Abstract
Long-lasting success in lung cancer therapy using tyrosine kinase inhibitors (TKIs) is rare since the tumors develop resistance due to the occurrence of molecularly altered subclones. The aim of this study was to monitor tumors over time based on the quantity of mutant plasma DNA and to identify early indications for therapy response and tumor progression. Serial plasma samples from lung adenocarcinoma patients treated with TKIs were used to quantify EGFR and KRAS mutations in circulating DNA by digital PCR. Mutant DNA levels were compared with the courses of responses to treatment with TKIs, conventional chemotherapy, radiotherapy, or combinations thereof. Variations in plasma DNA mutation levels over time were found in 15 patients. We categorize three major courses: First, signs of therapy response are associated with a fast clearing of plasma DNA mutations within a few days. Second, periods of stable disease are accompanied by either absence of mutations or fluctuation at low levels. Finally, dramatic increase of mutational load is followed by rapid tumor progression and poor patient survival. In summary, the serial assessment of EGFR mutations in the plasma of NSCLC patients allows conclusions about controlled disease and tumor progression earlier than currently available methods.
Lung cancer is the leading cause of cancer related mortality worldwide1. Non small cell lung cancer (NSCLC) is diagnosed in 85% of the patients, most of them in a locally advanced or metastatic stage2, which is associated with limited therapy options and poor prognosis. In recent years, cancer genome sequencing studies have revealed numerous molecular alterations in NSCLC, which have led to the reclassification of tumor subtypes and stratified therapies. For example, the presence of mutations in the epidermal growth factor receptor (EGFR) gene qualifies NSCLC adenocarcinoma patients for targeted therapy with tyrosine kinase inhibitors (TKIs), leading to improved overall survival3,4,5,6. However, survival times are limited due to the development of TKI resistance. Thus, recognizing impending resistance with a subsequent adoption of the therapy strategy would be highly desirable. In recent years, pilot studies have demonstrated the capability to detect mutations in circulating DNA in blood plasma, reflecting the landscape and heterogeneity of primary tumors and metastases7,8,9,10,11. Serial evaluation of mutant plasma DNA could provide a noninvasive assessment of therapy response and tumor progression, including the detection of resistance mutations or an increase of EGFR sensitizing mutations associated with clinical progression12,13,14,15,16,17. Most of these studies considered assessments within long time intervals (weeks or months) after initiation of treatment16,18,19,20. Here, we describe the analysis of serial plasma DNA samples from 16 NSCLC patients under TKI therapy. We quantified prominent mutations in EGFR and KRAS genes in cell free DNA using digital PCR assays and compared these to the clinical progression data of the same patients. The aim of the study was to derive patterns of mutant plasma DNA courses over time and to evaluate the potential of this liquid biopsy approach for monitoring tumor disease and predicting therapy response.
Results
The patient cohort comprised 16 adenocarcinoma patients under therapy at the Thoraxklinik Heidelberg from 2011 to 2016. All patients presented with stage III or IV disease and carried EGFR mutations as confirmed by molecular pathological analysis of tumor tissue (Table 1). One patient carried a second mutation in codon 12 of the KRAS gene (KRAS G12C). Eight patients harbored a sensitizing deletion of EGFR exon 19, six of the remaining eight carried an EGFR L858R mutation in exon 21. T790M mutation status of tumor tissue was unknown in 56.2% of the cases. The patients received TKI (erlotinib, gefitinib, or afatinib) therapy either as 1st-line or as treatment subsequent to surgery, chemotherapy or radiation therapy (Supplementary Table S1). At the end of the observation periods, all tumors had metastasized, and five patients had deceased with median overall survival times of nine months.
