Abstract
The integron is a bacterial recombination system that allows acquisition, stockpiling and expression of cassettes carrying protein-coding sequences, and is responsible for the emergence and rise of multiresistance in Gram-negative bacteria. The functionality of this system depends on the insertion of promoterless cassettes in correct orientation, allowing their expression from the promoter located upstream of the cassette array. Correct orientation is ensured by strand selectivity of integron integrases for the bottom strand of cassette recombination sites (attC), recombined in form of folded single-stranded hairpins. Here, we investigated the basis of such strand selectivity by comparing recombination of wild-type and mutated attC sites with different lengths, sequences and structures. We show that all three unpaired structural features that distinguish the bottom and top strands contribute to strand selectivity. The localization of Extra-Helical Bases (EHBs) directly favors integrase binding to the bottom strand. The Unpaired Central Spacer (UCS) and the Variable Terminal Structure (VTS) influence strand selectivity indirectly, probably through the stabilization of the bottom strand and the resulting synapse due to the nucleotide skew between the two strands. These results underscore the importance of the single-stranded nature of the attC site that allows such tight control over integron cassette orientation.
INTRODUCTION
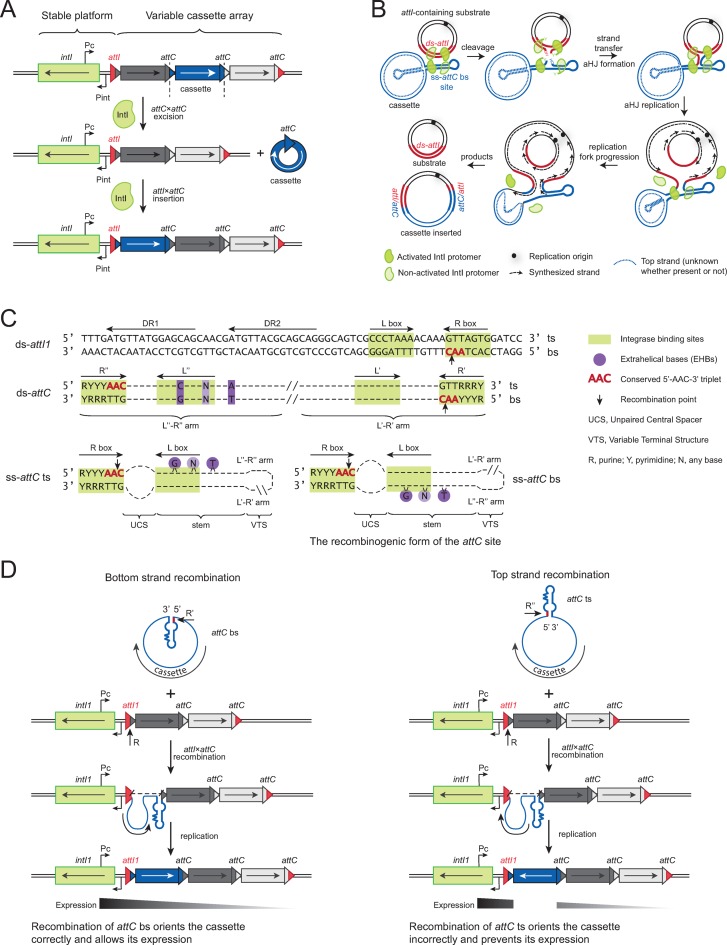
The integron is a bacterial recombination system that allows adaptation to environmental stresses and is largely responsible for the emergence and rise of antibiotic multiresistance in Gram-negative bacteria (1). The integron system is capable of capturing, stockpiling, excising and reordering mobile elements called cassettes. The stable platform of the integron contains an integrase gene (intI) under the control of its promoter Pint, as well as an integration site (attI) and a cassette promoter (Pc) driving the expression of the genes encoded in the cassette array located downstream (Figure 1A). The cassettes are promoterless coding DNA sequences (CDSs), each of them associated with a recombination site (attC).
Figure 1.
The integron. (A) Schematic representation of the integron. The stable platform consists of a gene coding for the integrase (intI) and its promoter Pint, a cassette promoter Pc, and an insertion site attI. Cassettes containing coding DNA sequences can be excised through an intramolecular attC×attC reaction mediated by the integrase. Circularized cassettes can be inserted through an attI×attC reaction mediated by the integrase, and constitute an array of variable size. (B) Mechanism of integron cassette insertion through recombination between ds-attI (red) and the bottom strand of ss-attC (blue). attC top strand is represented as a dotted line because the nature of the cassettes (ss or ds) is not known. After the first strand exchange an atypical Holliday junction (aHJ) is formed and further resolved through a replication step. (C) Sequence of a recombinogenic double-stranded (ds) attI1 site; schematic representation of a non-recombinogenic ds-attC site; schematic representations of a non-recombinogenic top strand (ts) and a recombinogenic bottom strand (bs) of a single-stranded (ss) attC site. Recombination occurs in the conserved 5′-AAC-3′ triplet, in the R box of the bs. (D) (Left panel) Recombination occurs in the bottom strand of the attC site (R′) and in the bottom strand of attI1, which leads to cassette insertion in correct orientation, meaning that the encoded gene can be expressed from the Pc promoter. (Right panel) Recombination occurs in the top strand of the attC site (R″) and in the bottom strand of attI1, which leads to cassette insertion in opposite orientation, meaning that the encoded gene cannot be expressed from the Pc promoter.
Sedentary chromosomal integrons (SCIs) are present in the chromosomes of ∼17% of sequenced bacterial genomes (2–4), while mobile integrons (MIs) are associated with mobile elements such as transposons and conjugative plasmids, and are responsible for the acquisition and dissemination of antibiotic resistance genes in Gram-negative bacteria (5). MIs and SCIs share the same architecture, recombination mechanisms and regulation of integrase expression by the SOS response (6). MIs usually contain few cassettes (up to 8 (7)), almost exclusively involved in antibiotic resistance, while SCIs can have arrays containing as many as 179 cassettes, as in the case of the Vibrio cholerae superintegron (8,9), where encoded genes play a broader yet largely unknown role in bacterial adaptation.
While attC sites located in the same SCI are often highly conserved (10), attC sites of MIs have a very low sequence similarity. Despite this paradoxical lack of sequence conservation, integron integrases (IntI) are capable of recognizing attC sites due to the conserved structure of their folded single strand (11,12). The recombinogenic form of attC sites is an imperfect hairpin formed by the bottom strand (13), with a core site produced by the pairing of the two arms (Figure 1C). The pairing of R′ and R″ forms the R box, while the pairing of L′ and L″ forms the L box. Recombination normally takes place in the R box, between the A and the C of the only sequence stretch conserved among all sites, the 5′-AAC-3′ triplet. This sequence is not important per se and is probably conserved for evolutionary reasons, ensuring universal micro-homology needed for strand transfer between all attC and attI sites (14). For the bottom strand of attC sites, the conserved 5′-AAC-3′ is located in the R′, and the consensus sequence extends beyond the triplet: the R′ consists of a conserved sequence 5′-RYYYAAC-3′ (R: purine; Y: pyrimidine), complementary to the inverse core site 5′-GTTRRRY-3′ in the R″ (15). The rest of the attC site consists of a hairpin with three unpaired features: the Extrahelical Bases (EHBs), the Unpaired Central Spacer (UCS) and the Variable Terminal Structure (VTS) (Figure 1C).
