Abstract
Porphyromonas gingivalis and Filifactor alocis are fastidious anaerobic bacteria strongly associated with chronic forms of periodontitis. Our understanding of the growth activities of these microorganisms in situ is very limited. Previous studies have shown that copy numbers of ribosomal-RNA precursor (pre-rRNA) of specific pathogen species relative to genomic-DNA (gDNA) of the same species (P:G ratios) are greater in actively growing bacterial cells than in resting cells. The method, so-called steady-state pre-rRNA-analysis, represents a novel culture-independent approach to study bacteria. This study employed this technique to examine the in situ growth activities of oral bacteria in periodontitis before and after non-surgical periodontal therapy. Sub-gingival paper-point samples were taken at initial and re-evaluation appointments. Pre-rRNA and gDNA levels of P. gingivalis and F. alocis were quantified and compared using reverse-transcriptase qPCR. The results indicate significantly reduced growth activity of P. gingivalis, but not F. alocis, after therapy. The P:G ratios of P. gingivalis and F. alocis were compared and a low-strength, but statistically significant inter-species correlation was detected. Our study demonstrates that steady-state pre-rRNA-analysis can be a valuable culture-independent approach to studying opportunistic bacteria in periodontitis.
Periodontitis is a globally pervasive disease characterized by progressive destruction of tooth-supporting structures and affects millions of people1. Periodontitis is also associated with several chronic systemic diseases2. The composition of the dental biofilm is diverse and plays an important role in the development of periodontitis3,4. Organisms including Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia belong to the ‘red complex’ of the dental biofilm and have been established as contributors to periodontitis5,6,7,8. Disproportionate overgrowth of opportunistic bacterial pathogens, including the red complex organisms, couples with the host immune response to potentiate inflammation and destruction of the periodontium.
P. gingivalis is proposed to be a “keystone pathogen” due to the microorganism’s prominent role in modulating the dynamics of the host immune response and composition/structure of the dental biofilm in order to persist in oral tissues9,10,11,12,13,14. Another emerging opportunistic pathogen, Filifactor alocis, has lately become a subject of great interest due to its association with periodontitis15,16,17,18,19,20,21,22 and peri-implantitis23,24. Our current understanding of the interaction between F. alocis and other periodontal pathogens is limited, however recent evidence suggests the pathogenicity of F. alocis is likely potentiated by P. gingivalis25,26,27,28. Thus, investigating the role of F. alocis in periodontitis is warranted.
Culture-independent technologies, such as the Human Oral Microbiome Identification using Next Generation Sequencing (HOMINGS), have allowed us to better understand periodontitis by profiling the microbial composition of the dental biofilm with high specificity29,30. Limitations to our understanding persist however, despite the availability of increasingly sensitive technologies such as HOMINGS. Use of additional culture-independent as well as physiologically relevant molecular-based techniques could further characterize periodontitis and may help define the etiological roles played by certain bacterial species, such as P. gingivalis and F. alocis.
Ribosomal RNA precursor (pre-rRNA) represents a large percentage of total RNA within cells31. As cell growth diminishes, pre-rRNA synthesis halts while maturation of the precursor molecules continues, resulting in substantial depletion of the pre-rRNA pool32,33. The pre-rRNA pool rapidly recovers upon nutritional stimulation34,35,36. Because pre-rRNA sequences are species-specific, it is possible to detect and quantify pre-rRNAs of specific bacterial species by applying reverse transcriptase quantitative PCR (RT-qPCR) to complex clinical and natural samples. Ratiometric pre-rRNA analysis, also termed molecular viability testing (MVT), has been shown to efficiently assess the viability of microbial pathogens in environmental and patient samples through detection of the synthesis of pre-rRNA upon brief nutritional stimulation34,35,37. MVT has also demonstrated enhanced sensitivity to the presence of viable microorganisms compared to DNA-targeted qPCR36.
Steady-state pre-rRNA analysis, a separate but related culture-independent method, assesses the growth activity of bacteria in samples without nutritional stimulation. Studies have shown that pre-rRNA copy number correlates with growth activity of bacterial cells (actively dividing cells have a high pre-rRNA copy number due to ongoing rRNA synthesis)32,33,38. Therefore, measurement of species-specific pre-rRNA normalized to genomic DNA (gDNA) of the same species (P:G ratio) can be used to assess ongoing proliferation of the targeted species in environmental or clinical samples. Due to the successful application of molecular methods similar to steady-state pre-rRNA analysis in other settings, we believe that this approach could be a valuable for understanding the growth activity/behavior of bacterial pathogens in periodontitis.
In this study we applied steady-state pre-rRNA analysis as a novel culture-independent approach to assess bacterial growth activity in periodontitis. We quantified pre-rRNA and gDNA of P. gingivalis and F. alocis from clinical chronic periodontitis samples before and after non-surgical periodontal therapy using RT-qPCR. P:G ratios were then generated to compare growth activity of P. gingivalis and F. alocis at baseline and after therapy. Clinical parameters including probing pocket depth (PPD), bleeding on probing (BOP), and presence of supra-gingival plaque were recorded before and after treatment to test for associations with growth activity as reflected in P:G ratio. Finally, the P:G ratios of P. gingivalis and F. alocis were compared to determine if any inter-species relationships could be found.
