Abstract
CRISPR/Cas9 nucleases have enabled powerful, new genome editing capabilities; however, the preponderance of non-homologous end joining (NHEJ) mediated repair events over homology directed repair (HDR) in most cell types limits the ability to engineer precise changes in mammalian genomes. Here, we increase the efficiency of isolating precise HDR-mediated events in mouse embryonic stem (ES) cells by more than 20-fold through the use of co-incidental insertion (COIN) of independent donor DNA sequences. Analysis of on:off-target frequencies at the Lef1 gene revealed that bi-allelic insertion of a PGK-Neo cassette occurred more frequently than expected. Using various selection cassettes targeting multiple loci, we show that the insertion of a selectable marker at one control site frequently coincided with an insertion at an unlinked, independently targeted site, suggesting enrichment of a sub-population of HDR-proficient cells. When individual cell events were tracked using flow cytometry and fluorescent protein markers, individual cells frequently performed either a homology-dependent insertion event or a homology-independent event, but rarely both types of insertions in a single cell. Thus, when HDR-dependent selection donors are used, COIN enriches for HDR-proficient cells among heterogeneous cell populations. When combined with a self-excising selection cassette, COIN provides highly efficient and scarless genome editing.
INTRODUCTION
The recent adaptation of RNA-guided nucleases for use in mammalian cells has yielded substantial benefits for stem cell and regenerative medicine research. The most frequently used nuclease, Cas9 from S. pyogenes, uses a structural guide RNA from a clustered regularly interspaced short palindromic repeat (CRISPR) locus to target its nuclease activity to a DNA sequence (1). The first 20bp of the guide RNA directs Cas9 nuclease activity to a DNA target by Watson–Crick base pairing, generating double strand breaks (DSB) at those sites (2,3). This, in turn, stimulates endogenous DNA repair pathways, which can be harnessed for local genome editing. In mammalian cells, an engineered single guide RNA (sgRNA) is frequently used in place of the processed CRISPR RNA (4,5). This process has been used in stem cells to generate new cell models of genetic disease and to repair pathologic mutations in patients’ stem cells for diverse human diseases including cancers, muscular dystrophy, HIV, and beta-thalassemia (6–11). The profound therapeutic potential of this system depends on its ability to engineer changes safely and efficiently into the genome.
In most mammalian cells, DSB are repaired predominantly through non-homologous end joining (NHEJ) pathways and not homology directed repair (HDR). This is problematic for precise genome editing because NHEJ repair can introduce small insertion/deletion (indel) mutations at the DSB, making those sites refractory to HDR-mediated precise editing. Recent studies have identified several enhancements for stimulating HDR repair of Cas9-generated DSB in pluripotent stem cells, particularly for use with single stranded oligonucleotide donors (ssODN) rather than dsDNA as donor DNA. A screen for small molecules identified two compounds (Brefeldin A and L755507) that stimulated a 9-fold increase for insertion of a point mutation by ssODN, as well as a 2- and 3-fold increase in the frequency of insertion of GFP into the Nanog locus using a dsDNA donor with large arms of homology (12). It remains unclear how this effect is caused by Brefeldin A, an inhibitor of ER to Golgi transport, or L755507, a β3-adrenergic receptor agonist. Delivering active Cas9 ribonucleoproteins (RNPs) directly into cells synchronized in the G2/M phase of the cell cycle also increased HDR events over 6-fold with a ssODN as the donor DNA. Use of a small linear dsDNA yielded a more modest 2-fold enrichment from timed RNP delivery (13). Several studies have tested inhibition of NHEJ activity as a method of enhancing HDR-dependent events using CRISPR/Cas9-mediated genome editing procedures. Inhibiting DNA Ligase IV with a small molecule (SCR7) or knockdown of KU70, KU80 or DNA Ligase IV proteins increased HDR-mediated genome editing by 4× to 19× in immortalized mammalian cell lines (14–16). Interestingly, different cell types display a wide range of sensitivity to SCR7, suggesting it may affect DNA repair activity in a cell-type specific manner (17); this differential activity provides a basis for its potential use as a cancer drug. Effects of SCR7 have not been reported for pluripotent cells.
To develop technologies and methods for making HDR-mediated genome engineering more efficient in ES cells, we focused on optimizing the use of dsDNA fragments as donor DNA. dsDNA can be inexpensively generated at kilobase lengths by de novo synthesis, by PCR, or through the generation of plasmids. Large dsDNA donors allow for increased insert and homology arm length relative to ssODN, broadening the range of applications. Our initial experiments elucidating effects of homology arm length and donor concentration on the frequency of HDR-mediated insertion revealed an unexpectedly high frequency of cells with insertions at both alleles when a single gene was targeted. Importantly, similar to the abundant bi-allelic events at a single genomic site, a coincidental insertion (COIN) effect also occurred when unlinked genes were simultaneously targeted by distinct sgRNA/donor DNA combinations. Thus, use of positive selection for an HDR event at one site provides substantial enrichment of HDR events at other genomic sites without the need for additional selectable markers.
MATERIALS AND METHODS
ES cell culture
For normal passage of cells, single cell suspension of 2 × 105 to 2 × 106 C57BL/6 mouse ES cells were plated onto individual wells of six-well plates (Falcon #353046) previously coated with 0.1% gelatin (Millipore #ES-006-B). Cells were grown in Knockout DMEM (GIBCO #10829-018) supplemented with the following: 15% Fetal Bovine Serum (GIBCO #10437-028), 2 mM l-Glutamine (GIBCO #25030-081), 1000 U/ml Pen Strep (GIBCO #15140), 1 mM HEPES (Thermo Scientific #SH30237.01), 1× MEM NEAA (GIBCO #11140), 55 uM 2-mercaptoethanol (GIBCO #21985-023), 100 U/ml LIF (Millipore #ESG1106), and 3 uM CHIR99021 (Sigma #SML1046). Cells were split 1:10 with 0.25% trypsin–EDTA (GIBCO #25200-072) every 2–3 days. For excision of pRIND, cells were treated with 1 uM 4-OH Tamoxifen for 3 days. For negative selection of pRIND, cells were treated with 2 uM Fialuridine for 5 days. Media were replaced daily for all experiments.
ES cell transfection and selection conditions
Cells were transfected shortly after plating with Lipofectamine 2000 (Life Tech #11668019) for ∼18 h. For each well, 10 ul of Lipofectamine and relevant DNAs were incubated separately in 250 ul OPTI-MEM (GIBCO #31985) for 10 min, then combined for 10 min prior to addition to cells. All transfections include 250ng pPGKpuro to allow for elimination of non-transfected cells. In addition, cells were transfected with 200 ng pX330 Cas9/sgRNA expression plasmid for each sgRNA target and 0.1–1 ug donor DNA. pPGKpuro was a gift from Rudolf Jaenisch (Addgene plasmid # 11349). pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from Feng Zhang (Addgene plasmid # 42230). For COIN experiments, -/+COIN arms were treated as similarly as possible and all plates were grown side-by-side for the same period of time (14 days) prior to quantification. The number of original clones were numerous and not quantified. Plates were split every 2–3 days at 1:10 dilutions to maintain ES cell viability. After 48 hours, cells were either selected for 4 days in 2 ug/ml puromycin to eliminate non-transfected cells then grown without selection for the remaining 10 days, or maintained in 200 ug/ml G418 for 14 days to isolate cells with genomic insertion of Neomycin resistance cassette.
PCR genotyping
Reactions were performed with Platinum Taq High Fidelity (Life Tech #11304) or Phusion High-Fidelity DNA Polymerase (NEB #M0530L) for Lef1, Rosa26::Neo, and Rosa26::pRIND. All PCR products are visualized on 1% agarose gel with Ethidium Bromide. For detection of on-target insertions, PCR products were cloned and sequenced verified.
DNA sequences used for PCR primers were: Lef1 WT and Lef1::PGK-Neo primers (Figure 2E): F1: GCCCTAAATGGAGCTTCCTC, R1: GAGAGCCCTCTCCCAATCTT, F2: AGGGCTTCACTCATAGCCAGT, R2: GCAGGTCGAGGGACCTAATA. Lef1::GFP primers (Figure 3D): F: GCCCTAAATGGAGCTTCCTC, R: GAGAGCCCTCTCCCAATCTT Rosa26::Neo primers (Figure 5D, Supplementary Figure S5A): F: AGGTCGACGGTATCGATAAG, R: TTTGCATTCCAAAAGGAACC. Rosa26::pRIND primers (Figure 5): External F: CCGGGCCTCGTCGTCTGATT, External R: GGCCCAAATGTGGAACACCACCTGA, Int/Ext F: TTCCTCTGGGGGAGTCGTTTT, Int/Ext R: AAGGGAGCTGACTTTCTACTGATTA, Internal F: CCATGGAGCACCCAGTGAAG, Internal R: CAGGGTGCTGGACAGAAATGT.
