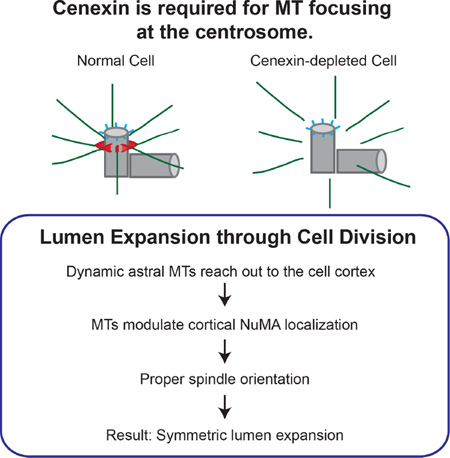
Summary
Establishing apical-basal polarity is instrumental in the functional shaping of a solitary lumen within an acinus. By exploiting micropatterned slides, wound healing assays, and 3-dimensional culture systems, we identified a mother centriole subdistal appendage protein, cenexin, as a critical player in symmetric lumen expansion through the control of microtubule organization. In this regard, cenexin was required for both centrosome positioning in interphase cells and proper spindle orientation during mitosis. In contrast, the essential mother centriole distal appendage protein, CEP164, did not play a role in either process, demonstrating the specificity of subdistal appendages for these events. Importantly, upon closer examination we found that cenexin depletion decreased astral microtubule length, disrupted astral microtubule minus-end organization and increased levels of the polarity protein, NuMA, at the cell cortex. Interestingly, spindle misorientation and NuMA mislocalization were reversed by treatment with a low dose of the microtubule-stabilizing agent, taxol. Taken together, these results suggest that cenexin modulates microtubule organization and stability to mediate spindle orientation.
Graphical Abstract
eTOC Blurb
By exploiting micropatterned slides, wound healing assays, and 3-dimensional culture systems, Hung et al. identify a mother centriole subdistal appendage protein, cenexin, as a critical player in symmetric lumen formation. In turn, lumen formation is mediated by spindle orientation through microtubule organization and stability.
Results and Discussion
In organs such as breast, kidney and intestine, polarized epithelial cells form acini, each with a single lumen (Figure 1E, left) [1]. However, in epithelial pre-invasive carcinomas, disruption of apical-basal polarity leads to multiple ectopic lumina (Figure 1E, right) [2]. Thus, a better understanding of the mechanisms responsible for multiple lumina formation will provide insight into the origin and progression of epithelial diseases.
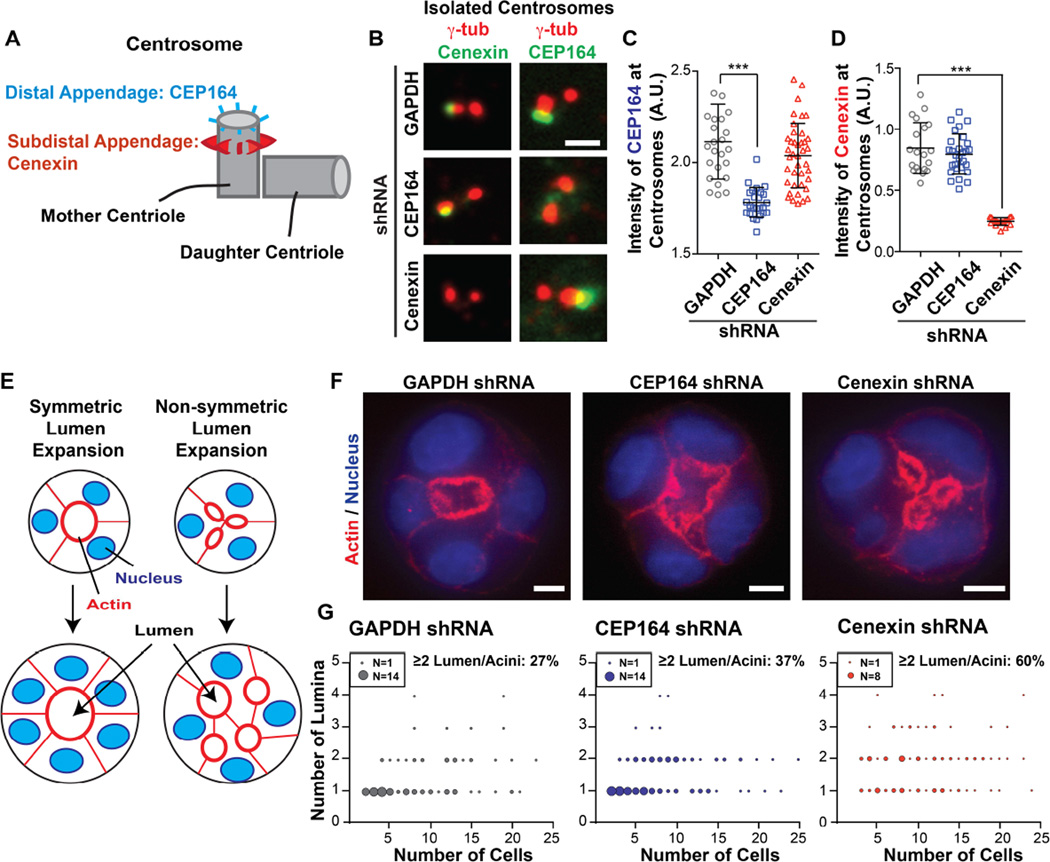
Figure 1. Depletion of the subdistal appendage protein, cenexin, leads to multiple lumina.
