Abstract
Members of the IS200/IS605 insertion sequence family differ fundamentally from classical IS essentially by their specific single-strand (ss) transposition mechanism, orchestrated by the Y1 transposase, TnpA, a small HuH enzyme which recognizes and processes ss DNA substrates. Transposition occurs by the ‘peel and paste’ pathway composed of two steps: precise excision of the top strand as a circular ss DNA intermediate; and subsequent integration into a specific ssDNA target. Transposition of family members was experimentally shown or suggested by in silico high-throughput analysis to be intimately coupled to the lagging strand template of the replication fork. In this study, we investigated factors involved in replication fork targeting and analysed DNA-binding properties of the transposase which can assist localization of ss DNA substrates on the replication fork. We showed that TnpA interacts with the β sliding clamp, DnaN and recognizes DNA which mimics replication fork structures. We also showed that dsDNA can facilitate TnpA targeting ssDNA substrates. We analysed the effect of Ssb and RecA proteins on TnpA activity in vitro and showed that while RecA does not show a notable effect, Ssb inhibits integration. Finally we discuss the way(s) in which integration may be directed into ssDNA at the replication fork.
INTRODUCTION
Transposable elements (TE) are genetic entities capable of moving from one location to another in a genome. Insertion sequences (IS) are the most compact and numerous TE present in bacteria. They are principal motors of genome remodelling and play an important role in horizontal gene transfer (1). Several transposition mechanisms have been described. Arguably the best documented involves double-strand DNA intermediates and employs transposases of the so-called DDE superfamily (DDE for Aspartate-Aspartate-Glutamate, residues constituting the catalytic site of these transposases) (2). In addition to coding for DDE transposases, classical IS also carry small terminal inverted repeats (IR) and often generate short flanking direct repeats (DR) of target DNA on insertion.
We have previously characterized a new transposition mechanism in which a single DNA strand is excised from a donor site and integrated into a single strand target. This mechanism is employed by IS200/IS605 family members which are widely distributed over the prokaryotic phylogenetic tree. These IS are quite different from classical IS. They do not carry terminal IR but have ends rich in secondary structures which are recognized and bound by their transposase, TnpA. Instead of the conserved active site amino acid DDE signature of classic Tpases, TnpA enzymes contain a motif composed of a single catalytic Tyrosine and a His-u-His amino acid triad (HuH, where ‘u’ represents a hydrophobic residue) involved in coordination of an essential metal ion. They are therefore called Y1 transposases. Our structural studies (3–6) revealed that these are members of a larger ‘HuH’ enzyme superfamily including RCR Rep proteins (involved in Rolling Circle plasmid and phage Replication), relaxases (involved in conjugal plasmid transfer) and Tpases of the Rolling Circle IS91/ISCR family of transposons. All use a catalytic tyrosine residue to attack the target phosphodiester bond creating a covalent 5′-phosphotyrosine enzyme-substrate intermediate.
Our in vitro and in vivo studies with IS608, initially isolated from Helicobacter pylori but active in Escherichia coli and ISDra2 from Deinococcus radiodurans, provided a detailed picture of the transposition pathway of this family (7–12). Transposition has an absolute requirement for single stranded (ss) DNA substrates and is strand-specific: the ‘top’ strand is recognized by the Tpase and undergoes strand cleavage and transfer while the ‘bottom’ strand is refractory to the enzyme and therefore inactive. Excision of the top strand as a transposon circle with joined left and right ends (transposon junction) is accompanied by rejoining of the DNA flanks (donor joint) (Figure 1A). The circle junction then undergoes TnpA-catalysed integration into an ssDNA target in a sequence-specific reaction. Insertion involves strand transfer of both the 5′ and 3′ ends of the single strand transposon circle junction into the single strand target. The left (5′) end always inserts 3′ to a tetra- or penta nucleotide target sequence specific for each element. For IS608 the tetranucleotide is TTAC (Figure 1A) and this sequence is also essential for subsequent transposition (8).
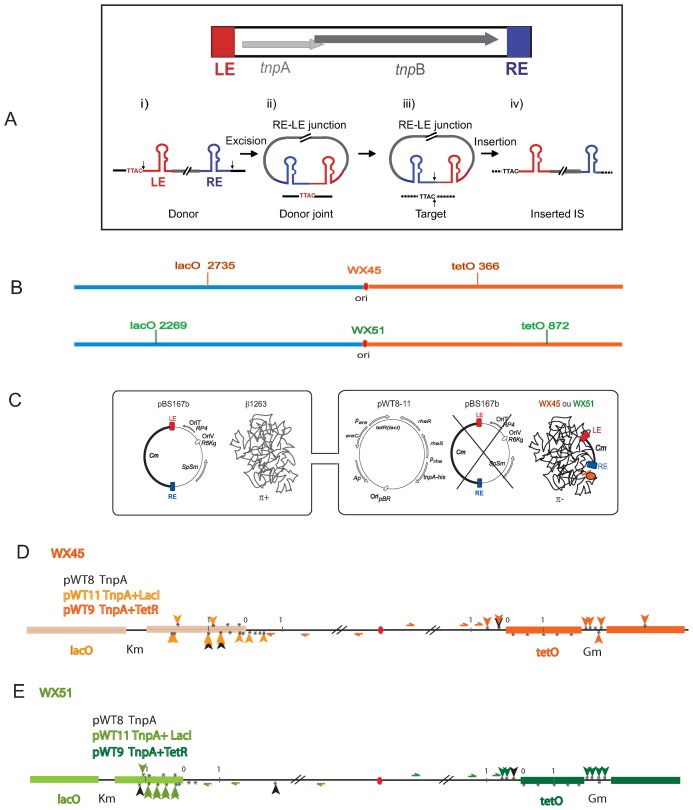
Figure 1.
IS608 insertion into blocked chromosome replication forks. (A) IS608 and its transposition pathway. IS608 organization. Top: the left (LE) and right (RE) IS ends are shown in red and blue respectively. Grey horizontal arrows: tnpA and tnpB; red and blue boxes, LE and RE ends (colour code retained throughout). Transposition pathway. Bottom (i) schematized single-stranded IS608 showing secondary structures of LE and RE, the flanking TTAC and cleavage positions at the IS ends (vertical black arrows). (ii) Excision and formation of circular ssDNA with an RE–LE transposon junction and a sealed donor joint (black line) retaining the TTAC. (iii) TnpAIS608 brings together the transposon junction with a new target DNA (dotted black line) carrying the required target TTAC. The two black arrows indicate points of cleavage and strand transfer. (iv) Insertion of IS608 into the target flanked on the LE by TTAC. (B) Position of the lacO and tetO arrays in Escherichia coli WX45 and WX51: the replication origin, ori, is shown as a red ellipse and the Left and Right replicores in blue and orange respectively. E. coli WX45 and WX51 contain arrays at different locations. (C) Experimental system: the cartoon represents the suicide mating strategy used to isolate insertions. The left side of the figure shows plasmid pBS167b, used as the IS608 donor plasmid. It is replication proficient in the donor strain by virtue of a copy of the π replication gene inserted into the donor chromosome. pBS167b is unable to replicate following conjugal transfer to the transposition test strain since this strain lacks the π gene. This is shown in the recipient strain on the right side of the figure. Three alternative plasmids were used in the recipient strain: pWT8 (carrying tnpA alone under control of the prha promoter); pWT11 (a pWT8 derivative including lacI under control of the para promoter); or pWT9 (a pWT8 derivative including tetR under control of the para promoter). Transposase and either LacI (pWT11) or TetR (pWT9) were induced in the recipient prior to mating. Following mating, the cognate inducers were added to release LacI or TetR respectively from the operator arrays and relieve the replication barrier permitting cells to resume growth before plating. (D and E) Insertions into E. coli WX45 and WX51: details of the lacO array (light orange or light green rectangles) on the left replicore and of the tetO array (orange or green rectangles) on the right replicore. Black vertical arrows: insertions obtained in the absence of LacI or TetR. Green or orange vertical arrows: insertions obtained in the presence of LacI or TetR in several independent experiments. The positions of the oligonucleotides (not to scale) used to localize the insertions are shown with half arrowheads. The kanamycin and gentamycin resistance cassettes used in the construction and insertion of the lac and tet operator arrays are also shown. * represents potential TTAC target sequences present in the region, 0 represents the beginning of arrays and 1 is distance in kb.
The single strand nature of IS200/IS605 family transposition raised the question of the source of ssDNA in the host cells. This can be generated in several ways. ISDra2 transposition in D. radiodurans is induced by irradiation which generates large amounts of single strand DNA during the process of post-irradiation repair and genome reconstitution (10). We also showed that the replication direction through IS608 and ISDra2 plays an important role in the excision step and that it is favoured when the active strand is located on the lagging-strand template at the replication fork. Furthermore, this lagging strand template appeared an attractive target for IS608 in E. coli and for ISDra2 in D. radiodurans as well for other IS200/IS605 family members (13).
In addition, a significant number of IS608 insertions into the E. coli chromosome were localized in the highly transcribed rrn genes (13), suggesting that high transcription levels might increase accessible ssDNA by affecting replication fork progression (14–17). This hypothesis was further supported using a plasmid target carrying the replication termination Tus/Ter system. The Tus–Ter complex forms a barrier that blocks progression of the replicative helicase, DnaB, when a fork arrives in the non-permissive direction (18–20). Tus binding to the Ter sites causes a transient pause, targeting IS608 insertions close to the Ter site on the lagging strand template of the replication fork (13).
Here we address how IS608 specifically targets the replication fork, the role of the transposase, TnpAIS608 and of the structural features of target DNA in target recognition. We investigated targeting of IS608 insertions into the E. coli chromosome using an operator/repressor replication fork roadblock system (21,22) and demonstrated that they were recruited to the lagging strand template both upstream of, and within, the stalled forks. We also used fluorescence microscopy to localize a fluorescent TnpAIS608 derivative and show that the protein co-localizes to the same repressor/operator stalled forks which attract IS608 insertions. We also demonstrate that TnpAIS608 interacts with DnaN, the β sliding clamp replisome processing factor and recognizes DNA resembling replication fork structures. This suggests that interaction of TnpAIS608 with these factors may play an important role in IS608 targeting.
In spite of the single strand nature of IS200/IS605 transposition, we also observed sequence-independent binding of TnpAIS608 to double strand DNA, a property which may have biological relevance. Here we show that the presence of dsDNA adjacent to an ssDNA IS608 end facilitates TnpAIS608-catalysed cleavage of these substrates in vitro. We also investigated the effect of dsDNA adjacent to single strand target and observed different effects depending on whether the ds DNA located 5′ or 3′ to the TTAC tetranucleotide.
ssDNA is rarely naked in vivo but it is protected by various proteins, mainly by the single strand binding protein Ssb and RecA (23–25). In addition to their general roles in DNA metabolism, both Ssb and RecA are also localized at the replication fork. In particular, RecA by binding to the ssDNA region, is intimately involved in processing stalled replication forks (26). We therefore also investigated the effect of Ssb and RecA on excision and insertion of IS608 in vitro. We show here different effects of these proteins on IS608 excision and integration and discuss how IS integration is influenced by these proteins.
MATERIALS AND METHODS
Bacterial and yeast strains and media
Strains used in this study are listed in Supplementary Table S1.
For bacteria, LB grown cultures were supplemented, where necessary, with appropriate antibiotics.
For yeast, minimal synthetic dropout medium (SD) deplete of certain amino acid or nucleotide for selection were used. According to transformed cell auxotrophy, l-histidine (10 mg/l), adenine (10mg/l) or X-α-Gal (20 μg/ml) were added as needed.