Figure 2.
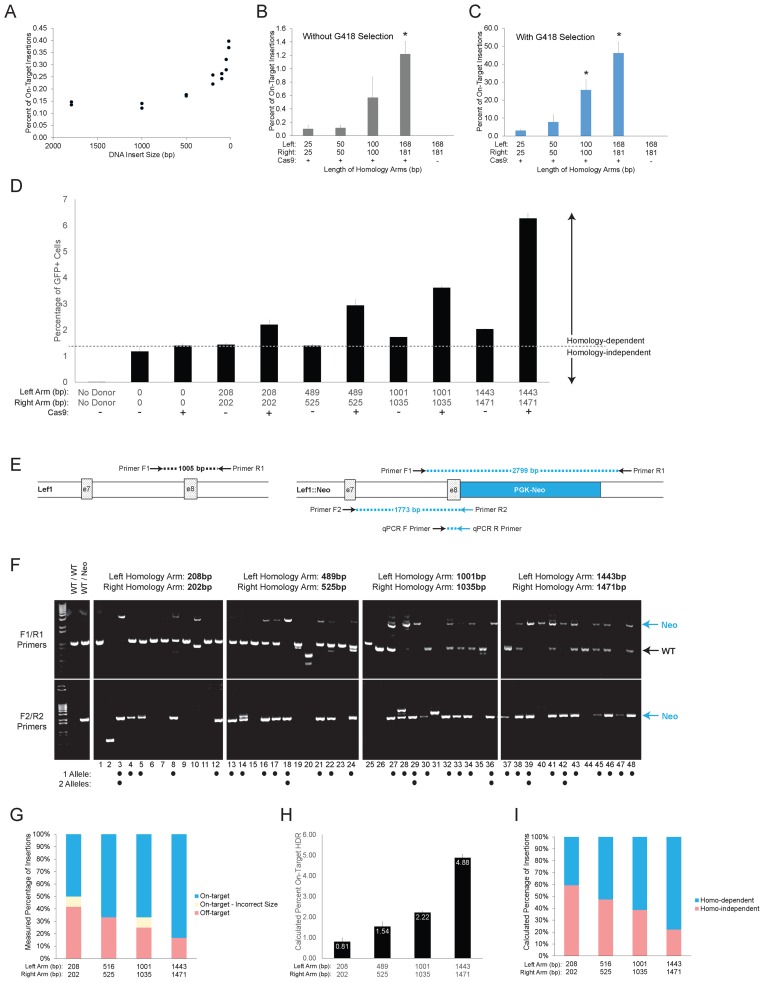
Effect of homology arm length on frequency of HDR-mediated donor DNA insertion. (A) Effect of insert size was determined by qPCR for inserts from 21 up to 1794 bp with a constant homology arm length (181/202 bp). DNA donors are shown in Supplementary Figures S2A,B. (B and C) Effect of homology arm length on targeting efficiency for nearly symmetrical arms from 25 to 181 bp was determined by qPCR for all transfected cells (B) or only those surviving G418 selection (C). Bars represent mean ± SD for biological and technical duplicates. DNA donor schematic and PCR products are shown in Supplementary Figure S2C and D. One-way ANOVA with post-hoc Tukey for multiple comparisons was performed. * represents P < 0.05 for all comparisons with the indicated bar. (D) Effect of homology arm length on targeting efficiency for nearly symmetrical arms from 202 to 1471 bp was determined by integration of CMV-EGFP expression cassette. Donor DNA constructs were co-transfected into ES cells ± Cas9 and gRNA targeting Lef1. After 18 days, the percentage of GFP+ cells was quantified by flow cytometry. Homology-independent integration was determined for donor constructs with no homology arms (third column and dashed line). Bars represent mean ± SD for biological and technical duplicates. Linear regression analysis, comparison of fits and F-test are shown in Supplementary Figure S2E. (E) Schematic of the parental Lef1 locus surrounding exon 7/8 (left) and Lef1::PGK-Neo allele after on-target insertion of Lef1-PGK-Neo DNA donor (right). PCR primer locations and corresponding PCR products are illustrated. Blue color denotes specificity for the Lef1::PGK-Neo allele. (F, G) PCR genotyping (F) and quantification (G) for 48 G418-resistant clones obtained using donor DNA with indicated homology arm lengths. A clone was identified as having a monoallelic on-target insertion if F1/R1 and F2/R2 primer pairs generated 1.0 and 1.7 kb bands, respectively. Biallelic clone were identified by F1/R1 primers producing a 2.8 kb band only and F2/R2 generating a 1.7 kb band. Clones with a single band from F2/R2 primers size were counted as incorrect size when the band was not 1.7 kb (#2,31). DNA donors are shown in Supplementary Figure S2C and D. (H and I) For each homology arm length in Figure 2D, the percent of on-target HDR (H) and the ratio of homology-dependent to homology-independent integrations (I) were calculated. Estimated values were obtained by subtracting the percent integration of donor with no homology arms (Figure 2D dotted line) from the total percentage of GFP+ cells.
Figure 3.
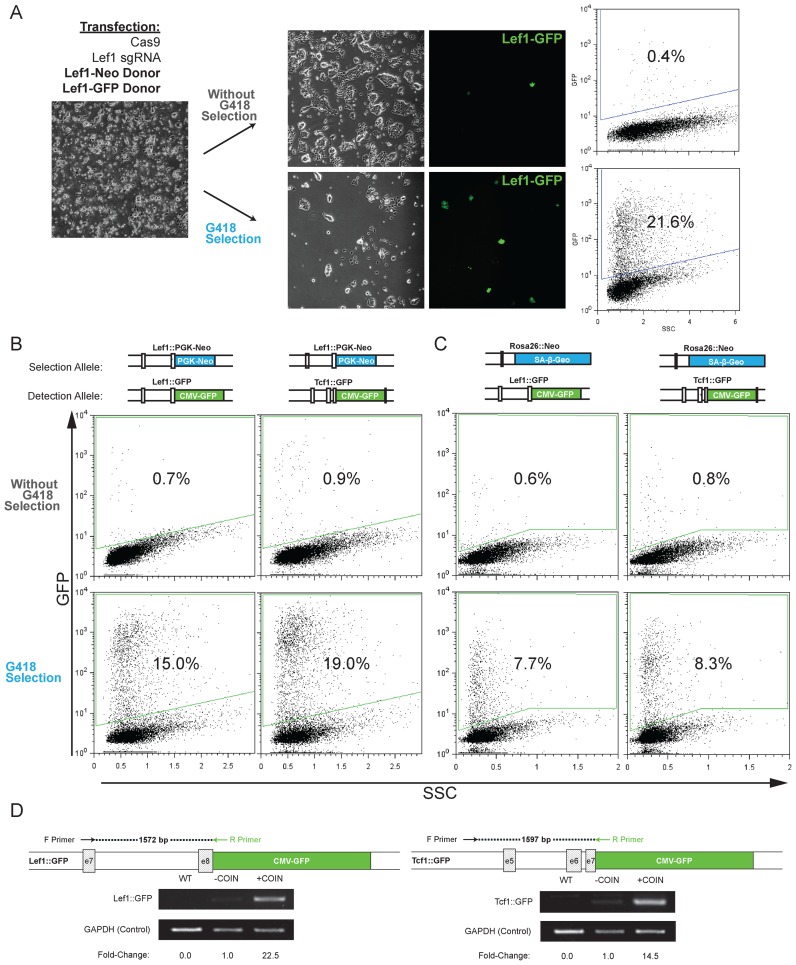
High frequency of coincidental insertion of distinct donor DNA targeting the same gene and independent genes. (A) Experimental approach showing transfection of donor DNA for selection (Lef1-PGK-Neo) and detection (Lef1-CMV-GFP), splitting cells into G418-containing media or non-selective media, and detection of Lef1::GFP frequency by microscopy and flow cytometry to determine percentage of GFP+ cells. Each donor DNA had 208/202bp homology arms. The insertion percentage is determined by flow cytometry by measuring the total GFP+ cells after gating on a transfected, GFP-negative control population. (B and C) Similar experimental set as shown in (A); however, sgRNA and donor DNA target insertion at distinct genes (Lef1, Tcf1, Rosa26). The combination of selection and detection donor DNA are noted at the top of each column. The insertion percentage is measured by counting total GFP+ cells by flow cytometry after gating for a transfected, GFP-negative control population. Representative microscopy images are shown in Supplementary Figure S3A. (D) Semi-quantitative PCR analysis of COIN for Lef1 and Tcf1 loci was performed to confirm that coincidental insertion increases on-target integrations. For each locus, two primers were designed; an external primer specific to the gene locus outside of the homology arms and an internal primer specific to the insert. Densitometry of the PCR products is used to compare the relative frequency of integration in a population of cells with and without COIN.