Table 1. Patient characteristics.
| Patient | Gender | Age [y] | Smoking status | TNM | Tumor tissue | Plasma | |||
|---|---|---|---|---|---|---|---|---|---|
| Mutation | EGFR T790M | No. samples | EGFR sens. mut. (No. pos. samples) | EGFR T790M (No. pos. samples) | |||||
| 1 | m | 56 | non smoker | cT4 cN0 cM1b | EGFR c.2235_49del | unk | 10 | 9 | n.d. |
| 2 | f | 54 | fmr. smoker; 35 pys | cT4 cN3 cM1b | EGFR L861Q | neg | 9 | 9 | 1 |
| 3 | m | 68 | fmr. smoker; 35 pys | cT2b cN2 cM1a | EGFR L858R | neg | 11 | 6 | 4 |
| 4 | f | 61 | cur. smoker; 20 pys | cT4 cN2 cM1b | EGFR c.2236_50del | unk | 6 | 1 | 0 |
| 5 | f | 55 | fmr. smoker | cT2A cN0 cM1b | EGFR c.2236_50del | unk | 3 | 1 | 1 |
| 6 | f | 73 | non smoker | pT1 pN1 pM0 R0 | EGFR L858R | unk | 4 | 4 | 0 |
| 7 | f | 57 | fmr. smoker; 28 pys | pT3 pN9 pM1a R0 | EGFR L858R | neg | 6 | 0 | 0 |
| 8 | f | 68 | non smoker | cT2A cN1 cM1b | EGFR L858R | unk | 10 | 5 | 1 |
| 9 | f | 73 | non smoker | cT4 cN2 cM1a | EGFR c.2235_49del | unk | 11 | 6 | 0 |
| 10 | f | 60 | non smoker | cT3-4 cN3 cM1b | EGFR c.2236_50del | neg | 8 | 2 | 1 |
| 11 | f | 73 | non smoker | cT2-3 cN2 cM1a | EGFR L858R | unk | 9 | 7 | 0 |
| 12 | m | 56 | cur. smoker; 25 pys | cT1a cN2 cM1b | EGFR c.2129 A > G; KRAS G12C | unk | 5 | 5 (KRAS) | 0 |
| 13 | f | 60 | non smoker | pT1B pN1 pM0 R0 | EGFR c.2236_50del | neg | 3 | 3 | 3 |
| 14 | m | 50 | fmr. smoker; 2 pys | cT4 cN3 cM1b | EGFR L858R | pos | 2 | 2 | 2 |
| 15 | f | 45 | non smoker | pT4 pN2 cM0 R1 | EGFR c.2235_49del | pos | 5 | 4 | 3 |
| 16 | f | 59 | cur. smoker; 43 pys | cT4 cN3 cM1b | EGFR c.2235_49del | unk | 5 | 2 | 1 |
(No.: number; y: years; sens.: sensitizing; pos: positive; m: male; f: female; fmr.: former; cur.: current; pys: packyears; unk: unknown; neg: negative; n.d. = not done).
Evaluation of DNA quantity and integrity
Plasma samples were collected between July 2014 and February 2016. The concentrations of circulating DNA in the plasma samples ranged between 7.4 and 4,768 ng/ml (median 30.7 ng/mL) and were not correlated to gender or clinical parameters such as tumor stage, therapy response, or outcome (data not shown). Size distributions of the DNA fragments varied among the samples. Frequently, DNA laddering with a prominent peak at around 166 bp could be observed (Supplementary Figure S2). Here, the smallest peaks are harboring the majority of tumor-derived DNA21,22,23. Variable amounts of high molecular weight DNA were frequently associated with higher cfDNA concentrations. Eight samples were excluded from further analysis due to low quality in the Bioanalyzer test.
Determination of mutant allele fractions in plasma DNA
Following quality control, 107 plasma samples were included into the mutation analysis. Sensitizing EGFR mutations were detected in 61.7% (61/102) of the samples (Table 1; Supplementary Table S2). The KRAS mutation was present in five samples from one patient. Overall, in 15 out of 16 patients (93.7%), at least one plasma sample was positive for the known mutation. The T790M resistance mutation appeared in 17.5% (17/97) of samples, which comprised 60% (9/15) of the patients.
The digital PCR method was able to detect between 0.033 and 0.1% mutant alleles in wt background (data not shown), which is in line with previously reported sensitivities17,24. The results of the mutation analysis were compared to clinical data, including treatment plan, disease progression, and other events. In most cases, mutant allele frequencies corresponded to the tumor burden, irrespective of individual differences in therapy and disease courses. Three predominant patterns in plasma DNA levels were observed in the majority of the patients and are illustrated in the following paragraphs.