(A) A model depicting CEP164 localization to distal appendages (blue) and cenexin localization to subdistal appendages (red) on the mother centriole.
(B) Isolated interphase centrosomes in U2OS cells expressing GAPDH-, CEP164-, or cenexin-shRNAs. Mother centrioles were decorated with antibodies against the distal appendage protein, CEP164 (right column, green), the subdistal appendage protein, cenexin (left column, green), and centrioles (γ-tubulin, red). Scale bar, 1 µm.
(C–D) The integrated intensity of CEP164 (C) or cenexin (D) at centrosomes measured in U2OS cells depleted of GAPDH, CEP164 and cenexin. Data are represented as mean ± SD of >20 centrosomes in each group. ***, p-value <0.001. See also Figure S1A.
(E) A model depicting symmetric lumen expansion where acini expand around the central lumen (left), and non-symmetric lumen expansion where cells expand while creating multiple unorganized lumina (right).
(F) 3D-cultured MDCK acini were fixed and stained for actin (phalloidin, red) and nuclei (DAPI, blue). A single confocal z-section at the center of the acinus is displayed. Scale bar, 5 µm.
(G) Scatter plot illustrating the number of lumina in different acini. The number of acini with a given number of lumen in the population correlates to the size of each point (refer to legend for each graph). Each graph represents >100 acini scored over 3 independent experiments. See also Figure S1H.
Lumen formation requires apical membrane/lumen establishment and symmetric lumen expansion [1]. Lumen generation requires a single epithelial cell to undergo the first cell division. After division, both mother and daughter cell centrosomes/spindle poles reorient to a position where the newly forming apical membrane will emerge [3]. In subsequent cell divisions, spindle orientation must be tightly regulated to complete apical domain formation at the center of a developing acinus [1]. Specifically, dividing cells must orient their spindles parallel to the apical lumen to expand the already existing central lumen.
Spindle orientation requires proteins at mitotic spindle poles and polarity proteins at the lateral cell cortex. Spindle pole proteins are involved in nucleating and anchoring microtubules whereas, polarity proteins (NuMA/LGN/Gαi) assist in astral microtubule capture at the cell cortex [4]. However, the molecular interface between mitotic spindle poles, astral microtubules, and cortical capture of astral microtubules is poorly understood.
The centrosome contributes to cell polarity. In luminal epithelial cells, the centrosome is involved in polarity formation in two distinct ways: 1) during division it organizes and orients the mitotic spindle ensuring single lumen expansion [5], and 2) in interphase it repositions itself toward the apical membrane [3]. More specifically, during mitosis the pericentriolar material proteins, pericentrin and CEP215, contribute to spindle orientation through their interaction with the mother centriole subdistal appendage proteins, centriolin and ninein [6]. In interphase, the subdistal appendage protein, cenexin, anchors both centriolin and ninein to subdistal appendages [7–9]. Cenexin also provides the structural integrity of subdistal appendages [10]. Thus, we hypothesize that these mother-centriole-specific substructures (Figure 1A), and the molecular components associated with them, play a role in spindle orientation and centrosome positioning. In this study, we dissect the role of subdistal appendages versus distal appendages in lumen formation.
To determine which appendage type was required for lumen formation, we created cell lines stably depleted of CEP164 (distal appendage protein) and cenexin (subdistal appendage protein) (Figure 1B–1D, S1A–S1C). Depletion of both was confirmed by loss of primary cilia (Figure S1D and S1E) as shown previously [11–13]. In addition, subdistal appendage proteins, centriolin [8, 9] and CEP128 [14, 15] were lost after cenexin depletion (Figure S1F and S1G). Importantly cenexin depletion did not disrupt CEP164 localization to distal appendages, and CEP164 depletion had no effect on cenexin localization to subdistal appendages (Figure 1B–1D) [10, 16]. This result demonstrated that cenexin depletion targeted subdistal appendages specifically.
Based on these findings, we tested lumen formation following depletion of CEP164 or cenexin (Figure 1F and 1G). At early stages of acinus formation (acini with ≤5 cells), the majority of control cells (94%, GAPDH-depleted) formed acini with a single lumen. Similar results were obtained with CEP164-depleted cells (81%). In contrast, only 55% of cenexin-depleted cells formed acini with a single lumen. As acini expanded with multiple cell divisions (acini containing >5 cells), the percentage of acini with multiple lumina was low in control (27%) and CEP164-depleted cells (37%), compared to 60% in cenexin-depleted cells (Figure 1G and S1H). These findings suggest a role for the subdistal appendage protein, cenexin, in symmetric lumen formation and expansion. Due to the significant increase of acini with multiple lumina in cenexin-depleted cells, we hypothesized that cenexin governs lumen formation by regulating centrosome positioning and spindle orientation during various stages of the cell cycle.
During interphase, centrosome positioning near the cell centroid is crucial for directional cell migration. Pathways involving cell polarity proteins and regulation of microtubule dynamics are emerging as common regulators of centrosome positioning in a variety of contexts [17, 18]. Thus, we first investigated whether cenexin depletion disrupts apical-basolateral polarity in epithelial cells. Neither cenexin depletion nor CEP164 depletion disrupted the adherens junction protein, E-cadherin, or the apical membrane polarity proteins, PAR3 and PKCζ localization (Figure S2A) [19], suggesting that cenexin and CEP164 are not involved in polarity establishment.
