Abstract
The compaction of nucleosomal structures creates a barrier for DNA-binding transcription factors (TFs) to access their cognate cis-regulatory elements. Pioneer factors (PFs) such as FOXA1 are able to directly access these cis-targets within compact chromatin. However, how these PFs interplay with nucleosomes remains to be elucidated, and is critical for us to understand the underlying mechanism of gene regulation. Here, we have conducted a computational analysis on a strand-specific paired-end ChIP-exo (termed as ChIP-ePENS) data of FOXA1 in LNCaP cells by our novel algorithm ePEST. We find that FOXA1 chromatin binding occurs via four distinct border modes (or footprint boundary patterns), with a preferential footprint boundary patterns relative to FOXA1 motif orientation. In addition, from this analysis three fundamental nucleotide positions (oG, oS and oH) emerged as major determinants for blocking exo-digestion and forming these four distinct border modes. By integrating histone MNase-seq data, we found an astonishingly consistent, ‘well-positioned’ configuration occurs between FOXA1 motifs and dyads of nucleosomes genome-wide. We further performed ChIP-seq of eight chromatin remodelers and found an increased occupancy of these remodelers on FOXA1 motifs for all four border modes (or footprint boundary patterns), indicating the full occupancy of FOXA1 complex on the three blocking sites (oG, oS and oH) likely produces an active regulatory status with well-positioned phasing for protein binding events. Together, our results suggest a positional-nucleosome-oriented accessing model for PFs seeking target motifs, in which FOXA1 can examine each underlying DNA nucleotide and is able to sense all potential motifs regardless of whether they face inward or outward from histone octamers along the DNA helix axis.
INTRODUCTION
Eukaryotic DNA wraps around histone octamers to generate nucleosome cores, where arrays of nucleosome cores are further organized into higher-order chromatin structures within the cell nucleus. This compaction creates a barrier for DNA-binding transcription factors (TFs) to access their cognate cis-regulatory elements buried within nucleosome particles (1,2). However, a large collection of enzymes including chromatin modifiers, chromatin remodelers and chaperone molecules (3–5), are capable of altering chromatin architecture through covalently modifying histone tails or repositioning, reconfiguring or ejecting nucleosomes to overcome these barriers. This dynamic balance between controlling genome packaging and regulatory element accessibility ultimately dictates transcription regulation in eukaryotic systems (6–8). A group of TFs called pioneer factors (PFs) including FOXA1 and GATA2 have the capability to engage with silent chromatin and compete with nucleosomes for direct access to cis-regulatory elements on DNA strands (9–11). In many cases, this access to the DNA occurs prior to transcriptional activation. For example, in one study the FOXA and GATA factors were characterized as pioneer factors occupying a specific enhancer of the Alb1 gene, and were required for the subsequent induction of Alb1 expression in early stages of mouse liver development (12). In another study, FOXA1 acted as a pioneer factor to facilitate the androgen receptor (AR) signaling pathway for hormone circulation in prostate cancer cells (13). It is believed that these pioneer factors can recruit the various remodeling enzymes mentioned above to establish an ‘open’ chromatin environment, which facilities DNA accessibility for other transcription factors that then initiate subsequent transcription events (8,9). However, a detailed mechanism of how these pioneer factors interplay with nucleosome particles to open up local histone–DNA complex and locate their cis-targets amidst the tangle of chromatin remains to be elucidated (9,11).
Understanding the mode of action of pioneer factors requires deciphering the molecular mechanisms that govern the selectivity, affinity and specificity for DNA binding. A number of techniques have been applied to evaluate the DNA-binding recognition and specificity of transcription factors. In particular, ChIP-seq technology has greatly facilitated the characterization of many transcription factor binding sites across the entire geneome (14). Efforts like the ENCODE project have generated a huge number of ChIP-seq datasets to reveal the genomic distribution of various TFs and histone modifications within several cell lines (15), and through mining these data it is now possible to determine complex mechanistic relationships between transcription factor binding, specific chromatin modifications, and nucleosome positioning. However, the resolution of ChIP-seq (which is several hundred base pairs) limits the precise definition of TF binding specificity (14,16). ChIP-exo is a novel protocol in which lambda exonuclease is introduced into the ChIP system (17). This exonuclease degrades double-stranded DNA in a 5′-3′ direction to within a few nucleotides of TF binding sites. The exonuclease-treated 5′-ends (or exo-5′-ends) are then sequenced, and the resulting high concentration of exo-5′-end sequencing reads tend to accumulate at one location representing a footprint boundary protected by protein binding, which is also often referred as a ‘border’ in ChIP-exo analysis (17). This new technique greatly increases the resolution of binding sites to a single base pair, and has been increasingly used in studying TF binding patterns (18–20). Several computational tools have been developed to process the ChIP-exo data, such as MACE (21), GEM (22) and ExoProfiler (23). However, all of these are focused on detecting borders (or footprint boundaries) for motif enrichment analysis with high accuracy, but have thus far ignored how these footprint boundaries themselves are structurally organized. Thus, we believe that the most critical novelty from ChIP-exo has not been fully evaluated and explored. For instance, how are these footprint boundaries or borders distributed within each individual binding site, and are they able to converge into fixed patterns across the whole genome? Would these fixed patterns be able to reflect structural properties of the underlying binding complex? Undoubtedly, high-resolution binding positions from ChIP-exo can also provide an opportunity to study how pioneer factors interplay with the underlying chromatin on a precise scale.
An unexplored application of the ChIP-exo method is to examine the underlying structural properties of TF footprint boundaries. Accordingly, we recently developed a modified ChIP-exo protocol in which both strands of digested DNA fragments are sequenced in a strand-specific paired-end fashion, i.e. both the exo-digested 5′-end (exo-5′-end) and sonicated 3′-end (son-3′-end) reads are collected from the sequencing (18,24). For the purpose of clarity, we refer to this technique as ChIP-ePENS (ChIP-exo paired-ends sequencing) throughout this paper. While we utilized this technique to redefine four Androgen Response Elements (AREs) with distinct motif compositions, the computational algorithm employed was not designed to take into account the paired-ends and strand-specific reads information (18). Thus in this study, we have developed a novel algorithm, ePEST (ChIP-exo paired-end sequencing processing toolkit), which leverages the statistical power of r-scan (25) for detecting binding peaks using son-3′-end reads, and the Chernoff inequality (26) for identifying precise footprint boundaries (or borders) from exo-5′-end reads, respectively. The detected borders are sequentially modeled as graphical components and classified into distinct border patterns based on their orientation and spacing. We applied our approach to analyze the FOXA1 ChIP-ePENS data in LNCaP cells (18), and have identified numerous FOXA1 motif-containing sites composed of precisely positioned borders. Interestingly, we found four primarily distinct modes of border-composition within these sites. Surprisingly, three fundamental nucleotide positions (oG, oS and oH) emerged as major determinants for blocking exo-digestion and combinatorically forming these four distinct border modes. Moreover, the oH site represents an obvious ‘well-positioned’ pattern relative to the FOXA1 motif across most binding sites. By integrating with histone MNase-seq datasets, we further uncovered that the configuration of nucleosome positioning is associated with the formation of these border modes. In addition, a ChIP-seq survey of eight chromatin remodelers also demonstrated that, the more well-positioned FOXA1 binding sites require higher concentrations of these remodeling molecules for their establishment or maintenance. Finally, we suggest a positional-nucleosome-oriented accessing model to explain the interplay between the pioneer factor FOXA1 and its immediate nucleosome for recognition of the buried cis-motif.
MATERIALS AND METHODS
Experimental protocol of ChIP-ePENS
The detailed protocol for ChIP-ePENS was published in our previous study (18). A key aspect is that ChIP-ePENS data generate sequence reads of fragments containing both exo-digested and sonicated ends (see Supplementary Figure S1). Similar to the original ChIP-exo, we added a ligation adaptor A2 to the both ends of sonicated DNA fragments after ChIP pull down, then lambda exonuclease (exo) is used to digest the unbound double-stranded DNA in the 5′-3′ direction, forming a hanged single stranded DNA, up to the point where it is protected by the binding protein complex (border or footprint boundary). After reverse cross-linking, A2 extension generates double-stranded DNA to get back from single strand, A1 adapters will then be ligated to the exo-digested ends, then sequencing is performed in a pair-end manner on both the 5′-exo-digested end (R1 reads) and 3′- sonicated-end (R2 reads).
