Abstract
We describe a case of hemi-atrophy in a young adult male, with a positive family history of three maternal uncles with Duchenne muscular dystrophy (DMD). The patient showed progressive weakness localized to the left side, an abnormal electromyography, and creatine kinase levels >3000 IU/l. Muscle biopsy showed both dystrophin-positive and -negative myofibers. An out-of-frame duplication variant in DMD, that is, c.(93+1_94-1)_(649+1_650-1)dup(p.?) resulting in duplication of exons 3–7 was inherited, but the muscle biopsy showed dystrophin mRNA with and without the duplication. Dystrophin quantification using mass spectrometry showed 25% normal dystrophin protein levels in the muscle biopsy from the stronger right side. Sex chromosome aneuploidy was ruled out. We conclude that the patient inherited the duplication variant, but early in development an inner cell mass underwent a somatic recombination event removing the duplication and restoring dystrophin expression. To our knowledge, this is the first report of a reversion leading to somatic mosaicism in DMD.
Introduction
Duchenne muscular dystrophy (DMD; MIM#310200) is a severe X-linked recessive disease characterized by progessive muscular degeneration. It is caused due to out-of-frame mutations in the DMD gene encoding for dystrophin protein, whereas in-frame mutations result in a milder Becker's muscular dystrophy phenotype.1, 2 Small areas of isolated somatic revertant fibers (ie, dystrophin-positive fibers) in muscle of DMD patients as well as in mouse models have been observed.3, 4, 5 In rare cases, somatic mosaicism has been observed for DMD, with patients presenting with cardiomyopathy and further molecular studies detect dystrophin-positive and dystrophin-negative cells.6, 7
Here we report what we believe is the first case of somatic mosaicism due to a reversion variant, leading to a hemi-atrophy phenotype in a patient with a family history of DMD.
Case report