We further tested if cenexin was involved in centrosome positioning by exploiting micropatterned surfaces to generate cells with nearly identical shapes and sizes (refer to Figure 2A, left) [20]. Centrosomes in nearly all control cells (GAPDH-depleted) localized within a 7 µm radius (dashed circle, Figure 2B) of the cell centroid. Strikingly, in cenexin-depleted cells, there was a six-fold increase in the percentage of centrosomes outside the centroid (30% compared to 5% in control cells and 3% in cells depleted of CEP164; Figure 2B and 2C). Moreover, cenexin depletion did not induce overt defects in microtubule nucleation at the centrosome (data not shown, [21]), but did cause defects in microtubule focusing at this site (Figure 2A, [21]). This suggested that defects in microtubule focusing disrupt centrosome positioning. To examine the contribution of microtubule organization and stability in centrosome positioning, we took two approaches: 1) Depletion of two well-established centrosome-localized modulators of microtubule nucleation, anchoring, and/or organization, PCM1 [22] and pericentrin (PCNT) [23–25] and by 2) treatment with the microtubule-destabilizing drug, nocodazole.
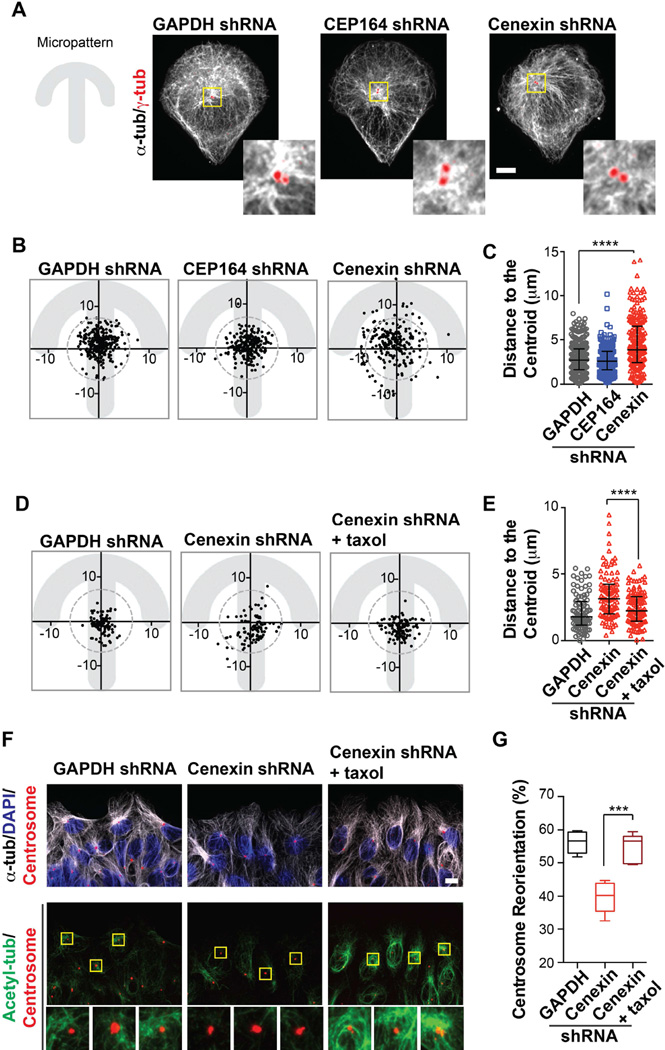
Figure 2. Cenexin is required for interphase centrosome centering and centrosome reorientation during cell migration.
(A) Left, Fibronectin-coated-crossbow micropattern on glass coverslips was used to mimic a migrating cell with a uniform cell shape and size. Right, U2OS cells were seeded onto micropatterns and stained for microtubules (α-tubulin, white) and centrosomes (γ-tubulin, red). Scale bar, 10 µm. Insets at the right of main images depict a 4×-magnified projection of microtubule anchoring at the centrosome. Cenexin-depleted cells show fewer microtubules focused at centrosomes.
(B) Interphase centrosome positions in cells grown on crossbow micropatterns. 270 centrosomes were plotted for the control cells, and 200 centrosomes were plotted for CEP164- depleted or cenexin-depleted cells. The centroid position in the pattern was determined and a circle with the radius of 7 µm was drawn. All points that fall outside of the circle were considered non-centered. Data were collected from >3 independent experiments.
(C) Distances between centrosomes and the cell centroid shown in (B) for GAPDH-, CEP164-, and cenexin-depleted cells. Data are represented as median ± interquartile range. ****, p-value<0.0001.
(D) As in (B), interphase centrosome positions in cells grown on crossbow micropatterns were treated with or without taxol as labeled; 100 centrosomes were plotted in each group.
(E) Distances between centrosomes and the cell centroid shown in (D). Data are represented as median ± interquartile range. ****, p-value<0.0001.
(F) U2OS cells (GAPDH-depleted, cenexin-depleted, and cenexin-depleted cells treated with taxol) were fixed 6 hours after applying a scratch wound. Top row, cells were stained with centrosomes (pericentrin, red), microtubules (α-tubulin, white), and nuclei (DAPI, blue). Bottom row, cells were stained for acetylated microtubules (green), and centrosomes (pericentrin, red). Insets, below main images, depict a 4×-magnified projection of acetylated microtubules at the centrosome. Scale bar, 10 µm.
(G) The percentage of centrosomes that orient towards the front quadrant (F, modeled in Figure S2K) at 6 hours after applying a scratch wound. Data are represented as a box-and-whisker plot with max and min of >300 cells over >3 regions in each group. Representative of n=2 experiments. ***, p-value<0.001.