Computational algorithm for ChIP-ePENS data
The computational algorithm, ePEST (ChIP-exo paired-end sequencing processing toolkit) is composed of four sequential steps (Supplementary Figure S2). In the first step, bad quality and duplicated reads are filtered out by scanning the distribution of 3′-sonicated end reads (R2). In the second step, the r-scan statistical model (25) is applied to identify binding regions (or peak-calling) using 3′-sonicated end reads (R2) too. In the third step, the Chernoff bound inequity (26) is used to evaluate the significance of the 5′-exo-digested R1 reads against background to identify borders (footprint boundaries) within a peak-pair bounded regions identified from the previous step. In the last step, all borders are connected into a directional linking graph based on their orientation and relative spacing among them, and an iterative re-sampling strategy to remove outlier links is applied to break the graph into individual isolated component and different border-composed binding sites are thus defined. A more detailed procedure is described in the Supplementary Information Note, and source codes for the implementation of this algorithm can be accessed from http://compbio.uthscsa.edu/ePEST/.
For the ChIP-exo FOXA1 data from MCF7 cell line (20), since all reads are generated from single exo-digestion ends, it is hard to directly apply our algorithm on it. We have pruned our pipeline as follows: we did not perform duplication filtering, we used the ChIP-seq data of FOXA1 in MCF7 for peak-calling in Step 2, and we then adapted the border-calling and border-matching procedures for the remaining process.
Motif analysis and asymmetric kurtosis calculation for borders
Firstly, all sites within the genomic blacklist have been removed (15). A flank DNA sequence was retrieved from each border-composed site to include borders and extensions from both sides to 45 or 60 bp. De novo motif discovery was performed by STEME online (27), and motif scanning was evaluated by FIMO (28) with P< 0.005. To measure the degree of sharpness for each border, we extended each border to both the upstream and downstream sides by 10 bp along the same strand, and calculated the kurtosis value using R1reads density by in-house python scripts, i.e., the ratio of the fourth moment and variance squared for the R1reads distribution on the relevant site.
To compile a control set of FOXA1 motif sites as a background, we collected all reliable sites in the above motif analysis to re-compile a FOXA1 motif PWM, then used this PWM to scan the whole reference genome hg19 by FIMO with more strict P< 0.00001, and removed those sites identified by our ChIP-ePENS analysis. For the remaining sites, we sorted them by the score from FIMO, and selected the top 20% as the final control set of FOXA1 motif sites as a ‘null’ background, which contains 20 773 sites.
Nucleosome positioning estimating on MNase-ChIP-seq data
MNase-ChIP-seq of H3K4me2 data was downloaded from GSM503903 in vehicle LNCaP (29), and only the 5′-end of uniquely mapped reads are used for the further analysis. Two Bigwig files of occupancy have been generated by separating reads into plus and minus strands respectively, then the excessive signal was calculated by subtracting the plus Bigwig file from the minus one, and a 15-bp window smoothing procedure was applied. To estimate nucleosome positioning, we firstly break the whole aggregative curve into three segments which presumably coincide with the three nucleosome particles, then to each segment we estimate the optimal boundaries and dyad position by a robust linear fitting method (RANSAC) using an in-house python script (30).
ChIP-seq of eight chromatin modifiers
ChIP-seq was performed as previously described (18). Briefly, vehicle-treated LNCaP cells were grown to 70%-80% confluence, and crosslinked with 1% formaldehyde for 10 min at room temperature. After washing twice with cold PBS, cells were collected and resuspended in lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris (pH 8.0), and 1x protease inhibitors). After sonication, the soluble chromatin was diluted in 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris (pH 8.0), 1× protease inhibitors), and incubated with 4 μg of antibodies overnight. Protein A Dynal magnetic beads were added and incubated for 1 h, and then washed using modified RIPA buffer (50 mM Tris–HCl pH 7.8, 1 mM EDTA, 0.25% Na Deoxycholate, 1% NP-40, 0.5 M LiCl) six times followed by Tris–HCl pH 8.0 twice. The eluted ChIP DNA was used for library generation with NEBNext ChIP-Seq Library Prep Master Mix Set according to the manufacturer's protocol. The library was amplified with 14 PCR cycles, and prepared with gel-based size selection (250 bp). The sequencing was performed using Illumina HiSeq 2500 at the OSUCCC sequencing core. Antibodies used in ChIP-seq were BRG1 (H-88), INOC1 (H-300), Mi2-α (C-16), CHD1 (H-210) from Santa Cruz Biotechnology (Santa Cruz, CA), CHD4 (ab72418), SNF2H (ab72499), SMARCA1 (ab37003), SMARCA2 (ab15597) from Abcam (Cambridge, UK).
Data access
The ChIP-seq data from this study has been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE72690. The main algorithm of ePEST software is implemented in Python, which can be downloaded from http://compbio.uthscsa.edu/ePEST/.
RESULTS
A computational approach for processing ChIP-ePENS data
Despite the advance of the ChIP-exo protocol in allowing single base pair resolution border definition, the traditional method suffers some sensitivity issues from background noise. For example, it is hard to discriminate the borders resulting from PCR-duplicated reads or from real exo-digested fragments, because both of them accumulate at a specific position to form border-like patterns (31). This essentially increases false positive ratio due to the creation of artificial borders. Our modified ChIP-ePENS protocol (Supplementary Figure S1), not only collects exonuclease-treated 5′-end (exo-5′-end) but also sonicated 3′-end (son-3′-end) together. Paired-end sequencing approaches provide information on both sides of the ChIP-exo library including the non-exo digested 3′-ends of fragments. Due to random sonication, these 3′-ends of fragments are expected to be relatively even distribution and therefore provide a distinguishing feature from duplicated fragments (Supplementary Notes). Based on these new characteristics, our algorithm ePEST is designed to only require a paired-complementary peaks from son-3′-end reads in peak-calling step, and allows multiple or even zero borders to occur within binding regions depending on statistical evaluation of Chernoff inequity on exo-5′-end reads (26), in which it is different from other approaches which impose a ‘border-paired-rule’ requiring that each binding site must have precisely two optimized borders paired on opposite strands (17,21,22). ePEST, summarized in Materials and Methods and Supplementary Figure S2, is essentially composed of four critical steps. In the first step we process and filter duplicated reads as mentioned above. In the second step, we apply the r-scan statistic method (25) to perform peak-calling using son-3′-end reads, analogous to ChIP-seq analysis. Notably, reads are processed on plus and minus strands separately, thus a pair of complementary peaks defines a binding boundary. Then in the third step, border-calling is conducted specifically within these binding regions bounded by peak pairs. At last, in the fourth step, borders will be assigned and composed into each individual binding site by a graph-based strategy for border matching procedure. As depicted in Supplementary Figure S2, son-3′ end reads provide a coarse-grained view of TF binding by mapping to the periphery of DNA-binding protein complexes, while exo-5′ end reads map throughout the footprint of such complexes and dissect binding sites into more refined components. Therefore, we can take advantage of both son-3′- and exo-5′-end reads available in our ChIP-ePENS data to combine peak-calling and border-calling in our pipeline. Doing so dramatically reduces background noise caused by irrelevant genomic regions and thereby enhances the accuracy of border detection.
In order to determine how various borders compose a binding site, we have adopted a graph-based strategy in the fourth step to make arrangements for each border and conduct border matching. Briefly, we first build a directional border-linking graph by connecting all borders according to their orientation and positions. We then use an iterative outlier detection method to break down abnormal outlier links (d>μ + 2.0σ) t o dissect the graph into small, isolated components that naturally correspond to each individual binding site. Thus, each component is actually a small sub-graph, and we call the bi-directional links connecting a plus border and a minus border as ‘backbone links’ in these sub-graphs. The rationale behind this strategy is (i) the distance (d) between two well-oriented borders within the same binding site is significantly smaller than the distance between borders from two different binding sites, and (ii) the distance between correctly linked borders assumes a fixed distribution allowing the few incorrectly assigned links to be regarded as outliers due to deviation of d from that fixed distribution (see more detailed description in Supplementary Information Note).
Four distinct border modes in ChIP-ePENS of FOXA1
Our previous study has generated two ChIP-ePENS datasets of FOXA1 in the prostate cancer cell line LNCaP under vehicle and dht-treated conditions, respectively (18). However, the data has only been used for demonstrating a co-regulatory role FOXA1 with respect to AR, and additional biological insights have not been fully elucidated. Thus, we applied our computational tool, ePEST, to re-analyze the data and identified a total of 119 291 border-composed sites (BCSs) in vehicle and 82 450 BCSs in dht-treated conditions. For simplicity, the following analysis focuses on vehicle data, though similar conclusions can also be made from the analyses of dht-treated data.