The EHBs are located on one of the arms of the site, in the vicinity of the L″ (Figure 1C). When the site folds, these bases do not have complementary bases on the other arm, and are extruded out of the helix formed by the hairpin. The crystal structure of the attC×attC synaptic complex with the V. cholerae integrase VchIntIA showed specific interactions between the integrase and two EHBs (16). Depending on the attC site, there can be either two or three EHBs: the base located within the L″ of the bottom strand (in position 16) is usually a G, and the furthermost EHB from the L″ (in position 23 or 24, depending on the site) is a conserved T (17). Some attC sites have an additional EHB in position 20. For the attC site to be efficiently recombined by IntI1, the spacing between the first and the last EHB has to consist of a 6 base pair stem (18).
The UCS separates the L and R boxes of the core site, its length on each arm is usually five to six bases and its sequence is not conserved.
The variability of attC site length (57–141 bp (19)) is mostly due to the differences in length of the region separating the R′-L′ and R″-L″ arms, consisting of a stem and a VTS (Figure 1C). attC sites of MIs usually have small VTSs, as short as three nucleotides. On the other hand, attC sites of SCIs often have longer VTSs (20), which can form complex branched structures.
attC sites are a highly unusual type of substrate for tyrosine-recombinases (Y-recombinases) (21). Apart from IntI, the only other known Y-recombinases capable of recombining both single-stranded and double-stranded substrates are the chromosome dimer resolution proteins XerC and XerD (22,23). IntI performs several types of recombination reactions between its ss-attC and ds-attI substrates (Figure 1A): the intramolecular attC×attC cassette excision reaction (24); the intermolecular attI×attC or attC×attC cassette insertion reaction (25); and the less frequent attI×attI reaction (26), which could be important when multiple integrons are present in the same cell (27). The recombination mechanism of IntI is similar to that of Y-recombinases (28), but the Holliday Junction (HJ) formed after the first strand exchange is atypical due to the single-stranded nature of the attC substrate. Unlike the classical resolution through a second cleavage observed for other Y-recombinases, these atypical HJs have to be resolved through replication, as proven for the attI×attC reaction (29) (Figure 1B).
Correct orientation of cassettes is essential to allow the encoded CDSs to be expressed from the promoter Pc and tested for possible selective advantage. Insertion of a cassette in the opposite orientation would position it facing the promoter, preventing its expression (Figure 1D). As most cassettes are promoterless (with only few exceptions (30–35)), their inversion would impede the role of the integron as a genetic platform for rapid adaptation. Correct orientation is ensured by the strand selectivity of the integrase toward the bottom strand. Indeed, the integrase cleaves the ss-attC site in the 5′-AAC-3′, which is located in the R′ for the bottom strand. Inversely, in the top strand the 5′-AAC-3′ is located in the R″. The recombination of the top strand would thus lead to the inversion of the cassette relative to the rest of the array (Figure 1D). Therefore, it is essential that the system is able to specifically recombine the bottom strand and avoid top strand recombination, especially in situations when only top strand of the attC site is present as a substrate (for instance upon conjugation, where the transferred ss-plasmid could be carrying the top strand of the attC site). As the sequence of attC sites is not conserved and the interaction of IntI with the folded attC site is mostly non-specific (16), the basis for this specific recognition of the bottom strand must lie in its structural features. Hairpins formed through the folding of the two complementary strands are inherently similar, but the three unpaired features are asymmetrical relative to the 5′-AAC-3′ triplet (Figure 1C). Because EHBs, UCS and VTS are the only features that distinguish the two strands, strand selectivity of the integrase for the bottom strand must be due to some of these features, or to the interplay of all three.
Previous studies have addressed the importance of attC site structural features in integrase binding (12) and in recombination by a cognate or heterologous IntI (18). We have previously addressed the question of strand selectivity by studying an attC site of V. cholerae superintegron, VCR2 (17). In the present study, we sought to determine if the current model of attC behavior, based on that of VCR2, can be applied to other attCs: as their sequences are variable, the conclusions made for one site might not be directly extrapolated to all attC sites. Moreover, apart from studying VCR2, an attC site from a SCI, we were particularly interested in MI sites, as their study is more relevant to understand the dynamics of cassette insertion in the context of antibiotic resistance gene dissemination. Also, we combined in vivo and in vitro approaches, as in vitro binding studies do not take into account factors involved in the stability of the synapse and the resolution of the recombination reaction.
Here, we present in vitro and in vivo data showing that the frequency and orientation of cassette insertion is ensured by the interplay of all three attC site structural features: EHBs, UCS and VTS. Moreover, the experimental evidence for the importance of the unpaired features is supported by the bioinformatics analysis that uncovered a strong nucleotide bias between the two strands, favoring the folding of the bottom strand.
MATERIALS AND METHODS
Bacterial strains and media
Detailed information on bacterial strains and media used in this study can be found in Supplementary Materials S1.
Plasmids
Wild-type and mutant attC sites were constructed by annealing of two or four overlapping phosphorylated oligonucleotides (Supplementary Table S1A), fully complementary except for the overhangs corresponding to EcoRI and BamHI restriction sites. The sites were then ligated into the p4116 plasmid for the delivery of the bottom strand, and into p4117 plasmid for the delivery of the top strand, both digested with EcoRI/BamHI. The plasmids were then transformed into the cloning strain Π1, a [Pir+] DH5α derivative that requires deoxythymidine to grow in LB media, and their sequence was verified by sequencing. The plasmids were then transformed into β2163 strain (Supplementary Table S2).
Suicidal conjugation assay
The suicidal conjugation assay (Supplementary Figure S1) for measuring the recombination frequencies of attC sites and strand selectivity is based on the previously described protocol (13). Detailed information can be found in Supplementary Materials S3–S5.
Electrophoretic Mobility Shift Assay (EMSA)
Each reaction contained 500 ng Poly[d(I-C)], 12 mM Hepes–NaOH pH 7.7, 12% glycerol, 4 mM Tris–HCl pH 8.0, 60 mM KCl, 1 mM EDTA, 0.06 mg/ml BSA, 1 mM DTT, 10% Tween 20, 0.01 nM specified 32P-labeled DNA oligonucleotide (Supplementary Table S2C) and the specified quantities of purified MBP-tagged IntI1Y312F (mutant unable to cleave DNA(12)), in a final volume of 20 μl. The samples were incubated at 30°C for 10 min without the probe followed by 20 min with the probe, then loaded to a 5% native polyacrylamide gels with 0.5x TBE as running buffer. The gels were visualized using a Molecular Dynamics intensification screen and a Typhoon FLA 9500 laser scanner.
