able to interact with each other (74). Our initial assays, however, show that the nuclear import of the S31.N25-(GA)5-3xyEGFP reporter is not affected in a kap123Δ strain (our unpublished data). We have also explored the timing of the nucle(ol)ar S31 assembly by analysing which pre-rRNAs stably co-purified with either S31-yEGFP or S31ΔN-yEGFP. Our data clearly indicate that wild-type or truncated S31, as practically all 40S r-proteins (118), are present in pre-40S particles containing the 20S pre-rRNA (see Supplementary Figure S11). Consistently, S31 could be apparently identified as properly positioned in late and likely cytoplasmic pre-40S r-particles (94). However, our results could not reveal whether S31 assembles into early 90S pre-ribosomal particles containing the 35S pre-rRNA. Indeed, it has been proven that, in contrast to early-acting assembly factors (i.e., see Pwp2-GFP in Supplementary Figure S11), SSU r-proteins interact particularly weakly with 90S pre-ribosomal particles (118). Thus, further experiments are required to address the precise timing of S31 assembly into pre-ribosomal particles, although it is clear that, at least the assembly of full-length, wild-type S31 r-protein seems to be a predominantly nucle(ol)ar event. Our results did neither directly elucidate whether S31 or especially S31ΔN could eventually assemble in the cytoplasm. This possibility is, however, unlikely since a S31ΔN protein fused in frame with a C-terminal 3xyEGFP, which hardly might passively enter the nucleus given its molecular mass of ca. 90 kDa, complements the sg phenotype of the ubi3Δ strain to a lesser extent than a similar construct fused in frame with a single C-terminal yEGFP (Supplementary Figure S6A). Taken together, our data support a scenario in which the bulk of S31 is efficiently delivered to the nucle(ol)us where it assembles prior to the nuclear export of pre-40S r-particles (summarized in Supplementary Figure S12A). On the other hand, only a small amount of S31ΔN could passively enter the nucleus and assemble into pre-40S r-particles, thus, a substantial fraction of nuclear pre-40S r-particles lacking S31ΔN would be exported to the cytoplasm (summarized in Supplementary Figure S12B). Upon arrival of these pre-40S r-particles in the cytoplasm, S31ΔN would be unable to efficiently assemble; thus, any excess of free S31ΔN would be subjected to rapid degradation, as described for other unassembled r-proteins (e.g. see (119–121)). Consistent with this model, it has been demonstrated that an ubi3Δ mutant is able to export pre-40S r-particles at an apparently similar rate to that of a wild-type strain (80). Moreover, we show that the sg and cs phenotypes and ribosome production defects of ubi3ΔN cells could be efficiently suppressed by overexpression of S31ΔN from a high-copy-number plasmid. The simplest explanation for the suppression data is that the elevated levels of S31ΔN may enhance the passive diffusion of the r-protein and hence increase its availability in the nucleus and the efficiency of its assembly. It should be stressed that overexpression of S31ΔN rescues the altered ribosome maturation of the ubi3ΔN strain as it reduces the large amounts of 20S pre-rRNA accumulating in this strain to practically wild-type levels. This observation implies that the growth and ribosome biogenesis defects present in the strain that genomically expresses the ubi3ΔN allele can be practically exclusively attributed to the presence of S31-deficient 40S r-subunits. In conclusion, the N-terminal extension of S31 is nearly dispensable for SSU maturation. This differs from the situation found for the C-terminal extension of yeast S14, which seems to directly influence D-site cleavage of 20S pre-rRNA (25). Strikingly, removal of the N-terminal tail of S31 results in a pronounced hypersensitivity against aminoglycosides, such as paromomycin and neomycin, even in strains where the ubi3ΔN allele is expressed from a high-copy number plasmid. Within SSUs, aminoglycosides bind to an internal loop of helix H44 of 18S rRNA, which is part of the A-site (96,99). Noticeably, the N-terminal tail of S31 also reaches the A-site of the SSU (7,122). Thus, this result suggests that the N-terminal extension of S31 might be important for the proper architecture of the A-site of wild-type ribosomes and hence for accurate decoding. In this sense, expression of NLS-S31ΔN fusion proteins containing a heterologous NLS, either the SV40 large T antigen NLS or the basic PY-NLS of Nab2, which are both known to act efficiently in yeast (115,116), in the ubi3Δ strain resulted in growth, pre-rRNA processing and aminoglycoside hypersensitivity phenotypes that were very similar to those of ubi3Δ [YCplac111-ubi3ΔN-HA] cells (Supplementary Figures S13-S15), suggesting that the substitution of the cognate N-terminal extension of S31 by a heterologous NLS sequence compromises 40S r-subunit assembly and function, likely through structural alterations of the rRNA region around the A-site. As abovementioned, mutations in different yeast SSU r-proteins that confer hypersensitivity to aminoglycosides often simultaneously display decreased translational accuracy (e.g. (103–105,110)). However, we have been so far unsuccessful in demonstrating a significant reduction in translation fidelity in the ubi3ΔN cells by quantitative lacZ-based assays of stop-codon read-through and +1 or −1 ribosomal frameshifting (our unpublished data).