Plasmids
pBS167b was constructed in two steps. First, a SpSm cassette replaced a Cm cassette in pSW23T to obtain pSW23SpSm. Second, the IS608 derivative transposon carrying Cm cassette was inserted into this intermediate plasmid to obtain pBS167b. pWT8 was constructed by cloning tnpA-his6 under control of Prha in pRha113 (Giacalone 2006). pWT11 was constructed by inserting a cassette expressing LacI-GFP under control of Para in pWT8. pWT9 was constructed by inserting a cassette expressing TetR-YFP under control of Para in pWT8.
pBS147 was constructed by cloning mCherry-tnpA fusion under control of Plac; pLL25 carries mCherry under control of Plac.
pDAG729 carries TetR-GFP fusion under control of Para (27).
pGBKT7-dnaN and pGADT7-dnaN were constructed by inserting the dnaN gene cassette into the NdeI and EcoRI site of pGBKT7 and pGADT7 cloning vector. pGADT7-tnpA was constructed by inserting the tnpA gene cassette into the NdeI and EcoRI site of pGADT7 cloning vector.
Recombinant pKT25, pUT18 and pUT18C carrying either tnpAIS608 or dnaN (NP_418156.1) were constructed as described by (28).
Mating out assays
The frequency of IS608 transposition was determined by a standard mating out assay (29).
Operator experiments
Fresh overnight cultures of donor (LB + DAP + SpSmCm) and recipient WX45 or WX51 cells (LB + GmKmAp + 0.5% Glucose) were diluted in LB medium without antibiotics for recipient and LB supplemented with diaminopimelic acid (DAP) for donor cells at 37°C. At OD600 of 0.5, the donor cells were incubated without agitation, the recipient was diluted to an OD600 of 0.15 and the transposase was induced with 0.5 mM rhamnose. After 60 min induction of TnpAIS608 expression followed by 30 min induction of LacI or TetR by 0.08% arabinose in the recipient, donor and recipient strains were mixed and incubated for 2 h. Cognate inducers IPTG or ATc (final concentration 1 mM and 10 μg/ml, respectively) were added to cultures and incubation continued for 30 min before concentrating and plating on Cm-containing LB agar plates. The assay is not quantitative, CmR colonies were collected from plates and bulk chromosomal DNA was isolated. The distribution of insertions in the population was analysed by polymerase chain reaction (PCR) using sets of forward primers (B243, B252, B237, B245 for WX45 and B240, B251, B258, B238 for WX51), left (LE) and right (RE) as reverse primers. Insertions in rrn operons were analysed with B247 as forward primer (Supplementary Data).
Insertion pattern analysis
PCR amplification was carried out with Phusion DNA Polymerase (Finnzymes) in GC buffer in the following conditions: 30 s 98°C, 35× (10 s 98°C, 30 s 64°C, 2 min 72°C). A total of 500 ng of bulk chromosomal DNA were used for each reaction.
Microscopy procedures
Overnight cultures grown from a single colony in LB supplemented with Sp, Sm and Ap at 37°C were diluted into LB (1/100). After 2 h, TetR expression was induced by adding 0.05% arabinose (during 30 or 60 min) and TnpAIS608 expression was induced by adding 50 or 100 μM of IPTG (during 30 or 60 min). Cultures were then stopped on ice, washed with M9 medium and plated on M9 agar-covered slide. Acquisitions were made with motorized and inversed Olympus IX81 microscope. Images were read with Metamorph (Molecular devices) and ImageJ (NIH) Softwares.
TnpAIS608 purification
TnpAIS608 was purified as described (9).
Oligonucleotide preparation
Oligonucleotide hybridizations were performed by incubating labelled DNA with cold oligonucleotides in Tris pH7.5 10 mM, NaCl 50 mM buffer at 95°C during 5 min (for denaturation) and left to slowly cool to 25°C. Hybridized DNA was then purified on 5% acrylamide gels, eluted overnight in elution buffer (Tris pH8 10 mM, ethylenediaminetetraacetic acid 1 mM, sodium dodecyl sulphate 0.2%, NaCl 300 mM). DNAs were then precipitated with ethanol and dissolved in 20 μl EB buffer (Qiagen).
TnpAIS608-DNA binding assays
Binding reactions set up
Radioactivity of each DNA construction was adjusted to 10 000 cpm per μl with water (∼15 nM). A total of 1 μl was then incubated with different concentrations of TnpAIS608 (60 min, 37°C, final volume 10 μl) in 20 mM HEPES (pH 7.5), 50, 150, 300 or 500 mM NaCl, 1 mM Dithiothreitol (DTT), 20 μg/ml bovine serum albumin and 20% glycerol (final volume: 10 μl for Electrophoretic Mobility Shift Assay (EMSA), 25 μl for dot blot).
Analysis of complexes
EMSA was carried out as described previously (12).
Dot blot: a standard dot-blot apparatus was modified so that the nitrocellulose and a second HybondN membrane, placed beneath the nitrocellulose, were sandwiched between two sets of rubber O-ring. Membranes were previously prepared as described by (30) and equilibrated with reaction buffer before and after sample loading.
Membranes were dried at room temperature and analysed by PhosphorImaging. Signals were quantified by Multi Gauge software and analysed as previously described (30). KD were calculated using GraphPad Prism software.
Cleavage and strand transfer assays: reaction conditions were as described (12).
TnpA and RecA/Ssb competition assays: RecA (in the presence of 2 mM adenosine triphosphate (ATP) or ATPγS) and Ssb were pre-incubated for 15 min in reaction buffer with substrates before addition of TnpA in standard conditions.
Yeast two-hybrid system
Plasmids expressing a protein fusion of a binding domain with a target protein (bait) and a fusion protein of an activating domain with interacting partner (prey) were transformed into the yeast host strain by the lithium acetate method (31). The transformants were selected in SD/-Leu/-Trp agar plate. The interaction between two testing proteins was tested by growth ability on SD/-Leu/-Trp/-His or SD/-Leu/-Trp/-Ade agar plates with X-α-gal as a color indicator.
BACTH complementation assay
After transformation, BTH101 cells harbouring a pair of the appropriate plasmids were plated on LB agar containing X-Gal and IPTG plus antibiotics and incubated at 30°C for 24–36 h. The efficiency of the interaction between two tested hybrid proteins was quantified by measuring the β-galactosidase activity in liquid cultures in a 96-well format. To be directly comparable to those obtained with the classical protocol (32), our specific activities were calculated for a volume of extract of 125 μl and for an OD600 nm of 1.
RESULTS
IS608 specifically targets blocked replication fork on the E. coli chromosome
To determine whether IS608 insertion targets blocked chromosomal replication forks as was observed for plasmids, we used transitory fork blockage resulting from lac and tet repressor binding in E. coli strains carrying arrays of lac and tet operators at known ectopic chromosome positions (21,22). Two recipient strains were used, each carried both lac and tet operators lacO and tetO arrays at different chromosomal locations (Figure 1B). For transposon delivery, we used an in vivo transposition system which relies on plasmid ‘suicide’ following conjugation into the recipient cell (13,33).
An IS608 derivative with both tnpA and tnpB genes (Figure 1A) replaced by a chloramphenicol resistance (CmR) cassette was introduced by conjugation using an RP4-based suicide plasmid to deliver one or other DNA strand (depending on the relative orientation of the IS to the origin of plasmid transfer). The transposon donor conjugative plasmid can replicate in the donor cell due to an essential plasmid replication gene, π, integrated into the chromosome. Since the recipient cell does not carry this gene, the plasmid cannot replicate once transferred (Figure 1C). Three alternative plasmids were used in the recipient strain: pWT8 (carrying tnpA alone under control of the prha promoter); pWT11 (a pWT8 derivative including lacI under control of the para promoter); or pWT9 (a pWT8 derivative including tetR under control of the para promoter) (Figure 1D). Prior to mating, transposase expression in the recipient was induced together with that of LacI (pWT11) or TetR (pWT9) to block active replication forks by formation of repressor/operator complexes (22). Following mating, the cognate inducers, IPTG or anhydrotetracyline (aTc) were added to release LacI or TetR respectively from the operator arrays and relieve the replication barrier to permit cells to resume growth before plating. Chromosomal DNA was extracted from a pool of CmR clones and the insertion distribution around the operator arrays was analysed by PCR with at least two primers specific for each locus (horizontal arrows in Figure 1D and E) and LE and RE as reverse primers (‘Materials and Methods’ section).
Without repressor, only a limited number of insertions were detected at the operator arrays (Figure 1D and E, black arrows). These occurred on the lagging strand template, in accordance with previous observations obtained with stalled plasmid replication forks and chromosome forks of actively growing E. coli cells (13).
However, with LacI and in spite of no notable change in insertion frequency (‘Materials and Methods’ section), we observed LacI-dependent IS608-specific targeting into lacO arrays. This was true for both recipient strains (Figure 1D and E, upper panels, coloured arrows). Insertions were localized to the replication fork-proximal region of the lacO arrays and up to ∼1 kb inside corresponding to the position of stalled forks observed previously (22). Again, the majority of insertions had occurred in the lagging strand although potential TTAC target sequences were also present in the leading strand.
The insertion pattern was slightly different with the tetO arrays (Figure 1D and E). No insertions were detected within the operator sequences themselves in the presence of TetR because, although the operator arrays carry several potential TTAC target sequences, these are all on the leading strand template and would therefore not be expected to favour IS608 insertion. However, some TetR-dependent insertions occurred on the lagging strand at TTAC sequences upstream of the operator array while others occurred in the Gm resistance cassette used in the array construction (21).
To rule out the possibility that the replication block regime changes the global insertion pattern in these strains, we examined insertions in the rrn genes previously observed in normally replicating E. coli (13) (Supplementary Figure S1A and B). PCR reactions were performed on clones grown with and without repressors using a primer complementary to the 5′ region of the rrn operons together with LE or RE primers. Insertions still occurred in the rrn operons whether or not repressors were present and the same insertion bias was observed (Supplementary Figure S1A and B) as seen previously (13).
Thus, IS608 insertion appears to be specifically attracted to repressor–operator blocked replication forks on the E. coli chromosome. Moreover, the orientation of these insertions is consistent with insertion into the lagging strand template.
TnpAIS608 is attracted to stalled replication forks in vivo
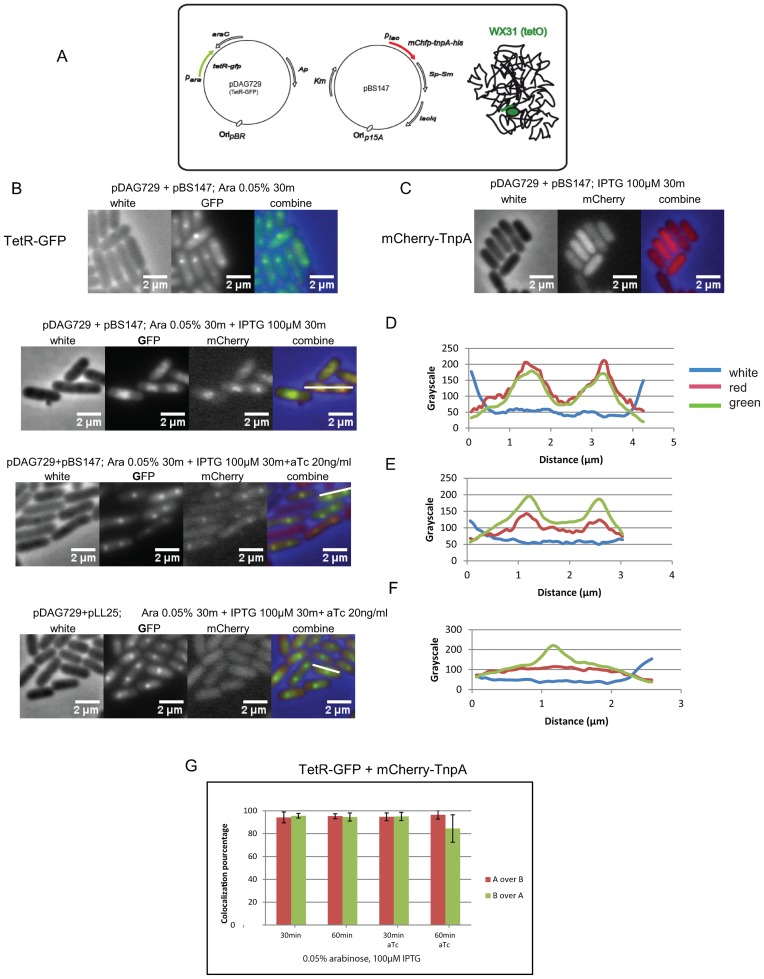
To examine whether specific IS608 targeting to repressor/operator blocked forks is mediated by TnpAIS608 itself, we developed a system to localize TnpAIS608 in vivo (Figure 2A). We constructed a functional mCherry-tagged transposase (mCherry-TnpA) whose position in the cell can be visualized by fluorescence microscopy. mCherry-TnpAIS608 was expressed under control of Plac in a p15a-based plasmid. A second compatible ColE1-based plasmid was used as a source of TetR-GFP (green fluorescent protein) fusion protein under control of the arabinose promoter. Inducer concentration and induction times were optimized (‘Materials and Methods’ section).