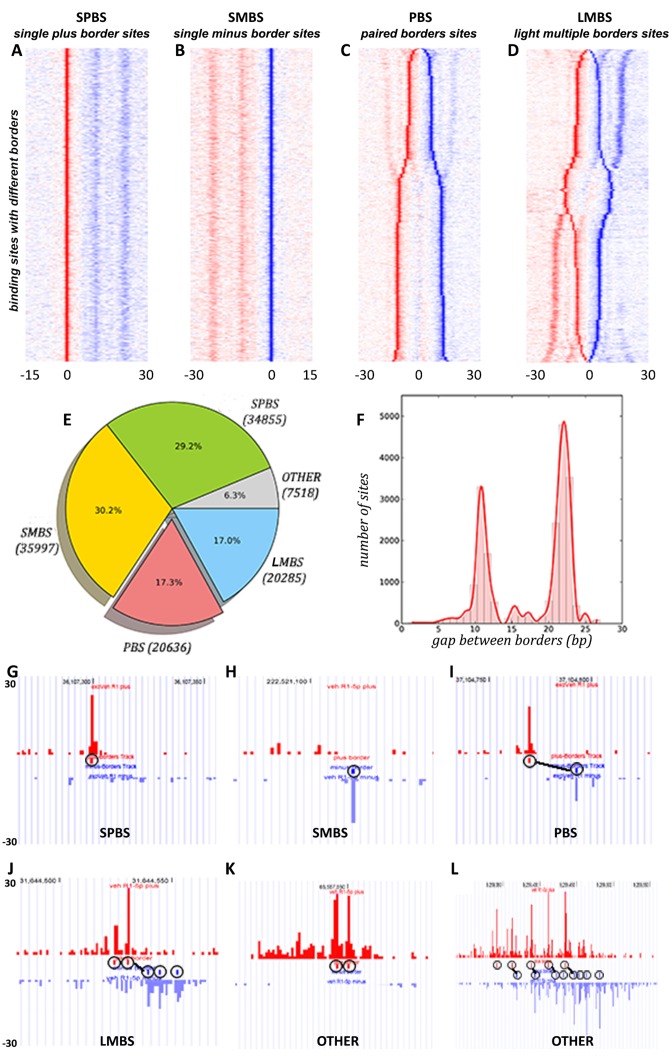
We categorized these BCSs into four distinct modes based on their deposition on the reference genome (plus strand or minus strand) and spacing of borders linked by our border matching algorithm in ePEST: single plus border site (SPBS)—only one border on the plus strand; single minus border site (SMBS)—only one border on the minus strand; paired-borders site (PBS) – two borders with one on plus and the other on minus strands; and light-multiple-borders site (LMBS)—three or more borders co-localized at the same binding site. For PBS and LMBS, exactly one backbone link is required to define these modes. The sorted heat-maps of binding sites comprising these four modes as well as the screenshots of examples in each of the four modes are shown in Figure 1A–D and Figure 1G–J, respectively. A fifth mode, ‘OTHER’, categorizes the remaining sites which do not fit into the above four modes. These sites consist primarily of two forms. The first exhibits two or more adjacent borders on the same strand at the same site (Figure 1K), and the second contains two or more backbone links connecting many borders at the same site (Figure 1L). We have excluded this mode from the further analysis since it represents only a very small portion of the total binding sites and does not produce clear border patterns when visualized as a heatmap. Of the 119 291 BCSs in the vehicle data, 34 855 (29.2%) are assigned to SPBS, 35 997 (30.2%) to SMBS, 20 636 (17.3%) to PBS, 20 285 (17.0%) to LMBS, as well as 7518 (6.3%) to other (Figure 1E). Interestingly, we observed that a big portion (almost 60%) of BCSs are single border sites (SPBS and SMBS), while a relatively small portion (34%) are PBS and LMBS. This unequal distribution in the numbers of BCSs in each of the four binding modes may imply FOXA1 shows conditional preference for one mode over another within specific genomic regions.
Figure 1.
Border patterns identified from FOXA1 ChIP-ePENS data in LNCaP cells. Each binding site was categorized by its border's conformation, and the density of reads occupancy on each site was normalized by a z-score method on plus (red) and minus (blue) strands separately. (A) SPBS—one single plus border on each site, aligned against the plus border only; (B) SMBS—one single minus border on each site, aligned against the minus border only; (C) PBS—two borders on plus and minus strands respectively on each site, aligned by the midpoint of the border pair and sorted by the spacing between them; (D) LMBS—three or more borders with one backbone on each site, aligned up by the midpoint of the backbone and sorted by the spacing of backbone as well as residual borders; (E) a pie chart showing the distribution of the numbers in each of the border modes; (F) a histogram of the spacing between the two borders in PBS; (G–L) Screenshots of examples of several sites with different border modes, and the links between the red border and blue border are backbone links.
Furthermore, we observed that the distance between the two paired borders in the PBS mode follows a typical bimodal distribution (Figure 1C and F), with one peak centralized at 12 bp and the other at 22 bp. Thus, we further classified this mode into two different sub-modes: 7873 shorter-PBS (st-PBS) with a distance between 0–15 bp and 12 763 longer-PBS (lg-PBS) with a distance between 16–30 bp. We further examined the LMBS mode, and found that most of these sites (∼64%) consist of three borders, where one border on the plus strand is frequently accompanied by two borders on the minus strand or vice verse (Figure 1D). Proximately, the LMBS mode seems like a combination of st-PBS and lg-PBS modes in terms of their paired border spacing. We speculated that the co-localization patterns in LMBS might result from transient occupancy of separate border positions in subsets of the cell population, or they could be due to a direct interaction of FOXA1 with these border positions simultaneously. Nevertheless, the limited number of distinct border modes within FOXA1 binding sites strongly suggests that there are a finite number of stable FOXA1 binding complex arrangements engaged in transcriptional regulation.
In order to investigate whether these four distinct border modes and their characteristic border patterns are a common characteristic for all TFs or particular to FOXA1, we examined AR ChIP-ePENS data from the same previous study (18). Surprisingly, while the same four modes were identified, border patterns within AR binding sites were drastically different from those of FOXA1 (Supplementary Figure S3B, D and E). Namely, there was no conspicuous bimodal distribution of paired border spacing in the PBS and LMBS modes. Our results prompted us to speculate that the patterns within the four distinct border modes may be intrinsically associated with the structural or functional role of the pioneer factor FOXA1. To confirm this speculation, we also looked into FOXA1 ChIP-exo data in the MCF7 cell line, which only contains exo-5′-end reads, by partially modifying our pipeline to adapt to the single-end ChIP-exo data (20), and found similar patterns within the four distinct border modes as seen in the LNCaP cell line (see Methods and Supplementary Figure S3A and C).