Acute and temporary changes in cfDNA mutation levels in response to TKI therapy
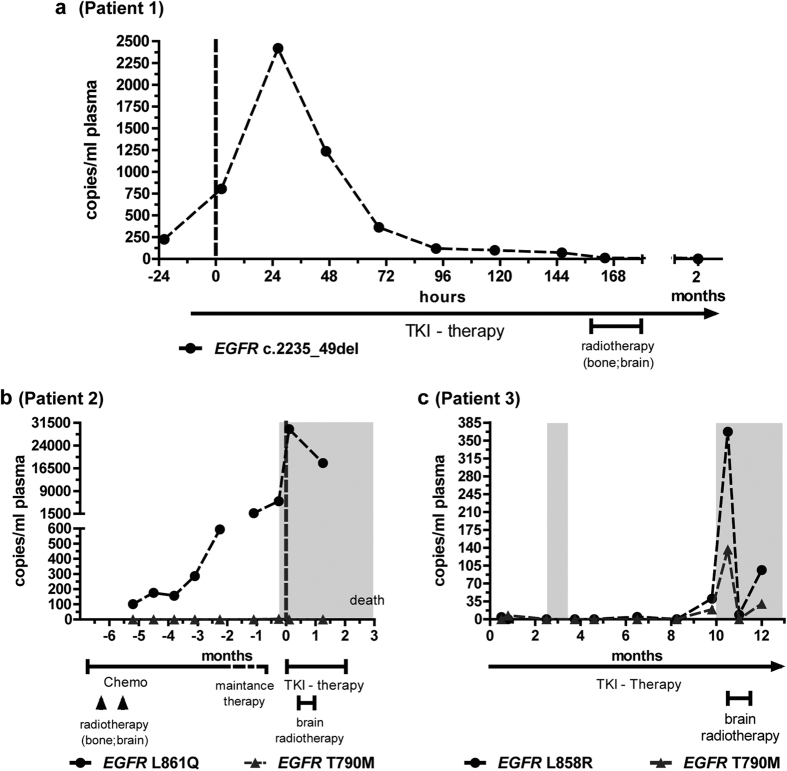
To assess the early time course of mutant cfDNA levels, changes within one week of initiation of TKI therapy were analyzed in two patients; one with daily tracking (Fig. 1A). Twenty-six hours after the first drug application (afatinib), an 11-fold increase of mutant DNA was measured, indicating DNA release from EGFR c.2235_49del-harboring tumor cells as a molecular response to TKI therapy25,26. The initial peak was followed by a continuous decline of mutant plasma DNA during the next two days. After 96 hours, mutation levels in plasma were lower than before therapy and further declined during the next three days. The clinical follow-up after two months was concurring with an objective therapy response and tumor remission. Concomitantly, EGFR sensitizing mutant alleles were no more detectable in plasma.
Figure 1. Occurrence of mutant EGFR cfDNA in response to therapy.
X-axis time frames, related to the start of TKI therapy (dashed vertical lines). Information about individual treatment schedules (chemo=chemotherapy; TKI-therapy; radiotherapy) are given below the x-axis. Y-axis: numbers of mutant alleles in plasma samples (copies/ml plasma), with sensitizing EGFR mutations labeled with black dots and T790M with grey triangles. White background: oncological stable state; grey background: disease progression.
In the second patient (Fig. 1B), the sensitizing EGFR mutation had developed over the course of five months prior to TKI therapy. The patient suffered a tumor progression after 1st-line chemotherapy, whereupon TKI therapy was introduced. A steep rise of mutant alleles, which is consistent with the eradication of susceptible tumor cells, was observed on the fifth day after TKI therapy. However, this was in contrast to patient 1, in whom mutant alleles had dropped below the pre-therapeutic level at this time point. Radiotherapy of brain metastases in patient 2 was initiated one day after TKI onset, and a further decline of mutant alleles in plasma was observed after five weeks. However, the number of mutant alleles remained higher than prior to therapy. Tumor progression instead of therapy response was diagnosed and the patient deceased two months later.