Figure 2.
Localization of mCherry-TnpAIS608 in vivo. (A) Experimental system:Escherichia coli WX31 carries tetO arrays (in green) on the chromosome and two compatible plasmids expressing TetR-GFP fusion under control of para (pDAG729) and mCherry-TnpAIS608 fusion under control of plac (pBS147) respectively. (B) TetR-GFP foci at blocked forks. Cultures of cells carrying pBS147 and pDAG729 were induced for TetR-GFP expression by addition of 0.05% arabinose for 30 min. (C) mCherry-TnpAIS608 homogenous localization in the cells. Cultures of cells carrying pDAG729 and pBS147 were induced for mCherry-TnpAIS608 expression (pBS147) by addition of 100 μM IPTG for 30 min. (D) TetR-GFP and mCherry-TnpAIS608 foci and their co-localization analysed by Metamorph. Left: cultures of cells carrying pDAG729 and pBS147 were induced for both TetR-GFP and mCherry-TnpAIS608 expression by addition of 0.05% arabinose and 100 μM IPTG for 30 min. Right: measurement of red and green fluorescence distribution over the cell cross-section shown in the Left section as a white bar. (E) TetR-GFP and mCherry-TnpAIS608 localization and addition of aTc. Left: cultures of cells carrying pDAG729 and pBS147 were induced for both TetR-GFP and mCherry-TnpAIS608 expression by addition of 0.05% arabinose and 100 μM IPTG for 30 min. The TetR inducer, aTc, was added to 20 ng/ml to reduce the amount of bound TetR. Right: measurement of red and green fluorescence distribution over the cell cross-section shown in the Left section as a white bar. (F) TetR-GFP and mCherry localization. Left: cultures of cells carrying pDAG729 and pLL25 (carrying the mCherry gene without TnpAIS608) were induced for both TetR-GFP and mCherry expression by addition of 0.05% arabinose and 100 μM IPTG for 30 min in the presence of aTc, (20 ng/ml). Right: measurement of red and green fluorescence distribution over the cell cross-section shown in the Left section as a white bar. (G) TetR-GFP and mCherry-TnpAIS608 co-localization. Analysis with Metamorph showing the percentage of TetR-GFP and mCherry-TnpAIS608 co-localization in different culture conditions.
The activity of the mCherry-TnpAIS608 derivative was compared to that of native TnpAIS608 in a series of mating out assays (29). These measure the frequency of transposition of a synthetic IS608 transposon carried by plasmid pBS102 (8) and composed of LE, RE and a CmR gene cassette which replaces tnpA and tnpB, into the conjugative plasmid target, pOX38Km. Wild-type- or mCherry-TnpAIS608 were supplied in trans from a compatible plasmid (‘Materials and Methods’ section). Under these conditions, mCherry-TnpAIS608 is only slightly less efficient than wild-type TnpAIS608 (Supplementary Figure S2).
When TetR-GFP was induced alone, discrete fluorescent foci were observed (Figure 2B) corresponding to TetR-GFP binding to the tetO operators. About 90% of the population showed foci after 30 min induction (0.05% arabinose). As expected for cells growing in LB medium with multiple replication forks, a majority of cells exhibited 2 foci (21). On the other hand, when mCherry-TnpAIS608 was induced alone (without TetR-GFP), no mCherry foci were observed but a similarly high percentage of cells showed a homogenously distributed red fluorescent signal, independently of the induction conditions (Figure 2C).
However, induction of both proteins resulted in red foci in about 20% of cells in two induction conditions (Figure 2D and E, left panels). With no anhydrotetracycline (aTc) inducer, TetR-GFP foci were quite large (Figure 2D). However, addition of sub-maximal levels of aTc to reduce TetR-GFP affinity for tetO resulted in a decrease in their size without significantly reducing their frequency in the cell population (Figure 2E). The mCherry-TnpAIS608 foci remained clearly visible. In a control experiment where mCherry-TnpAIS608 was replaced by mCherry alone (pLL25), fluorescence was homogenously distributed within the cells in spite of the TetR-induced blocked replication forks (Figure 2F, left panel), in a similar way to mCherry-TnpAIS608 itself when expressed alone without TetR induction (data not shown).
We further analysed the distribution of each protein by measuring the fluorescence level of each fluorophore in chosen longitudinal cell cross-sections (Metamorph software) (Figure 2D and E, right panels). Measurements were made on cell populations with and without sub-maximal aTc concentrations and with 30 and 60 min induction of the mCherry-TnpAIS608 (Figure 2G). The green columns in the histogram correspond to the percentage overlap of the green (Tet-GFP) signal with the red (mCherry-TnpAIS608). Clearly almost all the green signal co-localized with the red irrespective of culture conditions (time of induction, presence or absence of aTc). Reciprocally, the red columns showing the percentage of the red signal which co-localizes with the green indicate that almost all the red foci overlap with the green.
As expected, in a control experiment where mCherry-TnpAIS608 was replaced by mCherry alone the red fluorescent signal was distributed homogenously over the entire cell, only 20–30% of the red signal overlapped with the green, corresponding to the portion of mCherry localized in the vicinity of TetR-GFP foci (Supplementary Figure S3A).
These results therefore clearly show: that mCherry-TnpAIS608 foci strongly co-localized with TetR-GFP foci, that localization requires the TnpAIS608 moiety of mCherry-TnpAIS608 and that blocked replication forks are a prerequisite for visualization of mCherry-TnpAIS608 foci. Similar results were obtained with the lacO array and LacI-CFP fusion protein (Supplementary Figure S3B–D).
Targeting mechanisms
Interaction with β sliding clamp
A major question raised by the above results is how TnpAIS608 targets replication forks. A priori, this could occur in vivo by interaction with host-encoded proteins.
We first searched for host proteins permitting replication fork targeting by a physical protein-protein interaction approach ‘Tandem Affinity Purification’ (TAP-Tag). This general attempt was not successful. We then used in vivo two-hybrid approaches.
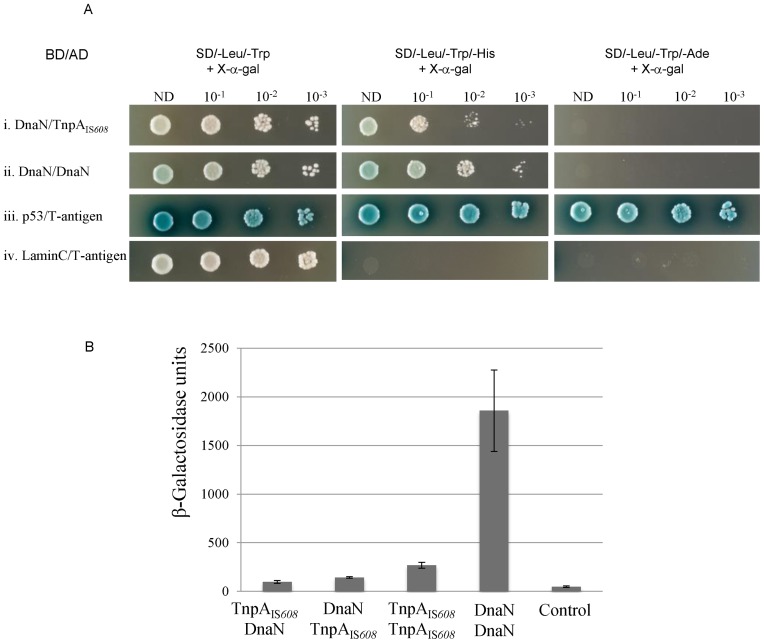
The β sliding clamp (DnaN) is known to interact with transposition proteins (34), we therefore tested the possibility that TnpAIS608 interacts with E. coli DnaN protein. DnaN/TnpAIS608 interactions could be readily detected on selective medium in a yeast two-hybrid system, (Figure 3Ai) in contrast to a negative control (Figure 3Aiv).
Figure 3.
Interaction of TnpAIS608 with DnaN in yeast and bacterial two-hybrid systems (BACTHs). (A) Yeast two-hybrid system. For each hybrid, dilutions (10−1, 10−2 and 10−3) of yeast cell cultures normalized (OD = 2) for each interaction and expressing both bait and prey constructs were spotted on several selective media. ND: non-diluted; BD: binding domain; AD: activation domain. Positive (p53/T-antigen) and negative (LaminC/T-antigen) controls are shown at the bottom of the figure. (B) BACTH. DnaN/TnpAIS608 and TnpAIS608/DnaN correspond to configurations with fused proteins T25-DnaN/TnpAIS608-T18 and T25-TnpAIS608/DnaN-T18 respectively (28). The efficiency of functional complementation between the indicated hybrid proteins was quantified by measuring β-galactosidase activities in Escherichia coli BTH101 carrying the corresponding plasmids as described in ‘Materials and Methods’ section. The figure represents the mean and standard deviation of at least six independent assays.
We further confirmed this interaction by a complementary bacterial two-hybrid system (BACTH) (28). The efficiencies of functional complementation between the different hybrids were quantified by measuring β-galactosidase activities in an E. coli reporter strain as described in ‘Materials and Methods’ section (Figure 3B). In this system, the positive control DnaN/DnaN interactions resulted in high β-galactosidase activity. DnaN/TnpAIS608interaction can be quantified in both configurations (permutation of hybrid partners, ‘Materials and Methods’ section) and conferred low but reproducible β-galactosidase activity, 2–3-fold above negative control, not very different from known TnpAIS608/TnpAIS608interactions (5-fold).
Thus the interaction of TnpAIS608with host β sliding clamp DnaN can be observed in two complementary two-hybrid systems. The relative low level of growth on selective plates and low β-galactosidase activity observed could either reflect weak interaction or may be due to the fact that the interaction interface is somehow influenced by fusion to respective binding and activation domains in these assays.