Motifs associated with distinct border modes
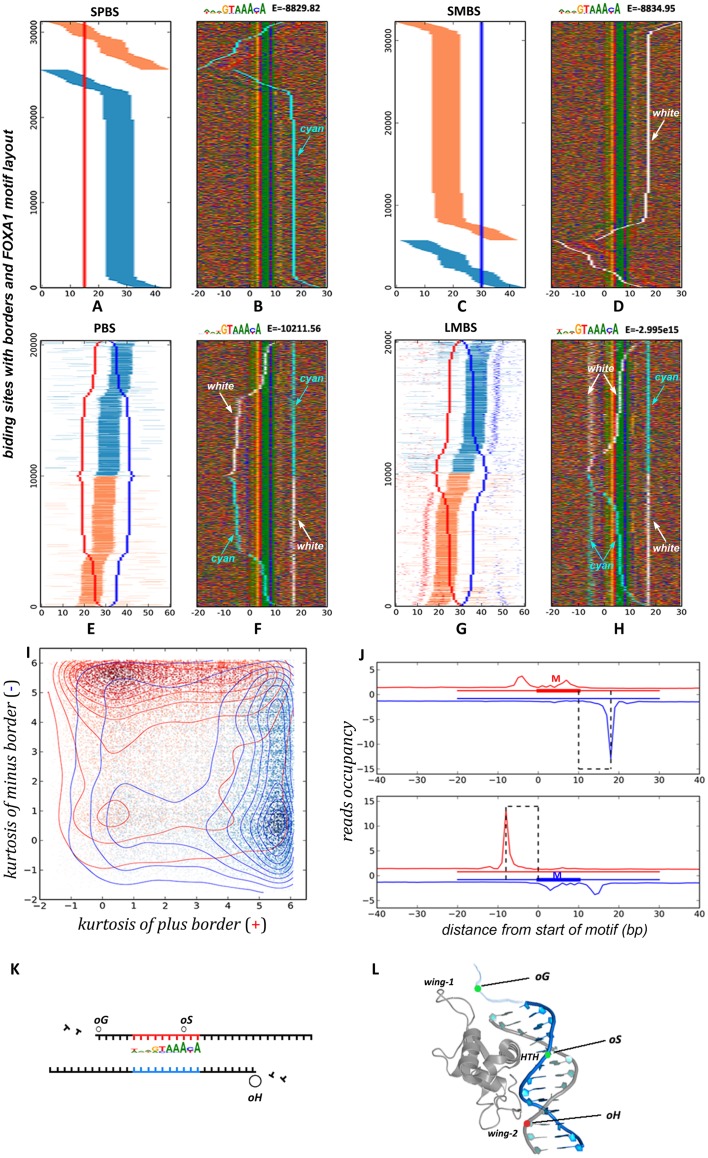
To examine if the identified BCSs contain the canonical FOXA1 binding motif and whether they include other possible TF binding motifs, we have performed a de-novo motif discovery by STEME (27) and motif scanning by FIMO (28). We defined the binding regions used for the motif discovery and scanning based on the following rules: for the SPBS mode, a 45 bp DNA sequence with 15 bp upstream and 30 bp downstream from the border position; for the SMBS mode, a 45 bp DNA sequence with 30 bp upstream and 15 bp downstream from the border position; for the PBS mode, a total of 60 bp extending 30 bp up- and downstream from the middle point of the two border positions; for the LMBS mode, similar to PBS mode, a 60 bp expanding 30 bp up- and downstream from the middle point of the two borders connected by the backbone link. As expected, we recovered a known FOXA1 motif as defined in the JASPAR database (32) with very stringent log10 E-values (−8829.82 in SPBS, −8834.95 in SMBS, −10211.56 in PBS and −3.00e15 in LMBS). We then used the position-weight-matrices (PWMs) of these motifs to re-scan and retrieve the occurrence positions using FIMO with a P-value <0.005. We further projected the motif positions over the four BCS modes (Figures 2A, C, E and G), where orange and blue line segments indicate FOXA1 motif occurrence on the plus and minus strand, respectively. Remarkably, the vast majority of binding regions are composed of at least one FOXA1 motif: 31 260 of 34 855 (89.7%) in SPBS (Figure 2A), and 32 319 of 35 997 (89.8%) in SMBS (Figure 2C) and 20 326 of 20 636 (98.5%) in PBS (Figure 2E), and 20 144 of 20 285 (99.3%) in LMBS (Figure 2G). Our results showed that the FOXA1 motif is indeed enriched in the identified border-composed sites, especially for the PBS and LMBS modes, which have a higher percentage of motif recovery at above 98%.
Figure 2.
Motif orientation associated with distinct border modes. The position of FOXA1 motif on each border-composed site has been determined by FIMO28, and labeled with orange or blue colors when it locates on plus or minus strands respectively. Further, all sites were re-aligned according to the identified FOXA1 motif in a 5′-3′ direction. DNA base content is indicated by four different colors, as well as ‘cyan’ and ‘white’ colors to indicate plus-border and minus-border positions respectively. (A–H) The left side shows relative position distribution of FOXA1 motifs adjacent to aligned border sites, the right side shows the base content matrix surrounding the aligned FOXA1 motifs. (I) Density-cloud plot of kurtosis values for the two borders in PBS. (J) Aggregated signals of exo-5′-end reads surrounding the FOXA1 motifs in PBS separated into two sets by their FOXA1 motif locations on plus strand or minus strand. (K) The schema of three basic blocking sites underneath borders. (L) The potential structural conformation of three blocking sites associated with the winged-helix domain of the FOXA1 protein.
Further, we aligned these motifs occurrences, and inspected how border positions are distributed relative to FOXA1 motifs in each mode. We displayed border positions using the ‘cyan’ color to indicates borders on the plus strand from original sites, the ‘white’ color to indicate borders on the minus strand, as well as other colors for DNA base identities (Figure 2B, D, F and H). Because the FOXA1 motif is not palindrome and defined in a strand-directional manner, regions with the FOXA1 motif on the minus strand (sites with horizontal blue line segments and vertical orange borders shown in Figure 2A) need to be mirrored for motif alignment. The upstream borders within these sites accordingly relocate downstream of the motifs (Figure 2B). Interestingly, after performing transformation of strand-directional replacement, we found that a clear, ‘well-positioned’ border configuration emerged, such that an almost vertical border line consistently appears at the downstream side of the FOXA1 motif in all the four modes (Figure 2B, D, F and H). Moreover, these downstream borders are precisely located 8 bp away from the motif on the opposite strand. Meanwhile, on the upstream side of these well-positioned sites, we observed various scenarios: no consistent border in SPBS and SMBS modes, one border in the PBS mode, and two primary borders in the LMBS mode (the two blue lines in upper part and two red lines in bottom part in Figure 2G). Interestingly, in the two PBS sub-modes, st-PBS sites show the upstream border directly overlapping the motif, while lg-PBS sites exhibit a border ∼10 bp upstream from the motif (Figure 2F). We noted that a mixture of these two border-spacing patterns exist in the LMBS mode, indicating the basic border composition is the same between PBS and LMBS, and the difference between these BCS modes is whether or not both upstream borders are simultaneously present within these sites in the whole cell population.
The asymmetric pattern of borders relative to motif orientation
By finely examining these binding sites with well-positioned borders, we observed an asymmetric pattern of borders with respect to 5′-3′-oriented motifs such that the downstream border on the opposite strand always shows higher occupancy than the upstream borders if they exist (no upstream borders are available for comparison in SPBS and SMBS, see Figure 2B, D and J). As an example, we used the PBS mode to elaborate on this pattern since it contains only two borders. For each linked border pair, we calculated the ‘kurtosis’ value of each border, which is a descriptor of the shape of a probability distribution, and used it to measure each border's sharpness compared to its neighboring background (33). Bigger ‘kurtosis’ values indicate greater sharpness of the border protruding from its surrounding bases. As displayed in Figure 2I (each point corresponds to a binding site in Figure 2E), the two borders on either the plus strand or minus strand have been transformed into a pair of ‘kurtosis’ values on the x-axis and y-axis, respectively. In addition, each point (or site) is labeled as red or blue to indicate FOXA1 motif occurrence on the plus or minus strand, respectively. A dot-density plot shows that a large number of binding sites are piled up at the left-top and right-bottom corners. The dense red region in the left-top corner indicates that when there is a motif on the plus strand (red point), the minus border generally exhibits a stronger sharpness (higher values on y-axis) and the plus border shows weaker sharpness (lower values on x-axis). The same pattern is visible for sites with a motif on the minus strand, as indicated by the dense blue region at the right-bottom corner.
We further separated all binding sites into two clusters according to the occurrence of FOXA1 motifs on the plus or minus strands, and then aggregated them to examine the exo-5′-end read occupancy signals (Figure 2J). This showed much higher reads occupancy on the minus strand downstream of the border position when the motif was on the plus strand (Figure 2J, top panel) or, conversely, higher occupancy on the plus strand upstream of the border position when the motif was on the minus strand (Figure 2J, bottom panel). We noted that these summits of reads occupancy at each border position correspond to the vertical lines (cyan and white color) in Figure 2F by collapsing them together. Our results clearly revealed an asymmetric pattern for the two borders surrounding the FOXA1 motif in the PBS mode. To further substantiate the notion that 5′-3′-oriented FOXA1 motifs exhibit a strong downstream border on the opposite strand, we also note that the SPBS and SMBS modes are essentially the same but exhibit the FOXA1 motif on the minus and plus strand, respectively. The asymmetric border pattern of both modes is identical upon proper orientation of their motif occurrences (Figure 2A–D).
To generalize the above observations, we propose a structural binding schema as depicted in Figure 2K, in which there exist three basic positions for blocking exo-traveling: from 5′-3′, the oG and oS sites are located on the same strand as FOXA1 motif, and the oH site is located downstream of the motif on the opposite strand. The oH site can strongly obstruct exo-digestion, while the oG and oS sites block digestion relatively weakly. These three blocking sites ultimately create the border-matching patterns we observed. BCSs with a well-positioned configuration, always possess a stronger border on the oH site, but exhibit different patterns over the oG and oS sites when a) no blocking on the oG and oS sites results in SPBS and SMBS modes, b) blocking on either the oG or oS site leads to one of two PBS sub-modes (st-PBS and lg-PBS), and c) blocking on both the oG and oS sites results in LMBS mode. The combination of these sites in parallel with different binding modes is depicted in details in Supplementary Figure S4. We re-grouped all well-positioned BCSs based on this proposed schema for further analysis, merging SPBS and SMBS into SBS, splitting PBS into st-PBS and lg-PBS, and keeping LMBS unchanged.