Results
Clinical findings after non-surgical periodontal treatment
Fifteen patients were enrolled in this study. Clinical diagnosis of periodontitis was completed using the guidelines of the American Academy of Periodontology39. Based on PPD and clinical attachment level (CAL), the periodontal pockets with the deepest PPD were selected as sample collection sites. A total of 45 sites were assessed for growth activity of P. gingivalis and F. alocis at initial and re-evaluation appointments using steady-state pre-rRNA analysis.
All sample sites had PPD ≥ 4 mm prior to treatment and 24 sample sites had PPD ≥ 4 mm upon re-evaluation. The PPDs were significantly reduced (one-tailed paired t-test, p = 2.5 × 10−13) from 5.82 ± 1.17 mm to 4.00 ± 1.45 mm after treatment. Mean improvement of CAL was 3.09 ± 2.65 mm after treatment (one-tailed paired t-test, p = 3.8 × 10−8), Table 1. Thirty-nine sample sites displayed BOP at baseline and 18 sample sites showed BOP at re-evaluation. The reduction in BOP after treatment was significant (McNemar’s test, p = 6.3 × 10−5). Forty sample sites had supra-gingival plaque at initial evaluation and 18 sample sites had supra-gingival plaque at re-evaluation. The reduction in plaque detected was significant (McNemar’s test, p = 4.5 × 10−6).
Table 1. Summary of clinical findings.
| Clinical parameter | Pre-treatment | Post-treatment |
|---|---|---|
| PPD ≥ 4 mm | 45 (100) | 24 (53) |
| Average PPD | 5.82 ± 1.17 mm | 4.00 ± 1.46 mm* |
| BOP | 39 (87) | 18 (40)§ |
| Plaque | 40 (89) | 17 (38)§ |
| Average CAL | 7.27 ± 2.16 mm | 4.18 ± 1.87 mm* |
Clinical parameters including the gender, number of pockets with PPD ≥ 4 mm, the average PPD, bleeding index (present/absent), and plaque index (present/absent) were assessed for all sites pre- and post-treatment. Values given are shown as number (percent of total sites). The significance in reduction of clinical signs of inflammation after treatment was assessed (*Paired t-test, p < 0.0001. §McNemar’s test, p < 0.0001).
Measurement of actively growing bacteria by steady-state pre-rRNA analysis
Because pre-rRNA and gDNA are not measured with equal efficiency (RT-qPCR is generally less efficient than qPCR), this value does not indicate that resting bacterial cells have on average, fewer than 0.5 or 0.1 pre-rRNA molecules per genome. The actual number of pre-rRNA molecules in resting cells is not known. However, by applying the same measurements at initial sample collection and re-evaluation appointments, it was possible to generate comparative reliable results.
Threshold values delineating growing vs. non-growing cell populations were derived from laboratory-based growth curve experiments, which identified the maximum P:G ratios that were seen in all samples of non-growing cultures (Fig. 1). Thresholds selected for this study were 0.5 for P. gingivalis and 0.1 for F. alocis. These values corresponded to the highest P:G ratios observed during the log phase of the bacterial growth curves from our nutrient-depletion experiment (see Methods). Use of alternative threshold values did not significantly alter results and interpretations.
Figure 1. Growth curves of P. gingivalis and F. alocis used to generate pre-rRNA:genomic-DNA (P:G) ratios.
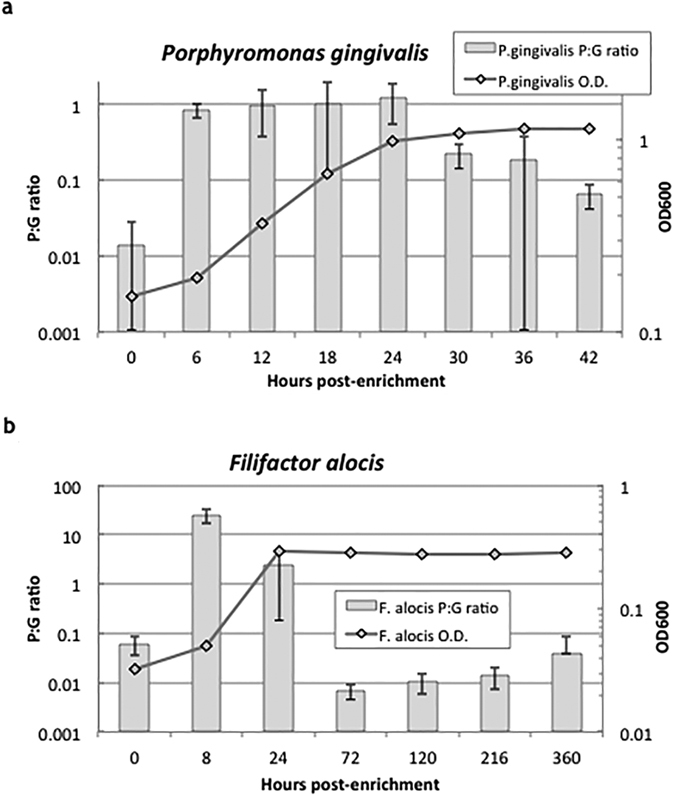
(a) Growth curve of P. gingivalis over 42 h time course. At inoculation and 6 h intervals, samples of the culture were taken and the P:G ratios were measured using RT-qPCR. The P:G threshold value of 0.5 was determined by identifying a point in the stationary phase that correlates with the highest P:G ratio. Thus, the threshold P:G ratio represents a value above which the bacteria can be conservatively viewed as actively dividing and metabolically active. (b) Growth curve of F. alocis over 360 h time course. P:G ratios were measured at time points and the threshold value of 0.1 was selected following the same strategies applied to P. gingivalis.