Bioinformatics analysis of attC site database
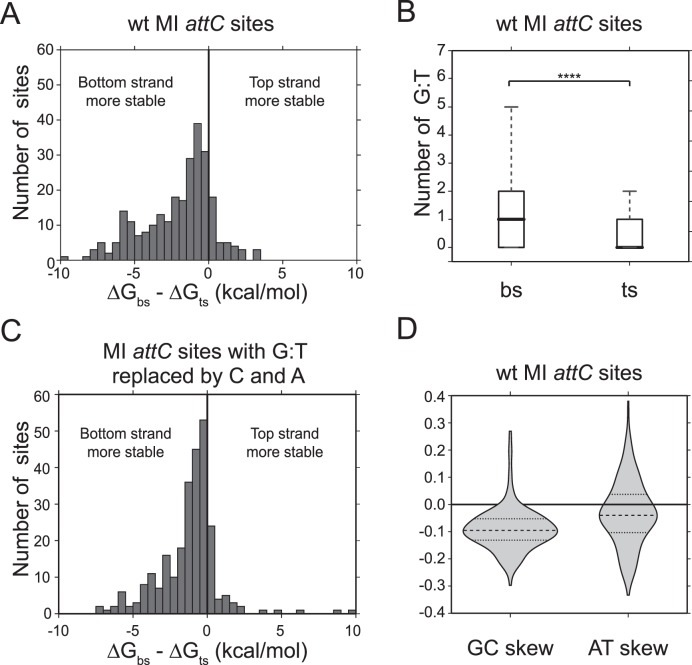
Sequences of 263 MI attC sites having less than a 95% identity were obtained from the authors of INTEGRALL database (36). We predicted attC site folding using the RNAfold program from the ViennaRNA2 package (37) (Supplementary Material S2). The mean ΔG difference between bottom and top strand folding was calculated from the differences in minimum free energy (MFE) ΔG values for each site. To investigate the bias of G:T base pairing, their number was calculated for each strand of each site, based on the predicted MFE structure. To assess the contribution of G:T base pairing to the ΔG bias, for each strand the occurring G:T pairs were replaced by non-pairing C and A. The folding of the modified sequences was predicted, and the mean ΔG difference between bottom and top strand folding was recalculated from the differences in MFE ΔG values for each site. GC skew measures the abundance of Guanines (G) compared to Cytosines (C) on the top strand: (G − C)/(G + C); AT skew measures the abundance of Adenines (A) compared to Thymines (T) on the top strand: (A − T)/(A + T).
RESULTS
The level of strand selectivity varies among attC sites
As a first step toward understanding which attC site features are responsible for the selectivity of the integrase for the bottom strand, we studied the recombination of several paradigmatic wild-type attC sites using a previously described suicidal conjugation assay (13) (Supplementary Figure S1 and Supplementary Materials S3–S5). Briefly, our assay consists of delivering a single strand of a plasmid, carrying a resistance marker and either a top or a bottom strand of an attC site into a recipient Escherichia coli strain containing a plasmid carrying an attI1 site and expressing IntI1. This assay is designed to measure the frequency of attI1×attC recombination by comparing the number of recombined cells having acquired the resistance marker, and the total number of recipient cells. The delivery of single-stranded substrates mimics the natural conditions in which the acquisition of cassettes occurs through horizontal gene transfer and has the advantage of delivering an already folded form of the attC site, decoupling the parameters influencing recombination itself from those related to prior folding (38). Also, the delivery of a single-stranded substrate allows studying the recombination of bottom and top strands separately. Upon the delivery of a single strand by conjugation, the complementary strand can be eventually resynthesized. Thus, even when the top strand is delivered, the recombination can happen in the bottom strand (after its resynthesis), instead of the top strand. In our assay, we are able to distinguish between top and bottom strand recombination by performing PCR reactions on the recombined products (Supplementary Figure S1 and Supplementary Material S5).
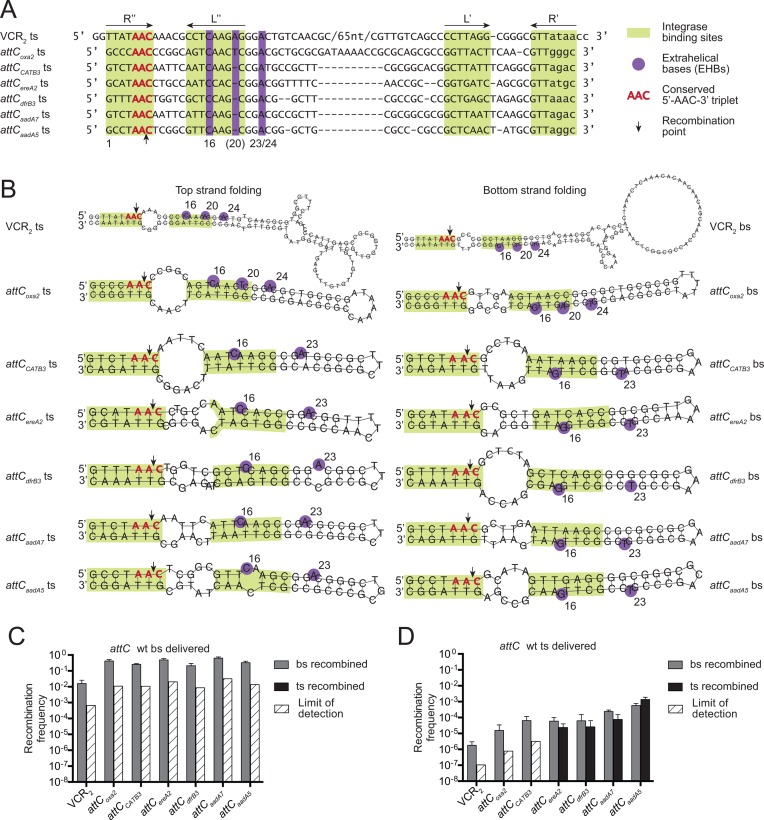
In this study we measured the frequencies of attI1×attC recombination performed by MI Class 1 integrase IntI1 on both SCI and MI attC sites in order to determine whether they share the same strand specificity determinants. On one hand, we tested VCR2 which is an attC site of the V. cholerae superintegron, but shares a 90% identity with VCR-like attCblaP3 found in MIs (2). On the other hand, we tested attCaadA7, attCaadA5, attCCATB3, attCdfrB3, attCereA2 and attCoxa2 from MIs associated with IntI1 (Figure 2A). Contrary to the 133bp long VCR2, these sites are much shorter: only 57–70 bp in length. We have selected these MI attCs to represent sites similar to the well-studied attCaadA7 by sequence and structure (e.g. attCCATB3), as well as sites presenting fewer similarities (e.g. attCereA2). Most of these small MI sites have only two EHBs, except for attCoxa2 that has three EHBs, like VCR2. The predicted structures of top and bottom strands show that they are asymmetrical due to the presence of the three unpaired features (Figure 2B).
Figure 2.
Wild-type attC site recombination. (A) Alignment of top strands of attC sites, based on CLUSTAL W 2.1 results, with manual adjustments to align the complementary L and R boxes and the EHBs. (B) Structures of top and bottom strands of attC sites, predicted by RNAfold program from the ViennaRNA2 package (37), with imposed constraints to pair the L and R boxes. (C and D) Recombination frequencies and recombination point localizations for different wild-type attC sites upon the delivery of the bottom strand (C) or the top strand (D). The bars correspond to recombination frequencies of each strain: grey bars for recombination of the bottom strand; black for recombination of the top strand. White cross-hatched bars correspond to the limits of detection by PCR (Supplementary Material S5), when recombination events of a particular strand were not observed. Values represent the mean of at least three independent experiments and error bars correspond to average deviations from the mean.
As expected, for all attC sites the delivery of the bottom strand leads to recombination at high efficiency (1.6 × 10−2 to 4.9 × 10−1, Figure 2C). Recombination took place exclusively in the delivered bottom strand, as required for cassette insertion in correct orientation.