This schema of structural blocking of exo-digestion proposed through our genomic analysis is remarkably consistent with the conclusion from a study on one specific enhancer locus of Alb bound by Hnf3 (a member of FOXA family) in mouse liver development, where Hnf3 binds to two sites eG and eH, which are separated by 20 bp at the core of the underlying nucleosome (34–36), very similar to oG and oH sites discovered here by ChIP-ePENS data analysis. Interestingly, as shown in Figure 2L, the FOXA1 protein contains three structural domains (wing-1, wing-2 and HTH) that might correspond to the three contact sites (oG, oS and oH) proposed in our schema (37,38). Each domain may bind to DNA with different affinities, and the oS site matches the recognized cis-FoxA1 motif contacted by the HTH domain penetrating into the major groove of the helical DNA strands (more in the Discussion).
Nucleosome positioning associating with the border patterns
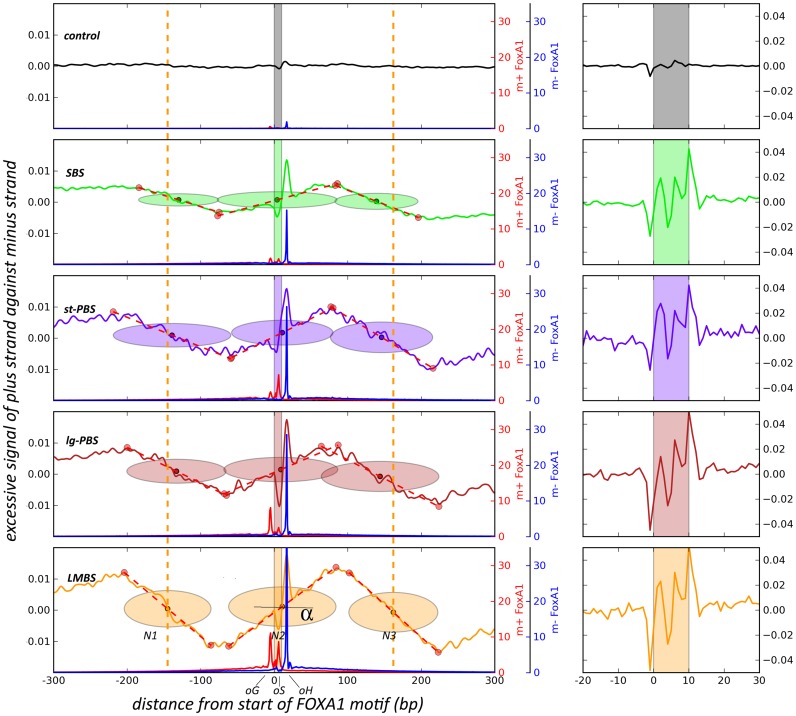
Since FOXA1 is a pioneer factor involved in many biological processes such as tissue development and disease progression (9,39), and has been shown to interact directly with nucleosome particles, it is worthwhile to further investigate the relationship between the underlying chromatin niche and the border patterns we have identified. Many recent reports found that nucleosome positioning impacts protein binding events (7,8,40). This prompted us to further analyze MNase-seq data to determine nucleosome positions surrounding BCSs. However, a big challenge of this correlation analysis is that most border sites locate in regions of enhancers, where nucleosomes might be apt to reposition, be evicted or be otherwise modified (41). In traditional methods for nucleosome position, reads from MNase-seq generally are shifted toward a supposed middle of nucleosome, and this shifting will lose the primeval information of status especially on enhancer sites. To overcome this shortage, we adopted a novel strategy to perform this analysis by using the MNase fragments pulled down by an H3K4me2 antibody in LNCaP cells (29). For an individual nucleosome particle, reads from single-5′-end sequencing of MNase-digested fragments will accumulate asymmetrically on either strand at both ends of the nucleosome, because MNase digestion of linker DNA creates an unequal probability for fragment sampling at these two terminal ends (see Supplementary Figure S5). Thus, we separated MNase reads onto the plus and minus strands respectively, and calculated the excessive signals by subtracting between them along chromosomal axis. This is a similar strategy adopted by another genome-wide study in mapping of nucleosomes (42). As shown in Figure 3, we identified an obvious three-nucleosome array surrounding the FOXA1 motif in all the four border modes (SBS, st-PBS, lg-PBS and LMBS), with an excessive ‘plus’ signal on the left (upstream) end and excessive ‘minus’ signal on the right (downstream) end of each neighboring nucleosomes (N1 and N3), but not in the control case due to fuzzy positioning of them. Expectedly, the middle nucleosome (N2) shows an abnormal reversal of signal accumulation from its left to right sides, which is different from the typical pattern of neighboring nucleosomes (34), indicating some potential destabilization on its body due to FOXA1 binding, and this fragile nucleosome may respond to MNase digestion in a different way as we revealed here. We should point out the H3K4me2-base method for pulling down nucleosomes will inherently enrich for active regions of the genome, while FOXA1 proteins also bind repressed targets on some conditions (43).
Figure 3.
Estimation of nucleosome position using a robust linear fitting method on strand-excessive MNase signals. For a canonical nucleosome (N1 or N3), more reads accumulate on the plus strand compared to the minus strand at the left edge of MNase-digested nucleosomes when regions are oriented with 5′-3′ FOXA1 motif direction, but fewer reads accumulate on the plus strand on the right edge. The middle N2 nucleosome presents a reverse pattern due to destabilization from FOXA1 binding. The slope or tangent α measures the stretching of well-positioned or fuzzy nucleosome at a fixed position. The red and blue lines indicate occupancy of exo-reads on plus and minus strands, respectively. A 15-bp window smoothing was applied on excessive signals in the left panel, but not in the right, zoomed-in panel.
The advantage of this analysis strategy for nucleosome positioning is that we can further utilize the asymmetric end signals to estimate nucleosome boundaries and dyad position by a robust linear fitting model (see Materials and Methods) (30). From the left panel in Figure 3, we can further see that FOXA1 engages at or very near to the exact dyad position of the underlying nucleosome (N2) regardless of the border mode, and this is likely because the regions represented in this panel are border sites with stable, well-positioned border configurations. This result is also quite consistent with the previous study showing that the Hnf3 winged-helix DNA-binding domain resembles the linker histone H1 and thereby binds DNA at the center of nucleosome cores (36). Iwafuchi-Doi et al. also recently reached a similar conclusion that FOXA binds to dyad of nucleosomes in which they combined low and high levels of MNase digestion in that study (44). Importantly, this result implies that the well-positioned configuration between the FOXA1 motif and the downstream border (oH) actually reflects an intrinsic structural property, and suggests the FOXA1-binding complex requires correct phasing between its cognate cis-motif and the dyad of the underlying nucleosome for a proper configuration or docking. As we know, the FOXA1 motif is a stationary cis-element on the DNA strand and only the reciprocation of the nucleosome can adjust the motif's position with respect to the floating dyad. To attain a proper conformation, the three basic sites (oG, oS and oH) revealed by our ChIP-ePENS analysis likely participate in this modulating motion, and might conditionally guide this process within different chromatin contexts as shown in the four modes here (SBS, st-PBS, lg-PBS and LMBS). In addition, we also noticed that there were almost no nucleosomes estimated in control regions. This does not mean that there are no nucleosomes within these sites, just that there are no excessive MNase signals for estimating the nucleosome position by segmental robust linear fitting. In other words, nucleosomes on these sites are quite delocalized with respect to FOXA1 motifs. This can be reflected by the slope of the dotted fitting lines in Figure 3. In LMBS, the angle α, indicating the slope, has the greatest value, which means that nucleosome positions are relatively fixed on these locations. In comparison, the slightly smaller slope in SBS indicates slightly poorer nucleosome positioning, and the slope of nearly zero indicates almost no fixed nucleosomes in the control case. Therefore, the distinct border modes resulting from the three blocking sites (oG, oS and oH) might be associated with differing degrees of proper phasing between nucleosomes and the FOXA1 motif. The right panel in Figure 3 shows the zoomed-in view of excessive MNase signals on FOXA1 motif sites, which are not much different among the four modes and relatively low in the control group.