Patient 3 had already received TKI therapy for more than 10 months before tumor progression became clinically evident with a brain metastasis, whereupon radiotherapy of the brain was initiated (Fig. 1C). One day after the first fraction, a steep increase of EGFR sensitizing and resistance mutations in cfDNA was observed, presumably caused by radiation-induced tumor cell death and DNA release through the blood brain barrier, which had become permeable due to radiation27,28. The identification of the T790M mutation in plasma suggested its origin from the irradiated metastasis, since T790M had not been found in the primary tumor. One day after termination of radiotherapy, cfDNA levels of the sensitizing mutation declined to almost zero, and the T790M mutation was no more detectable. MR imaging confirmed stability of the cranial metastasis. In the last sample, both mutations recurred, which was accompanied by progressive disease. These three examples suggested that plasma analysis immediately before and after therapy onset could be a valuable indicator of therapy response.
cfDNA mutations indicating therapy response and stable disease
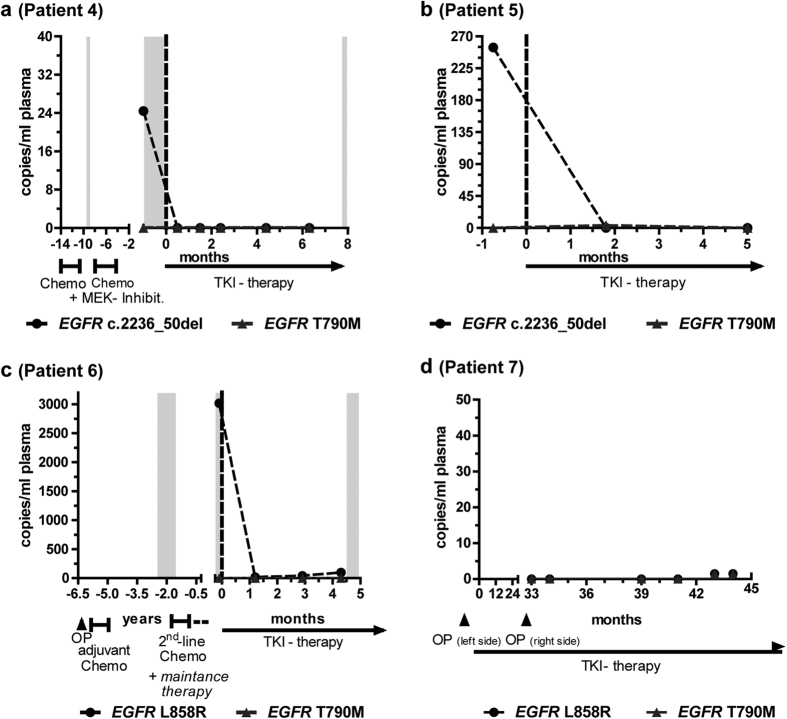
The examples illustrated above suggested that long-term response to TKI could be characterized by response immediately detectable in plasma, followed by a longer period of very low mutant cfDNA levels. These patterns could be equally highlighted in other plasma samples: Patients responding well to TKI therapy showed a strong decrease of EGFR mutant alleles from pre-therapeutic measurements to the next sample under therapy (Fig. 2). Interestingly, rising EGFR mutant cfDNA levels were observed in patient 6 two months prior to clinical progression (Fig. 2C). Overall, six (No. 1, 4, 5, 10, 11, and 16) out of seven patients with a clinical response to TKI therapy demonstrated such a complete absence of sensitizing mutations in the first assessment under EGFR TKI therapy. This was independent of prior treatments with chemotherapy or a MEK inhibitor. With continued stability of disease, EGFR mutant plasma levels remained low. In some cases, this trend sustained, indicating that TKI therapy could successfully be applied for even longer time periods: The monitoring of patient 7 started after 33 months of successful TKI therapy (Fig. 2D). Neither sensitizing nor T790M resistance mutations in EGFR could be detected in any of the six plasma samples collected over one year of TKI therapy. Patients 8 and 9 were monitored over 11 and 13 months, respectively (Supplementary Figure S3B,C). The plasma DNA mutation levels of these individuals showed slight variations on a low level, which is consistent with stable tumor disease.
Figure 2. Response to TKI therapy is accompanied by a quick drop of plasma mutant DNA levels.