TnpAIS608 binds to structured DNA and dsDNA
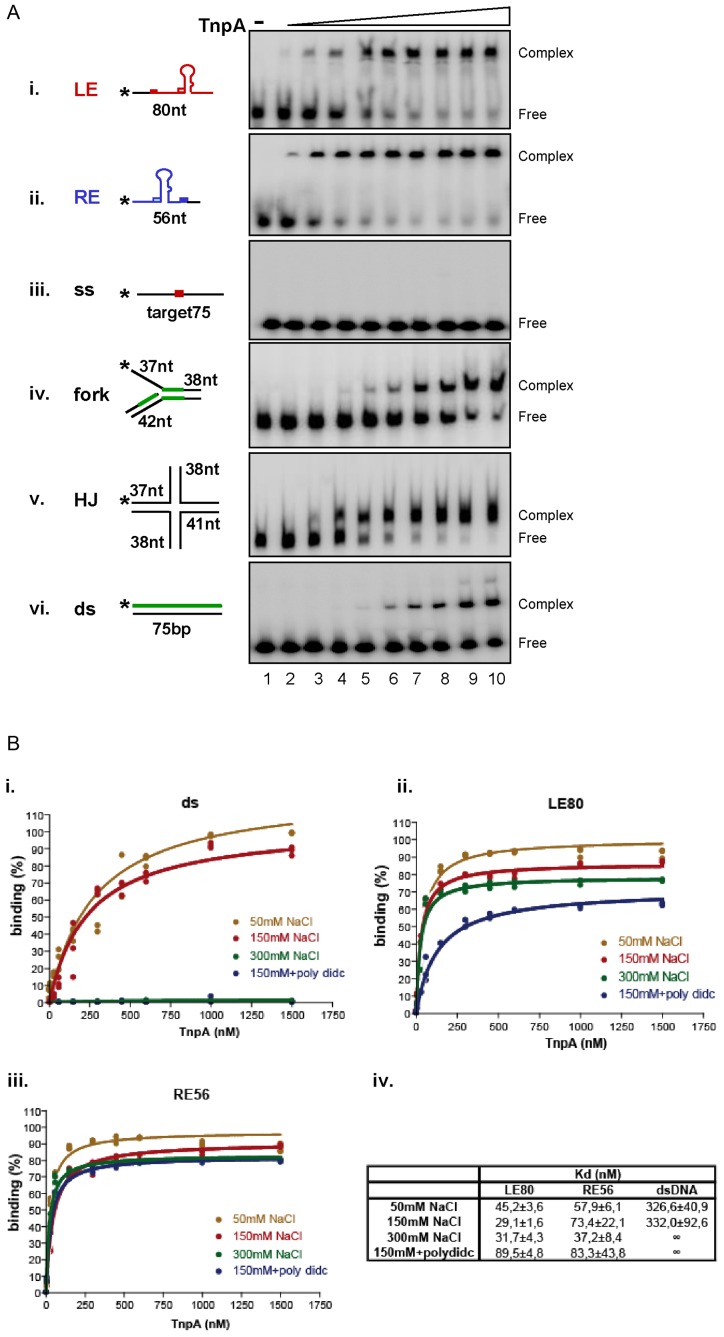
We then explored whether TnpAIS608 might also be targeted to replication forks by direct interaction with DNA structures mimicking a replication fork as reported for the TnsE protein of Tn7 (35). We therefore constructed several substrates and investigated TnpAIS608 binding properties by EMSA and ‘double filter retention’ (‘Materials and Methods’ section).
Although TnpAIS608 binds the ‘top’ strand of both LE and RE efficiently (Figure 4Ai and ii as observed previously (8,11), we were unable to detect complexes of TnpAIS608 by EMSA with ssDNA lacking the hairpin structure (Figure 4Aiii) even if this included the TTAC target sequence. In contrast, TnpAIS608 forms a complex with a branched DNA structure independently of TTAC target sequence (Figure 4Aiv). Copper-phenanthroline footprinting of the TnpAIS608-forked DNA complex showed that TnpAIS608 protects the region around the branch point (Supplementary Figure S4A, shown in green in the cartoons in Figure 4Aiv). It also formed a robust complex with a four-way holiday junction (Figure 4Av), a structure generated during restart of stalled replication forks (36). These results demonstrate that TnpAIS608 has a relatively strong binding affinity for DNA which imitates replication fork structures.
Figure 4.
TnpAIS608 DNA binding properties. (A) Binding to different substrates by EMSA. EMSA analysis of complexes formed between TnpAIS608 and different DNA substrates. The same conditions were used for all the substrates (i–vi). ‘-’ indicates no TnpAIS608-His6. Increasing TnpAIS608-His6 concentrations (6, 30, 60, 150, 300, 450, 600, 1000 and 1500 nM; lanes 2–10 respectively) are indicated by the triangle. Complexes were separated in native 5% polyacrylamide gels. i. ssLE; ii. ssRE; iii. ss target, iv. flap structured substrate, v. Holiday Junction substrate and vi. ds substrate. TnpAIS608 protected regions revealed by copper-phenanthroline footprinting are indicated in green (Supplementary Figure S4). (B) Double filter-binding analysis. Results of double filter-binding analysis at different ionic forces: 50, 150, 300 mM NaCl and 150 mM NaCl +dIdC. (i) TnpAIS608 binding to ds DNA; (ii) ssLE and (iii) ssRE substrates respectively. (iv) Corresponding Kd determined by Prism Graphpad software.
On the other hand, TnpAIS608 also forms a complex with dsDNA devoid of either LE or RE sequences (Figure 4Avi) although, as judged by the remaining uncomplexed DNA, this occurs with lower affinity. A similar complex was previously detected with dsRE and dsLE at high protein concentration, ds substrates being refractive to TnpAIS608-catalysed cleavage and strand transfer (8,9). Copper-phenanthroline foot-printing of this complex shows a homogenous partial protection along the dsDNA suggesting non-specific interactions (Supplementary Figure S4B, shown in green in the cartoons in Figure 4Avi).
The central challenge encountered by a protein that must bind a particular DNA sequence is the presence of vast amounts of DNA where non-specific binding can take place. This problem is resolved by regulatory proteins such as bacterial transcription factors, which recognize dsDNA substrates, by using several complementary mechanisms together called ‘facilitated diffusion’ on non-specific ds DNA (for review see (37)). The use of ssDNA substrates however sets TnpAIS608 apart from classical regulatory proteins which target dsDNA. TnpAIS608 might find its substrates by specific recognition of structured DNA per se, or act similarly to regulatory proteins by ‘diffusing’ along dsDNA before encountering for example, ssDNA situated between two Okazaki fragments on the replication fork.
The negatively charged dsDNA phosphate backbone interacts with positively charged amino acid residues within a protein. These electrostatic interactions generally occur at low but not at high ionic strengths. To examine whether TnpAIS608 binding to dsDNA is electrostatic and to obtain more accurate and quantitative data, we used the double filter binding technique (‘Materials and Methods’ section) to compare TnpAIS608 binding efficiencies between the dsDNA and IS608 ss ends (Figure 4B). TnpAIS608 binds dsDNA at 50 and 150 mM but no binding was detected at 300 mM NaCl. Moreover, with excess poly-dIdC competitor, no binding could be detected even at 150 mM NaCl (Figure 4Bi). However, both TnpAIS608-LE and TnpAIS608-RE complexes were resistant to high salt concentration (300 mM NaCl) and to poly-dIdC competitor (Figure 4Bii and iii). The binding constants, Kd, calculated from these experiments (‘Materials and Methods’ section), are shown in Figure 4Biv. Binding affinity of TnpAIS608for LE or RE is at least 5–10-fold higher than that for dsDNA under low salt condition, reflecting the binding selectivity. Furthermore, this technique permitted detection of TnpAIS608 binding to non-specific ssDNA and revealed that interaction with structured substrates is also sensitive to high salt concentration (Supplementary Figure S4C).
dsDNA neighbouring the target site affects integration
While TnpAIS608 binds ss IS ends with high affinity and shows some affinity for non-specific dsDNA, it requires an ssDNA target to catalyse integration (9). The presence of Okazaki fragments on the lagging strand results in transient alternating ds-ss-dsDNA. Binding to dsDNA may therefore facilitate TnpAIS608 access to the neighbouring ssDNA target on the lagging strand substrate. To determine whether this ‘non-specific’ dsDNA binding could influence TnpAIS608 activity on a neighbouring ssDNA IS608 target, we analysed cleavage and integration activity of ssDNA and partial dsDNA targets in vitro.
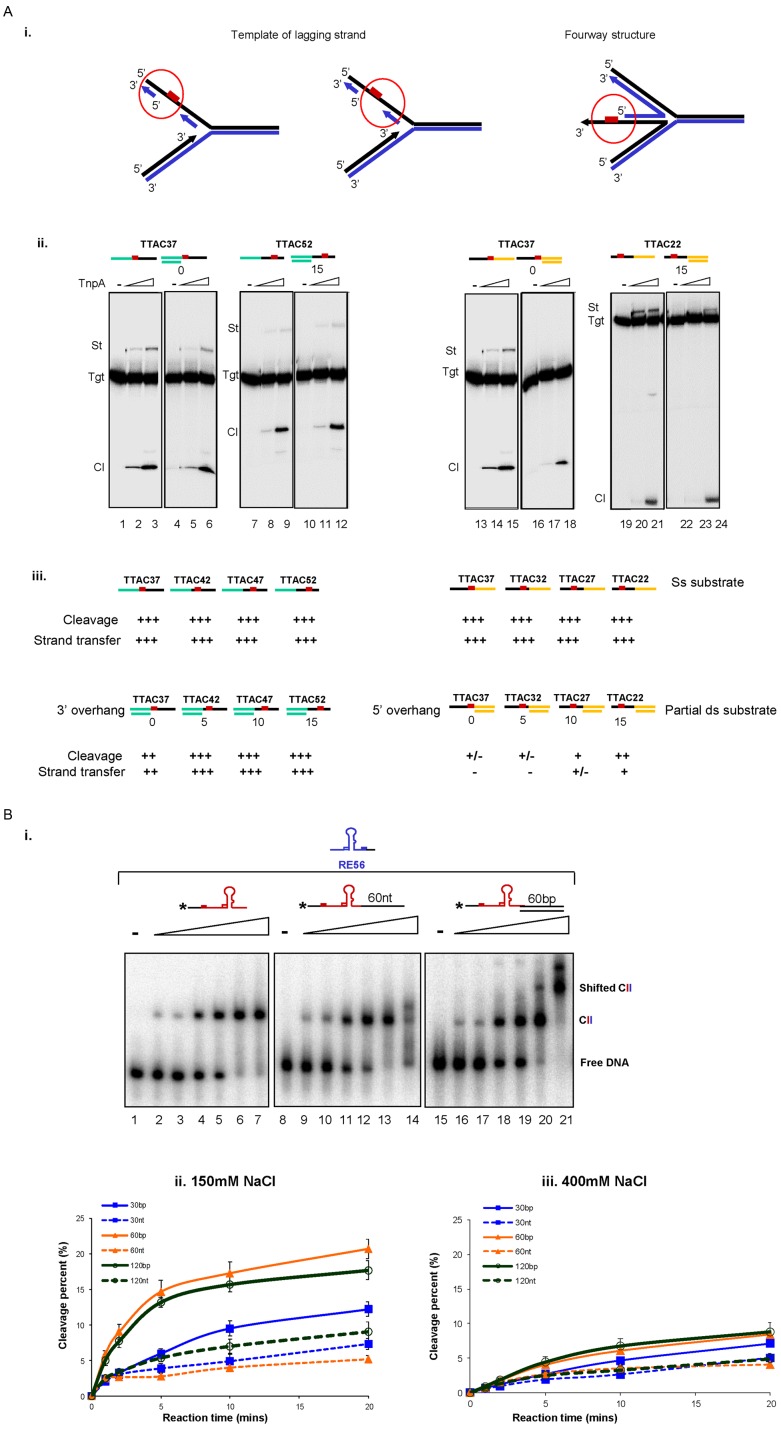
We used a series of labelled ssDNA target oligonucleotides carrying the TTAC tetranucleotide located at different positions (‘Materials and Methods’ section), together with LE80 in a standard integration reaction (9). In parallel, we compared the activity of these entirely ssDNA targets with a set of partial dsDNA derivatives which have a fixed double stranded end and a TTAC target tetranucleotide located in a single stranded DNA portion at 0, 5, 10 and 15 nt from the end of the dsDNA. These partial dsDNA substrates were generated by hybridization of different ssDNA substrates with short fixed oligonucleotides complementary to either the fixed 5′ or 3′ portions of the TTAC sequence (3′ or 5′ overhang derivatives, Figure 5Aiii, Supplementary Data). These mimic the situation on the lagging strand of a replication fork (Figure 5Ai). TnpAIS608 activities on two examples of these targets carrying variable ssDNA regions upstream or downstream of the TTAC are shown in Figure 5Aii (left and right panels respectively). The results are summarized in Figure 5Aiii.