Upon the delivery of the top strand, recombination frequencies decreased by several orders of magnitude, illustrating strand selectivity (Figure 2D). However, in terms of the DNA strand recombined (delivered top strand or resynthesized complementary bottom strand), attC sites behaved differently. For attCoxa2 and attCCATB3, we observed a similar effect as the one described for VCR2: even upon the delivery of the top strand, the recombination took place in the resynthesized bottom strand, though at a much lower rate than upon its delivery (Figure 2C versus Figure 2D). Such drop in recombination frequency is most likely due to the efficiency of resynthesis. Results obtained for VCR2, attCoxa2 and attCCATB3 confirm previous in vitro and in vivo observations showing that the integrase has no affinity for the top strand of wild-type attC sites, and is very selective toward the bottom strand (11,12).
Yet for other sites, such as attCereA2, attCdfrB3, attCaadA7 and attCaadA5, we observe less strand selectivity. Indeed, upon the delivery of the top strand of these sites (Figure 2D), the recombination in the delivered top strand occurs at frequencies similar to those of recombination in the resynthesized bottom strand. This means that upon the delivery of the top strand, the cassette has a similar probability of being inserted in either of the two orientations. However, the frequencies obtained in this case are several orders of magnitude lower than those obtained upon bottom strand delivery (Figure 2C), which reflects the fact that despite being capable of recombining the top strand, the integrase has a much lower affinity toward it.
These results show that the level of strand selectivity varies broadly among attC sites. This is not unexpected, given that the unpaired structural features that are likely responsible for strand selectivity also differ among sites. However, to exclude the possibility that these various levels of selectivity are due to differences in sequence rather than structure, we tested several mutants of VCR2 and attCaadA7, where we exchanged the sequences of integrase-binding sites (R boxes) between them (Supplementary Figure S2). Despite these changes, the integrase continued to show high strand selectivity for VCR2 and lower selectivity for attCaadA7 (even though the ratios of bottom and top strand recombination events varied for attCaadA7 mutants), confirming that strand selectivity is essentially ensured by structural features.
In order to understand how the unpaired structural features (EHBs, UCS or VTS) influence strand selectivity, we selected three attC sites for a more thorough investigation: VCR2 from the V. cholerae SCI and two short sites from MIs: attCoxa2 which has 3 EHBs like VCR2, and attCaadA7 which has 2 EHBs like most MI attC sites.
Localization of EHBs influences recombination frequency and strand selectivity
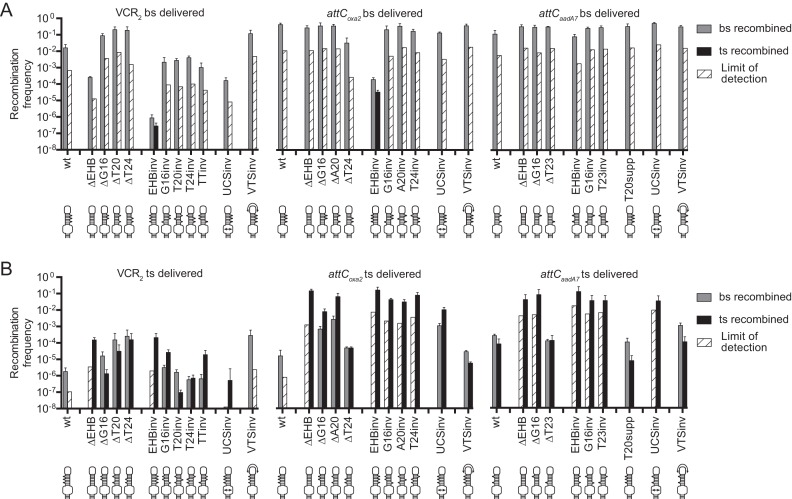
EHB deletions
First, we studied the effects of EHBs by the suicidal conjugation assay, either when these bases are deleted together, or one by one.
We found that the deletion of all EHBs at once decreases 50-fold the recombination of VCR2 when its bottom strand is delivered (Figure 3A, VCR2 bs ΔEHB), which reflects the importance of EHBs for efficient recombination. Upon the delivery of the top strand, EHB deletion has the effect of both increasing the recombination efficiency and inverting strand selectivity toward recombination in the top strand (Figure 3B, VCR2 ts ΔEHB). Interestingly, the deletion of EHBs of VCR2 one by one does not have the same effect as their combined deletion. When a bottom strand lacking only one of the EHBs is delivered, its recombination increases relative to the wild-type site, suggesting that any EHB by itself is not required for a high-efficiency recombination (Figure 3A, VCR2 bs ΔG16, ΔT20, ΔT24). The delivery of the top strand of VCR2 mutants missing one of the EHBs increases the recombination in both bottom and top strands relative to the wild-type site (Figure 3B, VCR2 ts ΔG16, ΔT20, ΔT24), suggesting that the absence of EHBs favors top strand recombination.
Figure 3.
Recombination frequencies and recombination point localizations for different attC site mutants upon the delivery of the bottom strand (A) or the top strand (B). The bars correspond to recombination frequencies of each strain: grey bars for recombination in the of the bottom strand; black for recombination of the top strand. White cross-hatched bars correspond to the limits of detection by PCR (Supplementary Material S5), when recombination events of a particular strand were not observed. Values represent the mean of at least three independent experiments and error bars correspond to average deviations from the mean. EHB, Extrahelical Bases; UCS, Unpaired Central Spacer; VTS, Variable Terminal Structure; inv, inversion; supp, supplementary. Schemes below the graphs represent the structure of corresponding attC site mutants.
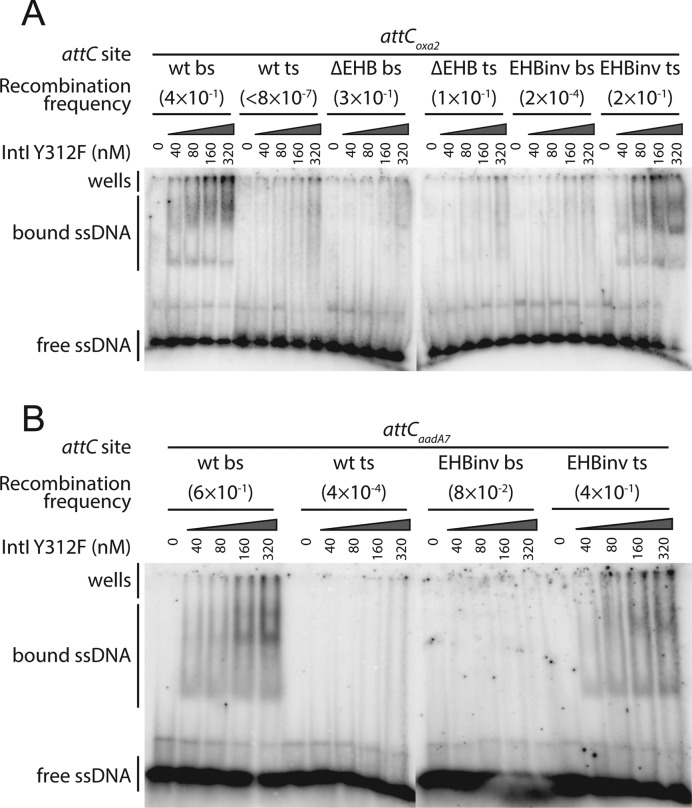
For the other two sites, the deletion of each one or all EHBs does not produce any noteworthy effect upon bottom strand delivery (Figure 3A, attCoxa2 bs ΔEHB, ΔG16, ΔA20, ΔT24, attCaadA7 bs ΔEHB, ΔG16, ΔT23), suggesting that for some attC sites EHBs are not essential for efficient recombination. Upon top strand delivery, EHB deletion from attCoxa2 or attCaadA7 has a general effect of favoring recombination in the top strand and in no case decreases the recombination efficiency (Figure 3B, attCoxa2 ts ΔEHB, ΔG16, ΔA20, ΔT24, attCaadA7 ts ΔEHB, ΔG16, ΔT23). In many cases, EHB deletions (either one by one, or combined) lead to almost identical recombination frequencies upon the delivery of either bottom or top strand. In order to determine whether this is a consequence of efficient IntI1 binding to both strands, we performed an Electrophoretic Mobility Shift Assay (EMSA) on bottom and top strands of attCoxa2 ΔEHB, and compared them to results obtained for wt attCoxa2. As previously demonstrated in EMSA experiments (11–13), the integrase binds only the bottom and not the top strand of wt attC sites, which we also observed for attCoxa2 (Figure 4A). This is consistent with the recombination results, where the bottom strand of attCoxa2 is recombined at the frequency of 4 × 10−1 and the top strand is recombined 5–6 orders of magnitude less efficiently (Figure 2C and D). However, for attCoxa2 ΔEHB the recombination results show highly efficient recombination of both bottom and top strands (3 × 10−1 and 1 × 10−1), whereas neither of these sites is efficiently bound by the integrase in the in vitro assay (Figure 4A), suggesting that binding affinity is not the only factor influencing recombination frequency.