Association between clinical parameters and actively growing P. gingivalis and F. alocis
The relationships between actively growing bacteria, as determined by P:G ratio, and clinical observations were determined using logistic regression. Actively growing P. gingivalis was positively correlated with PPD ≥ 4 mm before (p = 0.04) and after (p = 0.009) treatment. A significantly positive correlation (p = 0.0001) between actively growing F. alocis and PPD ≥ 4 mm was found only in sample sites after treatment.
Actively growing F. alocis was found to be positively correlated with BOP in sample sites at initial collection (p = 0.005), but not at re-evaluation. No significant associations between actively growing P. gingivalis and BOP or plaque score were found in our analysis. Positive associations between actively growing F. alocis and presence of plaque were found before (p = 0.003) and after treatment (p = 0.03).
Prevalence of actively growing P. gingivalis and F. alocis after non-surgical periodontal therapy
The number of sample sites with actively growing P. gingivalis and F. alocis was quantified at initial collection appointments and at re-evaluation (Fig. 2). At initial collection, 51% and 69% of sample sites had actively growing P. gingivalis or F. alocis, respectively (Fig. 2). After treatment, 33% and 53% of sample sites had actively growing P. gingivalis or F. alocis, respectively. The reduction in prevalence of P. gingivalis (Logistic regression, 95% CI: 0.2535, 0.7633) was statistically significant. The prevalence of actively growing F. alocis was also reduced, but the reduction was not significant (Logistic regression, 95% CI: 0.4750, 0.9315). Thirteen and twenty four sample sites with actively growing P. gingivalis or F. alocis, respectively, were observed at both initial collection and re-evaluation appointments.
Figure 2. Prevalence of actively growing periodontal pathogens in periodontitis sample sites.
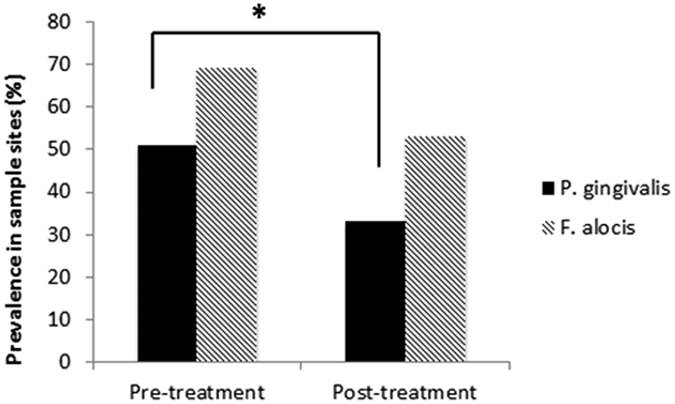
Percent of disease sample sites with actively growing P. gingivalis and F. alocis at initial and non-surgical periodontal therapy re-evaluation appointments (Logistic regression, *denotes statistical significance, p < 0.05).