Figure 5.
Effect of dsDNA neighbouring to ssDNA on integration and excision. (A) The dsDNA neighbouring to target site affected the integration activity. (i) Cartoon representing different ds-ss DNA regions at the replication fork. Red box represents the target TTAC. (ii) Integration activity on 3′ overhang (lanes 4–6 and 10–12) and 5′ overhang substrates (lanes 16–18 and 22–24) compared to ss substrates (lanes 1–3, 7–9 and 13–15, 19–21) respectively. (iii) Summary of TnpAIS608 relative cleavage and integration activities on ss and partial ds targets. (B) The presence of dsDNA adjacent to ss ends increases the rate of TnpAIS608-catalysed cleavage. (i) Binding of TnpAIS608 to ss0 (lanes 1–7), ss60 (lanes 8–14) and ds60 (lanes 15–21) substrates by EMSA. Increasing TnpAIS608-His6 concentrations (60, 150, 300, 450, 600 and 1000 nM) are indicated by the triangle. Complexes were separated in native 5% polyacrylamide gels. (ii) Cleavage kinetics in 150 mM NaCl with standard reaction conditions. (iii) Cleavage kinetics in 400 mM NaCl.
The sensitivity of integration as a function of proximity of the TTAC to the dsDNA region was compared with that obtained with the corresponding entirely ssDNA target. When 3′ overhang substrates were examined (TTAC37, TTAC42, TTAC47 and TTAC52 carry TTAC tetranucleotide situated at 37, 42, 47 and 52 nt from the 5′ end (Figure 5Aiii, left), cleavage and strand transfer were robust and significantly less sensitive to the length of the ssDNA region upstream of TTAC. Only in the case of the TTAC37 3′ overhang derivative in which the TTAC is abutted on its 5′ side by dsDNA, do the reactions have a slightly diminished activity.
In general, for 5′ overhang substrates (TTAC37, TTAC32, TTAC27, TTAC22 carry TTAC tetranucleotide situated at 37, 32, 27 and 22 nt from the 5′ end (Figure 5Aiii right) overall activity is reduced compared to the corresponding ssDNA target. In particular, integration is more affected than target cleavage (Figure 5Aiii, right). Activity was gradually restored as the length of the ssDNA region downstream of TTAC was extended. When this ssDNA region was extended to 15 nts (TTAC22 5′ overhang), cleavage became rather robust while strand transfer was only somewhat reduced. Similar results were obtained on replacing LE80 with the RE-LE junction (data not shown).
These results indicate that the adjacent dsDNA at the ss target site does not stimulate and even hampers IS608 integration.
dsDNA adjacent to IS608 ss ends increases the rate of TnpAIS608-catalysed cleavage
We also determined whether the capacity of TnpAIS608 to bind dsDNA ‘non-specifically’ could influence cleavage of a neighbouring ssDNA IS608 end (which occurs during IS excision). We thus measured the kinetics of TnpAIS608 cleavage of LE with or without adjacent dsDNA.
Firstly, the binding properties of TnpAIS608 to these LE substrates were analysed and compared to substrates with adjacent ssDNA or dsDNA by EMSA in an assay which detects synaptic complexes (CII complexes including TnpAIS608, LE and RE) using different labelled LE substrates and unlabelled RE. Incubating labelled LE and excess unlabelled RE favours formation of the TnpAIS608–LE–RE synaptic complex (11).
Figure 5Bi shows the synaptic complex (CII) obtained with fully single stranded LE (left panel). Addition of a 60-nt ssDNA tail to the 3′end of LE did not greatly affect its ability to form the synaptic complex (middle panel), although, with a high excess of TnpAIS608, the complex was somewhat destabilized and migrated as a smear. An LE substrate carrying an additional 60-bp ds DNA tail at the 3′end also generated robust synaptic complexes (Figure 5Biii) but, in addition, a super-shifted complex was detected at higher TnpAIS608 concentrations. This might correspond to a TnpAIS608–LE-RE synaptic complex with additional TnpAIS608 bound non-specifically to the dsDNA tail (right panel).
To determine whether the dsDNA tail influences cleavage efficiency of the adjacent LE, TnpAIS608 and the LE substrates with different lengths of adjacent dsDNA were incubated with Mg2+ (required for catalytic activity) and 150 mM NaCl (close to physiological salt concentration). The reaction was stopped after 1, 2, 5, 10 and 20 min and cleavage products were analysed in a denaturing gel and quantified. The presence of 60 or 120 bp dsDNA next to LE significantly increased the rate of TnpAIS608 catalysed LE cleavage, an effect which can still be detected with only a 30-bp dsDNA tail compared to ssDNA tails (Figure 5Bii, for relevant gels see Supplementary Figure S5). These differences were more pronounced at early time points of cleavage reaction (0–5 min, Figure 5Bii) and significantly reduced when the NaCl concentration was increased (Figure 5Biii, for the relevant gels see Supplementary Figure S5). This suggests that the assistance provided by the dsDNA results from electrostatic interactions with TnpAIS608. Similar results were obtained with the RE (Supplementary Figure S6).
Effect of Ssb and RecA on excision and integration of IS608 in vitro
Single strand DNA generated in vivo is rarely naked. The single strand binding protein, Ssb, binds ssDNA regions preventing premature annealing, protecting from nuclease digestion and removing ssDNA secondary structures (25). RecA is recruited to damaged DNA regions, binding to ssDNA and forming a nucleoprotein filament for DNA repair by homologous recombination (23). RecA is also involved in processing stalled replication forks (for review (38)). It is possible that these proteins influence TnpAIS608 activity. We therefore investigated the effect of RecA and Ssb on IS608 excision and insertion in vitro.
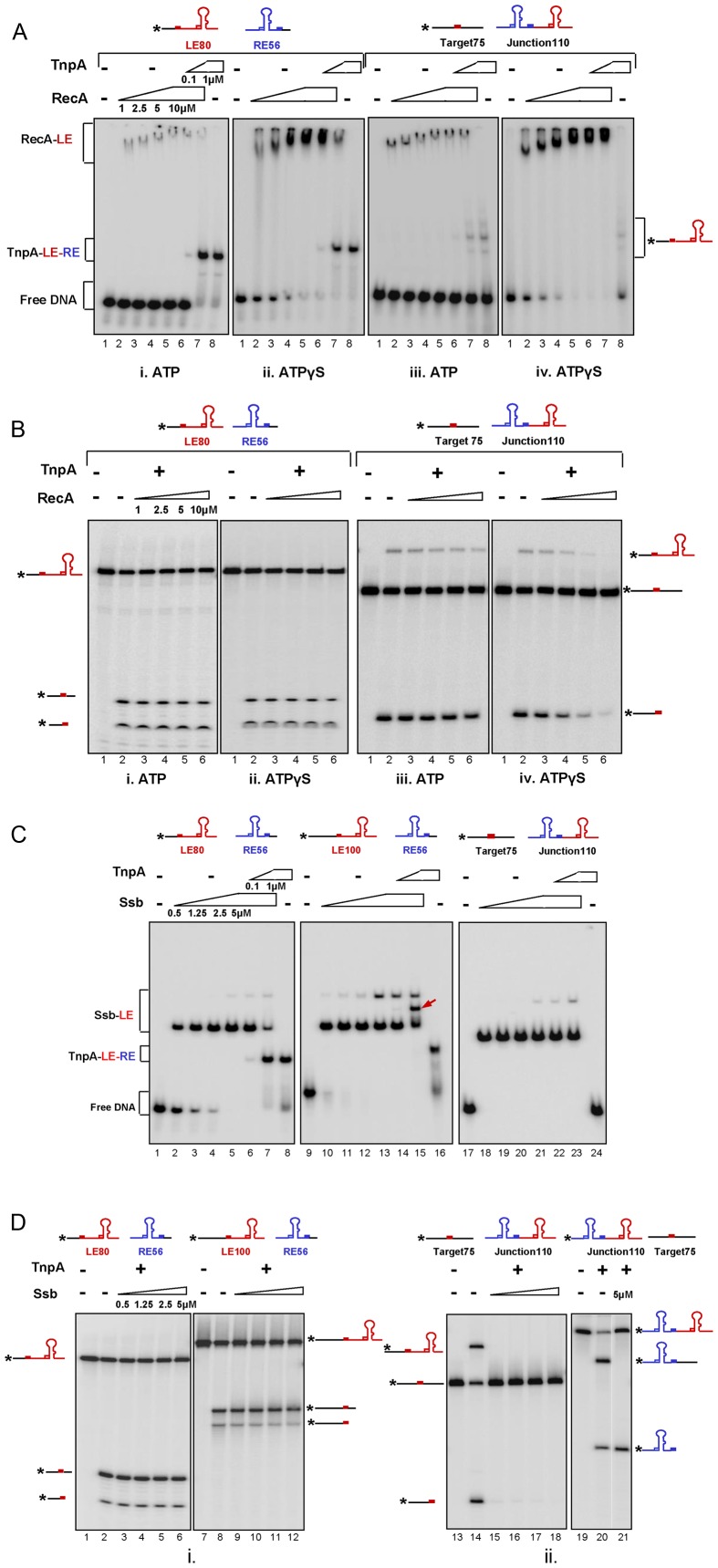
RecA: competition between RecA and TnpAIS608 for binding to LE and RE (representing the first step of excision) was tested using EMSA (Figure 6Ai and ii). We used the same conditions to form TnpAIS608–LE–RE synaptic complexes as described above (11) In the presence of Mg2+, ATP and labelled LE80, RecA formed a weak unstable complex whose mobility was increasingly reduced with increasing RecA concentration (Figure 6Ai, lanes 2–5). These complexes presumably correspond to the growing RecA nucleoprotein filament (39). Replacing ATP by ATPγS resulted in formation of more robust complexes as indicated by the loss of the naked DNA substrate (Figure 6Aii). The instability in the presence of ATP reflects turnover of the complex associated with ATP hydrolysis (23). However, after 15 min incubation of the substrates with RecA together with either ATP or ATPγS, the complex could be titrated by TnpAIS608 addition (Figure 6Ai and ii, lanes 6 and 7) to generate the same synaptic complex as observed without RecA (Figure 6Ai and ii, lane 8).
Figure 6.