Occupancy patterns of chromatin remodelers and histone modifications on border modes
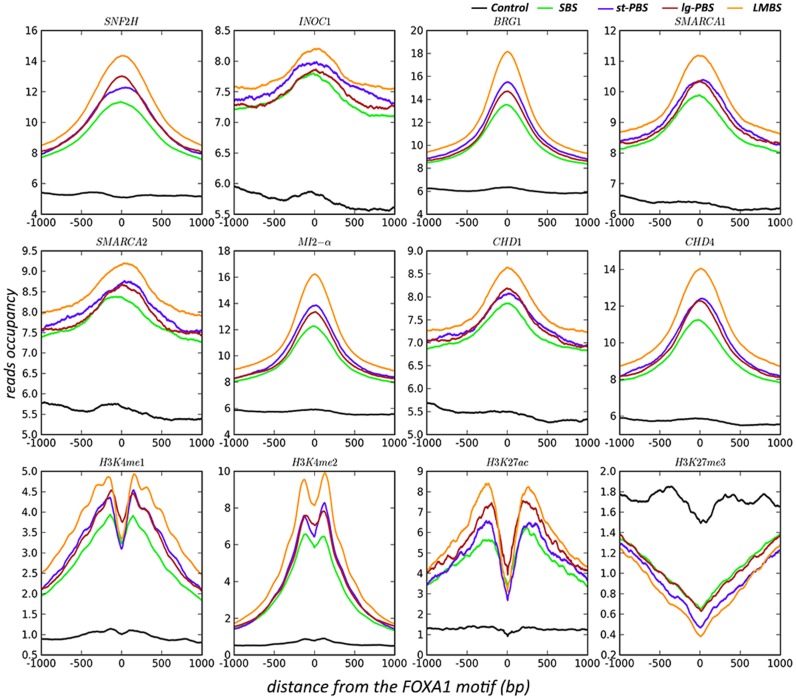
It is well recognized that nucleosome positioning is determined by many factors, including the actions between ATP-dependent chromatin remodelers and the thermodynamic forces from DNA sequence (45,46). For the FOXA1-binding complex wants to attain a well-positioned phase between its motif and the dyad of the underlying nucleosome for a proper configuration, it presumably requires collaboration with these forces to shift or alter the position and/or structure of the nucleosome (47). We therefore sought to investigate the connection between chromatin remodeling factors and the four distinct border modes associated with FOXA1. To do this, we have performed ChIP-seq data analyses on eight chromatin remodeler-associated molecules (SNF2H, INOC1, BRG1, SMARCA1, SMARCA2, Mi2-α, CHD1 and CHD4), which covered the four main families of chromatin remodeling complexes (SWI/SNF family, ISWI family, CHD family, and INO80 family) (48,49).
As shown in Figure 4, all well-positioned sites in the four border-mode groups showed much higher occupancy of reads centered on FOXA1 motif locations compared to control sites for each remodeling protein tested. In particular, the LMBS group shows the strongest signals compared to the other three groups, and SBS shows the weakest of the four groups. The st-PBS and lg-PBS groups fall in the middle, perhaps indicating a transitional status. This result is quite consistent with the results from the nucleosome positioning analysis on MNase data, where LMBS presents the most fixed configuration of nucleosomes, and SBS being the least fixed group. To investigate how additional histone modification marks correlate with these binding sites, we have further examined four marks including H3K4me1, H3K4me2, H3K27ac and H3K27me3. Similar to the chromatin remodelers, the three active marks (H3K4me1, H3K4me2 and H3K27ac) show the highest occupancy in LMBS and the lowest in SBS compared to the control case, while the repressive mark (H3K27me3) shows a reverse pattern with the highest occupancy in the control set. This indicated that the full occupancy of the FOXA1 complex on the three blocking sites (oG, oS and oH) likely produces more active status due to the well-positioned phasing between bound proteins.
Figure 4.
An aggregate plot of reads occupancy surrounding the FOXA1 motif sites from ChIP-seq of eight chromatin remodelers and four histone modifications.
DISCUSSION
ChIP-ePENS data analysis reveals intrinsic binding patterns
Lambda exonuclease is an enzyme which can digest double stranded DNA in 5′-3′ direction until it reaches an obstacle bound to the DNA, leaving to a single, 3′-5′ strand. ChIP-exo and ChIP-ePENS techniques utilize this feature of exonuclease to precisely digest free DNA up to the edge of a DNA-bound complex and then sequence the digested ends of the ChIP fragments. Aligned reads generated from digested ends accumulate to form sharp borders representing the boundary of transcription factor binding (17). However, beyond the crosslinking of the protein of interest, there are undoubtedly substrate structures that impede the 5′-3′ exonuclease activity. For instance, the exonuclease may pause in a strand specific and sequence dependent manner when it encounters ‘GGCGATTCT’ (50). Additionally, genome-wide factor binding is a complex process subject to spatial-temporal dynamics in a cell population, involving the association with additional recruited co-factors or other chaperones on or nearby an adjacent locus (51). Thereby the exonuclease digestion might be blocked by these accessory molecules. All these events have the potential to create diverse patterns of exonuclease boundaries throughout the genome.
Bearing this in mind, in designing our analysis method we did not impose the mandatory rule that there must be two paired borders on a single binding site as did other methods (17,21,22). Instead, we allowed for diverse border configurations to exist in binding sites to maximally maintain native border patterns. We firstly identify a concrete border configuration as a unique property at each binding site, then we aggregate the whole set of these individual binding sites in a genome-wide manner, which provides us an opportunity to distill intrinsic border patterns that might spontaneously emerge. Indeed, by applying our computational strategy on FOXA1 ChIP-ePENS data in LNCaP cells, we found four distinct modes of borders stably appearing on different loci. As a matter of fact, the diverse patterns of borders found in different binding loci are generated by the intrinsic properties a single target protein, FOXA1, indicating that this pioneer factor might be associated with different co-factors, or might adopt distinct structural conformations to adapt to site-specific environments. Therefore, these convergent modes, in principal, imply that the FOXA1 binding complex exists in a finite number of stable forms. Our unique graphical approach to border matching in the ePEST pipeline robustly preserves intrinsic border patterns on binding sites, and allows us to explore these latent properties further.
Three basic blocking sites (oG, oS and oH) underlying various borders
In contrast to ChIP-seq, ChIP-exo provides single base pair resolution for detecting the boundary of a DNA binding complex, and this leads to much higher precision in motif discovery near borders (11,18,19). However, in terms of motif analysis, much weight is placed on sequence fragments extracted from nearby borders while the shape or configuration of borders is wholly neglected. In reality, the borders themselves provide valuable information regarding the structural properties of the underlying binding complex. As we show, beneath the diverse border combinations, three basic sites (oG, oS and oH in Figure 2K) surrounding the FOXA1 motif are the primary locations for forming various borders. And more, the asymmetric pattern of ChIP-ePENS signal over these sites suggests a weaker occupancy over oG and oS, but stronger blocking on the oH site. This pattern illustrates a scenario in which there are two exonuclease molecules digesting opposing DNA strands in a 5′-3′ direction towards the center of a binding complex. One molecule would be consistently obstructed at the oH site, while the other molecule may or may not be impeded by an occupant of the oG and/or oS sites depending on specific conditions of the genomic region.
Previous studies on an albumin enhancer bound by Hnf3 revealed two sites, eG and eH, whose orientation and spacing are quite similar to our proposed oG and oH sites (34–36,52). Basically, the three sites (oG, oS and oH) are likely to be associated with the structural properties of the FOXA1 binding complex. It is well recognized that the ‘winged-helix’ DNA-binding domain of FOXA1 resembles linker histones such as H1 and H5 (36,53,54), binding to DNA as a monomer by using a recognition helix (HTH) flanked by two ‘wings’ (wing-1 and wing-2) that interact with one face of the DNA (see Figure 2L) (36–38). Cirillo and Zaret further verified the contacts of the two wings nicely adhere to DNA by hydroxyl radical foot-printing to map minor groove FOXA1-DNA contacts (55). We suggest that the three blocking sites here (oG, oS and oH) might reflect the three tertiary structures of FOXA1 (wing-1, HTH and wing-2) protruding to the DNA surfaces, and making intimate contacts that block exo-traveling. The HTH part contacts the FOXA1 motif sequence (GTAAACA) in a major groove of the DNA and the two wings touch DNA surfaces in the two flanking minor grooves. The spacing between oG, oS and oH is around 10 bp, which represents one turn of the DNA helix, so these sites would likely be present on the same side of DNA strand to allow simultaneous contact with each of the three FOXA domains. The question remains as to why the three sites display different binding affinities? The asymmetric pattern of border occupancy, with the highest signal on the oH site, implies a positional preference for initial FOXA1 contact as well as a directional, perhaps sequential, binding axis. We speculate that the three FOXA1 tertiary structures (wing-1, HTH and wing-2) perform different functional roles during the binding procedure, as we will further discuss in following sections.