Changes in bacterial load determined via genomic DNA quantification
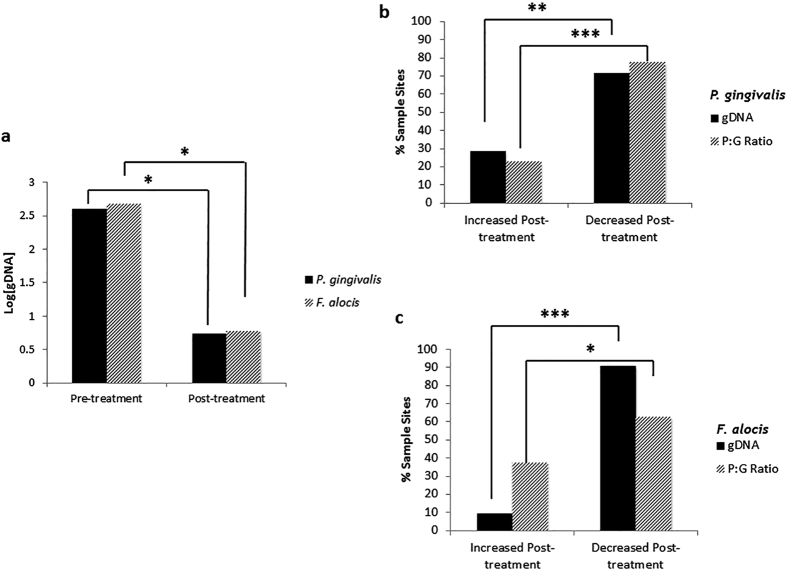
The bacterial load of P. gingivalis and F. alocis was indirectly determined via species-specific gDNA quantification before and after treatment (e.g. changes in [gDNA]) (Fig. 3). P. gingivalis and F. alocis load decreased significantly (Student’s t-test, p = 2.1 × 10−5 and p = 1.1 × 10−6, respectively) after treatment (Fig. 3a). We also evaluated the proportion of sites exhibiting growth activity and bacterial load changes for P. gingivalis (Fig. 3b) and F. alocis (Fig. 3c). There was a significantly higher proportion of sites in which the [gDNA] decreased for P. gingivalis (Two-sample Z-test, Z = −3.14, p = 0.002) and F. alocis (Two-sample Z-test, Z = −4.52, p = 6.3 × 10−6). Similarly, there was a significantly higher proportion of sites in which the P:G ratio decreased for P. gingivalis (Two-sample Z-test, Z = −6.48, p < 0.0001) and F. alocis (Two-sample Z-test, Z = −2, p < 0.05).
Figure 3. Changes in bacterial load and activity level of P. gingivalis and F. alocis.
(a) Levels of gDNA for P. gingivalis and F. alocis at initial and non-surgical periodontal therapy re-evaluation appointments. [gDNA] was measured in μg and was log-transformed for easier interpretation of data. (Student’s t-test, *denotes statistical significance at p < 0.001). (b) Percent of total sites in which P. gingivalis [gDNA] or P:G ratio increased or decreased after treatment. (Two-sample Z-test for proportions, *denotes statistical significance at p < 0.05; **denotes statistical significance at p < 0.01, ***denotes statistical significance at p < 0.0001) (c) Percent of total sites in which F. alocis [gDNA] or P:G ratio increased or decreased after treatment. (Two-sample Z-test for proportions, *denotes statistical significance at p < 0.05; **denotes statistical significance at p < 0.001).
Correlating growth activity between P. gingivalis and F. alocis
The growth activities of P. gingivalis and F. alocis relative to one another were assessed. Sample sites in which the P:G ratio of only one target organism was detected and outlier P:G ratios (greater than three standard deviations from the mean) were removed from this analysis (samples included in analysis n = 51). P:G ratios were log-transformed and compared using linear regression (Fig. 4). A small strength, but statistically significant correlation was observed (Pearson’s correlation coefficient r = 0.39, p = 0.005).
Figure 4. Correlation of growth activity of P. gingivalis and F. alocis.
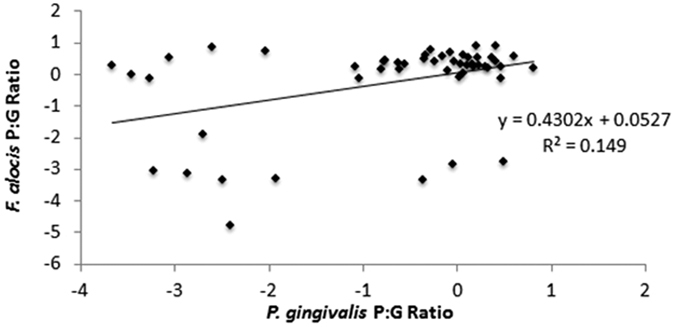
Correlation of P. gingivalis and F. alocis P:G ratios using linear regression (combined initial and non-surgical periodontal therapy re-evaluation appointments). Trendline equation: log(F. alocis P:G ratio) = 0.4302 * log(P. gingivalis P:G ratio) + 0.0527. R2 value: 0.14902. Outliers (greater than 3 standard deviations from the mean) and sample sites in which P:G ratios of only one target organism was generated were removed from analysis. Number of samples (sites with P:G ratios for both microorganisms) included in analysis = 51.
Discussion
This study investigated the use of steady-state pre-rRNA analysis to assess the growth activity of the periodontal pathogens P. gingivalis and F. alocis in patients with chronic periodontitis. Our results suggest that this novel culture-independent technique could be useful for directly understanding bacterial growth activities. This is the first study to evaluate the in situ growth activity of periodontal pathogens in patients with periodontitis through culture-independent techniques.
Non-surgical periodontal therapy provided to the study population resulted in improvement in clinical parameters associated with periodontitis and lowered the prevalence of actively growing P. gingivalis. Previous studies have shown a decrease in the relative abundance8,20,40 and number of viable cells41 of P. gingivalis following non-surgical periodontal therapy, thus our findings are consistent with existing literature.