Figure 5.
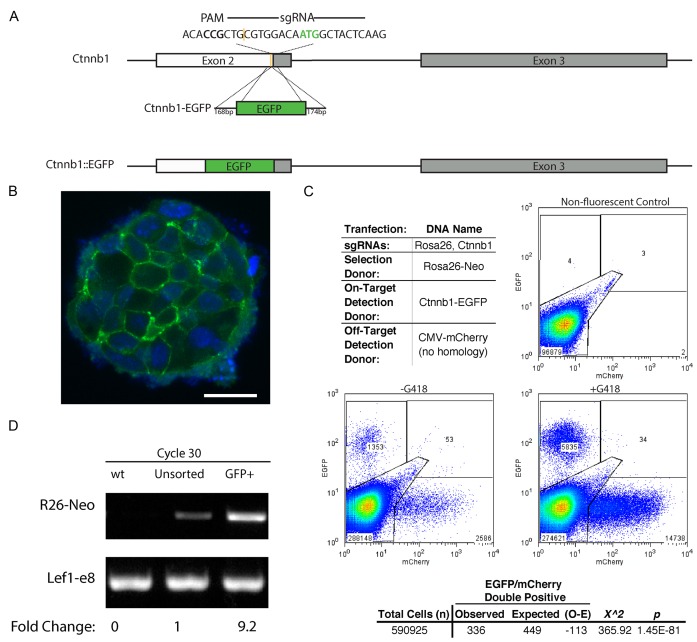
Segregation of on-target and off-target insertions among individual cells. (A) Schematic of mouse Ctnnb1 gene showing the sgRNA target sequence, the PAM site, the cut site (yellow) and the start codon (green ATG). Locations of exons 2 and 3 are noted by boxes. The size (bp) of homology arms and the Ctnnb1-EGFP donor with the in-frame EGFP insertion cassette are indicated. The expected result of editing is also indicated as the in-frame, Ctnnb1::EGFP allele. (B) A representative confocal image of colonies from GFP+ sorted, Ctnnb1-EGFP cells from the –G418 sample in (C). The nuclei are counterstained with DAPI. White scale bar represents 20 μm. (C) Representative flow cytometry dot plots using Ctnnb1-EGFP as the on-target detection donor DNA, CMV-mCherry (no homology) as the off-target detection donor, and Rosa26-Neo as the donor DNA for selection. Gates were drawn based on the non-fluorescent control. A Pearson chi square test was performed on the sum of events recorded for +G418 replicates. The expected was calculated as the frequency of mCherry+ events in the total population multiplied by the total EGFP+ events. (D) Semi-quantitative PCR analysis of COIN for on target Rosa26::Neo insertion (R26-Neo) was performed to confirm that coincidental insertion after selection for on-target insertion increases detection of on-target integrations. Product was amplified by an external primer specific to the gene locus outside of the homology arms and an internal primer specific to the insert. Lef1-e8 PCR product is a loading control for the PCR amplification. Densitometry of the PCR products is used to compare the relative frequency of integration in a population of cells before and after sorting for GFP+, Ctnnb1::EGFP cells. Reactions for cycles 22–28 are shown in Supplementary Figure S5.
Amplification was performed using an Eppendorf mastercycler pro with the following programs. Rosa26::pRIND Cycle Program: 98°—30 s; 30 cycles at 98°—10 s, 68.5°—30 s, 72°—1 min and 15 s; 72°—10 min. Lef1 Cycle Program: 94°—2 min; 30 cycles at 94°—30 s, 55°—30 s, 68°—4 min; 68°—5 min.
Flow cytometry
Single-cell suspensions were prepared by trypsinization and re-suspension in 1 ml of 1% BSA/PBS. Cells were analysed on a Beckman-Counter Cell Lab Quanta SC MPL flow cytometer or Beckman Coulter LSRFortessa. Data analysis was performed using FlowJo v9.3.2. Live cells were gated by forward scatter and side scatter area, singlets were gated by side scatter area and side scatter width. 5 × 104 to 5 × 105 singlet, live cells were counted for each sample (higher counts for more infrequent events). Live cell, green or red fluorescence events were quantified by gating the appropriate channel using fluorescence negative cells as control. Flow sorting was performed by the UIC Research Resources Flow Cytometry Service on the Beckman Coulter MoFlo.
Confocal microscopy
Cells were plated on 1 μ-Slide eight-well glass bottom slides (Ibidi #80827). ES cell colonies were fixed in 4% paraformaldehyde and counterstained with DAPI. Colonies were imaged on a Zeiss LSM 700 and analyzed using ImageJ software.
Generation of Cas9/sgRNA plasmids
All experiments use the pX330-U6-Chimeric_BB-CBh-hSpCas9 backbone. This plasmid was a gift from Feng Zhang (Addgene plasmid # 42230). For all targeted regions, sequences were analyzed for high quality guides using the CRISPR Design Tool (2). Selected target sequences + PAM are: Lef1: GCGCACGTAATAACGATGCG CGG, Tcf1: GTACTATGAACTGGCCCGCA AGG, Rosa26: GCTCCAGTCTTTCTAGAAGAT GGG (Note: additional 5′ G to satisfy U6 promoter requirement), Ctnnb1: AGTAGCCATTGTCCACGCAG CGG, Rosa26PB: AGTCTTCTGGGCAGGCTTAA AGG.
Generation of DNA donor templates
For Lef1, Tcf1 and Ctnnb1 targeting, linear donor templates were generated by PCR fusion of homology arms with the desired insert. For small homology arms (25–181 bp), long ssDNA Ultramers were purchased (Integrated DNA Technologies) with the desired homology length followed by a 20 bp sequence overlapping the insert. These were used as primers for PCR amplification of the insert. For large homology arms (>200 bp), 1500 bp arms for the left and right side were amplified from WT gDNA. A 20 bp overlap with the desired insert was added to the internal primer. 1.5 kb dsDNA products were then fused with the desired insert by PCR. Smaller arms were generated by nested PCR amplification using the 1.5 kb arm DNA as a template. Two different inserts were used for these reactions. The first insert was generated from a cassette encoding CMV-copGFP. CopGFP is a Copepod GFP variant that exhibits bright fluorescence (18). The PGK-Neo insert was amplified from PGKneolox2DTA, a gift from Philippe Soriano (Addgene plasmid #13443). Rosa26-SA-β-Geo donors were obtained from a previously generated Rosa26-SA-β-Geo plasmid. pRosa26-SA-β-Geo was generated from SA-β-Geo and pROSA26-1, both gifts from Philippe Soriano (Addgene plasmids #21709 and #21714). Linear donors with the largest arms (1087/4315 bp) were generated by linearizing the vector with SacII and KpnI. Smaller arms were generated by PCR amplification of internal segments.
Ctnnb1-EGFP donor was generated through amplification of EGFP from pSpCas9(BB)-2A-GFP (PX458), which was a gift from Feng Zhang (Addgene plasmid # 48138). CMV-mCherry (no homology) was generated by PCR of the CMV promoter, mCherry and poly-A from pmCherry-C1 (Clontech). pRIND donor plasmids were generated by isothermal assembly using pROSA26-1 as a base vector. ER tagged PBase sequence was PCR amplified from mPB-L3-ERT2 and PB transposon was amplified from pCyL50. Both plasmids were gifts from Allan Bradley obtained through Wellcome Trust (19,20). SA-PuroTK and SA-GFP fragments were constructed de novo by IDT. Full sequences of pRIND donor plasmids are included (pRIND-R26-puroTK and pRIND-R26-GFP).