PCM1- or PCNT-depletion caused only a subtle effect on centrosome centering (4%–10% not centered) (Figure S2B–S2D and S2E–S2G) compared to cenexin depletion (30%). Importantly, cenexin loss did not affect the presence of PCM1 or PCNT at the centrosome (Figure S2H) suggesting that cenexin, but not PCM1 or PCNT, has a significant role in centrosome positioning. PCM1 is involved in microtubule anchoring [22], but it is possible that cenexin anchors a separate subset of microtubules that predominate during centrosome positioning highlighting cenexin in centrosome centering and microtubule focusing.
In a second experiment, we directly tested if microtubule dynamics were important for centrosome positioning by treating cells with nocodazole (4 nM, 10 nM, and 20 µM) to destabilize microtubules (Figure S2I) [26]. Cells treated with low concentrations of nocodazole (4–10 nM) exhibited significant centrosome displacement from the cell’s centroid region (Figure S2J) similar to that seen in cenexin-depleted cells. Since cenexin depletion seemed to disrupt microtubules at the centrosome (Figure 2A), we attempted to rescue centrosome centering by stabilizing these microtubules with the microtubule-stabilizing drug, taxol (Figure 2D and 2E) [27]. Taxol treatment robustly reversed the effects of cenexin depletion allowing cells to re-center their centrosomes (Figure 2D and 2E) suggesting that stably focused microtubules at the centrosome are required for centrosome positioning.
We confirmed the role of cenexin in centrosome positioning using a physiologically-relevant wound-healing assay. The percentage of centrosomes that reorient to a position between the nucleus and the leading edge of the cell during migration was calculated. Two hours after inducing cell migration, 50% of control cells (GAPDH-depleted) reoriented their centrosome toward the leading edge, while fewer CEP164- and cenexin-depleted cells reoriented their centrosomes (43% for CEP164 depletion; 42% for cenexin depletion). Six hours after application of a scratch wound, ~60% of control (GAPDH-depleted) and CEP164-depleted cells reoriented their centrosomes (Figure S2K and S2L). However, centrosome reorientation was observed in only 40% of cenexin-depleted cells (Figure S2L), and could be rescued with an shRNA-resistant form of cenexin (Figure S2M). We conclude that cenexin is essential for centrosome reorientation during migration, while CEP164 may be dispensable in this process.
During migration, wound-edge migrating cells contain more centrosome-localized stable/acetylated microtubules [28]. After application of a scratch wound in control cells (6 hours), prominent acetylated microtubule arrays focused at the centrosome were noted (Figure 2F, GAPDH-depleted cells). In contrast, cenexin-depleted cells showed a significant reduction of acetylated tubulin at the centrosome concomitant with disorganized microtubules (Figure 2F), reminiscent of cenexin-depleted cells grown on micropatterns (Figure 2A). Taxol treatment of cenexin-depleted cells rescued microtubule organization (stabilized microtubules) at the centrosome, and centrosome reorientation. These findings (Figure 2F and 2G), together with those on centrosome centering (Figure 2A–2E), suggest that the subdistal appendage protein, cenexin, is critical for adjusting the position of the centrosome during migration via modulation of microtubule stability.
During mitosis, spindle pole integrity and function are crucial for spindle orientation. In fact, loss of function of spindle pole proteins can affect astral microtubule dynamics and spindle orientation [6, 29–31]. Moreover, proper control of division orientation is required for the symmetric expansion of a central lumen in a growing acinus and for the development of a variety of organs (modeled in Figure 1E, [1, 32]). For these reasons, we tested if appendages were present in spindle poles/mitotic centrosomes and if cenexin and CEP164 localized to these structures, as they did in interphase cells. Mitotic cells processed for transmission electron microscopy (TEM) showed that one of the two centrioles in mitotic centrosomes contained both subdistal and distal appendages (Figure 3A). We next determined that cenexin and CEP164 displayed conventional appendage-like localization at the subdistal and distal appendages of mitotic centrioles suggesting that both CEP164 and cenexin maintained their appendage localization during mitosis (Figure 3B). In support of previous studies [33], cenexin accumulated primarily at appendages of the older centriole (Figure 3C, top, and quantification of >30 cells in Figure 3D, left). Interestingly, the intensity difference of CEP164 between the two spindle poles was not as significant as cenexin (Figure 3C and 3D). These findings suggest that differential localization of cenexin to the poles might contribute to proper spindle orientation [34].