The proband presented with hemi-atrophy and weakness, largely limited to the left side (Figure 1a). Electromyography was consistent with a dystrophic myopathy, as were creatine kinase levels (>3000 IU/l; n<200 IU/l). Cytogenetic analyses were normal. Molecular testing was performed by quantitative dosage analysis of genomic DNA by a standardized test offered by Athena diagnostics (Marlborough, MA, USA). This test showed presence of a duplication c.(93+1_94-1)_(649+1_650-1)dup(p.?) in the DMD gene (NM_004006.2), with a dystrophinopathy phenotype predicted. As the duplication break points remain uncharacterized, the exon DNA change is represented as 2i_7i, and the RNA change results in duplication of exons 3–7, duplication of exons 3–7 is predicted to cause an out-of-frame mutation usually associated with DMD.
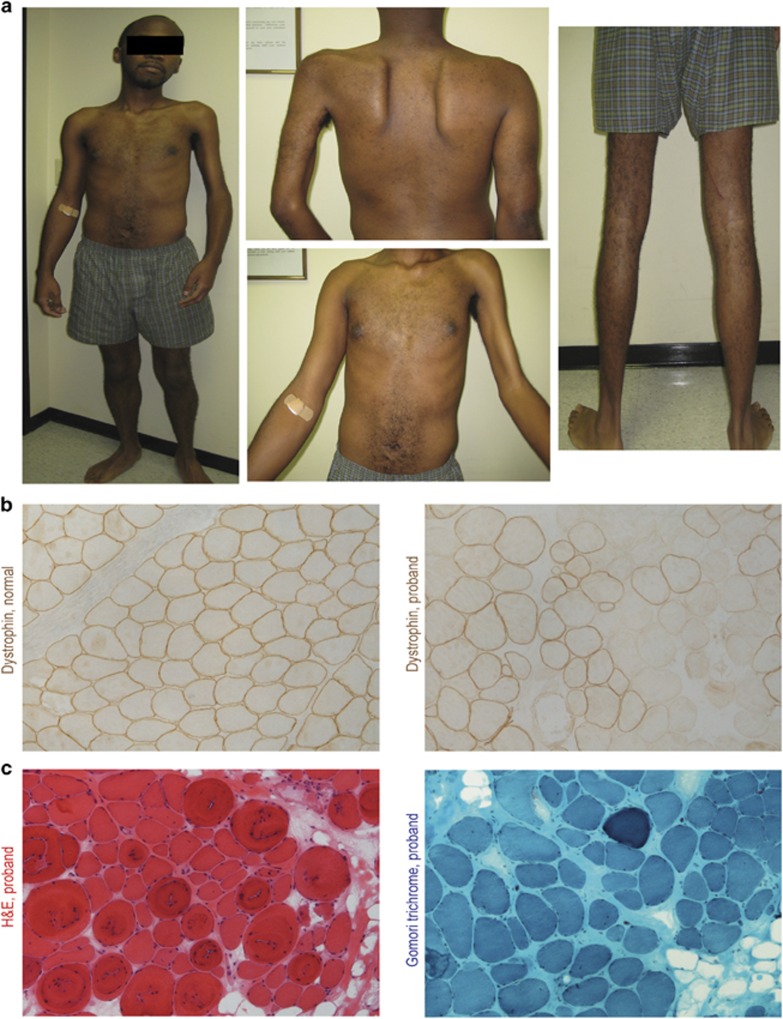
Figure 1.
Hemi-atrophy in a young adult associated with somatic mosaicism for dystrophin in muscle biopsy. (a) Marked muscle wasting and weakness is seen restricted to the left side. (b) Dystrophin immunostaining shows normal patterns at myofiber membranes in normal control, and mosaic patterns of dystrophin-positive and -negative fibers in the proband. (c) Histopathology of the proband by hematoxylin and eosin staining (left), and Gomori trichrome (right) shows myofiber size variation, variable endomysial fibrosis, hypercontracted fibers and central nuclei, consistent with a dystrophic myopathy. The full colour version of this figure is available at European Journal of Human Genetics online.
A muscle biopsy was taken from the stronger right deltoid when the patient was 22 years, and dystrophin immunostaining of cryosections showed a mosaic pattern with populations of both dystrophin-positive and -negative myofibers (Figure 1b). This pattern is typically seen with female carriers of DMD,8 however, the proband was male. Histopathology showed a remarkably variable dystrophic myopathy, with some regions showing normal histology, and others a marked dystrophic picture by both hematoxylin eosin and Gomori's-modified trichome methods (Figure 1c).
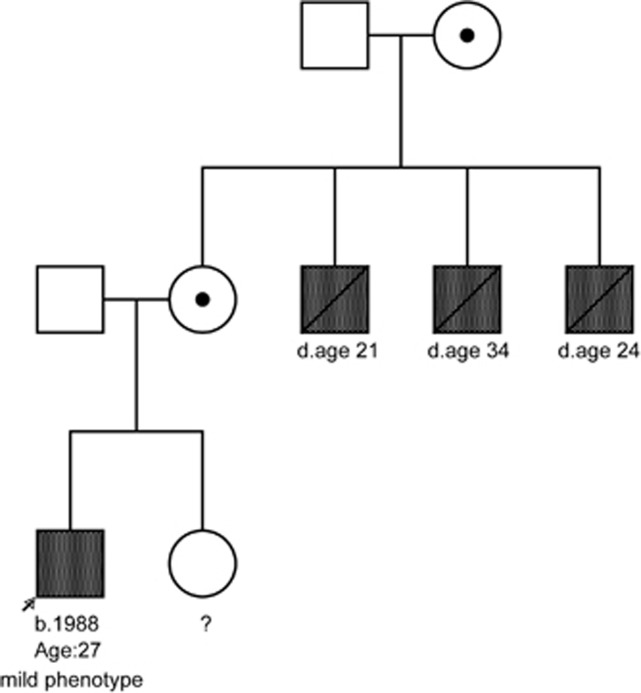
Family history was reported by the patient as positive for severe muscular dystrophy, with three maternal uncles with clinical pictures consistent with DMD (Figure 2). The maternal uncles died at 21, 24, and 34 years of age.