Figure 4.
Electrophoretic Mobility Shift Assay (EMSA). EMSA was performed as described in Materials and Methods in the presence of the indicated amounts of IntI1. (A) Oligonucleotides comprising 80–83 nt of either bottom or top strand of attCoxa2 wt, attCoxa2 ΔEHB and attCoxa2 EHBinv were labeled with 32P and used at a concentration of 0.01 nM in each binding reaction. (B) Oligonucleotides comprising 72–74 nt of either bottom or top strand of attCaadA7 wt and attCaadA7 EHBinv were labeled with 32P and used at a concentration of 0.01 nM in each binding reaction.
EHB inversions
To further investigate the role of EHBs, we tested mutants where one or all of these bases are inverted. The inversion of EHBs serves the purpose of making the mutated top strand spatially resemble the wild-type bottom strand: now, the EHBs on the top strand are located on the arm opposite the 5′-AAC-3′ triplet (Supplementary Figure S3), while on the bottom strand EHBs are now located on the same arm as the triplet.
For this set of mutations, the three sites behave in a slightly different way. The inversion of any EHB of VCR2 decreases the recombination frequency of the delivered bottom strand (Figure 3A, VCR2 bs G16inv, T20inv, T24inv), and the inversion of all three EHBs has a combined effect of drastically decreasing its recombination (>4 orders of magnitude), to an extent where we could detect recombination in the resynthesized top strand (Figure 3A, VCR2 bs EHBinv). EHB inversion on the top strand remarkably favors its recombination, especially when all EHBs are inverted (Figure 3B, Figure 3A, VCR2 ts EHBinv, G16inv, T20inv, T24inv). These results suggest a cumulative effect of the EHBs. To verify this hypothesis, we inverted both extrahelical T bases and indeed found an additive effect compared to the inversion of either extrahelical T alone (Figure 3A, VCR2 bs TTinv and Figure 3B, VCR2 ts TTinv).
For attCoxa2, when any one of its EHBs is inverted, both bottom and top strands recombine very efficiently (Figure 3A, attCoxa2 bs G16inv, A20inv, T24inv and Figure 3B, attCoxa2 ts G16inv, A20inv, T24inv), which is very different from the behavior of VCR2 mutants. However, just like for VCR2, the inversion of all three EHBs drastically decreases bottom strand recombination upon its delivery (by >3 orders of magnitude, Figure 3A, attCoxa2 bs EHBinv). Upon attCoxa2 top strand delivery, EHB inversion allows its more efficient recombination, raising its frequency up to the same order of magnitude as that of the wild-type bottom strand (1.6 × 10−1, Figure 3B, attCoxa2 ts EHBinv). In order to determine whether the inversion of EHBs changes integrase specificity from the bottom to the top strand, we performed EMSA on these mutants, and indeed found that IntI1 efficiently binds the top strand of attCoxa2 EHBinv, but not the bottom strand (Figure 4A).
attCaadA7 has only two EHBs, and the inversion of either or both of them allows equally efficient recombination of both bottom and top strands (Figure 3A, attCaadA7 bs EHBinv, G16inv, T23inv and Figure 3B, attCaadA7 ts EHBinv, G16inv, T23inv). In order to determine whether upon the inversion of EHBs the integrase is able to bind both attCaadA7 strands, we performed EMSA on these sites (Figure 4B). Surprisingly, we discovered that IntI1 was only able to efficiently bind the top strand. This observation, together with the fact that attCaadA7 bottom strand recombination did not decrease upon EHB inversion as it did for other attC sites, suggests that other unpaired features must influence strand selectivity and favor recombination of the bottom strand.
The number of EHBs
To test whether wild-type attCaadA7 behaves differently from VCR2 and attCoxa2 (Figure 2D) because it has only two EHBs, we added a supplementary extrahelical T in the position where the other two sites have their third EHB. The results show that this mutation increased strand selectivity toward the recombination of the bottom strand, which can be seen upon top strand delivery (Figure 3B, attCaadA7 ts T20supp). Together with the results of T20 deletion in VCR2 and attCoxa2, this leads us to conclude that the presence of a third EHB in position 20 has a cumulative effect in increasing strand selectivity.
UCS and VTS influence recombination frequency and strand selectivity
As suggested by our results, EHBs must not be the only structural feature influencing strand selectivity. We therefore sought to investigate the roles of the two other unpaired structures, UCS and VTS, by testing mutants in which these structures were inverted.
UCS inversion
Similarly to the inversion of EHBs, the inversion of UCS consists of exchanging its sequence between the two strands in a way that the mutated top strand resembles the wild-type bottom strand. Such inversion highly affects all three tested sites (Figure 3), and has different effects depending on the site. These differences were not unexpected, since UCS regions vary not only between the top and the bottom strands, but also between the sites.
For VCR2, UCS inversion decreases the recombination of the bottom strand 100-fold (Figure 3A, VCR2 bs UCSinv): to the same extent as the deletion of all EHBs. Upon the delivery of the top strand, UCS inversion changes strand selectivity toward top strand recombination (Figure 3B, VCR2 ts UCSinv). For attCoxa2, UCS inversion does not notably affect the recombination when the bottom strand is delivered (Figure 3A, attCoxa2 bs UCSinv), but upon top strand delivery recombination increases both in bottom and top strands (Figure 3B, attCoxa2 ts UCSinv). For attCaadA7, UCS inversion also does not notably affect the recombination when its bottom strand is delivered (Figure 3A, attCaadA7 bs UCSinv), but changes strand selectivity toward recombination in the top strand upon its delivery (Figure 3B, attCaadA7 ts UCSinv).