Quantitative PCR (qPCR) assay
A set of concentration standards for qPCR genotyping the Lef1::PGK-Neo insertion was generated by mixing genomic DNA (gDNA) obtained from a heterozygous Lef1::PGK-Neo line with WT gDNA. gDNA was mixed to generate the following percentages of Lef1::PGK-Neo: 50%, 25%, 10%, 2%, 0.5% and 0.1%. For a typical experiment, DNA was isolated from ∼2 × 106 cells by lysis with 0.5% SDS followed by ethanol precipitation. 50 ng of gDNA from each sample was combined with Perfecta SYBR Green Supermix (Quanta #95053) and primers specific for either the Lef1::PGK-Neo insert (Figure 1A) or a WT Lef1 (exon12) control ∼30 kb downstream. Primers used are: Lef1::PGK-Neo (F Primer): CTGCCCCTTTCCCTAACTG, Lef1::PGK-Neo (R Primer): CCGGATCCACTTTCATCATC, Lef1 (exon12) Forward: CTGCCCTGTGAAGTGTCTGA, Lef1 (exon12) Reverse: AATGAACTGCAAACGGGTTC. qPCR was performed on a C1000 thermal cycler and CFX96 Real Time System (Bio-Rad) with the following parameters: 95° for 2 min, then 40 cycles of 95° for 30 s and 60° for 30 s. Quantities were normalized to the Lef1 exon 12 control and cycle number was compared against the ladder for Lef1::PGK-Neo to quantify insert percentage. Controls to ensure signal was due to authentic on-target insertions include cloning and sequence verifying the qPCR reaction product from three independent experiments, and reactions using only one of the two primers to ensure signal was not due to concatemers of the donor DNA (Supplementary Figure S1A).
Figure 1.
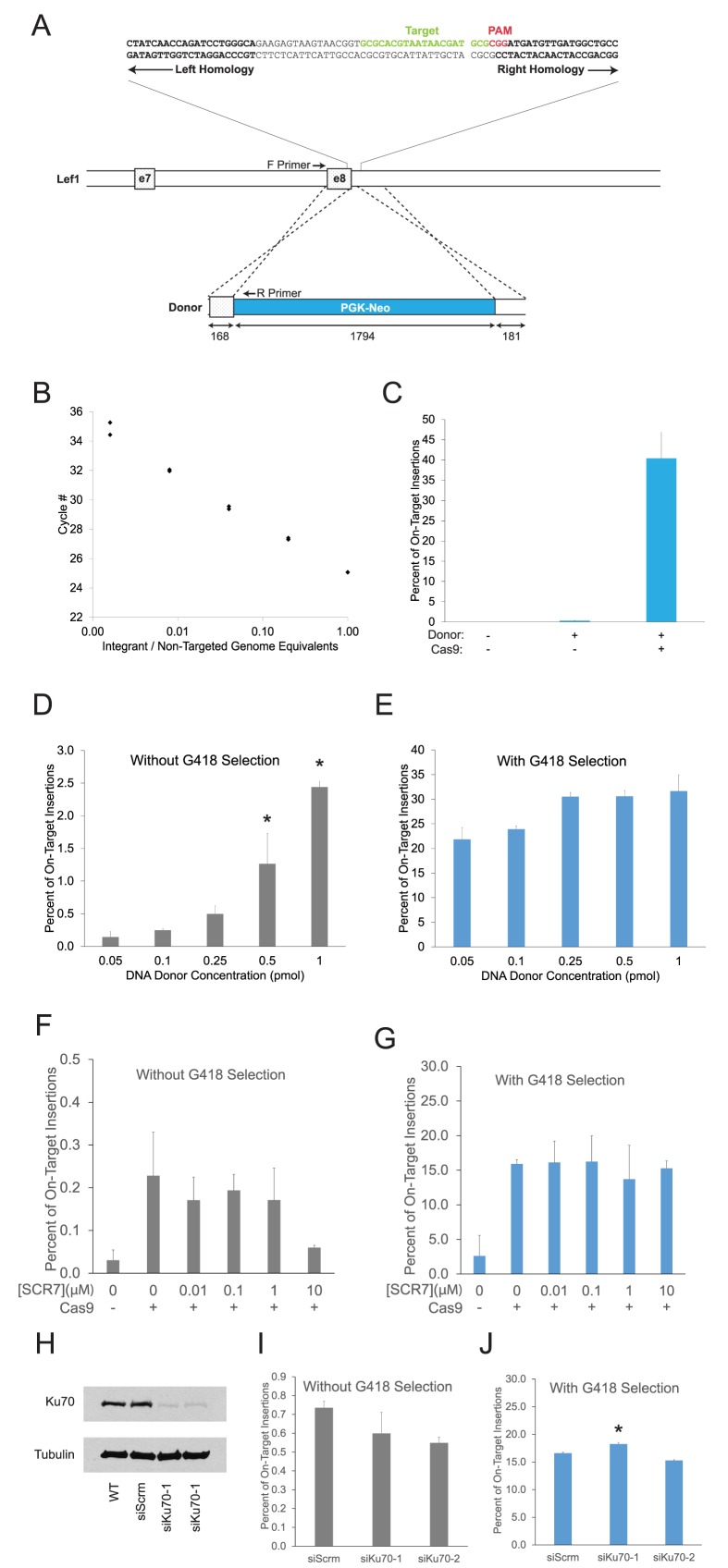
Quantitative PCR (qPCR) assay for optimization of HDR-mediated insertions. (A) Schematic of mouse Lef1 gene showing sgRNA target sequence (green), PAM site (red), and beginning of arms of homology (bold). Locations of exons 7 and 8 are noted by boxes. The size (bp) of homology arms and the Lef1-Neo DNA donor with a PGK-Neo selection cassette (blue) are indicated below each feature. Locations of primers used for quantitative PCR assay are noted. (B) qPCR detection of varying ratios of genomic DNA from non-targeted parental cells mixed with a heterozygous Lef1::PGK-Neo clone. (C) Percentage of on-target insertions following transfection of Cas9, donor DNA, and sgRNA from Figure 1A were determined using qPCR assay from Figure 1B after positive selection with G418 media. Bars represent mean ± SD for biological and technical duplicates. (D and E) qPCR analysis of increasing donor DNA concentration on on-target insertions among all transfected cells (D) or only those cells surviving G418 selection (E). Bars represent mean ± SD for biological and technical duplicates. One-way ANOVA with post-hoc Tukey for multiple comparisons was performed. * represents P < 0.05 for all comparisons with the indicated bar. (F and G) qPCR analysis of on-target insertions for cells grown in 0.01–10μM SCR7. Values shown for all transfected cells (F) and for cells surviving G418 selection. (E). Bars represent mean ± SD for biological and technical duplicates. One-way ANOVA with post-hoc Tukey for multiple comparisons was performed; no significant differences were observed. (H) Ku70 protein and tubulin control shown by western blot for cells transfected with two different siRNAs targeting Ku70, scrambled siRNA and wild-type control. (I and J) qPCR analysis of on-target insertions with Ku70 knockdown and scrambled siRNA control. Values shown for all transfected cells (I) and for cells surviving G418 selection (J). Bars represent mean ± SD for biological and technical duplicates. One-way ANOVA with post-hoc Tukey for multiple comparisons was performed. * represents P < 0.05 for all comparisons to control.
RESULTS
Inhibition of non-homologous end joining provides minimal benefit to genome editing in ES cells
To support experiments aimed at optimizing precise genome editing, we sought to develop a direct and sensitive method of measuring HDR insertions. We focused initial experiments on one genomic site in the Lef1 gene using a sgRNA that we previously validated for NHEJ-based indel mutagenesis (Figure 1A). The frequency of HDR insertion of a PGK-Neo selection cassette was determined with a quantitative real time PCR (qPCR) assay, which accurately determined the number of Cas9-dependent insertion events at a frequency as low as one insertion per 1000 genomes (Figure 1B), but was limited to homology arm lengths less than 200bp. The benefit of Cas9 and sgRNA was measured using the qPCR assay and exemplified by the 140-fold increased recovery of Lef1::PGK-Neo alleles following selection with G418 (Figure 1C).
The effect of donor DNA concentration was measured directly for on-target insertion at Lef1 and indirectly for off-target insertions at unknown sites. Genomic DNA was isolated from cells transfected with the Cas9/sgRNA expression plasmid plus a Lef1-Neo donor DNA (Figure 1D and E). On-target insertions were measured by qPCR (Figure 1B), and off-target insertions were determined by subtracting the percentage of on-target insertions from the total G418-resistant genome equivalents. While maintaining a constant 168/181 bp homology arm length, the number of on-target insertions was proportional to the amount of donor DNA transfected into cells (Figure 1D). The highest level of insertion (2.5% of transfected cells at 1 pmol donor DNA) did not appear to have reached saturation, but was limited by the amount of DNA that could be used without causing toxicity. By contrast, the fraction of on-target insertions among all G418 resistant cells was not substantially altered when more donor DNA was added (Figure 1E). Thus, increasing the concentration of donor DNA increases the frequencies of both on-target and off-target insertions.