Interestingly, we did not observe a significant reduction of actively growing F. alocis following non-surgical periodontal therapy. F. alocis has been previously implicated in cases of refractory periodontitis18 and non-surgical periodontal therapy without the use of antibiotics has shown limited effectiveness on reducing F. alocis42. Additionally, evidence suggests that the use of antibiotics may not have as great an effect on response to non-surgical periodontal therapy compared to the composition of the oral microbiota prior to treatment43. More studies are needed to determine the effectiveness of antibiotics on actively growing F. alocis in situ, and steady-state pre-rRNA analysis could be useful in such studies.
We compared the total load using [gDNA] of P. gingivalis and F. alocis at initial and re-evaluation appointments. Our results show a significant decrease in the abundance of these organisms. More comprehensive techniques, such as the Human Oral Microbiome Identification Microarray (the pre-cursor to HOMINGS), have also supported this result8,20. In order to assess the comparability of changes in bacterial growth activity and load, the increase/decrease in P:G ratios versus the [gDNA] was recorded. The two methods of measurement offered comparable results, however steady-state pre-rRNA analysis could provide a more detailed picture of bacterial growth in situ rather than relying solely on detecting nucleic acid.
One aim of this study was to identify any relationship between the growth activities of P. gingivalis and F. alocis before and after non-surgical periodontal therapy. The dental biofilm is a highly complex, multi-level ecosystem in which constituent bacterial species compete for nutrients and potentiate disease4. The specific interaction of P. gingivalis and F. alocis is a newly studied phenomenon and the growth of one relative to another may be a key indicator of conditions within the dental biofilm28. In this study a statistically significant, albeit weak, positive association was found between the P:G ratios of P. gingivalis and F. alocis. To date, only in vitro models have been used to investigate the potential interaction of P. gingivalis and F. alocis25,26,27. Thus, our study may offer a glimpse into a physiological and environmental relationship between these two opportunistic pathogens.
The relative amount of oral bacteria, including P. gingivalis and F. alocis, and their presence in patients with varying degrees of health and disease has become an active area of research over the past few years30,44,45. Our investigation originally aimed to identify healthy matched sample sites, however we faced challenges in identifying sample sites with no clinical signs of inflammation in this study population. Furthermore, the present study did not account for patient smoking status. In any future applications of steady-state pre-rRNA analysis in periodontitis research, it could be beneficial to include this demographic, as it is clinically and ecologically relevant46. We also included patients with varying degrees of chronic periodontitis and it may be interesting for further studies to select a more homogeneous patient population.
In conclusion, we applied steady-state pre-rRNA analysis as a culture-independent technique to assess growth activity of P. gingivalis and F. alocis in patients with chronic periodontitis. Our results demonstrate the potential value of this novel tool in the study of periodontitis. In future studies our approach can be expanded to larger patient populations using additional treatment modalities to further validate these new findings. Furthermore, it would be gainful to include other oral bacteria in future studies, with the aim to further characterize the dynamic nature of the dental biofilm in health and disease.
Materials and Methods
Study population
The clinical assessment and sample collection components of this longitudinal non-randomized cross-disciplinary study was performed on patients in the teaching clinics of the University of Florida College of Dentistry (UFCD) in Gainesville, Florida under the approved guidance of the Health Science Center Institutional Review Board (IRB, human subjects assurance number FWA 00005790) with all applicable federal regulations governing the protection of human subjects. Written informed consent was obtained from all subjects. The UFCD and the University of Washington IRBs also approved all experimental protocols. Molecular microbiological analyses were completed at the UFCD and University of Washington, in Seattle, Washington. Partially or fully dentate patients with at least twenty teeth were recruited. The patient age range for enrollment spanned the ages of 21 and 60 years, and the patient median age was 48 ± 11.5. Exclusion criteria for this study, in accordance with criteria outlined by the American Academy of Periodontology (AAP)39, included diagnosis with any systemic disease that could influence the progression and/or clinical characteristics of periodontal disease. Patients taking medications, which may affect the oral presentation of the periodontium or cause immune-modulating effects, were also excluded. These included antibiotics within three months prior to study enrollment, corticosteroids, chemotherapeutic agents, and bisphosphonates. Smoking status was not collected from patients. No exclusions were made on the basis of gender or race. However, our study population included 8 females and 7 males, for a total of 15 study participants, thus an approximately equal representation of the sexes was enrolled.
Clinical examination and sample site selection
Full periodontal charting of the patients’ dentition was recorded, including probing pocket depth (PPD, measurement from gingival margin to depth of pocket), clinical attachment level (CAL, measurement from cement-enamel junction to depth of pocket), plaque (presence or absence), and bleeding on probing (BOP, presence or absence). Measurements were preformed using UNC-12 periodontal probes (Hu-Friedy, Chicago, IL, USA). Diagnosis of the patients’ periodontal clinical presentation was made in accordance with criteria outlined by the AAP39. Ten study participants were diagnosed with generalized severe chronic periodontitis and one diagnosis each of generalized moderate or generalized slight chronic periodontitis was made. One diagnosis each of localized slight, localized moderate, and localized severe chronic periodontitis were also made. Two anterior, one pre-molar, and 11 molars (including one third molar) were selected as sample sites at initial evaluation appointments. The criteria used to select these sample sites included periodontal pockets with the deepest clinical PPD and worst CAL. Presence of BOP and plaque was not a requirement for sample site selection.