VTS inversion
VTS inversion as well can influence recombination frequency and strand selectivity of attC sites (Figure 3). For VCR2, after the inversion of its long VTS, recombination in the bottom strand increases upon the delivery of either strand (Figure 3A, VCR2 bs VTSinv and Figure 3B, VCR2 ts VTSinv). Conversely, upon the delivery of attCoxa2 top strand, VTS inversion increases recombination only in the top strand (Figure 3B, attCoxa2 ts VTSinv). For attCaadA7, VTS inversion does not lead to any important changes (Figure 3A, attCaadA7 bs VTSinv and Figure 3B, attCaadA7 ts VTSinv). We conclude that both UCS and VTS can influence attC site recombination frequency and strand selectivity, but the effects of their inversions is dependent on the site and the sequences of its UCS and VTS.
Nucleotide skew in attC sites
As the sequences of unpaired UCS and VTS are different between the two strands, the nucleotide content and distribution can also differ. We believe that this nucleotide skew might contribute to strand selectivity and explain the effects of UCS and VTS inversions seen in attC site mutants. Indeed, the analysis of 263 attC sites provided by the authors of the INTEGRALL database (36) showed that the ΔG of bottom strand folding is on average by 2.12 kcal/mol lower than the ΔG of top strand folding (7.6% of total ΔG), suggesting that folded bottom strands are more stable (Figure 5A).
Figure 5.
Analysis of 263 attC sites of Mobile Integrons from the INTEGRALL database (36). ΔG values were calculated with RNAfold program from the ViennaRNA2 package (37). (A) Difference in ΔG (kcal/mol) between bottom and top strands of wt attC sites. (B) Difference between the mean number of G:T mispairs on the bottom and top strand (t-test, P < 0.0001). (C) Difference in ΔG (kcal/mol) between bottom and top strands of sites where G:T mispairs were replaced by non-pairing C and A. (D) GC and AT skews calculated for the attC site top strand. Negative skews correspond to the enrichment of purines Guanine (G) and Adenine (A) on the bottom strand.
One of the reasons for this skew could be the presence of G:T mispairs on the bottom strand that form hydrogen bonds and stabilize it, compared to the top strand, which has non-pairing C and A in the corresponding positions. Indeed, the analysis of this attC site list showed that on average there are 1.24 G:T mispairs on the folded bottom strand and only 0.39 on the top strand, favoring bottom strand folding and recombination (Figure 5B).
However, the differential distribution of G:T mispairs does not account for all the ΔG difference between the two strands: even when the G:T pairings are replaced by the non-pairing C and A, the bottom strand is predicted to have on average a 1.28 kcal/mol lower energy of folding than the top strand, and thus to be more stable (Figure 5C). We suppose that this might be due to purine skew, as purines (guanines and adenines) have a higher self-stacking tendency than pyrimidines (39). Both GC and AT skews of MI attC sites are predominantly negative (Figure 5D), meaning that the bottom strand is enriched in purines, especially in guanines. This might explain why in most MI attC sites, the VTS of the bottom strand has a conserved 5′-GAA-3′ sequence, only composed of purines: as their presence in the unpaired regions stabilizes the structure, the sequence of the VTS must thus contribute to strand selectivity.
DISCUSSION
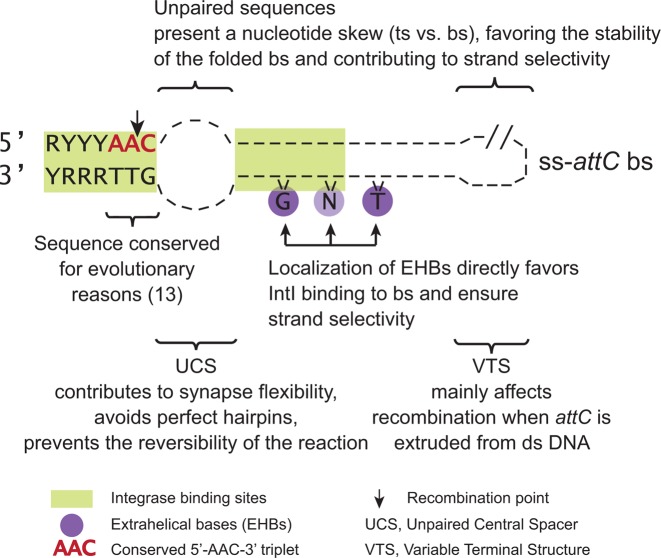
As previously mentioned, strand selectivity shown by the integrase in attI×attC recombination is essential for inserting cassettes in correct orientation relative to the Pc promoter. This allows expression of the encoded promoterless genes and thus ensures integron functionality. We have previously addressed the question of strand selectivity by studying the structural features of VCR2 (17). The conclusions of that article were that (i) the strand choice is mainly determined by the first EHB, while the other two EHBs increase recombination frequency; (ii) the structure of the UCS is critical for high-efficiency recombination but not for strand selectivity; and (iii) the VTS has no role in strand selectivity. Here, we conducted a more thorough study of VCR2 by retesting some of the mutants, as well as constructing and testing supplementary ones. This in-depth analysis allowed us to observe and quantify cases where recombination in bottom and in top strands occur at a comparable rate, which was not always detected in our previous study. Also, we have extended this study to attCs of MIs by testing six wild-type sites and submitting two of them to the same in-depth mutational analysis as VCR2. Moreover, our in vitro and in vivo results show that apart from integrase binding, other factors contribute to recombination efficiency and strand selectivity. This work allowed us to adjust previous statements (12,17), provide a more solid evidence for the roles of attC site structural features in recombination efficiency and strand selectivity, and uncover a strong nucleotide skew between the two strands, which could explain how UCS and VTS contribute to strand selectivity (Figure 6).
Figure 6.
Roles of attC site structural features during attI1×attC recombination.
EHBs are important, yet not the only determinants of strand selectivity
Results presented here confirmed that in all attC sites EHBs are important determinants of strand selectivity. This is accomplished by their localization on the same arm as the 5′-AAC-3′ triplet for the top strand, and opposite the triplet for the bottom strand. This leads to steric differences between the two strands: on the bottom strand, EHBs are located so as to lodge in the hydrophobic pocket of the integrase or within the reach of the interacting amino acids (16), whereas on the top strand they are positioned elsewhere and are most likely hindering integrase binding. Interestingly, we could not attribute this strand selectivity to the presence of any particular EHB, as it was previously suggested for G16 (16,17). Instead, after studying several attCs, and in particular small MI sites, we have seen that the extent to which the deletion and inversion of each EHB influences strand selectivity is different depending on the site, and that the effects are cumulative. For instance in attCoxa2, T20 seems to have an even greater importance for strand selectivity than G16. We believe that the difference between the two statements can be explained by the fact that previous works (16,17) corroborated their findings by the study of attC×attC recombination, whereas here we restrained our study to attI1×attC reactions, as we believe that the two reactions are not regulated identically.
For attC×attC recombination, the crystal structure of the corresponding synapse showed that the binding of T24 EHB by integrase monomers in cis prevents the nucleophilic attack in the L box, while the binding of G16 bases by integrase monomers in trans stabilizes the synapse. However, the ss-attC and ds-attI are structurally very different substrates. The crystal structure of the integrase bound to its substrates in an attI×attC synapse has not been resolved yet, mainly due to a lower affinity of the integrase for attI (11). Thus, we can only speculate about the overall structure of the synaptic complex formed by IntI1 during an attI×attC reaction and about the precise interactions of the integrase with each of the EHBs (Supplementary Figure S3). Hence, it is possible that the impact of EHBs on synapse assembly and recombination varies between complexes where the attC site is faced by another attC, or by an attI site. In fact, a previously described integrase mutagenesis and selection experiment already suggested that there is a balance between the high tolerance of the integrase toward such different substrates on one hand, and its capacity to efficiently assemble the synapse and perform recombination on the other (40).