Effect of RecA and Ssb on IS608 excision and integration of in vitro. (A) RecA binding and competition with TnpAIS608: Binding of increasing concentrations of RecA (1, 2.5, 5, 10 μM) to labelled ssLE and unlabelled ssRE in the presence of ATP (i, lanes 2–5) or ATPγS (ii, lanes 2–5). TnpAIS608 (0.1 and 1 μM) was added after 15 min of preincubation with RecA (i and ii, lanes 6–7). Synaptic complex TnpAIS608–LE–RE with 1 μM TnpAIS608 (i and ii, lane 8) is formed in the presence of ATP and ATPγS respectively. Binding of increasing concentrations of RecA (1, 2.5, 5, 10 μM) to labelled ss target and unlabelled ssRE-LE junction in the presence of ATP (iii, lanes 2–5) or ATPγS (iv, lanes 2–5). TnpAIS608 (0.1 and 1 μM) was added after 15mins of preincubation with RecA (iii and iv, lanes 6–7). Integration product with 1 μM TnpAIS608 (iii and iv, lane 8) is formed in the presence of ATP and ATPγS respectively. (B) Effect of RecA on excision and integration: excision and integration after 15 min of preincubation with increasing concentrations of RecA in the presence of ATP (i, lanes 2–6 and iii, lanes 2–6) or ATPγS (ii, lanes 2–6 and iv, lanes 2–6) respectively. (C) Ssb binding and competition with TnpAIS608: Binding of increasing concentrations of Ssb (0.5, 1.25, 2.5 and 5 μM) to labelled ssLE80 (lanes 2–5) or labelled ssLE100 (lanes 11–14) and unlabelled ssRE respectively. TnpAIS608 (0.1 and 1 μM) was added after 15 min of preincubation with Ssb (lanes 6–7). Synaptic complexes TnpAIS608–LE–RE with 1 μM TnpAIS608 are shown (lanes 8 and 16) respectively. Red arrow (lane 15) shows the position of co-complex TnpAIS608–LE–RE-Ssb. Binding of increasing concentrations of Ssb (0.5, 1.25, 2.5 and 5 μM) to labelled ss target and unlabelled ssRE-LE junction (lanes 18–21). TnpAIS608 (0.1 and 1 μM) was added after 15 min of preincubation with Ssb (lanes 22–23). Lane 24 includes 1 μM TnpAIS608 and labelled target and unlabelled junction. (D) Effect of Ssb on excision and integration: excision with labelled LE80 or LE100 and unlabelled RE56 after 15 min of preincubation with increasing concentrations of Ssb (lanes 2–6) and (lanes 8–12) respectively. Integration of cold RE–LE junction into labelled ss target in the absence (lane 14) and after 15 min of preincubation with increasing concentrations of Ssb (lanes 15–18). Integration of labelled RE–LE junction into cold ss target in the absence (lane 20) or in the presence of 5 μM Ssb (lane 21).
Competition between RecA and TnpAIS608 for binding to target DNA and to the IS608 transposon junction (representing the first step of integration) was also tested using EMSA (Figure 6Aiii and iv). Similarly, unstable nucleoprotein filaments were observed using labelled single strand target in the presence of ATP (Figure 6Aiii) and these were also more stable in the presence of ATPγS (Figure 6Aiv). However, these reactions generate low levels of additional intermediate bands with TnpAIS608although no TnpAIS608-junction-target co-complex could be detected by EMSA. Since these reactions were carried out in Mg2+, we attribute the additional bands to integration products (strand transfer between the LE copy in the unlabelled transposon junction and the labelled ssDNA target).
We then analysed the catalytic activity of TnpAIS608 in excision after 15 min of pre-incubation with RecA. RecA did not affect excision (Figure 6Bi) and permitted integration (cleavage or strand transfer) in the presence of ATP (Figure 6Biii). Only under conditions in which the RecA nucleoprotein filament was stabilized by eliminating the ATP hydrolysis, was integration inhibited at high RecA concentrations (Figure 6Biv).
These results suggest that TnpAIS608 is capable of efficiently excluding RecA.
Ssb: competition between Ssb and TnpAIS608 for binding to LE and RE and the IS608 target was also tested (Figure 6C). Ssb is known to bind ssDNA in several alternative modes, occupying 65 or 35 nt depending on reaction conditions (40). Without TnpAIS608, Ssb formed a complex with labelled LE80 (lanes 2–5) suggesting that it removes the LE hairpin structure since it is known to bind ssDNA by wrapping the DNA around an Ssb tetramer covering about 65 nt. This wrapping mode is favoured at salt concentrations of ≥200 mM, as used in our binding assay (40). Lane 5 reveals an additional but faint complex when the concentration of Ssb was increased. This complex could be due to binding of a second Ssb moiety.
TnpAIS608 was added 15 min later to a preformed Ssb–LE complex and was observed to titrate LE from this complex to form a TnpAIS608–LE–RE synaptic complex (lanes 6 and 7) as observed without Ssb (lane 8). A similar result was obtained with another LE (LE100, 100 nt) (lane 9–16). This included a 20-nt longer 5′flanking ssDNA segment which is not required for synaptic complex formation (11). In this case, the faint additional, slowly migrating, Ssb complex observed with LE80 was more pronounced presumably because the increased length of this LE allows more stable binding of additional Ssb. Moreover, an additional complex was observed once TnpAIS608 was added to the preformed Ssb–LE complex (lane 15, shown by an arrow). Since the 5′flanking ssDNA is not complexed with TnpAIS608, allowing access to Ssb, this is probably a co-complex of TnpAIS608–LE–RE–Ssb.
We also investigated integration using a labelled single strand target with a TTAC sequence and an unlabelled transposon junction. Ssb formed a complex with the labelled single strand target (Figure 6C, lanes 18–21). In contrast to Ssb-LE complexes, which can be titrated by TnpAIS608, this complex was resistant to TnpAIS608 (lanes 22 and 23), an observation consistent with the absence of detectable TnpAIS608-junction-target co-complexes in the absence of Ssb (lane 24).
The catalytic activity of TnpAIS608 on its substrate and target DNA in the presence of Ssb was then tested. Figure 6D, lanes 1–6 and 7–12 show that Ssb did not affect TnpAIS608 cleavage or strand transfer of LE80 or LE100 in the presence of RE. However, both target cleavage and strand transfer of an LE–RE junction were inhibited in the presence of Ssb (lanes 13–18). Although no TnpAIS608-junction-target co-complex could be detected by EMSA (Figure 6C, lanes 24), target cleavage and the strand transfer product were observed (Figure 6D, lane 14) in the absence of Ssb. When the labelling was inversed from the target to the junction, no effect of Ssb on junction cleavage was observed (lane 21).
These results suggest that Ssb might not affect the excision step of IS608 but could affect the integration step by hampering ss target site cleavage.
Integration opportunity and the kinetics of Okazaki fragment synthesis
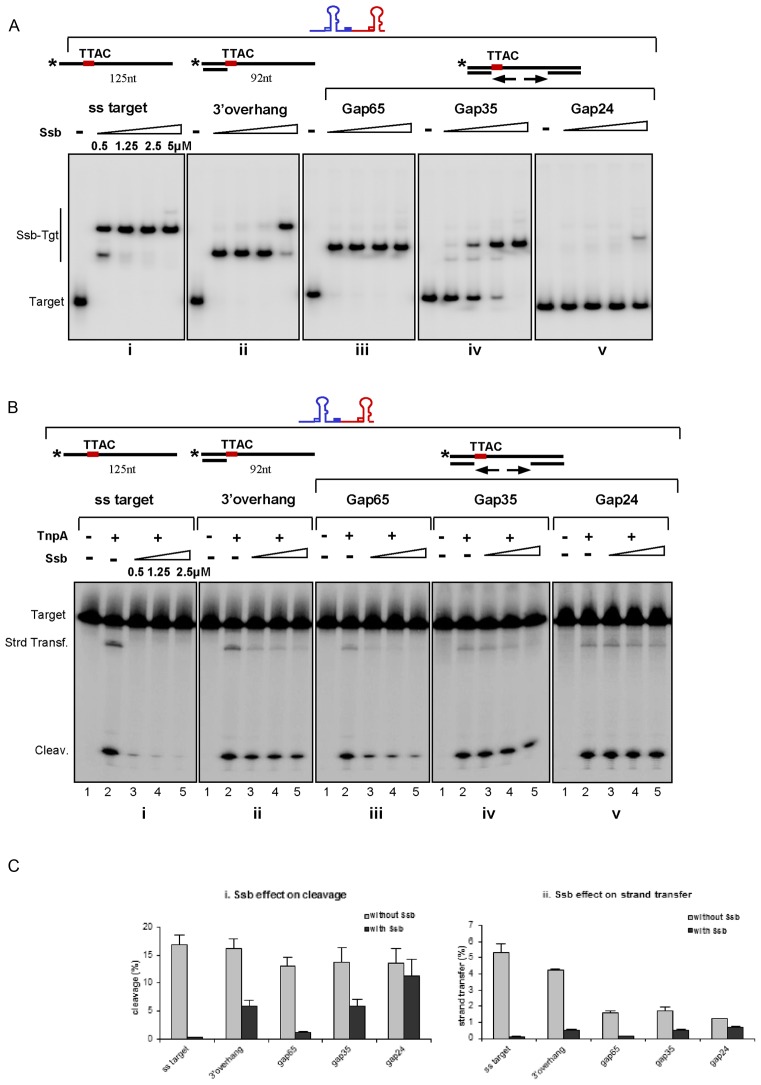
An important question raised by the above observations is how TnpAIS608 overcomes Ssb-mediated inhibition of ssDNA target cleavage (Figure 6D) to complete integration. Although the average length of ssDNA between Okazaki fragments at the replication fork is about 1.5–2 Kb (41) providing a robust matrix for Ssb efficient binding (Ssb requires at least 35 nt), extension of Okazaki fragments during the replication process will rapidly shorten this distance. Moreover, the presence of Okazaki fragments on the lagging strand results in transient ds-ss (partial ds DNA) or ds-ss-ds (gapped) DNA regions. We therefore determined the effect of Ssb on targets carrying partial ds or gapped DNA. For this we constructed several target DNAs with a TTAC tetranucleotide in the ssDNA region and tested integration activity in vitro.
These target DNAs were first tested by EMSA for their ability to bind Ssb. The entirely ssDNA target (125 nt) formed at least two complexes with Ssb (Figure 7Ai) while the partial ds (3′overhang 92 nt) target formed a single complex at low Ssb concentrations and two at higher concentrations (Figure 7Aii). Only one complex was formed with the gapped DNA bearing an ss DNA region of 65nt at all Ssb concentrations tested (gap65, Figure 7A iii). Reducing the gap to 35 nt (gap35) significantly reduced the affinity for Ssb as judged by the remaining uncomplexed DNA (Figure 7Aiv). Almost no complex was detected with a single strand gap shorter than 35 nt (gap24 in Figure 7Av and gap19, data not shown). These observations are consistent with the known minimal occupation binding properties of Ssb.
Figure 7.
Integration opportunity. (A) Binding of Ssb to different DNA target in presence of unlabelled RE–LE junction. Increasing concentrations of Ssb are used in the binding reaction with ss (i); 3′ overhang (ii); gap65 (iii); gap35 (iv) and gap24 (v) targets respectively. (B) Integration of cold RE–LE junction into different targets. TnpAIS608 (1 μM) was added after 15 min of preincubation with Ssb (i); 3′ overhang (ii); gap65 (iii); gap35 (iv) and gap24 (v) targets with increasing concentrations of Ssb respectively. (C) Comparison of target cleavage and integration efficiencies in absence and in presence of Ssb. Effect of 2.5 μM Ssb on target cleavage (i) and integration (ii).
The results of integration assays are shown in Figure 7B. In each case, the strand containing the TTAC integration site was 5′ end-labelled and incubated with TnpAIS608 and the LE–RE transposon junction in the presence of Mg2+. Without Ssb, robust cleavage and strand transfer occurred with the ssDNA target (Figure 7Bi, lane 2). Both activities were preserved for the 3′overhang target (Figure 7Bii, lane 2) as observed above (Figure 5A). In concordance with negative effect of a dsDNA portion downstream of the ss TTAC (Figure 5A, 5′ overhang substrates), the strand transfer activities of gap65, gap35 and gap24 DNA decreased significantly (Figure 7Biii, lane 2, 7Biv lane 2 and 7Bv lane 2) whereas cleavage was less affected.