Several studies have reported that inhibition of NHEJ-based repair enhanced on-target insertions in cancer cell lines and in mouse zygotes (14,15,21). To determine whether such approaches are effective in enhancing precise genome editing and reducing off-target insertion in ES cells, we inhibited NHEJ using two distinct methods. First, we tested a range of concentrations of SCR7, a DNA ligase IV inhibitor previously shown to stimulate HDR frequency (14,15,17,21). None of the concentrations of SCR7 caused a significant increase of on-target Lef1::PGK-Neo insertions (Figure 1F) or altered the ratio of on:off target insertions in ES cells (Figure 1G, Supplementary Figure S1A,B). Second, we inhibited Ku70, a DNA end binding protein required for an early step in NHEJ, by siRNA knockdown in ES cells (Figure 1H). Reduction of Ku70 by each of two independent siRNA did not consistently alter the frequency of on-target Lef1::PGK-Neo insertions among all transfected cells (Figure 1I) or among G418-resistant cells (Figure 1J). Inhibition of DNA-PK with NU7026 similarly did not increase on-target insertion (Supplementary Figure S1C). These data indicate a lack of benefit from inhibiting NHEJ for performing precise CRISPR-Cas9 genome editing procedures in ES cells, and they may reflect a larger cell-specific responsiveness to manipulation of partially redundant end joining pathways (22,23).
Opposing effects of homology arm length and insert size for on-target insertion at Cas9-generated DSB
Although decades of gene targeting experiments without RNA guided nucleases has led to the general consensus that longer arms of homology and smaller sized inserts between those arms are beneficial for gene targeting, we wanted to systematically measure the magnitude of change for each of these donor DNA characteristics using Cas9 in ES cells. To determine their effects on HDR-mediated insertion, we performed a series of experiments altering homology arm length and insert size for generating Lef1::PGK-Neo and Lef1::CMV-GFP insertions. Keeping homology arms constant at 168/181 bp, the distance between homology arms was varied from 1794 to 21 bp (Supplementary Figure S2A and B). Insertion frequency did not significantly change as insert size was reduced from 1794 to 500 bp, but the frequency increased logarithmically at insert sizes below 500 bp (Figure 2A). Effects of homology arm length were tested with a series of donor DNA consisting of a 1794 bp PGK-Neo insert flanked by homology arms ranging from 25 to 1471 bp (Supplementary Figure S2C and D). Combined with Cas9 and sgRNA expression, it was possible to detect on-target Lef1::PGK-Neo insertions with as little as 25 bp homology arms (Figure 2B and C). Increasing the length of homology arms substantially increases on-target insertion, with a notable jump at arm lengths >100 bp (Figure 2B). Importantly, in contrast to increasing donor DNA concentration (Figure 1D and E), increasing homology arm length increased the frequency of on-target insertions (Figure 2C), resulting in an overall rise in ratio of on:off-target ratio among transfected cells.
To determine the effects of homology arms larger than 200 bp, a combination of flow cytometry using a CMV-GFP insertion cassette and PCR analysis of clonal DNA samples was performed. The analysis of CMV-GFP donors was performed alongside a control donor DNA lacking homology arm sequence (Figure 2D). The CMV-GFP cassette does not require HDR-mediated on-target insertion for generating GFP expression in cells, making it useful for examining both the on- and off-target insertion frequencies in transfected cells. Based on the absence of HDR-mediated on-target Lef1::PGK-Neo insertions without Cas9 (Figures 1C and 2B, C), we considered the GFP+ cells recovered from cells without addition of Cas9 to be exclusively off-target insertions (Figure 2D). Similarly, GFP+ cells recovered from cells transfected with a CMV-GFP donor lacking any arms of homology were by definition considered to represent homology-independent insertion events (dotted line in Figure 2D, Supplementary Figure S2D). As with smaller homology arms, increasing the length of homology arms while keeping the molar concentration of donor DNA constant increased the frequency of all insertion events measured by GFP+ cells (Figure 2D). Adding 1kb arms of homology to each end of the donor DNA increased insertion without Cas9 expression; however the benefit of adding homology was substantially greater for Cas9 expressing cells for all donors examined (Figure 2D). Similar to effects with small homology arms, SCR7 did not affect insertion frequency using donors with 1443/1471bp homology arms (Supplementary Figure S2A).
We hypothesized that the increased frequency of insertions was due to on-target events while off-target insertions remained constant; this hypothesis is illustrated by the dotted line in Figure 2D. To measure directly the ratio of on:off target insertions generated from the various lengths of homology arms, DNA from individual G418-resistant clones was isolated and analysed for presence of Lef1::PGK-Neo on-target insertions with PCR and gel electrophoresis (Figure 2E). Clones with a 1.8 kb band from F2/R2 primers were scored on-target, and clones with only the 1.0 kb wild-type band from F1/R1 primers were scored as off-target (Figure 2F). Two clones (#2, 20) generated PCR fragments with sizes not corresponding to either wild-type or on-target insertions; these were scored as mis-targeted (Figure 2F). The percentage of each outcome for donor DNAs was consistent with longer arms of homology stimulating HDR mediated insertion (Figure 2G).
To test the hypothesis that off-target insertion frequencies were not affected by homology arms, the observed Lef1::PGK-Neo insertion frequencies were compared to the predicted frequencies of on-target and off-target insertions from CMV-GFP donor (Figure 2D). The predicted on-target insertions (Figure 2H) and ratio of on/off-target (Figure 2I) frequencies closely matched the observed frequencies (Figure 2G). In addition, when corrected for off-target insertions by subtracting the homology-independent insertion frequency (dotted line in Figure 2D), the observed frequencies of Lef1-CMV-GFP insertion fit a linear regression test forcing an intercept at zero (green line; Supplementary Figure S2D). By contrast, using the observed frequencies without subtraction of a constant off-target value results in a linear regression fit wherein a portion of the GFP+ cells would have to have an on-target insertion even without addition of homology arms (red line; Supplementary Figure S2D). Altogether, these data indicate that an increase in homology arm length from 25 bp (Figure 2B and C) up to 1500 bp (Figure 2D) is matched by an equivalent linear increase in on-target insertions. In contrast, the frequency of off-target insertions was constant and appeared to be unaffected by varying homology arm length.
Frequent co-occurrence of independent HDR-mediated insertions at Cas9-generated DSB
Analysis of individual clones revealed an unexpected relationship between the frequencies of monoallelic and biallelic Lef1::PGK-Neo insertions (Figure 2F). A priori, one would expect the two alleles to be targeted independently, and thus, biallelic insertions are expected to occur with a frequency equal to the product of two monoallelic insertions. However, a large portion (19%; 6 of 31) of on-target clones displayed biallelic insertions (Figure 2F). Interestingly, other groups have noted exceptionally high frequencies of biallelic events relative to monoallelic events following Cas9-generating DSB. Byrne et al. reported a high frequency of biallelic insertions using donor DNAs with large arms of homology (>2 kb) in human induced pluripotent stem cells (24). Canver et al reported frequent biallelic deletions between two Cas9-generated DSB in murine erythroleukemia (MEL) cells (25). Given the occurrence of biallelic events in these diverse experimental systems, we reasoned that the phenomena could be caused by a common mechanism, and that elucidating the mechanism could lead to a significant advance in genome engineering.
To determine the cause of frequent biallelic insertions, we tested whether the two insertions occurred independently or were caused by one allele acting as a template for gene conversion to generate the second allele. We transfected cells with two different Lef1 donor DNA; one expressing GFP used for detection and quantitation of insertion events (detection donor), and one expressing G418 resistance used for positive selection of cells (selection donor). Each donor DNA had 208/202bp arms of homology for the Lef1 target site. We reasoned that if gene conversion caused biallelism, then homologous biallelic insertions (Neo/Neo or GFP/GFP) should occur more frequently than heterologous biallelic insertions (Neo/GFP). Treatment with G418 was used to enrich for cells with Lef1::PGK-Neo alleles, and flow cytometry and fluorescence microscopy were used to measure the frequency of Lef1::CMV-GFP alleles (Figure 3A). Flow cytometry showed that the frequency of GFP+ cells was increased from 0.4% in cells without selection up to 21.6% in G418-resistant cells (Figure 3A). Thus, by performing selection for one insertion, the frequency of a second, heterologous insertion was dramatically increased. These data are not consistent with gene conversion causing biallelic insertions. We suggest that the high frequency of biallelism was caused by independent, but coincidental insertion (COIN) of each donor DNA after Cas9-mediated DSB.