Present work shows that EHBs are not the only determinants of strand selectivity, as it was previously thought (17). Indeed, the fact that recombination frequencies of top and bottom strands are not inverted upon EHB inversion, and that attC sites can be efficiently recombined even though they are not bound by IntI1 in vitro, clearly indicate that binding is not limiting in vivo and that other features strongly contribute to recombination efficiency and strand selectivity.
The UCS and the VTS contribute to strand selectivity by stabilizing the folded bottom strand
Our results show that UCS and VTS not only influence recombination frequency, but also play an important role in strand selectivity. Even though these unpaired structural features are not bound by the integrase (11,16) and do not determine strand selectivity in in vitro binding experiments (12), they most likely influence the recombination indirectly, by stabilizing the folded bottom strand or influencing the stability of the synapse. Indeed, this would explain the discrepancies between our in vitro results where the location of EHBs alone influences IntI1 binding through specific protein-DNA interactions, and our in vivo results where additional factors affecting synapse stabilization and recombination must be involved.
It has been suggested that the UCS, located between two integrase binding sites, might provide the flexibility necessary for the formation of the synapse. It could also avoid the formation of perfect hairpins that could possibly be deleterious for the cell, and contribute to the unidirectionality of the recombination reaction (16,17). Indeed, after the first strand exchange, the reverse reaction would likely be more favored in the case of a perfect hairpin, where the complementarity between the R′-L′ and R″-L″ regions would facilitate their pairing and the reconstitution of initial DNA substrates. The role of the UCS in synaptic flexibility is supported by an integrase mutagenesis and selection experiment (40), in which the selection of IntI1 mutants performing high-efficiency attI1×attC recombination of the VCR2 with a paired UCS yielded a predominant P109L mutant, which was predicted to have a higher flexibility in accommodating the substrates than the wild-type IntI1. Moreover, the P109L mutant showed lower recombination efficiency in attC×attC reactions, supporting the hypothesis that the two types of synapses could have different structural requirements for efficient recombination, and that attC site mutations could influence the two reactions in different ways.
The VTS plays a regulatory role in attC site folding, especially during cruciform extrusion from ds-DNA. In this process the formation of a hairpin on each strand of a symmetric and paired ds-DNA molecule is favored by DNA supercoiling (38,41). When the attC site has to be extruded from a ds-DNA, the length of its VTS is inversely correlated with the recombination frequency (38). This is in accordance with the results of in vitro single-molecule studies of cruciform extrusion (42). Apart from its length, the sequence of the VTS can also influence its recombination frequency by promoting hairpin stability (12) and changing the propensity of attC sites to form non-recombinogenic structures (38). We have previously shown that these two parameters only matter in the case of cruciform formation (38). Here, we have seen that the VTS can modulate recombination frequency and strand selectivity even in conjugative conditions and thus play a role in ensuring cassette insertion in the correct orientation, especially when the recombinogenic strand has to be resynthesized and then extruded from the ds-DNA.
The unpaired nature of EHBs, UCS and VTS is extremely important for their roles, as it allows the discrimination of the two strands. Indeed, between the bottom and top strands, these structures differ not only in their localization relative to the 5′-AAC-3′ triplet, but also in their nucleotide composition, as we have seen from the bioinformatics analysis. It is not entirely clear whether the observed nucleotide skews have been selected for their impact on strand selectivity, or if they appeared as a consequence of another phenomenon that favors the enrichment of purines on the bottom strand. However, the fact that these skews are so conserved among the MI attC sites reflects their biological importance.
Different levels of strand selectivity among attC sites
We have discovered that wild-type attC sites can in principle be recombined in the top strand when it is delivered, even though this event happens at around 1000–10 000 times lower frequency than the recombination in bottom strand upon its delivery. This suggests that the insertion of cassettes in wrong orientation due to recombination in the top strand is highly unlikely. However, when one compares the recombination frequency of the attCaadA5 top strand (Figure 2D) and the recombination frequency of the VCR2 bottom strand (Figure 2C), the insertion of attCaadA5 cassette in the wrong orientation happens only at a 12 times lower frequency, meaning that such events, though very rare, should still occur. Nonetheless, cassettes inserted in the orientation facing the promoter have never been observed in MIs nor SCIs.
We believe that this can be mainly explained by the fact that in natural conditions, the availability of bottom and top attC strands for attI×attC reaction must not be equimolar. Indeed, in a biological context, the circularized cassettes would mostly contain the bottom strand of attC sites due to the selectivity of the integrase for the bottom strand during the attC×attC excision reaction that generates these cassettes. It is not clear if, upon excision, circularized cassettes are maintained in their single-stranded form or if the complementary strand is resynthesized. However, even if the second is true and prior to recombination, the attC site has to be extruded from the double-stranded circularized form, the bottom strand would still be favored because of its lower ΔG that facilitates extrusion. Thus, the observed cassette insertion exclusively in the correct orientation can be explained by two consecutive selections of the bottom strand: first upon its excision and then upon its insertion. Moreover, the absence of cassettes facing the promoter in the sequenced integrons, especially from clinical isolates, could also be explained by the lack of selective advantage of such recombinants. Indeed, integron recombination is induced by the SOS response (6) in conditions such as the presence of antibiotics. The newly inserted cassette would thus be immediately tested for adaptive value, and only systems having integrated the antibiotic resistance cassette in correct orientation would be selected. In SCIs, the function of most cassettes is unknown but is also likely to be involved in adaptation to stressful conditions and be selected for, especially as cassette expression can also be dependent on the SOS response (43).
In conclusion, we have shown here the roles of attC site structural features during an attI1×attC insertion reaction (Figure 6), which result in preferential recombination of the bottom strand and lead to cassette insertion in correct orientation, allowing the expression of encoded genes from the Pc promoter of the integron platform. Strand selectivity is thus ensured by the interplay of the three unpaired structural features, due both to their dissimilarities in structure and differences in nucleotide composition. Such complex regulation is only possible because of the single-stranded nature of the attC substrate.
The understanding of factors controlling strand selectivity could lead to the development of approaches targeting integron cassette exchanges and potentially disrupting the spread of antibiotic resistance genes. Also, a synthetic integron has been developed as a tool for metabolic pathway optimization (44). For such biotechnological applications, correct orientation of cassettes is essential. The high level of attC site modularity opens the possibility of generating synthetic sites with customized sequences, but it is crucial to understand the features that need to be preserved, especially if one wants such synthetic systems to recombine efficiently and maintain cassette expression.