Pre-incubation with Ssb significantly inhibited both cleavage and strand transfer to the ssDNA target (Figure 7Bi lanes 3–5) as previously noted and quantified in Figure 7C. Surprisingly, both activities were partially recovered with the 3′overhang target (Figure 7Bii lanes 3–5 and Figure 7Ci and ii). This was particularly pronounced for the cleavage reaction perhaps because Ssb can diffuse bi-directionally along the ssDNA region liberating the TTAC integration site located directly adjacent to the 3′ dsDNA end from time to time and providing an opportunity for integration.
The gap65 DNA substrate exhibited an intermediate behaviour between ss and partial ds targets (Figure 7Biii, lanes 3–5). And, as expected, activities were less affected with the gap35 and gap24 (Figure 7Biv, lanes 3–5 and 7Bv lanes 3–5) or gap19 (data not shown) DNA substrates. For instance, almost of 80% of the cleavage activity and of 65% of strand transfer activity with gap24 was restored on addition of Ssb.
Thus, activities on gapped DNA are consistent with the Ssb binding efficiencies on corresponding substrates whereas the gain of activity on partial ds target suggests diffusion of Ssb on this substrate.
DISCUSSION
Members of the IS200/IS605 IS family are clearly distinct from classical IS by their organization, their transposase and their single strand transposition pathway. The replication fork is an important source of ssDNA in vivo for these ISs. Because of the discontinuous nature of lagging strand replication, the replication fork is rich in ssDNA including two regions of ssDNA on the lagging-strand template. The first, between the replicative helicase and Okazaki fragment, is prolonged before initiation of a new primer. The second, between two primers, is reduced in size as the DNA polymerase progressively extends the corresponding Okazaki fragments (42). Moreover, a growing body of evidence illustrates that three DNA polymerase copies are operational in a replisome which can potentially generate supplemental stretch of ssDNA (43,44).
Previous studies showed that transposition of two family members, IS608 and ISDra2, is intimately coupled to replication: excision and integration occurred mainly at the lagging strand template of the replication fork. In silico high-throughput analysis also revealed that other family members insert largely in the orientation corresponding to transposition into the lagging-strand template in various bacterial hosts (13,45). In this study, we analysed the link between IS608 transposition and the chromosomal replication fork in more detail. We also tried to identify factors involved in replication fork targeting and investigated DNA-binding properties of the transposase which can assist localization of ss substrates on the replication fork.
Replication and transposition
Transposition of a number of IS and transposons is intimately coupled to, or depends on, host replication. Replication can impinge on a number of different aspects of transposition. These include: the different replicative transposition modes themselves, repair of strand transfer products or the activity of components such as the transposon ends, the DNA target or the production of transposition proteins.
In the replicative transposition pathway used by bacteriophage Mu and some other transposons, the strand transfer product, the Shapiro intermediate, is processed by replication to generate a characteristic co-integrate (46).
IS911 and IS3 family members transpose via ‘copy out-paste in’ mechanism where replication is essential for conversion of the first strand transfer product, a single strand bridge between both IS ends, to a ds circular transposition intermediate prior to insertion (47–49).
Replication is also involved in repairing short target gaps introduced by many transposons on either side of the insertion to generate DR, a hallmark of classic transposition. Recent studies on bacteriophage Mu demonstrated that this step also involves the replisome and this mechanism can be generalized to other transposons (50).
Tn7 provides an interesting example. The lagging-strand template at the replication fork is actively targeted for transposition (51,52). In the ‘random’ transposition pathway employing transposon-encoded target selector protein TnsE, Tn7 is preferentially directed into discontinuously replicating DNA during conjugative plasmid transfer or into the lagging strand of the replication fork (53). Two factors assure TnsE-mediated targeting: specific DNA–protein interactions of TnsE with DNA structures carrying 5′ overhang ends (35) may favour insertion into the dsDNA carrying an Okazaki fragment (for review (54)); and insertion is facilitated by protein–protein interaction between TnsE and DnaN (34).
For IS10/Tn10 (55) and IS50/Tn5 (56,57), transposition is sensitive to replication fork passage. This not only results in a burst of transposase synthesis from the newly replicated hemi-methylated transposase promoter but also provides hemi-methylated IS ends which are considerably more efficient than fully methylated ends for transposition.
Finally, a recent large scale genomic analysis revealed a potential interplay of transposition with host replication fork in numerous bacterial IS families (58). However, exact mechanisms involved remain to be elucidated for each group.
TnpAIS608 DNA binding properties and replication fork targeting
In spite of the single strand nature of IS200/IS605 transposition, in vitro DNA binding studies demonstrated that TnpAIS608 also recognizes branched DNA structures mimicking replication forks or four-way Holiday Junctions which could be generated from stalled forks. TnpAIS608 binds with high affinity to ss LE and RE ends and binds dsDNA non-specifically at high protein concentration (Figure 4). Structural studies have shown that extrahelical T37 in the RE hairpin, a principal actor in strand specificity and synaptic complex stability, is positioned in a hydrophobic pocket of the protein where it is stacked against F75 and contacts L51 and R52 residues (3). Either mutation/deletion of this flipped out base (11) or mutation of this pocket (F75AR52A) seriously affect the formation/stability of excision synaptic complex CII (Supplementary Figure S4D).
Many DNA binding proteins localize their specific binding sites faster than expected from simple 3D diffusion inside the cell (37,59). This can be achieved by facilitated diffusion which includes several components such as 1D-sliding, dissociation/re-association and intersegmental transfer on non-specific dsDNA. This scenario could be relevant even for proteins using ssDNA as substrate since ssDNA invariably neighbours dsDNA in the cell.
Here we show that a portion of dsDNA adjacent to ssDNA IS ends increases TnpAIS608 cleavage kinetics in vitro. This implies that interaction between TnpAIS608 and dsDNA has a biologically relevant function on TnpA targeting to ssDNA end substrates. For example, diffusion of TnpAIS608 on ds Okazaki fragments would facilitate localization and processing of ss IS ends on lagging strand templates.
However, dsDNA adjacent to a single strand TTAC target did not stimulate and even hampered integration. The fact that integration activity is preserved on the 3′ overhang targets whereas cleavage and joining are inhibited on the 5′ overhang targets (Figure 5A) might be explained by asymmetry in the integration complex. The TTAC ss target sequence in the integration complex is thought to be recognized by the left guide sequence, AAAG, in the transposon junction 5′ to the LE hairpin foot in the same way as this sequence is recognized in the LE flank prior to IS excision. This involves a complex network of base interactions at the hairpin foot (4). Although TnpAIS608 structural studies identified a protein region which binds the hairpin structure (3), this region may also be able to interact non-specifically with other ds DNA. Indeed, apart from some specific contacts responsible for strand discrimination, TnpAIS608 establishes numerous non-sequence-specific interactions with the hairpin via the phosphate backbone. Different effects of flanking dsDNA on excision and integration might be due to occupation of the two DNA binding domains of the TnpAIS608 dimer by the two hairpins (of LE and RE) at the transposon junction. This could prevent non-specific binding to dsDNA. This also implies that the two steps involve different mechanisms.
Interaction with host proteins/the β sliding clamp
We detected interaction of full-length TnpAIS608 with the E. coli β sliding clamp using the in vivo yeast two-hybrid system. This interaction was confirmed by a complementary BACTH.
In addition to providing processivity to DNA polymerases, the β sliding clamp is a ubiquitous and essential component of DNA metabolic machineries. It serves as a mobile platform that interacts with a large variety of proteins to assure a dynamic regulatory network linking DNA replication, recombination, repair and other cellular processes. In view of the importance of the sliding clamp in cell physiology, it is not surprising that interaction with a transposase should not to be strong enough to compete with proteins involved in vital cellular processes.
In a recent survey undertaken with a large number of sequenced bacterial genomes (58), a clear orientation bias in relation to chromosome replication was observed for some IS families, raising the possibility that their transposition apparatus interacts with host replication. These authors also selected transposases from IS with different transposition mechanisms and all appeared to interact with purified E. coli DnaN protein in an in vitro binding assay using 20 aa synthetic peptides carrying the potential interaction motif Qxx/xL. While the interaction between the β sliding clamp and the corresponding full-length transposases remains to be formally demonstrated, it is possible that the interaction is a widespread mechanism of dialogue with the host to coordinate transposition with replication and probably other cellular processes.
The transposases used in the study by Gómez et al. 2014 (45) were chosen because they all included a potential interaction region (60). It is important to note, however, that in many cases, this signal is not always present in transposases of closely related members of the corresponding IS families.
We also noticed a short potential DnaN-interaction motif (Q151TKAL155) in the TnpA C-terminus. However, its mutation (Q151ATKAL155A) or deletion did not affect the TnpA/DnaN interaction in the yeast two-hybrid system or the excision frequency (data not shown). In these cases, it is possible that there are other potential β clamp interaction motifs elsewhere in the proteins or that these transposases interact with other, as yet unidentified components of the replication fork. Preliminary screening of the TnpAIS608 interaction network against an E. coli genomic bank including fragments of ∼0.5–1.5 kb of E. coli chromosomal DNA cloned in one of vectors of the ‘Bacterial adenylate cyclase two-hybrid system’ (28) has indeed revealed other interesting potential partners, proteins of known functions involved in DNA metabolism or associated with the replication apparatus (data not shown). The nature and impact of these interactions remain to be addressed.
Interplay of ss transposition with Ssb and RecA proteins
The single strand transposase must confront ssDNA binding proteins such as Ssb and RecA at the replication fork. These are ubiquitous and important players in host DNA metabolism. Ssb is believed to function mainly on the lagging strand during DNA replication: it binds to ssDNA in several modes and can diffuse along ssDNA molecules (for recent review (61)). RecA is essential for homologous recombination and actively involved in the repair of stalled replication forks (62).
We have shown that TnpAIS608 can efficiently compete with Ssb and RecA for hairpin structures at single strand ends in vitro: IS608 excision is not affected upon pre-incubation with Ssb or RecA prior to transposase addition. Similar effects of these proteins on integron recombination activity has also been observed in reactions occurring on folded single-stranded recombination sites (63).
While integration into a RecA-covered ss target DNA could be readily observed, Ssb was found to inhibit integration to different extents. While target cleavage and integration (strand transfer) are highly sensitive to Ssb, cleavage was significantly restored on a ssDNA target with a partial ds 3′ overhang. It is possible that this kind of substrate permits Ssb diffusion (64) rendering a small stretch of target-containing DNA transiently accessible for TnpAIS608-catalysed reactions to occur.
Integration can also occur in gapped substrates which do not allow stable binding of Ssb. Recently, an additional mode of ultrafast redistribution of Ssb has been observed on long ssDNA (65), in contrast to its behaviour on short ssDNA oligonucleotides. While the behaviour of Ssb on short ss substrates is consistent with low integration efficiency observed in vivo, it will be important to examine the competition effect of Ssb using longer DNA substrates.
It is important to note that we examined here DNA substrates where gaps are flanked by DNA portions on both sides while the gaps on a lagging strand template in vivo would be flanked transiently by the RNA primers on one side. After Okazaki fragments have been synthesized, RNA primers are removed by 5′-3′ flap exonuclease activity of DNA Pol I simultaneously with the gap-filling by its polymerase activity (for review see (66)). We do not therefore exclude that TnpA might behave differently on this kind of substrate with an RNA segment.
It is well established that RecA binding to ssDNA generally occurs in two phases: slow nucleation followed by fast filament extension. In some conditions, Ssb can inhibit RecA filament formation (67). The Ssb barrier to RecA nucleation gives rise to a need for protein mediators, such as RecFOR, that bypass the barrier and facilitate the nucleation process. RecFOR can specifically recognize the junction of ssDNA and dsDNA or RNA–DNA hybrid, at the stalled replication fork and then facilitate the assembly of RecA and displacement of Ssb (24). Thus, integration could occur since we know that a RecA filament does not significantly inhibit target cleavage and integration, consistent with the observation that integration is more favourable into stalled replication forks.