The independence of each insertion event suggested that the COIN effect may also affect insertions targeted to distinct loci on different chromosomes, similar to co-incidental insertions at the Lef1 locus on two homologous chromosomes. To test this possibility, insertions were examined for three genes (Lef1, Tcf1, Rosa26) in different combinations (Figure 3B and C, Supplementary Figure S3A). Insertion of a Neomycin-resistance cassette was targeted by Cas9 to either the Lef1 or Rosa26 genes (selection allele). A second insertion of a GFP-expression cassette was targeted by Cas9 to either Lef1 or Tcf1 genes (detection allele). As with the previous experiment, the frequency of the GFP+ insert was evaluated by flow cytometry with or without G418 selection. Each combination of selection and detection donors generated a substantial COIN effect, ranging from 10-fold for Rosa26-Neo/Tcf1-GFP to 22-fold for Lef1-Neo/Tcf1-GFP combination (Figure 3B and C). Semi-quantitative PCR measuring the on-target insertion of CMV-GFP at Lef1 and Tcf1 with the selection donor, Rosa26-Neo, verifies that the COIN effect increased frequency of on-target insertions (Figure 3D). These results demonstrate the utility of COIN to enrich for insertions at unlinked sites in the genome.
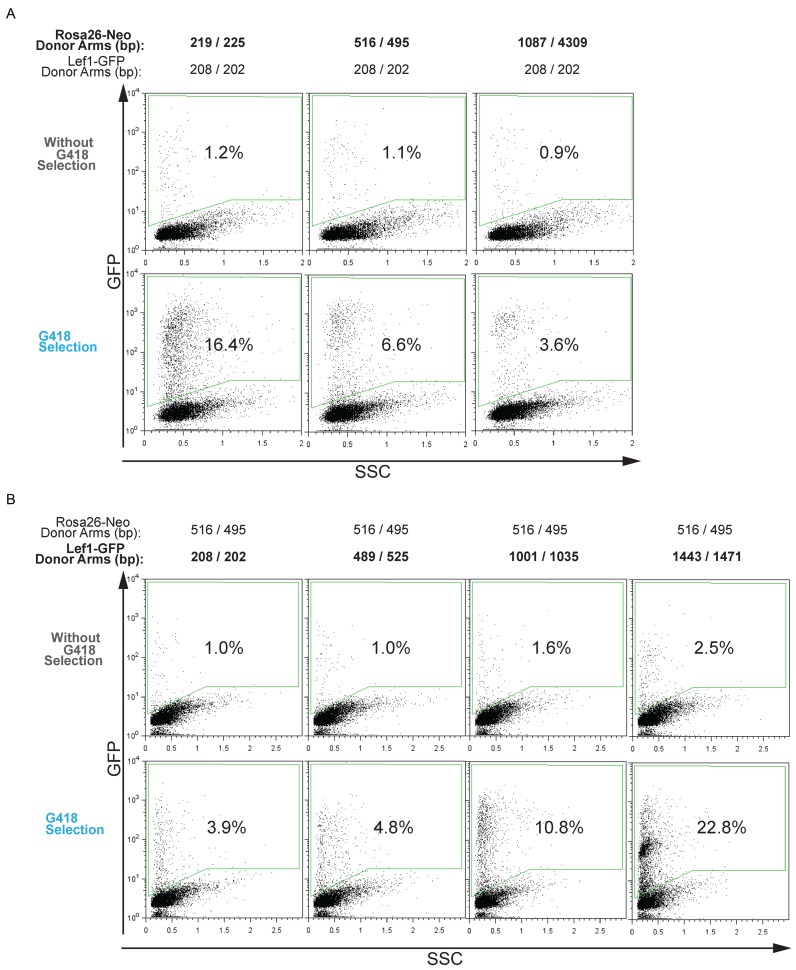
To elucidate the mechanism underlying the COIN effect, we posited that it works through co-occurrences that allows selecting for one event (e.g Rosa26::Neo) to enrich for cells more likely to have completed the second, independent event (e.g. Tcf1::GFP). Conceptually, heterogeneity of HDR activity among individual cells in a culture could provide the basis for co-occurring HDR insertion events. This hypothesis predicts that selection donor DNAs that are highly effective at HDR insertion will produce the weakest COIN effect, because they provide the least stringent selection for HDR activity. Conversely, using relatively weak DNA donors for selection should produce the greatest COIN effect. We tested this possibility by comparing the COIN effect on Lef1::GFP insertion with positive selection provided by Rosa26-Neo donor DNAs with various homology arm lengths (Figure 4A). The weakest COIN effect came from the selection donor with the longest homology arms (1087/4039 bp), as it provided only 5-fold enrichment of Lef1::GFP insertions (Figure 4A). Selection donors with the shortest homology arms (219/235bp) provided the greatest COIN effect with 16.4% GFP+ cells following G418 selection compared to 3.6% GFP+ cells with the longest homology arms (1087/4039 bp) (Figure 4A). These results are consistent with COIN enriching for HDR-mediated insertions by positive selection for the most HDR-proficient cells.
Figure 4.
Homology arm length alters the frequency of co-incident insertion. (A) Flow cytometry analysis using Lef1-GFP as the donor DNA for detection and Rosa26-Neo as the donor DNA for selection. Percentage of GFP+ cells was determined for all transfected cells (top) and for cells following G418 selection (bottom). As indicated at the top of each column, homology arm length was varied for the Rosa26-Neo donor DNA and kept constant at 208/202bp for Lef1-GFP. Insertion percentage is measured by flow cytometry after gating on a transfected, GFP-negative control population. Representative microscopy images are shown in Supplementary Figure S4A. (B) Similar to (A), except homology arm length was varied for Lef1-GFP and kept constant for Rosa26-Neo at 516/495 bp. Insertion percentage is measured by flow cytometry after gating on a transfected, GFP-negative control population. Representative microscopy images are shown in Supplementary Figure S4B.
To determine if the COIN effect can enrich for both relatively frequent and infrequent HDR events, the homology arm length of the Lef1-GFP donor DNA was varied. The homology arm length of the selection donor DNA (Rosa26-Neo) was kept constant at 516/495 bp, a length that provided an intermediate COIN enrichment effect (Figure 4A). As expected, the addition of longer arms of homology increased baseline insertion frequency without COIN, from 1.0% for 208/202bp arms to 2.5% for 1443/1471 arms (Figure 4B). COIN increased insertion frequencies for all Lef1-GFP donors (from 3.9% to 22.8%), with the COIN effects rising with increasing baseline detection donor insertion rates. Taken together, these data suggest that COIN works by positive selection of cells from within a heterogeneous population of cells.
Although enriching cells with high HDR activity provided a plausible mechanism underlying the COIN effect, the selection and detection donors used in the previous experiments were not specific for on-target insertions. To distinguish between co-insertion at on-target and off-target sites at an individual cell level, we used three different donor DNA: (i). CMV-mCherry dsDNA lacks arms of homology to the mouse genome. The CMV promoter will drive mCherry expression when the donor is inserted at sites throughout the genome. Due to the absence of homology arms, mCherry+ cells are by definition the result of an off-target insertion. (ii) Ctnnb1-EGFP dsDNA provides for insertion of EGFP into the Ctnnb1 gene (Figure 5A). Due to the absence of a promoter or splice acceptor site in the Ctnnb1-EGFP donor, it requires an in-frame insertion into the Ctnnb1 gene encoding β-catenin to generate GFP+ fluorescence via a β-catenin-EGFP fusion protein. All GFP+ cells resulting from this donor DNA displayed GFP fluorescence localized to the plasma membrane in a characteristic β-catenin pattern (Figure 5B). As such, activity of the Ctnnb1-EGFP donor is specific to on-target insertions. (iii) Rosa26-Neo dsDNA includes arms of homology to the Rosa26 locus and a splice acceptor sequence upstream of the β-geo cassette. Therefore, the Rosa26-Neo donor can confer resistance to G418 following either an on-target insertions or an off-target insertion within an active gene. The on-target insertions of Rosa26-Neo can be measured using semi-quantitative PCR (Supplementary Figure S5).