Supplementary Material
Acknowledgments
We would like to thank Thomas Jové for sharing the list of MI attC sites from the INTEGRALL database, and Jean Cury for his advice on bioinformatics analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut Pasteur, the Centre National de la Recherche Scientifique, the French Government's Investissement d'Avenir program Laboratoire d'Excellence ‘Integrative Biology of Emerging Infectious Diseases’ [ANR-10-LABX-62-IBEID]; French National Research Agency [ANR-12-BLAN-DynamINT; Paris Descartes University – Sorbonne Paris Cité and École Doctorale Frontières du Vivant (FdV) – Programme Bettencourt (to A.N.); Marie Curie Intra-European Fellowship for Career Development [FP-7-PEOPLE-2011-IEF, ICADIGE to J.A.E.]. Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1.Escudero J.A., Loot C., Nivina A., Mazel D. The integron: adaptation on demand. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0019-2014. doi:10.1128/microbiolspec.MDNA3-0019-2014. [DOI] [PubMed] [Google Scholar]
- 2.Mazel D., Dychinco B., Webb V.A., Davies J. A distinctive class of integron in theokok cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 3.Rowe-Magnus D.A., Guerout A.M., Ploncard P., Dychinco B., Davies J., Mazel D. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. U.S.A. 2001;98:652–657. doi: 10.1073/pnas.98.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cambray G., Guerout A.-M., Mazel D. Integrons. Annu. Rev. Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 5.Stokes H.W., Hall R.M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 6.Guerin E., Cambray G., Sanchez-Alberola N., Campoy S., Erill I., Da Re S., González-Zorn B., Barbé J., Ploy M.-C., Mazel D. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 7.Naas T., Mikami Y., Imai T., Poirel L., Nordmann P. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 2001;183:235–249. doi: 10.1128/JB.183.1.235-249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidelberg J.F., Eisen J.A., Nelson W.C., Clayton R.A., Gwinn M.L., Dodson R.J., Haft D.H., Hickey E.K., Peterson J.D., Umayam L., et al. DNA sequence of both chromosomes of the cholera pathogen : Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe-Magnus D.A., Guerout A.-M., Mazel D. Super-integrons. Res. Microbiol. 1999;150:641–651. doi: 10.1016/s0923-2508(99)00127-8. [DOI] [PubMed] [Google Scholar]
- 10.Rowe-Magnus D.A., Guerout A.-M., Biskri L., Bouige P., Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francia M.V., Zabala J.C., la de Cruz F., García Lobo J.M. The IntI1 integron integrase preferentially binds single-stranded DNA of the attC site. J. Bacteriol. 1999;181:6844–6849. doi: 10.1128/jb.181.21.6844-6849.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson C., Kamali-Moghaddam M., Sundström L. Integron integrase binds to bulged hairpin DNA. Nucleic Acids Res. 2004;32:4033–4043. doi: 10.1093/nar/gkh730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouvier M., Demarre G., Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frumerie C., Ducos-Galand M., Gopaul D.N., Mazel D. The relaxed requirements of the integron cleavage site allow predictable changes in integron target specificity. Nucleic Acids Res. 2010;38:559–569. doi: 10.1093/nar/gkp990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall R.M., Brookes D.E., Stokes H.W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald D., Demarre G., Bouvier M., Mazel D., Gopaul D.N. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 17.Bouvier M., Ducos-Galand M., Loot C., Bikard D., Mazel D. Structural features of single-stranded integron cassette attC sites and their role in strand selection. PLoS Genet. 2009;5:e1000632. doi: 10.1371/journal.pgen.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larouche A., Roy P.H. Effect of attC structure on cassette excision by integron integrases. Mob. DNA. 2011;2:3. doi: 10.1186/1759-8753-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokes H.W., O'Gorman D.B., Recchia G.D., Parsekhian M., Hall R.M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 20.Cury J., Jové T., Touchon M., Néron B., Rocha E.P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016;44:4539–4550. doi: 10.1093/nar/gkw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunes-Düby S.E., Kwon H.J., Tirumalai R.S., Ellenberger T., Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Val M.-E., Bouvier M., Campos J., Sherratt D., Cornet F., Mazel D., Barre F.-X. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Midonet C., Barre F.-X. Xer site-specific recombination: promoting vertical and horizontal transmission of genetic information. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0056-2014. doi:10.1128/microbiolspec.MDNA3-0056-2014. [DOI] [PubMed] [Google Scholar]
- 24.Collis C.M., Hall R.M. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 1992;6:2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 25.Collis C.M., Grammaticopoulos G., Briton J., Stokes H.W., Hall R.M. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 26.Hansson K., Sköld O., Sundström L. Non-palindromic attl sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol. Microbiol. 1997;26:441–453. doi: 10.1046/j.1365-2958.1997.5401964.x. [DOI] [PubMed] [Google Scholar]
- 27.Escudero J.A., Loot C., Parissi V., Nivina A., Bouchier C., Mazel D. Unmasking the ancestral activity of integron integrases reveals a smooth evolutionary transition during functional innovation. Nat. Commun. 2016;6:1–12. doi: 10.1038/ncomms10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grindley N.D.F., Whiteson K.L., Rice P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 29.Loot C., Ducos-Galand M., Escudero J.A., Bouvier M., Mazel D. Replicative resolution of integron cassette insertion. Nucleic Acids Res. 2012;40:8361–8370. doi: 10.1093/nar/gks620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tolmasky M.E., Crosa J.H. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 31.Szekeres S., Dauti M., Wilde C., Mazel D., Rowe-Magnus D.A. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 32.Guerout A.-M., Iqbal N., Mine N., Ducos-Galand M., Van Melderen L., Mazel D. Characterization of the phd-doc and ccd toxin-antitoxin cassettes from Vibrio superintegrons. J. Bacteriol. 2013;195:2270–2283. doi: 10.1128/JB.01389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bissonnette L., Champetier S., Buisson J.P., Roy P.H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biskri L., Mazel D. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. Agents Chemother. 2003;47:3326–3331. doi: 10.1128/AAC.47.10.3326-3331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Fonseca É.L., Vicente A.C.P. Functional characterization of a Cassette-specific promoter in the class 1 integron-associated qnrVC1 gene. Antimicrob. Agents Chemother. 2012;56:3392–3394. doi: 10.1128/AAC.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moura A., Soares M., Pereira C., Leitão N., Henriques I., Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 37.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loot C., Bikard D., Rachlin A., Mazel D. Cellular pathways controlling integron cassette site folding. EMBO J. 2010;29:2623–2634. doi: 10.1038/emboj.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigel A., Operschall B.P., Sigel H. Comparison of the π-stacking properties of purine versus pyrimidine residues. Some generalizations regarding selectivity. J. Biol. Inorg. Chem. 2014;19:691–703. doi: 10.1007/s00775-013-1082-5. [DOI] [PubMed] [Google Scholar]
- 40.Demarre G., Frumerie C., Gopaul D.N., Mazel D. Identification of key structural determinants of the IntI1 integron integrase that influence attC x attI1 recombination efficiency. Nucleic Acids Res. 2007;35:6475–6489. doi: 10.1093/nar/gkm709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bikard D., Loot C., Baharoglu Z., Mazel D. Folded DNA in action: hairpin formation and biological functions in prokaryotes. Microbiol. Mol. Biol. Rev. 2010;74:570–588. doi: 10.1128/MMBR.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramreddy T., Sachidanandam R., Strick T.R. Real-time detection of cruciform extrusion by single-molecule DNA nanomanipulation. Nucleic Acids Res. 2011;39:4275–4283. doi: 10.1093/nar/gkr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krin E., Cambray G., Mazel D. The superintegron integrase and the cassette promoters are co-regulated in Vibrio cholerae. PLoS ONE. 2014;9:e91194. doi: 10.1371/journal.pone.0091194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bikard D., Julie-Galau S., Cambray G., Mazel D. The synthetic integron: an in vivo genetic shuffling device. Nucleic Acids Res. 2010;38:e153. doi: 10.1093/nar/gkq511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.