A subsequent time interval of low mutant EGFR levels indicates a phase of stable disease. For designations, please refer to legends of Fig. 1. OP: operation/tumour resection; MEK-Inhibit.: Selumetinib therapy.
Association of EGFR and KRAS mutations with therapy failure and tumor progression
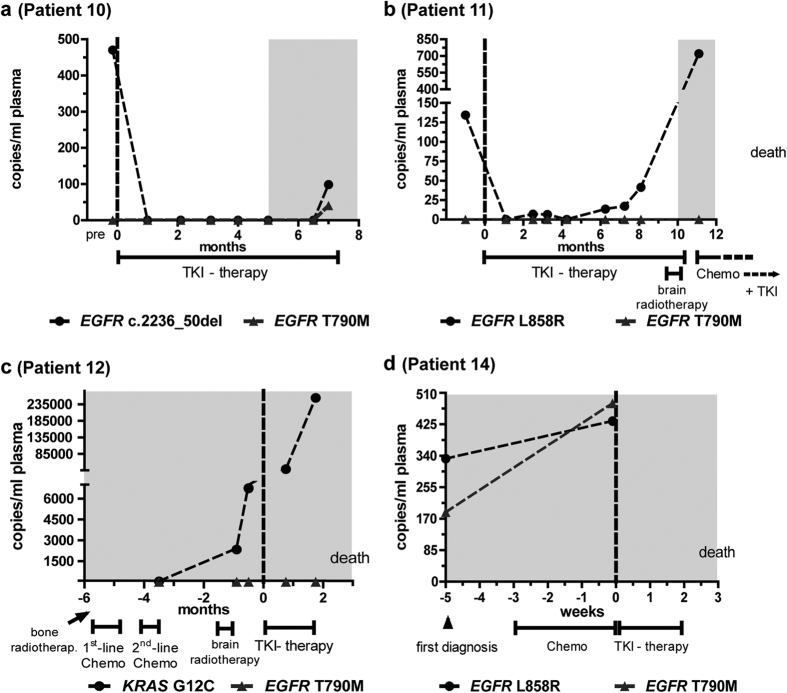
In contrast to response and controlled tumor disease, various forms of therapy failure, such as tumor relapse after initially stable disease, progressive disease despite therapy or changes of the treatment plan, were observed in six patients (Fig. 3, Supplementary Figure S4). Tumor progression and therapy failure were associated with a detectable T790M mutation in the cfDNA of five patients (No. 3, 10, 13, 14, and 15), with variable time points of occurrence during tumor progression. In these patients, the resistance mutation exhibited a course similar to the sensitizing mutation, albeit to a lower extent and somewhat lower level. For example, in patient 10 (Fig. 3A), where T790M had not been found in tissue, tumor progression concurred with the first detection of T790M in cfDNA. Three further patients (No. 2, 3, and 13; Fig. 1B,C; Supplementary Figure S4A) exhibited detectable plasma T790M levels, although this mutation had not been detected in the primary tumor tissues and metastasis biopsies.
Figure 3. Different manifestations of tumor progression.
Four different manifestations of progressive disease, all having in common that rise in plasma course reflected an objective aggravation of cancer disease. For designations, please refer to legends of Fig. 1. pre TKI: plasma sample collected prior to TKI onset.
Patient 15 was studied during the late 1st-line TKI therapy with gefitinib, tumor progression, and 2nd line treatment with erlotinib (Supplementary Figure S4B). The patient had a detectable T790M resistance mutation during 1st-line TKI treatment, which might have contributed to inefficient therapy. Initial decreases of EGFR sensitizing and resistance (T790M) mutations could be seen four weeks after erlotinib administration, indicating a therapeutic benefit. However, both mutations recurred only eight weeks later in the cfDNA. The disease progressed after initiation of systemic chemotherapy, and the patient deceased two months later.
T790M occurrence was associated with acquired resistance to TKI therapy but also pre-existing in TKI-naïve patients (No. 14 and 16; Fig. 3D, Supplementary Figure S3A). For example, patient 14 (Fig. 3D) exhibited high plasma levels of sensitizing and resistance EGFR mutations in two pre-therapeutic samples. He showed no objective response to TKI initiation, but a steady progression of tumor disease and deceased two months after the first diagnosis. No plasma samples under TKI therapy were available, but the T790M mutation had been confirmed in the biopsy of primary tumor tissue.