In summary, we have tackled the characterization of the eukaryote-specific extension of the SSU r-protein S31. Our results demonstrate that yeast cells expressing S31ΔN from a single genomic locus or few extrachromosomal ubi3ΔN copies display slow growth and cold-sensitivity due to impaired SSU biogenesis and function. Overexpression of S31ΔN almost completely restores growth and ribosome production, but likely not the translation defects, to the wild-type extent. It is unclear whether mutations in human S27A are linked to ribosomopathies, such as Diamond-Blackfan anemia (123). Dinman and co-workers have reported that the yeast rpl10[R98S] mutation, which mimics the most commonly identified ribosomal mutation linked to T-cell acute lymphoblastic leukemia, impairs cell growth and LSU production (124). Interestingly, this mutant acquires spontaneous extragenic suppressors at an elevated rate that map in the NMD3 gene. These suppressor mutations only restore the growth and the ribosome biogenesis defects of rpl10[R98S] cells, however, they do not suppress the translational defects associated with L10[R98S]-containing ribosomes. In light of these results, these authors have put forward the interesting hypothesis that error-prone translation by defective ribosomes, present in mutant cells that have recovered the growth and ribosome production status of normal cells due to secondary suppressor mutations, is the primary cause of the so-called ribosomopathy paradox: the transition from a ribosome-based hypo- to hyper-proliferative disease, particularly cancer (124). Our results implicate gene amplification as another potential mechanism to achieve this unfortunate transition.
Supplementary Material
Acknowledgments
We thank those colleagues cited in the text who supplied materials, M. Carballo of the Biology Service from the University of Seville for help with the phosphorimager, and M. Pastor-Herrera of the Proteomics and K. Levitsky of the Microscopy Service units from IBiS. We are grateful to F. Espinar-Marchena for handling structures of ribosomes. We specially thank Patrick Linder and Costa Georgopoulos for their continuous support.