To test co-occurrence of on-target versus off-target insertions, ES cells were transfected with sgRNA, Cas9, and the three different donor DNA as indicated in Figure 5C. Flow cytometry of transfected cells showed the mCherry+ cells displayed a wide range of fluorescence intensity, whereas EGFP+ cells displayed a narrow range of fluorescence intensity (Figure 5C). These results are consistent with a variety of insertion sites and position effects on expression levels for CMV-mCherry and a single on-target insertion site for Ctnnb1-EGFP. Interestingly, the frequency of mCherry/EGFP double positive cells, a population with both on and off-target insertions, remained low in all replicates. Indeed, when examined using a Pearson chi-square test, the observed number of double positives was significantly lower than expected based on frequencies of single positive cells (Figure 5C). Applying G418 selection increased the frequency of EGFP+ (on target) and mCherry+ (off-target), consistent with the Rosa26-Neo donor's lack of specificity for on- or off-target insertions. Conversely, when on-target EGFP+ cells were isolated by cell sorting, and their genomic DNA was analysed, the EGFP+ cells displayed a substantially higher frequency of on-target Rosa26::Neo insertions relative to the overall unsorted population (Figure 5D, Supplementary Figure S5). Together, these data support mutual exclusivity of on-target insertions versus off-target insertion among individual cells. They also indicate that selection donors that are specific for HDR (on-target) insertions provide a better enrichment for HDR-mediated genome edits at a second site. These observations are consistent with the mechanisms of COIN being the enrichment of cells with high HDR activity from a heterogeneous population.
Seamless genome editing with PBase-Removable Insertion DNA (pRIND)
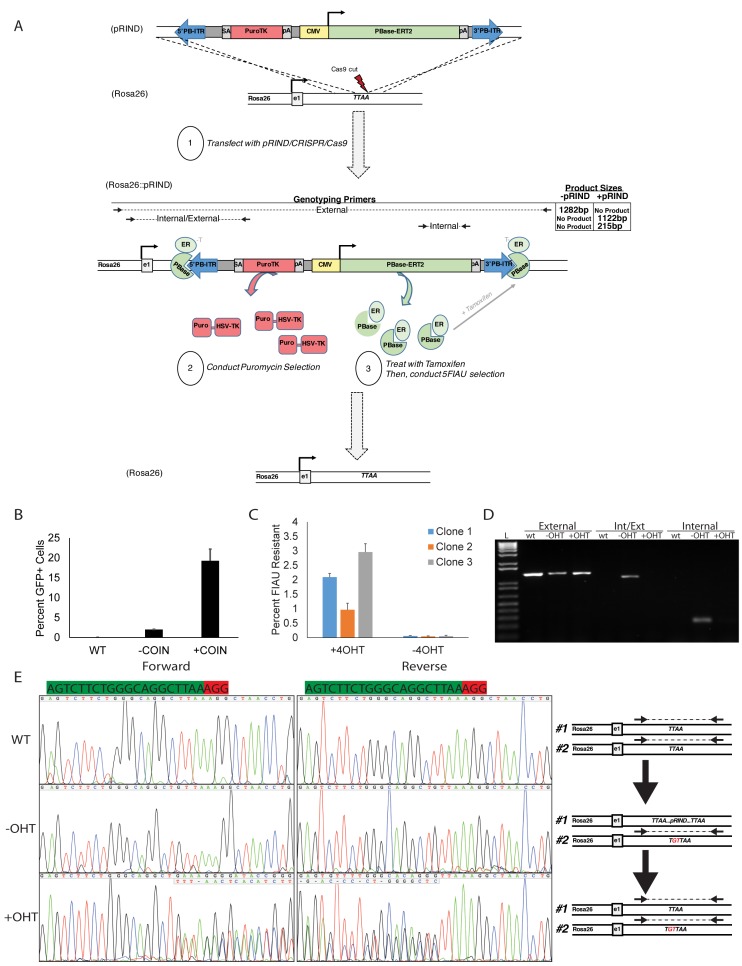
One potential limitation of COIN is the presence of heterologous DNA sequence from the insertion of a selection cassette into the genome of the edited cell. Although the selection cassette can be inserted into a safe-harbor site, such as the Rosa26 gene, its permanent existence could pose a barrier for rigorous applications. In addition, it would also preclude using the same reagents for a second round of COIN to generate additional mutations in the same cell line. Therefore, we sought to develop a method that enables COIN-enhancement of efficiency without a permanent insertion of a selectable marker in the genome of edited cells. Previous research has demonstrated the utility of PiggyBac based approaches for editing the genome of stem cells in a scarless or seamless manner. A defining characteristic of the PiggyBac transposase (PBase) is its ability to excise DNA flanked by inverted terminal repeats (ITR) and leaving behind only the last four bases (TTAA) of an ITR in the genome (26). When inserted into a genomic region that already contains the TTAA sequence, PBase effectively restores the site back to the original wild-type sequence (26). The method we describe below is based on combining the CRISPR-COIN and PiggyBac methods within a specialized selection cassette named pRIND, for PBase-Removable Insertion DNA.
The Rosa26-pRIND donor DNA was engineered to contain every element needed to achieve a scarless genome edit via COIN in a single cassette (Figure 6A). Homology arms were added to the pRIND cassette to target insertion to a TTAA sequence in the first intron of the Rosa26 gene (Figure 6A). A complementary Cas9/sgRNA plasmid was used to generate a DSB at this TTAA. Within the pRIND cassette a splice acceptor enables expression of Puro-ΔTK fusion protein (27) from the endogenous Rosa26 promoter following on-target insertion. Positive selection is provided by Puromycin resistance, while ΔTK provides negative selection by converting normally inert nucleoside analogues, such as Fialuridine (FIAU), into toxic metabolites (27). The final components of pRIND enable conditional removal of the cassette from the genome. Inverted terminal repeats (ITR) were placed at the ends of the cassette, and a CMV promoter drives expression of PBase from within the pRIND to stimulate its own excision. To prevent immediate removal, PBase is fused to a modified estrogen receptor (ERT2) (19). In the absence of 4-Hydroxy Tamoxifen (4OHT), PBase-ERT2 is sequestered away from the nucleus. When excision is desired, 4OHT is added to the media to stimulate nuclear translocation of PBase (Figure 6A).
Figure 6.
PBase-removable insertion DNA (pRIND) can be used as a selection donor for COIN and excised in an inducible and scarless manner. (A) Schematic representation of pRIND-PuroΔTK donor construct and primers used to assay for insertion. (B) PuroΔTK-SDS constructs were targeted to the Rosa26 locus by the inclusion of 500bp homology arms and used to stimulate integration of a GFP expression cassette at the Lef1 locus. The percent of GFP+ cells was measured by flow cytometry after 18 days. (C) Three clones treated with or without 4-OHT were treated with FIAU and percent surviving clones is shown. (D) PCR using the External, Int/Ext, and Internal primers denoted in A to measure insertion and excision of the pRIND cassette for a representative clone. (E) Sequencing of the Cas9-sgRNA cut site generated using the External Primer amplicons from D using nested sequencing primers. The scarless removal of pRIND can be traced by sequencing before and after excision by tamoxifen (OHT) treatment. The schematic at right demonstrates the anticipated amplicons available for sequencing analysis.
Functionality of pRIND for COIN was tested using 500bp homology arms for Rosa26, and provided a tenfold enrichment for isolating Lef1-GFP+ cells (Figure 6B). After 4-OHT, the excision of pRIND by PBase-ERT2 was detected by the loss of sensitivity to FIAU in 1–3% of cells in three independently derived Rosa26::pRIND cell lines (Figure 6C). Individual FIAU-resistant cells were clonally expanded from 4-OHT-treated Rosa26::pRIND clonal lines. Analysis of genomic DNA isolated from clones of cells after steps 1–3 (Figure 6A) were performed with PCR and demonstrated the insertion of pRIND and subsequent 4-OHT stimulated excision of pRIND from the Rosa26 locus (Figure 6D). Sanger sequencing of the site of insertion illustrates the homozygous sequence prior to step 1 (Figure 6E, top), a 2bp insertion in the non-targeted Rosa26 allele prior to step 2 (Figure 6E, middle), and the scarless excision of pRIND to return the sequence of the targeted allele back to wild-type after step 2 (Figure 6E, bottom). Thus pRIND constructs can be used to take advantage of the COIN effect in genome editing experiments and provide a simple method of seamless excision following selection.
DISCUSSION
The adaptation of the CRISPR/Cas9 system for use in mammalian cells provided a simple and powerful method to edit the genome of nearly any cell type, including pluripotent stem cells (3–5). Currently, the high frequency of mutagenic NHEJ-driven repair relative to HDR processing of Cas9-mediated DSB constitutes a barrier to engineering precise changes (5,28). Given the potential therapeutic uses for pluripotent cells, overcoming barriers to editing their genomes with sequence precision has potential for significant impact.