Figure 2.
Pedigree showing three maternal uncles affected with severe muscular dystrophy. Proband shows a mild dystrophinopathy phenotype.
As the proband showed an asymmetric clinical picture and positive family history, we hypothesized that a somatic reversion event early during the patient's development may have corrected the exon 3–7 DMD duplication in a subset of myogenic cells.
Methods
Proband was recruited under protocol 2405 approved by the Office for the Protection of Human Subjects at Children's National Medical Center, Washington, DC, USA. Details of the duplication variant identified have been uploaded to the Leiden open variation database: http://www.LOVD.nl/DMD (patient ID #0033321).
Total RNA was extracted from muscle biopsy and converted to cDNA using a protocol described earlier.9 RT-PCR primers (Supplementary Table 1) were designed using Primer3 software10 with NM_004006.2 as reference sequence; exon numbering was based on NG_012232.1 and White et al.11 RT-PCR was carried out using Accuprime Super Mix II (Life Technologies, Carlsbad, CA, USA). RT-PCR products were run on an agarose gel and extracted, purified using QIAquick gel extraction kit (Qiagen, Germantown, MD, USA) and sequenced. Dystrophin quantification using a stable isotope standard and mass spectrometry was performed on the muscle biopsy as described previously.12
Results
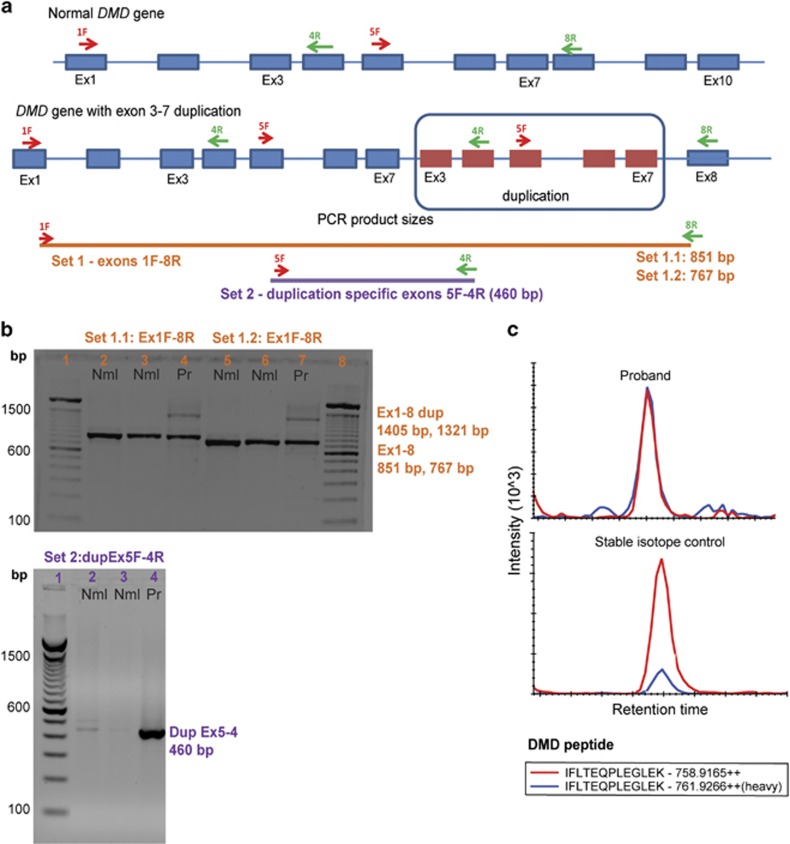
RT-PCR primer sets were designed to determine the structure of the dystrophin mRNA in the proband's muscle tissue vs normal controls (Figure 3a). As the proband's DNA duplication variant involved exons 3–7, RT-PCR primers designed in exons 1 and 8 were predicted to amplify both the normal dystrophin transcript as well as the duplicated transcript. As expected, two normal controls showed the expected dystrophin transcript using two different exon 1 primers, with a shared exon 8 primer (851 and 767 bp; Figure 3b). The proband showed two bands, corresponding to both the normal transcript, as well as a novel band corresponding to the duplication (1405 and 1321 bp; Figure 3b).