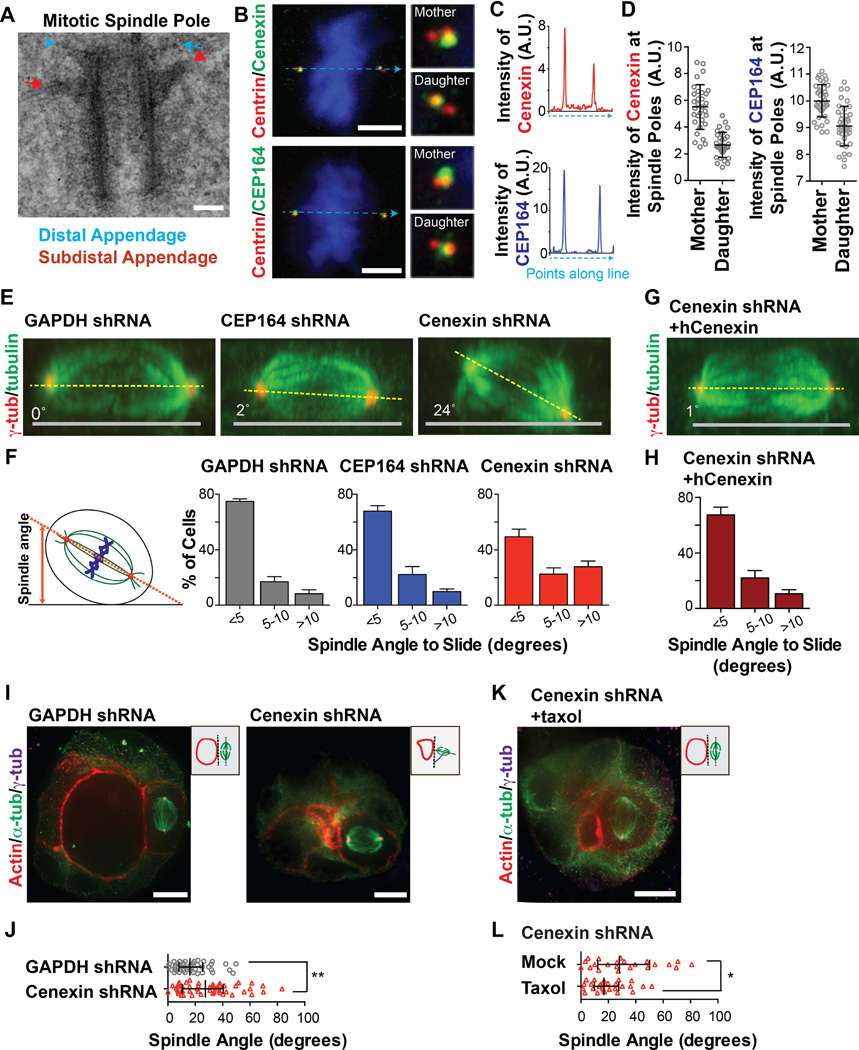
Figure 3. Cenexin depletion causes spindle orientation defects in mitosis that contribute to multiple lumina generation in acini cultures.
(A) Mitotic cells were collected by mitotic shake-off and processed for transmission electron microscopy. Shown is a representative mother centriole from a mitotic spindle. Scale bar, 0.1 µm. The distal appendage is highlighted by a blue arrowhead, and the subdistal appendage is highlighted by a red arrow.
(B) Immunofluorescence staining of U2OS cells at metaphase; centrioles (centrin, red), CEP164 or cenexin (green). Dashed line (blue) depicts where line-scan measurements were obtained for (C). Scale bar, 5 µm. Insets depict 4×-magnification of mother and daughter spindle poles.
(C) Line scans through two spindle poles of images in (B). Upper image, demonstrates the presence of more cenexin on one pole (the mother/older spindle pole) than the other. Bottom image, depicts similar amounts of CEP164 intensity across two spindle poles.
(D) The integrated intensity of cenexin (left) or CEP164 (right) at the mother and daughter spindle poles were measured in >30 mitotic cells (U2OS). Data are represented as mean ± SD of >30 cells in each graph.
(E and G) Orthogonal view (x–z) of metaphase MDCK cells stained for microtubules (α-tubulin, green), and spindle poles (γ-tubulin, red) depleted of GAPDH, CEP164 or cenexin (E), and subsequently rescued with shRNA-resistant human cenexin (G). Grey line depicts the culture substrate, and yellow-dashed lines portray the orientation of spindles to substrates. See also Figure S3A and S3C.
(F and H) Quantification showing a significant increase (>10°) in spindle angles from cells depleted of cenexin (3-fold) compared to control cells (GAPDH shRNA), while no significant change was seen in CEP164-depleted cells (F). Data are represented as mean ± SEM of 3 independent experiments with >25 cells measured in each treatment/experiment. Collective raw spindle angles are shown in Figure S3A and S3C.
(I and K) 3D-cultured MDCK acini were stained for actin (red), centrosomes (γ-tubulin, magenta), and microtubules (α-tubulin, green). From the left to right are control cells (GAPDH-depleted), cenexin-depleted cells (I), and cenexin-depleted cells treated with taxol (K). Scale bar, 10 µm.
(J) Raw spindle angles measured from acini in (I). Spindle angles were measured in GAPDH-depleted cells (n=43), and cenexin-depleted cells (n=50). **, p-value<0.01.
(L) Raw spindle angles measured from cenexin-depleted acini treated with taxol (n=27) or without taxol (n=22) for 2 hours (J). *, p-value<0.05.
CEP164 or cenexin was depleted and tested for changes in spindle positioning during mitosis (Figure 3E). Only cenexin-depleted cells showed variability in the orientation of their spindle to the substratum (Figure 3E, 3F, and S3A). More specifically, 50% of cenexin-depleted cells demonstrated a spindle angle >5°, whereas most control spindles were parallel to the substratum (<5°, 75% for control). CEP164-depleted cells displayed a modest difference compared to control (<5°, 68% for CEP164; Figure 3F and S3A). Importantly, spindle orientation defects could be rescued by expressing an shRNA-resistant form of cenexin (Figure 3E–3H and S3B–S3C). We observed similar defects in U2OS cells (Figure S2D) or when additional shRNAs were used (Figure S2E) further demonstrating that the subdistal appendage protein, cenexin, but not the distal appendage protein, CEP164, is essential for modulating spindle orientation.