The courses of plasma DNA in two patients (11 and 12; Fig. 3B,C, respectively) indicated that huge increases of mutant alleles were associated with particularly poor prognosis and survival times of less than 6 months. Interestingly, in patient 11, progressive disease could already be detected in plasma several months before clinical progression. Mutant DNA levels increased 15-fold over pre-therapeutic levels within three months. Patient 12, harboring two mutations (in EGFR and in KRAS), showed no response to any therapy and died ten months after the first diagnosis. Here, disease progression was accompanied by considerably high amounts of cfDNA and a vast number of KRAS mutant alleles.
Discussion
In accordance with previous reports11,16,20,29,30, we demonstrate that serial mutation analysis of cfDNA in plasma is feasible for therapy monitoring. Despite variances in treatment or tumor manifestation, changes in mutant cfDNA levels were detected in almost all patients, in accordance with clinical course of disease. Three major categories could be attributed: evidence for therapy response, periods of stable disease, and impending tumor progression.
While several previous studies demonstrated the feasibility of serial assessment of mutations in cfDNA, albeit at longer intervals16,18,19,20, our data suggest that it is worthwhile to examine the first time periods before and immediately subsequent to therapy onset. Therapy response based on cfDNA mutations was observed after only 26 h after first drug application and mutational load decreased in the following days, indicating that the impact of TKI therapy is largest within the first few days. This was supported by repeated plasma analysis and clinical follow-up for more than two months. In addition, the data from patient 3 show that response in plasma is not only seen during the first days after TKI application but also when radiotherapy of distant regions (brain) is applied. Unfortunately, this patient progressed only two weeks afterwards due to the deterioration of extracerebral manifestations, which was again reflected by elevated mutation levels in cfDNA. Marchetti et al.13 reported decreased plasma DNA levels on the fourth day of TKI therapy in the majority of responding patients but did not analyze the developments of cfDNA mutations in the first hours after drug application. They further noticed varying decreases in association with shorter progression free survival for some patients, defining them as slow responders13. Using instantaneous assessment of cfDNA levels after surgical resection of colorectal cancer, Diehl and colleagues could make a first prediction for therapy success31. Our results suggest that this strategy could be adopted for EGFR sensitizing mutations in TKI-treated NSCLC patients. In addition, EGFR T790M mutant alleles in TKI-naïve patients20,32,33 could provide further insights into incipient development of therapy resistance.
Our data also suggest that longer periods of controlled disease after initial therapy response are paralleled by either absence of sensitizing mutations or fluctuations at low levels. Thus, as long as this pattern is found, the mutations in cfDNA suggest sustained therapy response. However, since most of these NSCLC tumors will inevitably progress, it is important to find evidence for this ongoing process as early as possible. Our data suggest that impending tumor progression is characterized by a continuous increase of EGFR sensitizing (and, in several cases, resistance) mutations up to four months before clinical symptoms were observed. Thus, regular tumor monitoring using cfDNA in short time intervals would be useful to determine progression earlier than currently possible and to potentially adapt the treatment strategy accordingly. Finally, vigorous tumor progression is paralleled by the occurrence of vast numbers of mutant alleles in the plasma (EGFR mutations in patients 2, 11, 13, and 14, as well as, KRAS mutations in patient 12). This finding was associated with very short patient survival times.
Concerning minimal invasiveness of diagnostic procedures, the liquid biopsy approach is superior to the analysis of tissue biopsies, which can be hardly implemented in regular, short time intervals. Furthermore, liquid biopsies can provide extra information: For example, the frequent T790M mutation in EGFR had not been identified in the tissues (patients 3, 10, and 13) but was detected in the cfDNA samples. However, much more knowledge is required to corroborate the current findings before they can be translated into the clinical setting. This includes the analysis of larger and systematically controlled patient cohorts, early and more frequent sampling as well as the standardization of pre-analytical processes34,35. Furthermore, a solid understanding of the mechanisms, including spatial and temporal patterns of tumor DNA release into the bloodstream is required.