Figure 3.
mRNA studies of proband muscle show loss of familial duplication mutation in dystrophin transcript. (a) RT-PCR primer sets were designed to cover regions present within or outside of the exon 3–7 duplication. Primer sets 1.1 and 1.2 cover DMD exons 1–8. Primer set 2 is specific for the duplicated region with exon 5F as forward primer and exon 4R as reverse primer. (b) Agarose gel image showing RT-PCR products from primer sets using cDNA from two normal samples (Nml) and proband (Pr). Primer sets 1.1 and 1.2 cover DMD exons 1–8 with expected sizes of 851 and 767 bp, respectively. The proband shows both a normal band (Ex1-8) similar to the normal samples, as well as presence of a larger duplicated transcript (Ex1-8dup) with sizes of 1405 and 1321 bp for the primer sets, as expected. Set 2 duplication-specific RT-PCR primers detected the duplicated mRNA transcript in the proband only (DupEx5-4) with an expected size of 460 bp, which was observed. All bands were sequenced and confirmed predicted exonic structures. Faint bands seen in normal controls with the DupEx5-4 primers were also Sanger sequenced and did not align to the duplication-specific PCR region (non-specific products). (c) Quantification of dystrophin protein in proband muscle (top panel) and normal control (bottom panel). Five peptides were quantified and averaged (single-peptide exemplar shown). The proband dystrophin level was quantified at ~25.7% of normal. The full colour version of this figure is available at European Journal of Human Genetics online.
To confirm the structure of the proband's mRNA containing the duplication predicted by the DNA variant data, a forward primer was used for exon 5 and reverse primer for exon 4: this would be predicted to only amplify across the novel duplication break point (Figure 3; set 2). As expected, a 460-bp RT-PCR product was seen in the proband, but not normal controls. Sequence analysis of each RT-PCR band confirmed the predicted exonic arrangements. This data indicated that the patient was producing two distinct dystrophin mRNA transcripts; one normal transcript and one duplicated transcript. The most likely event was a somatic recombination event early in development, leading to reversal (reversion) of the duplication and somatic mosaicism for normal and DMD myofibers.
Proband dystrophin levels in the stronger right side muscle biopsy were quantified at ~25.7% of normal levels using a mass spectrometry assay (Figure 3c).
Discussion
We report the first case of a reversion of a familial DMD variant, leading to somatic mosaicism and a hemi-atrophy phenotype. Our lab previously reported somatic mosaic dystrophinopathy in an isolated case of exertional heart failure (cardiomyopathy), where a de novo mutation occurred during early development lead to a mixture of normal and mutant cells.6 The cardiomyopathy patient showed high levels of dystrophin-positive cells in muscle, and did not show significant muscle symptoms. About half of DMD patient muscle biopsies show somatic revertant myofibers, where rare somatic mutations lead to rare (<1%) dystrophin-positive myofibers in otherwise dystrophin-negative muscle.3, 4, 5 In the case presented here, a reversion event likely happened very early in the development of the myogenic lineage, where the inherited duplication was lost, reverting to the normal sequence. Through subsequent embryonic development, the right side of the patient showed ~25% dystrophin-positive cells precluding most symptoms; whereas the left side likely showed a much lower percentage of dystrophin-positive myofibers (an additional biopsy from the affected side was not possible). RT-PCR, sequencing and quantitative dystrophin protein analyses confirmed this model. An alternative model of Klinefelter's syndrome (XXY carrier state for DMD with X-inactivation) was ruled out by cytogenetic studies.