The interplay of FOXA1 with the underlying nucleosome on the three blocking sites
FOXA1 binding to its cognate site undoubtedly creates a strong obstacle for exonuclease traveling, confirmed by the persistently strong border signals on the oH site residing at downstream side of the FOXA1 motif. However, the interesting question is what causes the relatively weak borders on the oG and oS sites upstream of and overlapping the FOXA1 motif, respectively. What conditions result in the lack of a border on these sites leading to the single border mode (only oH), and what determines the conditional formation of the st-PBS and lg-PBS modes in which borders appear over the oS (∼12 bp upstream of oH), and oG (∼22 bp upstream of oH) sites, respectively. In theory, if there were only one stable FOXA1 binding complex conformation, there would be one rather than several stable border patterns detected by our analysis. A reasonable speculation is that accessory molecules may involve in FOXA1 binding contact these three sites (oG, oS and oH) in a context-specific manner.
Considering an alternative explanation, FOXA1 does not bind to free DNA in nuclei, but instead, mostly binds to nucleosomal DNA wrapped around histone octamers (9,12,52). As we revealed in Figure 3, FOXA1 binding events always occurred at the exact or very near center of nucleosome cores. Prior to FOXA1 binding, the three sites (oG, oS and oH) on DNA are occupied by nucleosomal histone proteins or other chaperone molecules. FOXA1 needs to compete with these proteins for the three sites. Previous studies have shown that FOXA1 resembles a ‘winged helix’ structure as linker histones (36,53,54), and competes with them to ‘open’ chromatin rather than compact the nucleosome particles as H1 and H5 (36,52). And recent experiments using single-base resolution •OH foot-printing also show that the globular domain of histone H1 interacts with the DNA minor groove located at the center of the nucleosome and contacts a 10-bp region of DNA localized symmetrically with respect to the nucleosomal dyad (56,57). Coincidentally, the oG, oS and oH blocking sites revealed by our analysis are located near to or exactly on the dyad position encompassing the FOXA1 motif. Moreover, these sites are separated by ∼10 bp, and would thus be expected to mediate the competition between linker histones (H1 or H5) and FOXA1. Based on these observations, we conjecture that the three blocking sites are initially occupied by linker histones on the dyad site. FOXA1 attacks the oH site first with very strong affinity, and then gradually competes for occupancy of the other two sites as indicated by the relatively weak and dynamic reads occupancy over the oG and oS sites due to variable competition and transitions in FOXA1 binding status. This hypothesis is quite consistent with a study showing dynamic binding of histone H1 to chromatin in living cells (58). While oS is the point for cis-motif recognition on the dyad position, we found no evidence of conserved nucleotides at the oH site (Figure 2B, D, F and H). Thus, the precise position of the oH site with respect to the FOXA1 motif is likely constrained by a spatial requirement rather than nucleotide composition. oH is therefore likely to provide an important scaffolding guide for subsequent contact with oG and oS during the docking of FOXA1 with its underlying nucleosome.
Attaining a well-positioned configuration between the FOXA1 motif and the nucleosome dyad
By competing with H1 to bind the dyad site, FOXA1 can take charge of the underlying nucleosome, and initiate subsequent events. However, FOXA1 also needs to bind to its stationary cis-target site, which is not necessarily in proximity to the dyad. The interesting question then comes as to how a single monomer of FOXA1 is able to engage two disparate sites simultaneously? The reasonable explanation is that the two sites (cis-motif and dyad) assume proper phasing to establish a favorable spatial relationship, and our results clearly demonstrated this pattern. As we know, the cis-motif has a fixed position on the DNA strand, while only the dyad position can be changed by nucleosome sliding along chromatin axis (59). However, this sliding requires energy and is generally facilitated by many chromatin remodelers with ATPase motor (48,49,60). Indeed, our ChIP-seq data of eight remodeler-associated proteins show much higher occupancy of these molecules over well-positioned FOXA1 motif sites, especially in the LMBS mode (Figure 4). Rather than being recruited in a DNA sequence-specific manner to the cis-FOXA1 motif, these factors are recruited to the dyad position to assist in nucleosome positioning, leading the coincidental overlap of remodeler occupancy signals with the cis-FOXA1 motifs. This co-localization phenomenon has also been found in other studies (61,62), for example, the catalytic subunit BRG1 of BAF complexes co-localizes to GATA1-bound distal sites to shift and reorganize nucleosomes during hematopoietic stem cells (HSCs) differentiation (61). In addition to the ∼60 bp of extra-nucleosomal DNA adjacent to the entry/exit site of nucleosomes that is important for the binding of remodelers such as ISW2, it is also well recognized that another hot-spot for chromatin remodeling enzyme access exists near the dyad position (63). Studies show ISW2 can bind to the DNA minor groove of two helical turns flanking the dyad axis just ∼20 bp away (64), which partially overlap the three competition sites (oG, oS, and oH) between H1 and FOXA1. The detailed mechanism for nucleosome movement remains to be fully elucidated though the DNA twist model and the bulge propagation model have been proposed (59,60). Our results of the co-localization of FOXA1 motifs and eight remodelers on dyad loci strongly suggest an intrinsic relationship between FOXA1 and chromatin remodeling complexes, as well as the underlying nucleosome.
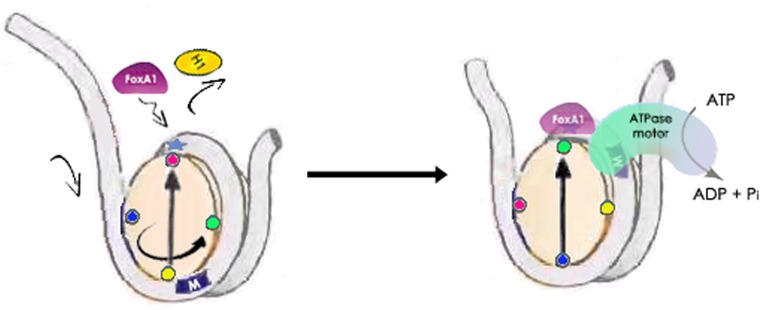
Therefore, in order to attain the well-positioned configuration between the cis-FOXA1 motif and the nucleosome dyad position, we proposed a positional-nucleosome-oriented binding model for this process (Figure 5). Initially, when the FOXA1 motif is randomly positioned out of phase with the dyad of the underlying nucleosome (like the control case in Figure 3, and left panel in Figure 5), the FOXA1 protein enters the dyad locus from one side to compete with and replace H1. Then with assistance from chromatin remodeling complexes and ATPase motor, FOXA1, coupled to the dyad of the nucleosome core, translates along the nucleosomal DNA in search of its cognate cis-motif target until the correct phasing of the motif and dyad is achieved and a stable FOXA1 complex configuration is formed. In this model, FOXA1 does not initially search for its cis-target buried within the nucleosome, but instead locates the dyad position by competing with histone H1. Thereafter, in association with the remodeler complexes it locates the connate motif locally, where each nucleotide is examined no matter if it faces inward or outward from the histone octamer. Alternatively, Soufi et al. recently suggested a partial motif recognition strategy in the study of four reprogramming OSKM factors accessing silent chromatin (65), in which TFs may target a part of their canonical motif displayed on the nucleosome surface. However, in this model, TFs cannot fully examine each DNA base wrapped in the super-helical turns around the histone octamer, especially those positions facing inward to the octamer surface. Thus, TFs are only able to sample partially exposed motifs by stochastic collision with nucleosome surfaces. Therefore, different strategies might exist for different pioneer factors to access variable chromatin, and other layers of modulating access remain for further exploration. Further applications using our ChIP-ePENS assay integrated with chromatin features should extend and shed light on our understanding the mechanistic features of pioneer factor actions.
Figure 5.
A proposed model of the positional-nucleosome-oriented binding pattern for the pioneer factor FOXA1. To compete with H1, the FOXA1 protein occupies the dyad position, and changes its translational position together with the dyad due to the activity of ATPase-dependent chromatin remodelers, to search for its cognate cis-motif by examining every nucleotide, and ultimately arriving at a well-positioned phase between the cis-motif and dyad position.
ACCESSION NUMBERS
The ChIP-seq data from this study has been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE72690.
Supplementary Material
Acknowledgments
We are grateful to Dr Ken Zaret at Institute for Regenerative Medicine, University of Pennsylvania for reading the manuscript and providing suggestive comments, and to all members of the Jin and Wang laboratories for valuable discussion.