Footnotes
Present address: Institut Rondeaux, 6 rue Franz Heller, F- 35700 Rennes, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness (MINECO) and FEDER [BFU2013-42958-P to J.d.l.C.]; Swiss National Science Foundation [PP00P3_123341, PP00P3_146344/1 and 31003A_156764/1 to D.K.]; A.F.-P. was recipient of a fellowship from the University of Seville and of a bridge contract from the Internal Research Plan from the University of Seville; S.M.-V. is a recipient of a FPI fellowship from the MINECO. Funding for open access charge: MINECO and FEDER [BFU2013-42958-P to J.d.l.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 2.Yusupova G., Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 3.Brodersen D.E., Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 4.Wilson D.N., Nierhaus K.H. Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 2005;40:243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- 5.Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015;22:336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 8.Timsit Y., Acosta Z., Allemand F., Chiaruttini C., Springer M. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int. J. Mol. Sci. 2009;10:817–834. doi: 10.3390/ijms10030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 10.Leulliot N., Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 11.Meskauskas A., Dinman J.D. Ribosomal protein L3: gatekeeper to the A site. Mol. Cell. 2007;25:877–888. doi: 10.1016/j.molcel.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 13.Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L., Lindahl L., Liljas A., Lipton J.M., et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayerle M., Woodson S.A. Specific contacts between protein S4 and ribosomal RNA are required at multiple stages of ribosome assembly. RNA. 2013;19:574–585. doi: 10.1261/rna.037028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Changchien L.M., Schwarzbauer J., Cantrell M., Craven G.R. On the role of protein S4 N-terminal residues 1 through 30 in 30S ribosome function. Nucleic Acids Res. 1978;5:2789–2799. doi: 10.1093/nar/5.8.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoang L., Fredrick K., Noller H.F. Creating ribosomes with an all-RNA 30S subunit P site. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12439–12443. doi: 10.1073/pnas.0405227101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calidas D., Lyon H., Culver G.M. The N-terminal extension of S12 influences small ribosomal subunit assembly in Escherichia coli. RNA. 2014;20:321–330. doi: 10.1261/rna.042432.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zengel J.M., Jerauld A., Walker A., Wahl M.C., Lindahl L. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA. 2003;9:1188–1197. doi: 10.1261/rna.5400703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey D., Oleinikov A.V., Traut R.R. The hinge region of Escherichia coli ribosomal protein L7/L12 is required for factor binding and GTP hydrolysis. Biochimie. 1995;77:925–930. doi: 10.1016/0300-9084(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 20.Diaconu M., Kothe U., Schlunzen F., Fischer N., Harms J.M., Tonevitsky A.G., Stark H., Rodnina M.V., Wahl M.C. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Guillier M., Allemand F., Graffe M., Raibaud S., Dardel F., Springer M., Chiaruttini C. The N-terminal extension of Escherichia coli ribosomal protein L20 is important for ribosome assembly, but dispensable for translational feedback control. RNA. 2005;11:728–738. doi: 10.1261/rna.7134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaman S., Fitzpatrick M., Lindahl L., Zengel J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol. Microbiol. 2007;66:1039–1050. doi: 10.1111/j.1365-2958.2007.05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lumsden T., Bentley A.A., Beutler W., Ghosh A., Galkin O., Komar A.A. Yeast strains with N-terminally truncated ribosomal protein S5: implications for the evolution, structure and function of the Rps5/Rps7 proteins. Nucleic Acids Res. 2009;38:1261–1272. doi: 10.1093/nar/gkp1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galkin O., Bentley A.A., Gupta S., Compton B.A., Mazumder B., Kinzy T.G., Merrick W.C., Hatzoglou M., Pestova T.V., Hellen C.U., et al. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA. 2007;13:2116–2128. doi: 10.1261/rna.688207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakovljevic J., de Mayolo P.A., Miles T.D., Nguyen T.M., Léger-Silvestre I., Gas N., Woolford J.L., Jr The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 2004;14:331–342. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- 26.Moreland R.B., Nam H.G., Hereford L.M., Fried H.M. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc. Natl. Acad. Sci. U.S.A. 1985;82:6561–6565. doi: 10.1073/pnas.82.19.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrov A., Meskauskas A., Dinman J.D. Ribosomal protein L3: influence on ribosome structure and function. RNA Biol. 2004;1:59–65. doi: 10.4161/rna.1.1.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meskauskas A., Dinman J.D. A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site. Nucleic Acids Res. 2010;38:7800–7813. doi: 10.1093/nar/gkq641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meskauskas A., Dinman J.D. Ribosomal protein L3 functions as a ‘rocker switch' to aid in coordinating of large subunit-associated functions in eukaryotes and Archaea. Nucleic Acids Res. 2008;36:6175–6186. doi: 10.1093/nar/gkn642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Gómez J.J., Fernández-Pevida A., Lebaron S., Rosado I.V., Tollervey D., Kressler D., de la Cruz J. Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3. PLoS Genet. 2014;10:e1004205. doi: 10.1371/journal.pgen.1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gamalinda M., Woolford J.L., Jr Deletion of L4 domains reveals insights into the importance of ribosomal protein extensions in eukaryotic ribosome assembly. RNA. 2014;20:1725–1731. doi: 10.1261/rna.046649.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillet B., García-Gómez J.J., Pausch P., Falquet L., Bange G., de la Cruz J., Kressler D. The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear pre-60S assembly site. PLoS Genet. 2015;11:e1005565. doi: 10.1371/journal.pgen.1005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stelter P., Huber F.M., Kunze R., Flemming D., Hoelz A., Hurt E. Coordinated Ribosomal L4 Protein Assembly into the Pre-Ribosome Is Regulated by Its Eukaryote-Specific Extension. Mol. Cell. 2015;58:854–862. doi: 10.1016/j.molcel.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moradi H., Simoff I., Bartish G., Nygard O. Functional features of the C-terminal region of yeast ribosomal protein L5. Mol. Genet. Genomics. 2008;280:337–350. doi: 10.1007/s00438-008-0369-7. [DOI] [PubMed] [Google Scholar]
- 35.Deshmukh M., Stark J., Yeh L.C., Lee J.C., Woolford J.L., Jr Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J. Biol. Chem. 1995;270:30148–30156. doi: 10.1074/jbc.270.50.30148. [DOI] [PubMed] [Google Scholar]
- 36.Kressler D., Bange G., Ogawa Y., Stjepanovic G., Bradatsch B., Pratte D., Amlacher S., Strauss D., Yoneda Y., Katahira J., et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012;338:666–671. doi: 10.1126/science.1226960. [DOI] [PubMed] [Google Scholar]
- 37.Sulima S.O., Gulay S.P., Anjos M., Patchett S., Meskauskas A., Johnson A.W., Dinman J.D. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2014;42:2049–2063. doi: 10.1093/nar/gkt1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrov A.N., Meskauskas A., Roshwalb S.C., Dinman J.D. Yeast ribosomal protein L10 helps coordinate tRNA movement through the large subunit. Nucleic Acids Res. 2008;36:6187–6198. doi: 10.1093/nar/gkn643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bussiere C., Hashem Y., Arora S., Frank J., Johnson A.W. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J. Cell Biol. 2012;197:747–759. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodin M.H., Dinman J.D. A flexible loop in yeast ribosomal protein L11 coordinates P-site tRNA binding. Nucleic Acids Res. 2010;38:8377–8389. doi: 10.1093/nar/gkq711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhodin M.H., Rakauskaite R., Dinman J.D. The central core region of yeast ribosomal protein L11 is important for subunit joining and translational fidelity. Mol. Genet. Genomics. 2011;285:505–516. doi: 10.1007/s00438-011-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pratte D., Singh U., Murat G., Kressler D. Mak5 and Ebp2 act together on early pre-60S particles and their reduced functionality bypasses the requirement for the essential pre-60S factor Nsa1. PLoS One. 2013;8:e82741. doi: 10.1371/journal.pone.0082741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaap P.J., van't Riet J., Woldringh C.L., Raue H.A. Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J. Mol. Biol. 1991;221:225–237. doi: 10.1016/0022-2836(91)80216-h. [DOI] [PubMed] [Google Scholar]
- 44.Underwood M.R., Fried H.M. Characterization of nuclear localizing sequences derived from yeast ribosomal protein L29. EMBO J. 1990;9:91–99. doi: 10.1002/j.1460-2075.1990.tb08084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos C., Ballesta J.P.G. The highly conserved protein P0 carboxyl end is essential for ribosome activity only in the absence of proteins P1 and P2. J. Biol. Chem. 1995;270:20608–20614. doi: 10.1074/jbc.270.35.20608. [DOI] [PubMed] [Google Scholar]