Reverse somatic mosaics in other monogenic diseases have been seen when there is a selection bias for revertant (normal) cells, such as in blood disorders with clonal expansion of cell subsets in the bone marrow.13 However, reversions of monogenic disorders are otherwise rare.14, 15 The nature of the patient's duplication variant facilitated reversion with homologous recombination leading to loss of the duplication early in development.
Acknowledgments
We would like to thank the patient for participating in this study. This work was supported by grants from the National Institutes of Health 3R01NS29525 (EPH). JP is a predoctoral student in the Molecular Medicine Program of the Institute for Biomedical Sciences at the George Washington University. This work is from a dissertation to be presented to the above program in partial fulfillment of the requirements for the PhD degree.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Hoffman EP, Brown RH, Kunkel LM: Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 1987; 51: 919–928. [DOI] [PubMed] [Google Scholar]
- Beggs AH, Hoffman EP, Snyder JR et al: Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am J Hum Genet 1991; 49: 54–67. [PMC free article] [PubMed] [Google Scholar]
- Fanin M, Danieli GA, Vitiello L, Senter L, Angelini C: Prevalence of dystrophin-positive fibers in 85 Duchenne muscular dystrophy patients. Neuromuscul Disord 1992; 2: 41–45. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Morgan JE, Watkins SC, Partridge TA: Somatic reversion/suppression of the mouse mdx phenotype in vivo. J Neurol Sci 1990; 99: 9–25. [DOI] [PubMed] [Google Scholar]
- Yokota T, Lu QL, Morgan JE et al: Expansion of revertant fibers in dystrophic mdx muscles reflects activity of muscle precursor cells and serves as an index of muscle regeneration. J Cell Sci 2006; 119 (Pt 13): 2679–2687. [DOI] [PubMed] [Google Scholar]
- Kesari A, Neel R, Wagoner L, Harmon B, Spurney C, Hoffman EP: Somatic mosaicism for Duchenne dystrophy: Evidence for genetic normalization mitigating muscle symptoms. Am J Med Genet A 2009; 149A: 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deburgrave N, Daoud F, Llense S et al: Protein- and mRNA-based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Hum Mutat 2007; 28: 183–195. [DOI] [PubMed] [Google Scholar]
- Rajakulendran S, Kuntzer T, Dunand M et al: Marked hemiatrophy in carriers of Duchenne muscular dystrophy. Arch Neurol 2010; 67: 497–500. [DOI] [PubMed] [Google Scholar]
- Kesari A, Pirra LN, Bremadesam L et al: Integrated DNA, cDNA, and protein studies in Becker muscular dystrophy show high exception to the reading frame rule. Hum Mutat 2008; 29: 728–737. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ: Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds): Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press, 2000, pp 365–386. [DOI] [PubMed] [Google Scholar]
- White SJ, den Dunnen JT: Copy number variation in the genome; the human DMD gene as an example. Cytogenet Genome Res 2006; 115: 240–246. [DOI] [PubMed] [Google Scholar]
- Brown KJ, Marathi R, Fiorillo AA et al: Accurate quantitation of dystrophin protein in human skeletal muscle using mass spectrometry. J Bioanal Biomed 2012; Suppl 7: 001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M: Mutations to the rescue. Nat Med 2011; 17: 405–407. [DOI] [PubMed] [Google Scholar]
- Hall JG: Review and hypotheses: somatic mosaicism: observations related to clinical genetics. Am J Hum Genet 1988; 43: 355–363. [PMC free article] [PubMed] [Google Scholar]
- Wahn V, Stephan V, Hirschhorn R: Reverse mutations—spontaneous amelioration or cure of inherited disorders? Eur J Pediatr 1998; 157: 613–617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.