We found that spindle misorientation in cenexin depleted cells was the likely cause of multi-lumina formation in acini (as in Figure 1F and 1G). In control acini (GAPDH depletion), the spindle was nearly parallel to the apical lumen in most cells (on average 18°, Figure 3I and 3J), suggesting that spindle alignment contribute to normal symmetric lumen expansion. However, in cenexin-depleted cells, a higher degree of spindle angle variability and an overall increase in spindle angle towards the apical lumen was observed (Figure 3I and 3J). Taken together, these results indicate that cenexin is required for control of spindle orientation and single lumen formation. Importantly, taxol treatment rescued spindle misorientation in cenexin-depleted cells, as there was a significant decrease in the spindle angle in relation to the lumen under these conditions (Figure 3K and 3L), This suggests that cenexin regulates microtubule stability that influences centrosome positioning in both non-dividing and mitotic cells.
To better understand the role of cenexin in spindle positioning we examined the overall morphology of the mitotic spindle in cenexin-depleted cells. Astral microtubule arrays in cells depleted of cenexin were disrupted when compared to control cells (GAPDH-depleted, Figure 4A). The plus-end microtubule binding protein EB1 was used to visualize astral microtubules and revealed a significant decrease in both astral microtubule number (30% of control, Figure 4C) and length (17% of control, Figure 4D) in cenexin depleted cells. Having observed defects in the astral microtubule plus ends (EB1 staining), we examined the minus ends at the poles by visualizing the minus-end binding protein, Kif2a [35]. Interestingly, we observed a significant defocusing of Kif2a at spindle poles in cenexin-depleted cells compared to controls (Figure 4B), while the presence of Kif2a at the spindle poles was unchanged (Figure S4A). This suggests that cenexin loss causes disorganization of microtubule minus-ends, which can lead to defects in anchoring, stability, and/or growth of astral microtubules. Cenexin depletion did not affect spindle pole localization of canonical spindle pole proteins involved in microtubule nucleation and spindle orientation, namely γ-tubulin, pericentrin, and the pericentrin binding protein, CEP215 (Figure S4B) [6, 25] nor did we observe any significant difference in cell cycling, mitotic progression, chromosome congression, kinetochore alignment or kinetochore-fiber formation under these conditions (Figure S4C–S4F, [36]). Thus, we conclude that spindle microtubules, as well as microtubule-nucleating and - depolymerizing activity at the spindle pole are not grossly affected in cenexin-depleted cells, whereas cenexin is required for proper organization of astral microtubules.
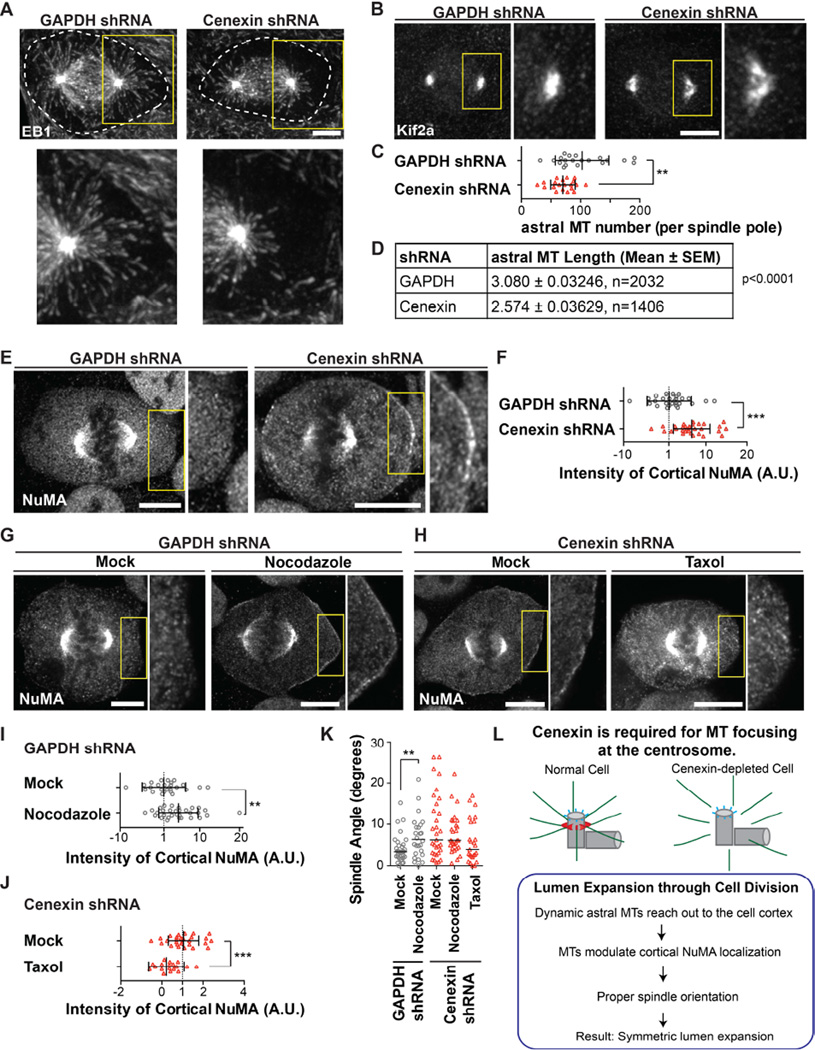
Figure 4. Cenexin depletion produces astral microtubule defects in mitosis causing NuMA mislocalization and spindle tilt.
(A–B) Metaphase spindles in MDCK cells were decorated with a plus-end microtubule marker, EB1 (A), or the minus-end microtubule marker, Kif2a (B). Dashed white line outlines edges of the cell (A). (A–B) Insets taken from yellow box and magnified 2× to illustrate the shortening of astral microtubules (A) or the defocusing of Kif2a (B). Scale bar, 5 µm.