- 46.Francisco-Velilla R., Remacha M., Ballesta J.P. Carboxy terminal modifications of the P0 protein reveal alternative mechanisms of nuclear ribosomal stalk assembly. Nucleic Acids Res. 2013;41:8628–8636. doi: 10.1093/nar/gkt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melnikov S., Ben-Shem A., Yusupova G., Yusupov M. Insights into the origin of the nuclear localization signals in conserved ribosomal proteins. Nat. Commun. 2015;6:7382. doi: 10.1038/ncomms8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pausch P., Singh U., Ahmed Y.L., Pillet B., Murat G., Altegoer F., Stier G., Thoms M., Hurt E., Sinning I., et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat. Commun. 2015;6:7494. doi: 10.1038/ncomms8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kenney S.P., Meng X.J. Identification and fine mapping of nuclear and nucleolar localization signals within the human ribosomal protein S17. PLoS One. 2015;10:e0124396. doi: 10.1371/journal.pone.0124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carter C.J., Cannon M. Maturation of ribosomal precursor RNA in Saccharomyces cerevisiae. A mutant with a defect in both the transport and terminal processing of the 20S species. J. Mol. Biol. 1980;143:179–199. doi: 10.1016/0022-2836(80)90197-7. [DOI] [PubMed] [Google Scholar]
- 51.O'Connor M., Gregory S.T., Dahlberg A.E. Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res. 2004;32:5750–5756. doi: 10.1093/nar/gkh913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khairulina J., Graifer D., Bulygin K., Ven'yaminova A., Frolova L., Karpova G. Eukaryote-specific motif of ribosomal protein S15 neighbors A site codon during elongation and termination of translation. Biochimie. 2010;92:820–825. doi: 10.1016/j.biochi.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 53.Santos C., Rodriguez-Gabriel M.A., Remacha M., Ballesta J.P. Ribosomal P0 protein domain involved in selectivity of antifungal sordarin derivatives. Antimicrob. Agents Chemother. 2004;48:2930–2936. doi: 10.1128/AAC.48.8.2930-2936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Cruz J., Karbstein K., Woolford J.L., Jr Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lacombe T., García-Gómez J.J., de la Cruz J., Roser D., Hurt E., Linder P., Kressler D. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol. Microbiol. 2009;72:69–84. doi: 10.1111/j.1365-2958.2009.06622.x. [DOI] [PubMed] [Google Scholar]
- 56.Finley D., Bartel B., Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 57.Kaiser C., Michaelis S., Mitchell A. Methods in yeast genetics: a Cold Spring Harbor Laboratory Course Manual. NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 58.Babiano R., Gamalinda M., Woolford J.L., Jr, de la Cruz J. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol. Cell. Biol. 2012;32:3228–3241. doi: 10.1128/MCB.00539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Foiani M., Cigan A.M., Paddon C.J., Harashima S., Hinnebusch A.G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kressler D., Rojo M., Linder P., de la Cruz J. Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:4598–4608. doi: 10.1093/nar/27.23.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de la Cruz J., Kressler D., Rojo M., Tollervey D., Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kressler D., de la Cruz J., Rojo M., Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. NY: John Wiley & Sons, Inc; 1994. pp. 13.10.11–13.14.17. [Google Scholar]
- 65.Kressler D., de la Cruz J., Rojo M., Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodríguez-Mateos M., Abia D., García-Gómez J.J., Morreale A., de la Cruz J., Santos C., Remacha M., Ballesta J.P.G. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009;37:3514–3521. doi: 10.1093/nar/gkp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vilardell J., Warner J.R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petitjean A., Bonneaud N., Lacroute F. The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eucaryotic homolog of the eubacterial and archaebacterial L1 ribosomal protein. Mol. Cell. Biol. 1995;15:5071–5081. doi: 10.1128/mcb.15.9.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zanelli C.F., Maragno A.L., Gregio A.P., Komili S., Pandolfi J.R., Mestriner C.A., Lustri W.R., Valentini S.R. eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- 70.Rodríguez-Mateos M., García-Gómez J.J., Francisco-Velilla R., Remacha M., de la Cruz J., Ballesta J.P.G. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:7519–7532. doi: 10.1093/nar/gkp806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kouba T., Danyi I., Gunisova S., Munzarova V., Vlckova V., Cuchalova L., Neueder A., Milkereit P., Valasek L.S. Small ribosomal protein RPS0 stimulates translation initiation by mediating 40S-binding of eIF3 via its direct contact with the eIF3a/TIF32 subunit. PLoS One. 2012;7:e40464. doi: 10.1371/journal.pone.0040464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frey S., Pool M., Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- 73.Dieci G., Bottarelli L., Ottonello S. A general procedure for the production of antibody reagents against eukaryotic ribosomal proteins. Protein Pept. Lett. 2005;12:555–560. doi: 10.2174/0929866054395860. [DOI] [PubMed] [Google Scholar]
- 74.Schütz S., Fischer U., Altvater M., Nerurkar P., Pena C., Gerber M., Chang Y., Caesar S., Schubert O.T., Schlenstedt G., et al. A RanGTP-independent mechanism allows ribosomal protein nuclear import for ribosome assembly. Elife. 2014;3:e03473. doi: 10.7554/eLife.03473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerbasi V.R., Weaver C.M., Hill S., Friedman D.B., Link A.J. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 2004;24:8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.García-Gómez J.J., Lebaron S., Froment C., Monsarrat B., Henry Y., de la Cruz J. Dynamics of the putative RNA helicase Spb4 during ribosome assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 2011;31:4156–4164. doi: 10.1128/MCB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Old W.M., Meyer-Arendt K., Aveline-Wolf L., Pierce K.G., Mendoza A., Sevinsky J.R., Resing K.A., Ahn N.G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 78.Liu H., Sadygov R.G., Yates J.R., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 79.Zybailov B., Mosley A.L., Sardiu M.E., Coleman M.K., Florens L., Washburn M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 80.Ferreira-Cerca S., Pöll G., Gleizes P.E., Tschochner H., Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 81.Lecompte O., Ripp R., Thierry J.C., Moras D., Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Monia B.P., Ecker D.J., Finley D., Crooke S.T. A human ubiquitin carboxyl extension protein functions in yeast. J. Biol. Chem. 1990;265:19356–19361. [PubMed] [Google Scholar]
- 83.Ripmaster T.L., Vaughn G.P., Woolford J.L., Jr DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:7901–7912. doi: 10.1128/mcb.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sachs A.B., Davis R.W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 85.Wu K., Wu P., Aris J.P. Nucleolar protein Nop12p participates in synthesis of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:2938–2949. doi: 10.1093/nar/29.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Udem S.A., Warner J.R. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- 87.Kosugi S., Hasebe M., Tomita M., Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu P.C., Thiele D.J. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol. Biol. Cell. 2001;12:3644–3657. doi: 10.1091/mbc.12.11.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 90.Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomson E., Rappsilber J., Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007;13:2165–2174. doi: 10.1261/rna.747607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fassio C.A., Schofield B.J., Seiser R.M., Johnson A.W., Lycan D.E. Dominant mutations in the late 40S biogenesis factor Ltv1 affect cytoplasmic maturation of the small ribosomal subunit in Saccharomyces cerevisiae. Genetics. 2010;185:199–209. doi: 10.1534/genetics.110.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Milkereit P., Strauss D., Bassler J., Gadal O., Kuhn H., Schutz S., Gas N., Lechner J., Hurt E., Tschochner H. A Noc-complex specifically involved in the formation and nuclear export of ribosomal 40S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 94.Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L. III, Karbstein K., Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. Nuclear export of 60S ribosomal subunit depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garreau de Loubresse N., Prokhorova I., Holtkamp W., Rodnina M.V., Yusupova G., Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 97.Singh A., Ursic D., Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 98.Palmer E., Wilhelm J.M., Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 99.Ogle J.M., Carter A.P., Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 100.Lee W.-C., Zabetakis D., Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol. Cell. Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lafontaine D.L.J., Preiss T., Tollervey D. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis. Mol. Cell. Biol. 1998;18:2360–2370. doi: 10.1128/mcb.18.4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.King T.H., Liu B., McCully R.R., Fournier M.J. Ribosome structure and activity are altered in cells lacking snoRNPs that form psseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 103.Alksne L.E., Anthony R.A., Liebman S.W., Warner J.R. An accuracy center in the ribosome conserved over 2 billion years. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9538–9541. doi: 10.1073/pnas.90.20.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Synetos D., Frantziou C.P., Alksne L.E. Mutations in yeast ribosomal proteins S28 and S4 affect the accuracy of translation and alter the sensitivity of the ribosomes to paromomycin. Biochim. Biophys. Acta. 1996;1309:156–166. doi: 10.1016/s0167-4781(96)00128-5. [DOI] [PubMed] [Google Scholar]
- 105.Dresios J., Derkatch I.L., Liebman S.W., Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- 106.Chernoff Y.O., Vincent A., Liebman S.W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Loar J.W., Seiser R.M., Sundberg A.E., Sagerson H.J., Ilias N., Zobel-Thropp P., Craig E.A., Lycan D.E. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 2004;168:1877–1889. doi: 10.1534/genetics.104.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anand M., Chakraburtty K., Marton M.J., Hinnebusch A.G., Kinzy T.G. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 2003;278:6985–6991. doi: 10.1074/jbc.M209224200. [DOI] [PubMed] [Google Scholar]
- 109.Kim S.Y., Craig E.A. Broad sensitivity of Saccharomyces cerevisiae lacking ribosome-associated chaperone Ssb or Zuo1 to cations, including aminoglycosides. Eukaryot. Cell. 2005;4:82–89. doi: 10.1128/EC.4.1.82-89.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anthony R.A., Liebman S.W. Alterations in ribosomal protein RPS28 can diversely affect translational accuracy in Saccharomyces cerevisiae. Genetics. 1995;140:1247–1258. doi: 10.1093/genetics/140.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Archibald J.M., Teh E.M., Keeling P.J. Novel ubiquitin fusion proteins: ribosomal protein P1 and actin. J. Mol. Biol. 2003;328:771–778. doi: 10.1016/s0022-2836(03)00374-7. [DOI] [PubMed] [Google Scholar]
- 112.McIntosh K.B., Bhattacharya A., Willis I.M., Warner J.R. Eukaryotic cells producing ribosomes deficient in Rpl1 are hypersensitive to defects in the ubiquitin-proteasome system. PLoS One. 2011;6:e23579. doi: 10.1371/journal.pone.0023579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nakao A., Yoshihama M., Kenmochi N. RPG: the Ribosomal Protein Gene database. Nucleic Acids Res. 2004;32:D168–D170. doi: 10.1093/nar/gkh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jakel S., Mingot J.M., Schwarzmaier P., Hartmann E., Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rout M.P., Blobel G., Aitchison J.D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 116.Schlenstedt G., Smirnova E., Deane R., Solsbacher J., Kutay U., Görlich D., Ponstingl H., Bischoff F.R. Yrb4p, a yeast Ran-GTP–binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sydorskyy Y., Dilworth D.J., Yi E.C., Goodlett D.R., Wozniak R.W., Aitchison J.D. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 2003;23:2042–2054. doi: 10.1128/MCB.23.6.2042-2054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ferreira-Cerca S., Pöll G., Kuhn H., Neueder A., Jakob S., Tschochner H., Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 119.Warner J.R., Mitra G., Schwindinger W.F., Studeny M., Fried H.M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol. Cell. Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsay Y.F., Thompson J.R., Rotenberg M.O., Larkin J.C., Woolford J.L., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 1988;2:664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- 121.Maicas E., Pluthero F.G., Friesen J.D. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol. Cell. Biol. 1988;8:169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rabl J., Leibundgut M., Ataide S.F., Haag A., Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 123.Gazda H.T., Sheen M.R., Vlachos A., Choesmel V., O'Donohue M.F., Schneider H., Darras N., Hasman C., Sieff C.A., Newburger P.E., et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sulima S.O., Patchett S., Advani V.M., De Keersmaecker K., Johnson A.W., Dinman J.D. Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5640–5645. doi: 10.1073/pnas.1400247111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.