To optimize the frequency of precise insertions into the ES cell genome, we systematically tested characteristics of donor DNA used as a substrate for HDR. We use these empirical observations to suggest several guidelines. Varying the size of the donor insert rather than the homology arms demonstrated an inverse relationship between targeting efficiency and insert size. Interestingly, this relationship was not linear, and when flanked by 0.2kb homology arms reduction <0.5 kb of insert caused steep increases in on-target insertion. As expected, the length of homology arms was critical for frequency of on-target insertion. Perhaps unexpectedly, the length of homology arms did not affect the frequency of off-target insertions, consistent with the concept that on- and off-target insertions are mediated by distinct mechanisms. Conversely, increases in donor DNA concentrations increased the frequency of both on- and off-target insertion. Thus both mechanisms are limited by donor availability, and it may be beneficial to use the lowest possible concentration.
Recent rational approaches to the off-target insertion problem have aimed either to enhance on-target rates by stimulating HDR activity, or to reduce off-target integration by inhibiting NHEJ activity on the donor DNA (14,15,29,30). Additional recent advances from a small molecule screen identified two compounds that increased insertion of a Nanog-GFP donor DNA by 2- to 3-fold (12). Electroporation of active Cas9 RNPs into G2/M synchronized cells similarly increased the frequency of a 12 bp insertion from a 250 bp dsDNA donor (13). Although attempting to control cellular repair activity has produced variable benefits, the most promising results demonstrated a 4× to 19× in HDR-mediated editing by inhibition of NHEJ with a small molecule (SCR7) or knockdown of KU70, KU80 or DNA Ligase IV proteins (14,15). Unfortunately, these benefits do not appear to translate to ES cells, because NHEJ inhibition with either SCR7 or knockdown of KU70 led to no increase in HDR efficiency or the ratio of on:off target insertion. Moreover, this approach is frequently cytotoxic and potentially mutagenic, posing a significant problem for their use for rigorously engineering cell lines (17).
In contrast, substantially greater overall efficiencies were attained by strategies to eliminate unwanted cells through positive selection. Importantly, we describe the effect of COIN, whereby selection for one insertion increases the frequency of a second, unlinked insertion in individual cells. COIN increased the frequency of isolating HDR-mediated insertion by up to 42-fold when selection and detection dsDNA donors were targeted to a single site, and up to 22-fold when the two donors were targeted to distinct sites. Unlike cell cycle synchronization or NHEJ inhibition, these enrichments were not caused by an increase in the actual frequency of HDR-mediated insertions among transfected cells. Instead, the increased efficiency was caused by the elimination of cells that were incapable of performing HDR-mediated insertions. Intriguingly, one can imagine combining these separate mechanisms in amenable cell types. In ES cells, the COIN effect reduces the number of clones one must screen by 90–95% in order to obtain an on-target insertion. The increased efficiency facilitates ease in design of effective donor DNAs by reducing the length of homology arms needed to less than 0.2 kb. It also removes the need to include a selection marker cassette within the donor DNA at the site of interest. When considering the inverse logarithmic relationship of insert length and targeting efficiency, this benefit can be substantial. Finally, by using dsDNA as donors, COIN allows changes greater than 1kb to be inserted at a frequency similar to that of using ssODN, which are limited to ∼50 bp of insert size.
In order to enable broad application of COIN, we generated a specialized donor DNA, Rosa26-pRIND. In combination with a matched sgRNA/Cas9 vector, this system enabled proper targeting followed by seamless excision without additional transfections. Because the targeted Rosa26 allele is returned to the original sequence, repeated retargeting is possible using the same sgRNA and Rosa26-pRIND donor. Although not demonstrated here, the pRIND cassette can also be used independently of COIN procedures as a direct selectable marker system. For sites near a genomic TTAA sequence, homology arms can be attached to the pRIND cassette using PCR or isothermic Gibson-type reactions. Similar piggyBac based strategies have been used for seamless editing of several genes in human pluripotent stem cells (31) and subsequently combined with CRISPR/Cas9 (6,9). Like pRIND, the previous results demonstrated the utility for precise removal of the selection cassette. Unlike pRIND, previous uses of piggyBac required separate steps to screen and expand clones of cells, introduce PBase expression by transfection, and subsequently screen and expand the final clones (6,9,31). By including the inducible pBase-ERT2 element within pRIND, a single transfection is required, and the resources and time are reduced by about half for obtaining the final edited cell line.
Finally, while we use COIN specifically for the promotion of HDR in mouse ES cells, we expect the principles to be broadly applicable. High levels of co-occurrence in terms of bialleleic events have been reported for genome editing procedures in a variety of mammalian systems, including human pluripotent stem cells (24,25,28,32). Recently, efficient precise editing of human pluripotent cells was described via a method named CORRECT, which uses Cas9 and ssODN to generate small homozygous mutations (33). Although the high level of biallelic editing was not examined mechanistically in that study, it can be simply explained by the COIN effect. We suggest that COIN is an example of an important and broadly applicable principle to consider when performing genome engineering experiments. The principle is based on the co-occurrence of independent events wherein heterogeneity exists among genetically homogenous cells. The rare cells that are proficient at completing one genomic manipulation have an increased probability of completing a second, independent genomic manipulation, provided the two manipulations are mediated by sufficiently similar mechanisms. Currently, the implementation of COIN for precise genome engineering is limited by the availability of selection donors with specificity for an on-target insertion only, such as Ctnnb1-EGFP. Developing a variety of on-target specific selection donors will enable broad usage of the COIN technique for enhancing efficiency of precise genome engineering.
Supplementary Material
Acknowledgments
We thank Jenny Zhang and Maureen Regan for helpful discussions associated with the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01-HD081534 to B.J.M, T32-HL007829 to B.R.S.]; Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust [SCH-032 to B.R.S.]. Funding for open access charge: NIH [R01-HD081534].
Conflict of interest statement. None declared.
REFERENCES
- 1.Joung J.K., Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nat. Biotechnol. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 2.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science (New York, N.Y.) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J., et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science (New York, N.Y.) 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie F., Ye L., Chang J.C., Beyer A.I., Wang J., Muench M.O., Kan Y.W. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N.S., Cai W., Yang G., Bronson R., Crowley D.G., et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S., et al. Small Molecules Enhance CRISPR Genome Editing in Pluripotent Stem Cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 15.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vartak S.V., Raghavan S.C. Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. FEBS J. 2015;282:4289–4294. doi: 10.1111/febs.13416. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava M., Nambiar M., Sharma S., Karki S.S., Goldsmith G., Hegde M., Kumar S., Pandey M., Singh R.K., Ray P., et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 18.Shagin D.A., Barsova E.V., Yanushevich Y.G., Fradkov A.F., Lukyanov K.A., Labas Y.A., Semenova T.N., Ugalde J.A., Meyers A., Nunez J.M., et al. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol. Biol. Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 19.Cadinanos J., Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., Lin C., Lu D., Ning Z., Cox T., Melvin D., Wang X., Bradley A., Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh P., Schimenti J.C., Bolcun-Filas E. A mouse geneticist's practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu G., Duan J., Shu S., Wang X., Gao L., Guo J., Zhang Y. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc. Natl. Acad. Sci. U.S.A. 2016;113:1256–1260. doi: 10.1073/pnas.1521597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakuma T., Nakade S., Sakane Y., Suzuki K.T., Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 24.Byrne S.M., Ortiz L., Mali P., Aach J., Church G.M. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43:e21. doi: 10.1093/nar/gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canver M.C., Bauer D.E., Dass A., Yien Y.Y., Chung J., Masuda T., Maeda T., Paw B.H., Orkin S.H. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yusa K. piggyBac Transposon. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0028-2014. MDNA3-0028-2014. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y.T., Bradley A. A new positive/negative selectable marker, puDeltatk, for use in embryonic stem cells. Genesis. 2000;28:31–35. doi: 10.1002/1526-968x(200009)28:1<31::aid-gene40>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 28.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holkers M., Maggio I., Henriques S.F., Janssen J.M., Cathomen T., Goncalves M.A. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods. 2014;11:1051–1057. doi: 10.1038/nmeth.3075. [DOI] [PubMed] [Google Scholar]
- 30.Pinder J., Salsman J., Dellaire G. Nuclear domain 'knock-in' screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat. Protoc. 2013;8:2061–2078. doi: 10.1038/nprot.2013.126. [DOI] [PubMed] [Google Scholar]
- 32.Wan H., Feng C., Teng F., Yang S., Hu B., Niu Y., Xiang A.P., Fang W., Ji W., Li W., et al. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 2015;25:258–261. doi: 10.1038/cr.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.