Periodontal therapy
Non-surgical periodontal therapy was completed after initial sample collection in the UFCD periodontology clinics. This therapy included supra- and sub-gingival debridement via scaling, root planing, and oral hygiene instructions. Re-evaluation was completed at least 6 weeks post-treatment by the same provider/trained periodontist. The study patients received supra- and sub-gingival scaling and oral hygiene instructions and were recommended to return for 3-month supportive periodontal maintenance.
Bacterial growth curve generation using pre-rRNA upshift protocol
Ratios of pre-rRNA to gDNA (P:G ratios) were measured for both species to assess their respective growth activities. Threshold P:G ratio values to discriminate growing from non-growing populations were identified based on growth curves created for P. gingivalis and F. alocis (Fig. 1). P. gingivalis ATCC 33277 was cultured anaerobically for 36 h at 37 °C in trypticase soy broth (TSB) (Fisher Scientific, Waltham, MA, USA) supplemented with yeast extract (1 mg/mL) (Fisher Scientific), haemin (5 μg/mL) (Sigma-Aldrich, St. Louis, MO, USA) and menadione (1 μg/mL) (MP BIomedicals, Santa Ana, CA, USA). At 36 h, the culture of P. gingivalis was re-inoculated at 1:10 dilution into fresh supplemented TSB and grown at 37 °C for one week to reach late stationary phase and allow for drainage of the pre-rRNA pool. 100 μL of the carbon-depleted culture was harvested in triplicate and centrifuged at 6000 g and 4 °C for 10 min. The pellets were immediately placed onto ice and transferred to −80 °C for storage. The carbon-depleted culture was then re-inoculated at 1:10 dilution into pre-warmed fresh supplemented TSB and grown at 37 °C. At 6, 12, 18, 24, 30, 36, and 42 h post-inoculation 1 mL samples were harvested according to the methods described above.
The growth curve used to generate the threshold P:G ratio for F. alocis was made via the same approach to that of P. gingivalis. F. alocis ATCC 35896 was cultured anaerobically for 36 h at 37 °C in brain heart infusion (BHI) (Fisher Scientific) supplemented with yeast extract (0.5 mg/mL) (Fisher Scientific), L-cysteine (50 μg/mL) (Sigma-Aldrich), and 20% arginine (Fisher Scientific). At 36 h, the culture of F. alocis was re-inoculated at 1:10 dilution into fresh supplemented BHI and grown at 37 °C for one week to reach late stationary phase and allow for drainage of the pre-rRNA pool. Pelleted samples were collected according to the methods described above at 0, 8, 24, 72, 120, 216, and 360 post-carbon depletion.
Bacterial sampling protocol
Teeth with periodontal pockets selected for sampling were isolated with cotton rolls and air dried to prevent saliva contamination. Sub-gingival plaque collection was completed using the paper point collection technique, which has been found to be similar to curette sampling in its ability to detect specific bacteria via qPCR43. Paper points (Henry-Schein, Melville, NY, USA) were inserted into the periodontal pockets and held in situ for at least 20 seconds. The paper points were then placed in sterile Eppendorf tubes which were directly put in cooler boxes previously frozen at −80 °C (Nalgene, Fisher Scientific). Upon collection, frozen boxes with samples were immediately stored and kept at −80 °C for future processing.
Nucleic acid extraction and RT-qPCR
DNA and RNA of each individual sample were simultaneously extracted using the MasterPure Complete DNA and RNA Purification Kit (Epicentre, Madison, WI, USA). Paper points were suspended in the provided lysis buffer (300 μL and proteinase k solution (1 μL) prior to following the furnished protocols. The suspension was vortexed for 1 minute and heated at 65 °C for 15 minutes to lyse the cells. Total nucleic acid (TNA) was eluted in 30 μL TE, from which 10 μL was removed for DNA measurement. From the remaining 20 μL, RNA was purified by DNase I treatment and re-precipitated as directed by the included protocol, and resuspended in the same volume of RNAse-free water.
Pre-rRNA and purified DNA was directly measured by qPCR as follows. All primers were designed using Primer3 software. qPCR primers for P. gingivalis were F- CGAGGTGTACTACCTGATAAATCG and R- CCCTCGACTTGCATGTGTTA and the RT primer was GTTTCAACGGCAGGCTGA. qPCR primers for F. alocis were F- AACCGGAGCAAAACTGAGAA and R- CCGTCCGCCACTAACTTCTA and the RT primer was TACTGATCGTTGCCTTGGTG. As in previous studies35,36,37, RT-qPCR primers for amplifying pre-rRNA straddled the junction between the 5′ terminus of the mature rRNA (16S) and the pre-rRNA leader region (ETS1), such that intact pre-rRNA molecules were required as templates. An in silico analysis of the primer sequences using NCBI’s BLAST tool suggested that all four 16S paralogs would be detected for every published complete genome strain of P. gingivalis (n = 7) and F. alocis (n = 1). A similar analysis using NCBI’s BLAST tool against the non-redundant database predicted no non-target amplification.