In summary, we show that early and frequent mutation analysis in plasma of advanced NSCLC patients provides molecular data, which can be correlated with clinical information. Mutations in cfDNA can be identified prior to clinically detectable progression and therefore could be used to inform earlier decisions on revised therapy strategies.
Materials and Methods
Patient cohort and plasma preparation
The study was approved by the local ethics committee of the Medical Faculty Heidelberg (S048/2012) with amendment (July 31, 2014). All methods were carried out in accordance with the guidelines of the Translational Lung Research (TLRC) group in the German Center for Lung Diseases. Likewise, all experimental procedures were approved by the TLRC group in the German Center for Lung Diseases. Sixteen patients with advanced or recurrent NSCLC adenocarcinoma were recruited at the Thoracic Clinic in Heidelberg. Written informed consent was obtained from all patients in accordance with the guidelines of the ethics committee of the Medical Faculty Heidelberg. Serial blood sampling was implemented over the course of treatment with either TKI or other modalities. In eleven cases, samples could be drawn prior to TKI therapy. Between two and eleven samples of peripheral blood from patients were collected in EDTA tubes (Sarstedt, Nümbrecht, Germany) and processed within one hour. Blood samples were centrifuged at 2000 × g for 10 minutes at 10 °C, plasma was withdrawn and stored at −80 °C until use.
DNA extraction and quality control
cfDNA was extracted from 500 μL aliquots of frozen plasma with the QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) according to adapted manufacturer´s recommendations (Supplementary Methods). DNA quality and fragment sizes were examined using the Bioanalyzer 2100 with High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA).
DNA analysis using digital PCR
Absolute quantification of plasma cfDNA was performed using the Quantstudio 3D digital PCR System (Thermo Fisher Scientific, Waltham, MA, USA) and the validated Taqman assay for the reference gene TERT according to the manufacturer´s protocol (Supplementary Methods). The amount was calculated based on an external standard reference curve of fragmented genomic DNA (Roche Diagnostics, Mannheim, Germany). CfDNA from plasma samples was subjected to the measurement of known sensitizing EGFR and resistance (T790M), as well as KRAS mutations using digital PCR and TaqMan® Rare Mutation Assays (Supplementary Table S3 in supplemental methods). The chips were read using the Quantstudio 3D digital PCR instrument and the data were analyzed with QuantStudio™ 3D AnalysisSuite™ Software (example plot shown in Supplementary Figure S1). Three independent observers performed blinded analysis of the plots. Raw data for absolute quantification and rare allele detection were listed as copies/μl reaction volume. Pooled blood samples were used as controls to exclude background signals. The numbers of mutant alleles in the samples were reported as copies/mL plasma (Supplementary Methods).
Additional Information
How to cite this article: Riediger, A. L. et al. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci. Rep. 6, 33505; doi: 10.1038/srep33505 (2016).
Supplementary Material
Acknowledgments
Plasma samples were provided by Lung Biobank Heidelberg, a member of the BioMaterialBank Heidelberg (BMBH) and the biobank platform of the German Center for Lung Research (DZL). We thank Simon Ogrodnik for excellent technical assistance and Denis Roster for help with data analysis.
Footnotes
Author Contributions Conception and design: M.T., H.S. Sample collection and provision: T.M., M.M. and M.S. Acquisition of data: A.L.R., S.D., U.S. and I.H.-G. Analysis and interpretation of data: A.L.R., S.D., U.S., M.M., T.M., M.T. and H.S. Drafting of the manuscript: A.L.R., S.D. and H.S. Revision of the manuscript: A.L.R., S.D., M.M., T.M., M.T. and H.S. Supervision: H.S.