Members of the IS200/IS605 family are widespread in bacteria and in archaea and can occur in relatively high copy number. Although it is difficult to evaluate the impact of different DNA structures and host proteins on their transposition activity, the success of certain elements is clearly related to the favourable interplay between Y1 HuH transposase with these replication fork-associated factors.
Supplementary Material
Acknowledgments
We would like to thank Jean-Christophe Meile and Jerome Rech for advice on microscopy, Dave Sherratt and Xindan Wang for operator-carrying strains, Violette Morales for advice on substrate assembly.
Footnotes
Present address: Susu He, Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, NIH, Bethesda, MD, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural CNRS (to M.C., B.T.H.); Agence National de la Recherche (France) Mobigen [ANR-08-BLAN-0336 to M.C.]; Mobising [ANR-12-BSV8-0009-01 to B.T.H.]; Paul Sabatier University (to L.L.); ARC Foundation (to L.L.). Funding for open access charge: Mobising [ANR-12-BSV8-0009-01].
Conflict of interest statement. None declared.
REFERENCES
- 1.Siguier P., Gourbeyre E., Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler M., Mahillon J. Insertion Sequences Revisited. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA. Vol. 2. Washington D.C.: ASM press; 2002. pp. 305–366. [Google Scholar]
- 3.Ronning D.R., Guynet C., Ton-Hoang B., Perez Z.N., Ghirlando R., Chandler M., Dyda F. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell. 2005;20:143–154. doi: 10.1016/j.molcel.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 4.Barabas O., Ronning D.R., Guynet C., Hickman A.B., Ton-Hoang B., Chandler M., Dyda F. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell. 2008;132:208–220. doi: 10.1016/j.cell.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickman A.B., James J.A., Barabas O., Pasternak C., Ton-Hoang B., Chandler M., Sommer S., Dyda F. DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans. EMBO J. 2010;29:3840–3852. doi: 10.1038/emboj.2010.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He S., Guynet C., Siguier P., Hickman A.B., Dyda F., Chandler M., Ton-Hoang B. ) IS200/IS605 family single strand transposition: mechanism of IS608 strand transfer. Nucleic Acids Res. 2013;41:3302–3313. doi: 10.1093/nar/gkt014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guynet C. Thesis: Study of Transpositional Mechanism of the IS608 Bacterial Insertion Sequence. Toulouse: University Paul Sabatier; 2008. [Google Scholar]
- 8.Ton-Hoang B., Guynet C., Ronning D.R., Cointin-Marty B., Dyda F., Chandler M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 2005;24:3325–3338. doi: 10.1038/sj.emboj.7600787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guynet C., Hickman A.B., Barabas O., Dyda F., Chandler M., Ton-Hoang B. In vitro reconstitution of a single-stranded transposition mechanism of IS608. Mol. Cell. 2008;29:302–312. doi: 10.1016/j.molcel.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Pasternak C., Ton-Hoang B., Coste G., Bailone A., Chandler M., Sommer S. Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 2010;6:e1000799. doi: 10.1371/journal.pgen.1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He S., Hickman A.B., Dyda F., Johnson N.P., Chandler M., Ton-Hoang B. Reconstitution of a functional IS608 single-strand transpososome: role of non-canonical base pairing. Nucleic Acids Res. 2011;39:8503–8512. doi: 10.1093/nar/gkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He S., Guynet C., Siguier P., Hickman A.B., Dyda F., Chandler M., Ton-Hoang B. IS200/IS605 family single-strand transposition: mechanism of IS608 strand transfer. Nucleic Acids Res. 2013;41:3302–3313. doi: 10.1093/nar/gkt014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ton-Hoang B., Pasternak C., Siguier P., Guynet C., Hickman A.B., Dyda F., Sommer S., Chandler M. Single-stranded DNA transposition is coupled to host replication. Cell. 2010;142:398–408. doi: 10.1016/j.cell.2010.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivatsan A., Tehranchi A., MacAlpine D.M., Wang J.D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merrikh H., Machon C., Grainger W.H., Grossman A.D., Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470:554–557. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGlynn P., Savery N.J., Dillingham M.S. The conflict between DNA replication and transcription. Mol. Microbiol. 2012;85:12–20. doi: 10.1111/j.1365-2958.2012.08102.x. [DOI] [PubMed] [Google Scholar]
- 17.Mirkin E.V., Mirkin S.M. Mechanisms of transcription-replication collisions in bacteria. Mol. Cell. Biol. 2005;25:888–895. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulugu S., Potnis A., Shamsuzzaman, Taylor J., Alexander K., Bastia D. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9569–9574. doi: 10.1073/pnas.171065898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulcair M.D., Schaeffer P.M., Oakley A.J., Cross H.F., Neylon C., Hill T.M., Dixon N.E. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell. 2006;125:1309–1319. doi: 10.1016/j.cell.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan D.L., Bastia D. Mechanisms of polar arrest of a replication fork. Mol. Microbiol. 2009;72:279–285. doi: 10.1111/j.1365-2958.2009.06656.x. [DOI] [PubMed] [Google Scholar]
- 21.Lau I.F., Filipe S.R., Soballe B., Okstad O.A., Barre F.X., Sherratt D.J. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 22.Possoz C., Filipe S.R., Grainge I., Sherratt D.J. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 2006;25:2596–2604. doi: 10.1038/sj.emboj.7601155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox M.M. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 24.Morimatsu K., Wu Y., Kowalczykowski S.C. RecFOR proteins target RecA protein to a DNA gap with either DNA or RNA at the 5′ terminus: implication for repair of stalled replication forks. J. Biol. Chem. 2012;287:35621–35630. doi: 10.1074/jbc.M112.397034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shereda R.D., Kozlov A.G., Lohman T.M., Cox M.M., Keck J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusetti S.L., Cox M.M. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 27.Ah-Seng Y., Rech J., Lane D., Bouet J.Y. Defining the role of ATP hydrolysis in mitotic segregation of bacterial plasmids. PLoS Genet. 2013;9:e1003956. doi: 10.1371/journal.pgen.1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimova G., Pidoux J., Ullmann A., Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galas D.J., Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J. Mol. Biol. 1982;154:245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- 30.Wong I., Lohman T.M. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gietz R.D., Woods R.A. Genetic transformation of yeast. Biotechniques. 2001;30:816–820. doi: 10.2144/01304rv02. [DOI] [PubMed] [Google Scholar]
- 32.Miller J.H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, p. 25.5. NY: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 33.Bouvier M., Demarre G., Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks A.R., Li Z., Shi Q., Owens R.M., Jin M.M., Peters J.E. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009;138:685–695. doi: 10.1016/j.cell.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters J.E., Craig N.L. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 2001;15:737–747. doi: 10.1101/gad.870201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seigneur M., Bidnenko V., Ehrlich S.D., Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 37.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets. Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michel B., Boubakri H., Baharoglu Z., LeMasson M., Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z., Yang H., Pavletich N.P. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 40.Lohman T.M., Ferrari M.E. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 41.Johnson A., O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 42.Duderstadt K.E., Reyes-Lamothe R., van Oijen A.M., Sherratt D.J. Replication-fork dynamics. Cold Spring Harb. Perspect. Biol. 2014;6:1–18. doi: 10.1101/cshperspect.a010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georgescu R.E., Kurth I., O'Donnell M.E. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat. Struct. Mol. Biol. 2012;19:113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes-Lamothe R., Sherratt D.J., Leake M.C. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gómez MJ1 D.-M.H., González-Tortuero E., López de Saro F.J. Chromosomal replication dynamics and interaction with the β sliding clamp determine orientation of bacterial transposable elements. Genome Biol. E vol. 2014;6:727–740. doi: 10.1093/gbe/evu052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapiro J.A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. U.S.A. 1979;76:1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duval-Valentin G., Marty-Cointin B., Chandler M. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J. 2004;23:3897–3906. doi: 10.1038/sj.emboj.7600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandler M., Fayet O., Rousseau P., Ton Hoang B., Duval-Valentin G. Copy-out-Paste-in transposition of IS911: a major transposition pathway. Microbiol. Spectr. 2015;3:1–17. doi: 10.1128/microbiolspec.MDNA3-0031-2014. [DOI] [PubMed] [Google Scholar]
- 49.Siguier P., Gourbeyre E., Varani A., Ton-Hoang B., Chandler M. Everyman's guide to bacterial insertion sequences. Microbiol. Spectr. 2015;3:1–35. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 50.Jang S., Harshey R.M. Repair of transposable phage Mu DNA insertions begins only when the E. coli replisome collides with the transpososome. Mol. Microbiol. 2015;97:746–758. doi: 10.1111/mmi.13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolkow C.A., DeBoy R.T., Craig N.L. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 52.Parks A.R., Peters J.E. Tn7 elements: engendering diversity from chromosomes to episomes. Plasmid. 2009;61:1–14. doi: 10.1016/j.plasmid.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters J.E., Craig N.L. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 54.Fricker A.D., Peters J.E. Vulnerabilities on the lagging-strand template: opportunities for mobile elements. Annu. Rev. Genet. 2014;48:167–186. doi: 10.1146/annurev-genet-120213-092046. [DOI] [PubMed] [Google Scholar]
- 55.Roberts D., Hoopes B.C., McClure W.R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- 56.Yin J.C., Krebs M.P., Reznikoff W.S. Effect of dam methylation on Tn5 transposition. J Mol. Biol. 1988;199:35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]
- 57.Dodson K.W., Berg D.E. Factors affecting transposition activity of IS50 and Tn5 ends. Gene. 1989;76:207–213. doi: 10.1016/0378-1119(89)90161-3. [DOI] [PubMed] [Google Scholar]
- 58.Gómez M.J., Díaz-Maldonado H., González-Tortuero E., López de Saro F.J. Chromosomal replication dynamics and interaction with the β sliding clamp determine orientation of bacterial transposable elements. Genome Biol. Evol. 2014;6:727–740. doi: 10.1093/gbe/evu052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammar P., Leroy P., Mahmutovic A., Marklund E.G., Berg O.G., Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 60.Dalrymple B.P., Kongsuwan K., Wijffels G., Dixon N.E., Jennings P.A. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ha T., Kozlov A.G., Lohman T.M. Single-molecule views of protein movement on single-stranded DNA. Annu. Rev. Biophys. 2012;41:295–319. doi: 10.1146/annurev-biophys-042910-155351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Indiani C., Patel M., Goodman M.F., O'Donnell M.E. RecA acts as a switch to regulate polymerase occupancy in a moving replication fork. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5410–5415. doi: 10.1073/pnas.1303301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loot C., Parissi V., Escudero J.A., Amarir-Bouhram J., Bikard D., Mazel D. The integron integrase efficiently prevents the melting effect of Escherichia coli single-stranded DNA-binding protein on folded attC sites. J. Bacteriol. 2014;196:762–771. doi: 10.1128/JB.01109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roy R., Kozlov A.G., Lohman T.M., Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee K.S., Marciel A.B., Kozlov A.G., Schroeder C.M., Lohman T.M., Ha T. Ultrafast redistribution of E. coli SSB along long single-stranded DNA via intersegment transfer. J. Mol. Biol. 2014;426:2413–2421. doi: 10.1016/j.jmb.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurth I., O'Donnell M. Replisome dynamics during chromosome duplication. EcoSal Plus. 2009;3:1–26. doi: 10.1128/ecosalplus.4.4.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cox M.M. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.