RNA was converted to cDNA by reverse transcription (RT) with the ImProm II system (Promega, Fitchburg, WI, USA), using 4 μL template and 3 μM 16S-specific primers. Genomic DNA and cDNA were measured by qPCR (using the same primer/probe set) utilizing Power SYBR Green (Life Technologies, Grand Island, New York, USA). Each 20 μL reaction contained 2 μL template and 375 nM each of forward and reverse primers. Reactions (including 7-log standard curves) were run on an Applied Biosystems StepOnePlus under the following conditions: 95 °C for 10 minutes followed by 40 cycles of 95 °C for 15 seconds, 57 °C for 30 seconds, and 72 °C for 30 seconds. Thresholds were automatically set by the StepOnePlus software (Life Technologies, Grand Island, New York, USA) or manually adjusted in the exponential range when necessary.
Statistical analyses
Differences in microbiological and clinical parameters were assessed using Logistic regression, Linear regression, Student’s t-test, and McNemar’s test. Logistic regression was completed with Statistical Analysis System GENMOD procedure using generalized estimating equation models for both binary and continuous outcomes. 95% confidence intervals were generated based on frequency tables created to compare prevalence of actively growing P. gingivalis and F. alocis before and after treatment. Log-transformation of data was completed when necessary to allow for interpretation of data. Linear regressions were performed using Excel 2010.
Additional Information
How to cite this article: Spooner, R. et al. In Situ Anabolic Activity of Periodontal Pathogens Porphyromonas gingivalis and Filifactor alocis in Chronic Periodontitis. Sci. Rep. 6, 33638; doi: 10.1038/srep33638 (2016).
Acknowledgments
We would like to acknowledge the support of the NIDCR grant R21DE021510 and R01DE016593. We would also like to thank Dr. Huaihou Chen from the University of Florida Department Of Biostatistics for his statistical support and expertise and Dr. Chul Hee Choi with his valuable assistance with bacterial growth assays. The views expressed in this article are those of the author(s) and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, or the United States Government. “Author, Dr. Ralee Spooner is a military service member (or employee of the U.S. Government). This work was prepared as part of his official duties.” Title 17, USC, §105 provides that ‘Copyright protection under this title is not available for any work of the U.S. Government. Title 17, USC, §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Footnotes
Author Contributions R.E.S. analyzed data and wrote the paper. K.M.W. designed experiments, analyzed data, and wrote the paper. K.L. designed experiments. P.L.H. analyzed the data. G.A.C. and Ö.Y. designed the study, supervised experiments, and wrote the paper.
References
- Pihlstrom B. L., Michalowicz B. S. & Johnson N. W. Periodontal diseases. Lancet 366, 1809–1820 (2005). [DOI] [PubMed] [Google Scholar]
- Atanasova K. R. & Yilmaz O. Prelude to oral microbes and chronic diseases: Past, present and future. Microbes Infect. 17, 473–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles R., Teles F., Frias-Lopez J., Paster B. & Haffajee A. Lessons learned and unlearned in periodontal microbiology. Periodontol. 2000 62, 95–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo A. E. & Frias-Lopez J. Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes Infect. 17, 505–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky S. S., Haffajee a D., Cugini M. a., Smith C. & Kent R. L. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998). [DOI] [PubMed] [Google Scholar]
- Kumar P. S., Griffen A. L., Moeschberger M. L. & Leys E. J. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43, 3944–3955 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledder R. G. et al. Molecular analysis of the subgingival microbiota in health and disease. Appl. Environ. Microbiol. 73, 516–523 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo A. P. et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 80, 1132–1421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Darveau R. P. & Curtis M. a. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Hasegawa M. & Inohara N. The Role of Oral Pathobionts in Dysbiosis during Periodontitis Development. J. Dent. Res. 93, 539–546 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. H. et al. Porphyromonas gingivalis-nucleoside-diphosphate-kinase inhibits ATP-induced reactive-oxygen-species via P2X7 receptor/NADPH-oxidase signalling and contributes to persistence. Cell. Microbiol. 15, 961–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner R., Deguzman J., Lee K. L. & Yilmaz O. Danger signal adenosine via adenosine 2a receptor stimulates growth of Porphyromonas gingivalis in primary gingival epithelial cells. Mol. Oral Microbiol. 29, 67–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. et al. Porphyromonas gingivalis attenuates ATP-mediated inflammasome activation and HMGB1 release through expression of a nucleoside-diphosphate kinase. Microbes Infect. 17, 369–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter G. et al. Molecular analysis of bacteria in periodontitis: Evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149, 67–75 (2003). [DOI] [PubMed] [Google Scholar]
- Dahlén G. & Leonhardt Å. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol. Immunol. 21, 6–11 (2006). [DOI] [PubMed] [Google Scholar]