(C) Astral microtubule number per spindle pole was determined. Data are represented as mean ± SD of 20 spindle poles. **, p-value<0.01.
(D) Astral microtubule length was measured from spindle pole to microtubule tip labeled with EB1. n>10 cells/group. n-values listed in chart are the number of individual astral microtubules measured.
(E) Shown is a single z-section of NuMA staining in a metaphase U2OS cell depleted of GAPDH or cenexin. Scale bar, 5 µm. Inset taken from yellow box was magnified 2× to illustrate the increase in cortical NuMA after cenexin depletion compared to control (GAPDH shRNA).
(F) The cortical integrated intensity of NuMA was measured in metaphase cells depleted of GAPDH or cenexin. Data are represented as mean ± SD of >28 cells/treatment. **, p-value<0.01.
(G) GAPDH-depleted U2OS cells were treated with nocodazole and labeled for NuMA. Inset taken from yellow box was magnified 2× to illustrate the increase in cortical NuMA when disrupting astral microtubules with nocodazole treatment. Scale bar, 5 µm. See also Figure S4H and S4I.
(H) Cenexin-depleted U2OS cells were treated with taxol and labeled for NuMA. Inset taken from yellow box was magnified 2× to illustrate the decrease in cortical NuMA when stabilizing astral microtubules with taxol treatment. Scale bar, 5 µm. See also Figure S4J and S4K.
(I–J) The cortical integrated intensity of NuMA was measured in metaphase cells depleted of GAPDH (I), or cenexin (J). Data are represented as mean ± SD of >20 cells/treatment. **, p-value<0.01. ***, p-value<0.001.
(K) Spindle angles were measured in GAPDH- and cenexin-depleted metaphase cells treated with or without taxol or nocodazole (as depicted). Data are represented as median of >25 cells/treatment. **, p-value<0.01.
(L) A model portraying cenexin functions in microtubule focusing and mitotic spindle orientation.
The interaction between astral microtubules and the cell cortex has been shown to direct spindle orientation. This process involves the NuMA/LGN/Gαi complex (NuMA complex), which is transiently organized at the cell cortex during metaphase, to assist in astral microtubule capture [4]. Since we observed spindle misorientation (Figure 3E–3L) and astral microtubule defects in cenexin-depleted cells (Figure 4A–4D), we hypothesized that cenexin depletion could disrupt the distribution of the NuMA complex at the cell cortex. Interestingly, we found that cenexin depletion in two separate cell lines (MDCK and U2OS) increased cortical NuMA localization when compared to control cells (Figure 4E, 4F, and S4G). These findings suggested that loss of astral microtubules caused by cenexin depletion, did not impede NuMA recruitment to the cell cortex, but instead, increased NuMA localization at this site.
NuMA may not require astral microtubules for delivery to the cell cortex [37], but instead, it may require astral microtubules to dissociate from the cortex [38]. In fact, in our system, a second approach suggested the requirement for astral microtubules in cortical NuMA dissociation. Control cells treated with nocodazole to selectively destabilize astral microtubules (Figure S4H) [26] showed a 5-fold increase in NuMA at the cell cortex compared to vehicle-treated cells (Figure 4G and 4I). This closely mimicked the phenotype of cenexin depletion (Figure 4E and 4F).
To further confirm that cenexin depletion affects the stability/growth of astral microtubules and therefore impedes NuMA dissociation from the cell cortex, taxol was used to stabilize astral microtubules (Figure S4J) [27]. A significant reduction in cortical NuMA was observed under these conditions compared to mock treated cenexin depleted cells (Figure 4H and 4J). The taxol treatment rescued the spindle misorientation phenotype in cenexin-depleted cells (1.6-fold, Figure 4K and S4K), while low-dose nocodazole treatment caused spindle misorientation in control cells (Figure 4K and S4I). Thus, we argue that astral microtubules loss observed in cenexin-depleted cells causes NuMA mislocalization at the cell cortex and spindle misorientation.
We present a model wherein the cenexin-regulated subdistal appendages are required for microtubule organization at the mother centriole (Figure 4L) [39, 40]. In line with previous findings [8, 10], cenexin depletion causes subdistal appendage protein loss with no overt defects to distal appendages (Figure 1B–1D). We conclude that cenexin-regulated-subdistal appendages and not distal appendages are specifically required for centrosome positioning both in interphase cells for proper directed migration (Figure 2), and during mitosis for appropriate placement and orientation of the mitotic spindle (Figure 3). In addition, our findings suggest that cenexin affects a specific pool of microtubules, namely astral microtubules, that influences spindle orientation and modulates localization of NuMA at the cell cortex (Figure 4). We reason that this regulation is required for apical-basal axis orientation and epithelial lumen positioning (Figure 3E–3L; modeled in Figure 4L).
Supplementary Material
Highlights.
Cenexin controls centrosome positioning during cell migration.
Cenexin is required for spindle orientation.
Cenexin is essential for proper lumen formation.
Cenexin controls the above activities by modulating MT organization and stability.