Authors contributions: Z.Y. and V.X.J. conceived the project. Z.Y. and V.X.J. designed the research with input from Z.C. and Q.W. Z.Y. developed the algorithm. Z.Y. performed computational analyses with the help from Z.C. Z.C. and Q.W. designed the experiments and Z.C. performed ChIP-seq and ChIP-ePENS data experiments. Z.Y. and V.X.J. wrote the manuscript with input from Q.W., S.F., B.S., T.H.H. and revisions from all co-authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health (NIH) [R01 GM114142 to V.X.J., R01 CA151979 to Q.W. and U54 CA113001 (Integrative Cancer Biology Program, to T.H.H, Q.W., V.X.J.)]; NCI CCSG [P30 CA054174]; University of Texas System STARS award. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Workman J.L., Kingston R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Bell O., Tiwari V.K., Thomä N.H., Schübeler D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 3.Swygert S.G., Peterson C.L. Chromatin dynamics: Interplay between remodeling enzymes and histone modifications. Biochim. Biophys. Acta - Gene Regul. Mech. 2014;1839:728–736. doi: 10.1016/j.bbagrm.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen T., Dent S.Y.R. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurard-Levin Z.A., Quivy J.-P., Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- 6.Farnham P.J. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morse R.H. Getting into chromatin: how do transcription factors get past the histones. Biochem. Cell Biol. 2003;81:101–112. doi: 10.1139/o03-039. [DOI] [PubMed] [Google Scholar]
- 8.Voss T.C., Hager G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaret K.S., Carroll J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwafuchi-doi M., Zaret K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnani L., Eeckhoute J., Lupien M. Pioneer factors: Directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y.A., Yu J. Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis. 2015;2:144–151. doi: 10.1016/j.gendis.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furey T.S. ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat. Rev. Genet. 2012:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 17.Rhee H.S., Pugh B.F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z., Lan X., Thomas-Ahner J.M., Wu D., Liu X., Ye Z., Wang L., Sunkel B., Grenade C., Chen J., et al. Agonist and antagonist switch DNA motifs recognized by human androgen receptor in prostate cancer. EMBO J. 2015;34:502–516. doi: 10.15252/embj.201490306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim H., Uhlenhaut N.H., Rauch A., Weiner J., Hübner S., Hübner N., Won K.J., Lazar M.A., Tuckermann J., Steger D.J. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 2015;25:836–844. doi: 10.1101/gr.188581.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serandour A.A., Brown G.D., Cohen J.D., Carroll J.S. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol. 2013;14:R147. doi: 10.1186/gb-2013-14-12-r147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L., Chen J., Wang C., Uusküla-Reimand L., Chen K., Medina-Rivera A., Young E.J., Zimmermann M.T., Yan H., Sun Z., et al. MACE: model based analysis of ChIP-exo. Nucleic Acids Res. 2014;42:e156. doi: 10.1093/nar/gku846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y., Mahony S., Gifford D.K. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput. Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starick S.R., Ibn-Salem J., Jurk M., Hernandez C., Love M.I., Chung H.R., Vingron M., Thomas-Chollier M., Meijsing S.H. ChIP-exo signal associated with DNA-binding motifs provide insights into the genomic binding of the glucocorticoid receptor and cooperating. Genome Res. 2015;25:825–835. doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z., Lan X., Wu D., Sunkel B., Ye Z., Huang J., Liu Z., Clinton S.K., Jin V.X., Wang Q. Ligand-dependent genomic function of glucocorticoid receptor in triple-negative breast cancer. Nat. Commun. 2015;6:8323. doi: 10.1038/ncomms9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlin S., Brendel V. Chance and statistical significance in protein and DNA sequence analysis. Science. 1992;257:39–49. doi: 10.1126/science.1621093. [DOI] [PubMed] [Google Scholar]
- 26.Hagerup T., Rüb C. A guided tour of chernoff bounds. Inform. Process. Lett. 1990;33:305–308. [Google Scholar]
- 27.Reid J.E., Wernisch L. STEME: Efficient em to find motifs in large data sets. Nucleic Acids Res. 2011;39:e126. doi: 10.1093/nar/gkr574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant C.E., Bailey T.L., Noble W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He H.H., Meyer C.A., Shin H., Bailey S.T., Wei G., Wang Q., Zhang Y., Xu K., Ni M., Lupien M., et al. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischler M.A., Bolles R.C. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM. 1981;24:381–395. [Google Scholar]
- 31.Carroll T.S., Liang Z., Salama R., Stark R., de Santiago I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front. Genet. 2014;5:75. doi: 10.3389/fgene.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H., et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balanda K.P., Macgillivray H.L. Kurtosis: a critical review. Am. Stat. 1988;42:111–119. [Google Scholar]
- 34.McPherson C.E., Shim E.Y., Friedman D.S., Zaret K.S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 35.McPherson C.E., Horowitz R., Woodcock C.L., Jiang C., Zaret K.S. Nucleosome positioning properties of the albumin transcriptional enhancer. Nucleic Acids Res. 1996;24:397–404. doi: 10.1093/nar/24.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirillo L.A., McPherson C.E., Bossard P., Stevens K., Cherian S., Shim E.Y., Clark K.L., Burley S.K., Zaret K.S. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark K.L., Halay E.D., Lai E., Burley S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 38.Littler D.R., Alvarez-Fernández M., Stein A., Hibbert R.G., Heidebrecht T., Aloy P., Medema R.H., Perrakis A. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010;38:4527–4538. doi: 10.1093/nar/gkq194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang C., Pugh B.F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ay A., Arnosti D.N. Nucleosome positioning: an essential component of the enhancer regulatory code. Curr. Biol. 2010;20:R404–R406. doi: 10.1016/j.cub.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 42.Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watts J.A., Zhang C., Klein-Szanto A.J., Kormish J.D., Fu J., Zhang M.Q., Zaret K.S. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 2011;7:e1002277. doi: 10.1371/journal.pgen.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwafuchi-Doi M., Donahue G., Kakumanu A., Watts J.A., Mahony S., Pugh B.F., Lee D., Kaestner K.H., Zaret K.S. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struhl K., Segal E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes A.L., Rando O.J. Mechanisms underlying nucleosome positioning in vivo. Annu. Rev. Biophys. 2014;43:41–63. doi: 10.1146/annurev-biophys-051013-023114. [DOI] [PubMed] [Google Scholar]
- 47.Cairns B.R. Chromatin remodeling complexes: Strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 49.Bartholomew B. Regulating the Chromatin Landscape: Structural and Mechanistic Perspectives. Annu. Rev. Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins T.T., Dalal R. V, Mitsis P.G., Block S.M. Sequence-dependent pausing of single lambda exonuclease molecules. Science. 2003;301:1914–1918. doi: 10.1126/science.1088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calo E., Wysocka J. Modification of Enhancer Chromatin: What, How, and Why. Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaret K.S. Nucleoprotein architecture of the albumin transcriptional enhancer. Semin. Cell Biol. 1995;6:209–218. doi: 10.1006/scel.1995.0029. [DOI] [PubMed] [Google Scholar]
- 53.Ramakrishnan V., Finch J.T., Graziano V., Lee P.L., Sweet R.M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 54.Cerf C., Lippens G., Ramakrishnan V., Muyldermans S., Segers A., Wyns L., Wodak S.J., Hallenga K. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5. Biochemistry. 1994;33:11079–11086. doi: 10.1021/bi00203a004. [DOI] [PubMed] [Google Scholar]
- 55.Cirillo L.A., Zaret K.S. Specific Interactions of the Wing Domains of FOXA1 Transcription Factor with DNA. J. Mol. Biol. 2007;366:720–724. doi: 10.1016/j.jmb.2006.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Syed S.H., Goutte-Gattat D., Becker N., Meyer S., Shukla M.S., Hayes J.J., Everaers R., Angelov D., Bednar J., Dimitrov S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyer S., Becker N.B., Syed S.H., Goutte-Gattat D., Shukla M.S., Hayes J.J., Angelov D., Bednar J., Dimitrov S., Everaers R. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: Nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 2011;39:9139–9154. doi: 10.1093/nar/gkr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 59.Bowman G.D. Mechanisms of ATP-dependent nucleosome sliding. Curr. Opin. Struct. Biol. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flaus A., Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: Farewell to the tuna-can octamer. Curr. Opin. Genet. Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Hu G., Schones D.E., Cui K., Ybarra R., Northrup D., Tang Q., Gattinoni L., Restifo N.P., Huang S., Zhao K. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Côté J., Peterson C.L., Workman J.L. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kagalwala M.N., Glaus B.J., Dang W., Zofall M., Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zofall M., Persinger J., Kassabov S.R., Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 65.Soufi A., Garcia M.F., Jaroszewicz A., Osman N., Pellegrini M., Zaret K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.