- Kumar P. S. et al. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44, 3665–3673 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlafer S. et al. Filifactor alocis–involvement in periodontal biofilms. BMC Microbiol 10, 66 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen A. L. et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo A. P. V. et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J. Periodontol. 83, 1279–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine D. H. et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J. Clin. Microbiol. 51, 2850–2861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R. R. D. S. et al. Levels of Candidate Periodontal Pathogens in Subgingival Biofilm. J. Dent. Res., doi: 10.1177/0022034516634619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N., Ochi M., Miyakawa H. & Nakazawa F. Analysis of bacterial flora associated with peri-implantitis using obligate anaerobic culture technique and 16S rDNA gene sequence. Int J Oral Maxillofac Implant. 28, 1521–1529 (2013). [DOI] [PubMed] [Google Scholar]
- da Silva E. H., Feres M., Figueriredo L. C., Shibli J. A., Ramiro F. S. & Faveri M. Microbiological diversity of peri-implantitis biofilm by Sanger sequencing. Clin. Oral Implants Res. 25, 1192–9 (2014). [DOI] [PubMed] [Google Scholar]
- Aruni A. W., Roy F. & Fletcher H. M. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect. Immun. 79, 3872–3886 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wright C. J., Dingming H., Uriarte S. M. & Lamont R. J. Oral Community Interactions of Filifactor alocis In Vitro. PLoS One 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni A. W., Zhang K., Dou Y. & Fletcher H. Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways. Infect. Immun. 82, 3261–3274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni A. W. et al. Filifactor alocis - a new emerging periodontal pathogen. Microbes Infect. 17, 517–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes B. P. F. A., Berber V. B., Kokaras A. S., Chen T. & Paster B. J. Microbiomes of Endodontic-Periodontal Lesions before and after Chemomechanical Preparation. J. Endod. 41, 1975–1984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belstrom D., Paster B. J., Fiehn N. E., Bardow A. & Holmstrup P. Salivary bacterial fingerprints of established oral disease revealed by the Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) technique. J Oral Microbiol 8, 30170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerther D. B., Pernthaler J., Schramm A., Amann R. & Raskin L. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge wastewater treatment systems. Appl. Environ. Microbiol. 66, 2154–2165 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi G. A. & Brabant W. H. Depletion of pre-16S rRNA in starved Escherichia coli cells. J. Bacteriol. 179, 4457–4463 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Stroot P. G. & Oerther D. B. Reverse Transcription of 16S rRNA to Monitor Ribosome-Synthesizing Bacterial Populations in the Environment. Appl. Environ. Microbiol. 75, 4589–4598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi G. A., Weigel K. M., Lefthand-Begay C. & Meschke J. S. Molecular detection of viable bacterial pathogens in water by ratiometric pre-rRNA analysis. Appl. Environ. Microbiol. 76, 960–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel K. M. et al. Molecular Viability Testing of Bacterial Pathogens from a Complex Human Sample Matrix. PLoS One 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do J. S., Weigel K. M., Meschke J. S. & Cangelosi G. A. Biosynthetic enhancement of the detection of bacteria by the polymerase chain reaction. PLoS One 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi G. A. & Meschke J. S. Dead or alive: Molecular assessment of microbial viability. Applied and Environmental Microbiology 80, 5884–5891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroot P. G. & Oerther D. B. Elevated precursor 16S rRNA levels suggest the presence of growth inhibitors in wastewater. in Water Science and Technology 47, 241–250 (2003). [PubMed] [Google Scholar]
- Armitage G. C. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 4, 1–6 (1999). [DOI] [PubMed] [Google Scholar]
- Jünemann S. et al. Bacterial community shift in treated periodontitis patients revealed by Ion Torrent 16S rRNA gene amplicon sequencing. PLoS One 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonyi M. et al. Assessment of viable periodontal pathogens by reverse transcription quantitative polymerase chain reaction. J. Periodontal Res. 48, 671–676 (2013). [DOI] [PubMed] [Google Scholar]
- Jentsch H. F. R., März D. & Krüger M. The effects of stress hormones on growth of selected periodontitis related bacteria. Anaerobe 24, 49–54 (2013). [DOI] [PubMed] [Google Scholar]
- Bizzarro S. et al. Microbial profiles at baseline and not the use of antibiotics determine the clinical outcome of the treatment of chronic periodontitis. Sci. Rep. 6, 20205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco T. G. B. et al. Microbial signature profiles of periodontally healthy and diseased patients. J. Clin. Periodontol. 41, 1027–1036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belstrøm D. et al. Differentiation of salivary bacterial profiles of subjects with periodontitis and dental caries. J. Oral Microbiol. 7, 27429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzarro S., Loos B. G., Laine M. L., Crielaard W. & Zaura E. Subgingival microbiome in smokers and non-smokers in periodontitis: An exploratory study using traditional targeted techniques and a next-generation sequencing. J. Clin. Periodontol. 40, 483–492 (2013). [DOI] [PubMed] [Google Scholar]