Acknowledgments
We thank Christine Powers (UMMS) for the help of EM processing and imaging, Joshua van Kleef and Yu-Shan Hung (ANU) for the help of Matlab software, YoonJeung Chang and Wendy Zimmerman (UMMS, Doxsey Lab) for reading this manuscript. The following grants supported this work S10RR027897 (TEM, UMMS), R01 GM051994-14 (S.D.), and R00 GM107355 (H.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Overeem AW, Bryant DM, Ijzendoorn SCD Van. Mechanisms of apical – basal axis orientation and epithelial lumen positioning. Trends Cell Biol. 2015:1–10. doi: 10.1016/j.tcb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 2.McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21:727–735. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Fraticelli AE, Auzan M, Alonso MA, Bornens M, Martin-Belmonte F, Rodríguez-Fraticelli AE, Auzan M, Alonso MA, Bornens M, Martín-Belmonte F. Cell confinement controls centrosome positioning and lumen initiation during epithelial morphogenesis. J. Cell Biol. 2012;198:1011–1023. doi: 10.1083/jcb.201203075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotak S, Gönczy P. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr. Opin. Cell Biol. 2013;25:741–748. doi: 10.1016/j.ceb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 2008;183:625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C-T, Hehnly H, Yu Q, Farkas D, Zheng G, Redick SD, Hung H-F, Samtani R, Jurczyk A, Akbarian S, et al. A Unique Set of Centrosome Proteins Requires Pericentrin for Spindle-Pole Localization and Spindle Orientation. Curr. Biol. 2014;24:2327–2334. doi: 10.1016/j.cub.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soung N-K, Park J-E, Yu L-R, Lee KH, Lee J-M, Bang JK, Veenstra TD, Rhee K, Lee KS. Plk1-dependent and - independent roles of an ODF2 splice variant, hCenexin1, at the centrosome of somatic cells. Dev. Cell. 2009;16:539–550. doi: 10.1016/j.devcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr. Biol. 2012;22:1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 2003;161:535–545. doi: 10.1083/jcb.200301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MFB. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hehnly H, Hung H-F, Doxsey S. One among many: ODF2 isoform 9, a.k.a. Cenexin-1, is required for ciliogenesis. Cell cycle. 2013;12:1021. doi: 10.4161/cc.24330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang J, Seo SG, Lee KH, Nagashima K, Bang JK, Kim BY, Erikson RL, Lee K-W, Lee HJ, Park J-E, et al. Essential role of Cenexin1, but not Odf2, in ciliogenesis. Cell cycle. 2013;12:655–662. doi: 10.4161/cc.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg Ea. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrøder J, Rogowski M, Jakobsen L, Vanselow K, Geimer S, Pedersen L, Andersen J. Identification and characterization of two novel centriolar appendage component proteins. Cilia. 2012;1:44. [Google Scholar]
- 15.Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 2012;199:1083–1101. doi: 10.1083/jcb.201202126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elric J, Etienne-Manneville S. Centrosome positioning in polarized cells: common themes and variations. Exp. Cell Res. 2014;328:240–248. doi: 10.1016/j.yexcr.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Luxton GG, Gundersen GG. Orientation and function of the nuclear–centrosomal axis during cell migration. Curr. Opin. Cell Biol. 2011;23:579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 2014;15:225–242. doi: 10.1038/nrm3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Théry M, Racine V, Piel M, Pépin A, Dimitrov A, Chen Y, Sibarita J-B, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibi M, Zou P, Inoko A, Shiromizu T, Matsuyama M, Hayashi Y, Enomoto M, Mori D, Hirotsune S, Kiyono T, et al. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J. Cell Sci. 2011;124:857–864. doi: 10.1242/jcs.075705. [DOI] [PubMed] [Google Scholar]
- 22.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doxsey SJ, Steln P, Evans L, Calarco PD, Kirschnefi M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 24.Dictenberg JB, Zimmerman W, Sparks Ca, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gama-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasquez RJ, Howell B, Yvon aM, Wadsworth P, Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell. 1997;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yvon A-MC, Wadsworth P, Jordan MA. Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol. Biol. Cell. 1999;10:947–959. doi: 10.1091/mbc.10.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palazzo A, Ackerman B, Gundersen GG. Cell biology: Tubulin acetylation and cell motility. Nature. 2003;421:230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]
- 29.Delaval B, Bright A, Lawson ND, Doxsey S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nat. Cell Biol. 2011;13:461–468. doi: 10.1038/ncb2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hehnly H, Canton D, Bucko P, Langeberg LK, Ogier L, Gelman I, Santana LF, Wordeman L, Scott JD. A mitotic kinase scaffold depleted in testicular seminomas impacts spindle orientation in germ line stem cells. Elife. 2015;4:1–22. doi: 10.7554/eLife.09384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hehnly H, Doxsey S. Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev. Cell. 2014;28:497–507. doi: 10.1016/j.devcel.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blasky AJ, Mangan A, Prekeris R. Polarized protein transport and lumen formation during epithelial tissue morphogenesis. 2015:1–17. doi: 10.1146/annurev-cellbio-100814-125323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange BMH, Gull K. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 1995;130:919–927. doi: 10.1083/jcb.130.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita YM. The centrosome and asymmetric cell division. Prion. 2009;3:84–88. doi: 10.4161/pri.3.2.8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganem NJ, Compton Da. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasic I, Nerurkar P, Meraldi P. Centrosome age regulates kinetochore–microtubule stability and biases chromosome missegregation. Elife. 2015;4:1–15. doi: 10.7554/eLife.07909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotak S, Busso C, Gönczy P. NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 2014;33:1815–1830. doi: 10.15252/embj.201488147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Z, Wan Q, Liu J, Zhu H, Chu X, Du Q. Evidence for dynein and astral microtubule-mediated cortical release and transport of Gαi/LGN/NuMA complex in mitotic cells. Mol. Biol. Cell. 2013;24:901–913. doi: 10.1091/mbc.E12-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 40.Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, et al. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 2012;148:189–200. doi: 10.1016/j.cell.2011.10.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.