References
- Ferlay J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–E386, doi: 10.1002/ijc.29210 (2015). [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D. & Jemal A. Cancer statistics, 2016. CA Cancer J Clin 66, 7–30, doi: 10.3322/caac.21332 (2016). [DOI] [PubMed] [Google Scholar]
- Mitsudomi T. et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol 23, 2513–2520, doi: 10.1200/JCO.2005.00.992 (2005). [DOI] [PubMed] [Google Scholar]
- Mok T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361, 947–957, doi: 10.1056/NEJMoa0810699 (2009). [DOI] [PubMed] [Google Scholar]
- Rosell R. et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 13, 239–246, doi: 10.1016/S1470-2045(11)70393-X (2012). [DOI] [PubMed] [Google Scholar]
- Zhou C. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 12, 735–742, doi: 10.1016/S1470-2045(11)70184-X (2011). [DOI] [PubMed] [Google Scholar]
- Chan K. C. et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 59, 211–224, doi: 10.1373/clinchem.2012.196014 (2013). [DOI] [PubMed] [Google Scholar]
- Dawson S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368, 1199–1209, doi: 10.1056/NEJMoa1213261 (2013). [DOI] [PubMed] [Google Scholar]
- Murtaza M. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 6, 8760, doi: 10.1038/ncomms9760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112, doi: 10.1038/nature12065 (2013). [DOI] [PubMed] [Google Scholar]
- Newman A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 20, 548–554, doi: 10.1038/nm.3519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz L. A. Jr. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540, doi: 10.1038/nature11219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A. et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol 10, 1437–1443, doi: 10.1097/JTO.0000000000000643 (2015). [DOI] [PubMed] [Google Scholar]
- Marcq M., Vallee A., Bizieux A. & Denis M. G. Detection of EGFR mutations in the plasma of patients with lung adenocarcinoma for real-time monitoring of therapeutic response to tyrosine kinase inhibitors? J Thorac Oncol 9, e49–e50, doi: 10.1097/JTO.0000000000000134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536, doi: 10.1038/nature11156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard G. R. et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 20, 1698–1705, doi: 10.1158/1078-0432.CCR-13-2482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K. S. et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 21, 560–562, doi: 10.1038/nm.3854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. et al. Microdroplet digital PCR: detection and quantitation of biomarkers in archived tissue and serial plasma samples in patients with lung cancer. J Thorac Oncol 10, 212–217, doi: 10.1097/JTO.0000000000000349 (2015). [DOI] [PubMed] [Google Scholar]
- Narayan A. et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res 72, 3492–3498, doi: 10.1158/0008-5472.CAN-11-4037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D. et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 6, 20913, doi: 10.1038/srep20913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 102, 16368–16373, doi: 10.1073/pnas.0507904102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA 112, E1317–E1325, doi: 10.1073/pnas.1500076112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere F. et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 6, e23418, doi: 10.1371/journal.pone.0023418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One 9, e110780, doi: 10.1371/journal.pone.0110780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felip E. et al. A phase II pharmacodynamic study of erlotinib in patients with advanced non-small cell lung cancer previously treated with platinum-based chemotherapy. Clin Cancer Res 14, 3867–3874, doi: 10.1158/1078-0432.CCR-07-5186 (2008). [DOI] [PubMed] [Google Scholar]
- Gong Y. et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med 4, e294, doi: 10.1371/journal.pmed.0040294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Avella D. et al. Radiation-induced blood-brain barrier changes: pathophysiological mechanisms and clinical implications. Acta Neurochir Suppl 71, 282–284 (1998). [DOI] [PubMed] [Google Scholar]
- Qin D. et al. Improved efficacy of chemotherapy for glioblastoma by radiation-induced opening of blood-brain barrier: clinical results. Int J Radiat Oncol Biol Phys 51, 959–962 (2001). [DOI] [PubMed] [Google Scholar]
- Siravegna G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21, 795–801, doi: 10.1038/nm.3870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen B. S. et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 120, 3896–3901, doi: 10.1002/cncr.28964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F. et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 14, 985–990, doi: 10.1038/nm.1789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S. et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 359, 366–377, doi: 10.1056/NEJMoa0800668 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell R. et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 17, 1160–1168, doi: 10.1158/1078-0432.CCR-10-2158 (2011). [DOI] [PubMed] [Google Scholar]
- Bronkhorst A. J., Aucamp J. & Pretorius P. J. Cell-free DNA: Preanalytical variables. Clin Chim Acta 450, 243–253, doi: 10.1016/j.cca.2015.08.028 (2015). [DOI] [PubMed] [Google Scholar]
- van der Vaart M. & Pretorius P. J. Is the role of circulating DNA as a biomarker of cancer being prematurely overrated? Clin Biochem 43, 26–36, doi: 10.1016/j.clinbiochem.2009.08.027 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.