Abstract
Cancer patients participating in studies involving experimental or diagnostic next-generation sequencing (NGS) procedures are confronted with the possibility of unsolicited findings. The Center for Personalized Cancer Treatment (CPCT), a Dutch consortium of cancer centers, is offering centralized large-scale NGS for the discovery of somatic tumor mutations with their germline DNA as reference. The CPCT aims to give all cancer patients with advanced disease stages access to tumor DNA analysis in order to improve selection for experimental therapy. In this article, our experiences at the CPCT will serve as an example to discuss the ethical and practical aspects regarding the management of unsolicited findings in personalized cancer research and treatment. Generic issues, relevant for all researchers in this field are discussed and illustrated by description of three patients faced with an unsolicited DNA finding, while they intended to be candidate for future anticancer treatment by participating in a trial that included NGS of both somatic and germline DNA. As options for DNA analysis expand and costs decrease rapidly, more and more patients are offered large-scale NGS testing. After reviewing current recommendations in literature, we conclude that classical informed consent procedures need to be adapted to become more explicit in asking patients if they want to be informed about unsolicited findings and if so, what level of detail of genetic risk information exactly they want to be returned after the analysis.
Introduction
In the era of personalized cancer treatment, large-scale genetic analysis of tumors is considered to be key for a better selection of patients for an appropriate anticancer therapy. Owing to this development, cancer treatment is moving onward from only organ based, one size fits all medicine, to specific anticancer treatments based on specific somatic genetic mutations.
With the rapid development of next-generation sequencing (NGS), it is now possible and affordable to sequence individual genomes in a short period of time to identify somatic genetic alterations.1
Being a powerful diagnostic tool, the introduction of NGS is accompanied by ethical challenges. As it is still important to sequence germline DNA, as well as tumor DNA, to identify true somatic tumor DNA mutations in an individual patient, one of these challenges is how to deal with genetic risk information that is inevitably generated by these tests and which may have potential medical, psychological, financial and social consequences.2 These genetic findings are also challenging for laboratories performing whole-genome sequencing, because many variants are not (yet) considered to be clinically relevant. Only a minority of variants is of direct clinical importance for patients and their family members.3
In some cases, the returning of genetic risk information after a NGS procedure is essential because of the possible impact and challenges for patients and their relatives. How this should be done is the subject of an ongoing debate. Like others,4, 5 we are convinced that we have the responsibility to offer research participants the option to be notified of findings that potentially affect a person's health or may prevent significant harm.
The additional genetic information that is found in the search for better selection of antitumor treatment has many different annotations. Here we prefer the use of 'unsolicited' findings, because this describes that this finding was discovered unintentionally, as a by-product of a research question.6 However, the terms ‘secondary' findings or ‘incidental' findings are also widely used.
The Center for Personalized Cancer Treatment (CPCT) is a Dutch consortium including the Netherlands Cancer Institute (NKI), 8 University Hospitals and over 10 large teaching hospitals in the Netherlands. The mission of this consortium is to improve treatment outcome and patient care in the field of oncology and avoid unnecessary exposure to side effects. In particular, the CPCT aims to give all cancer patients with advanced disease stages access to tumor and germline DNA analysis in order to improve selection for therapy. CPCT offers biopsies and sequencing in patients undergoing standard of care targeted treatment to generate a database. From 2011 until August 2015, we have taken tumor biopsies and blood samples of over 600 late-stage cancer patients over the past 3 years and NGS has been performed in >370 tumor samples. The occurrence of unsolicited findings has proven to be not hypothetical, which was shown recently by findings in three patients who urged us to revisit the CPCT policy regarding disclosure of genetic risk information. In this article, our experiences at the CPCT will serve as an example to discuss the ethical and practical aspects regarding the management of unsolicited findings in personalized cancer research and treatment. This may also guide other consortia when setting up NGS testing.
NGS at the CPCT
The CPCT offers large-scale NGS-based tumor diagnostics as of 2011. First, a comprehensive test for ‘actionable' mutations based on the IonTorrent Personal Genome Machine (Life Technologies, Carlsbad, CA, USA) and Illumina MiSeq (Illumina, Hayward, CA, USA) has been adapted, which covers hotspot mutations in oncogenes and complete coding sequences of tumor suppressors but also allows for the detection of relevant copy number amplifications. This was offered as a diagnostic test. Second, we offered a so-called targeted mini-cancer genome sequencing panel, involving approximately 2000 cancer-related genes. As sequencing output is increasing and running costs are decreasing, we have recently moved toward whole-exome analysis. For correlation purposes to identify true somatic mutations, both tumor and germline DNA are sequenced on the same panels. In the near future, whole-genome sequencing will be implemented.
Methods
Unsolicited findings and the CPCT disclosure policy
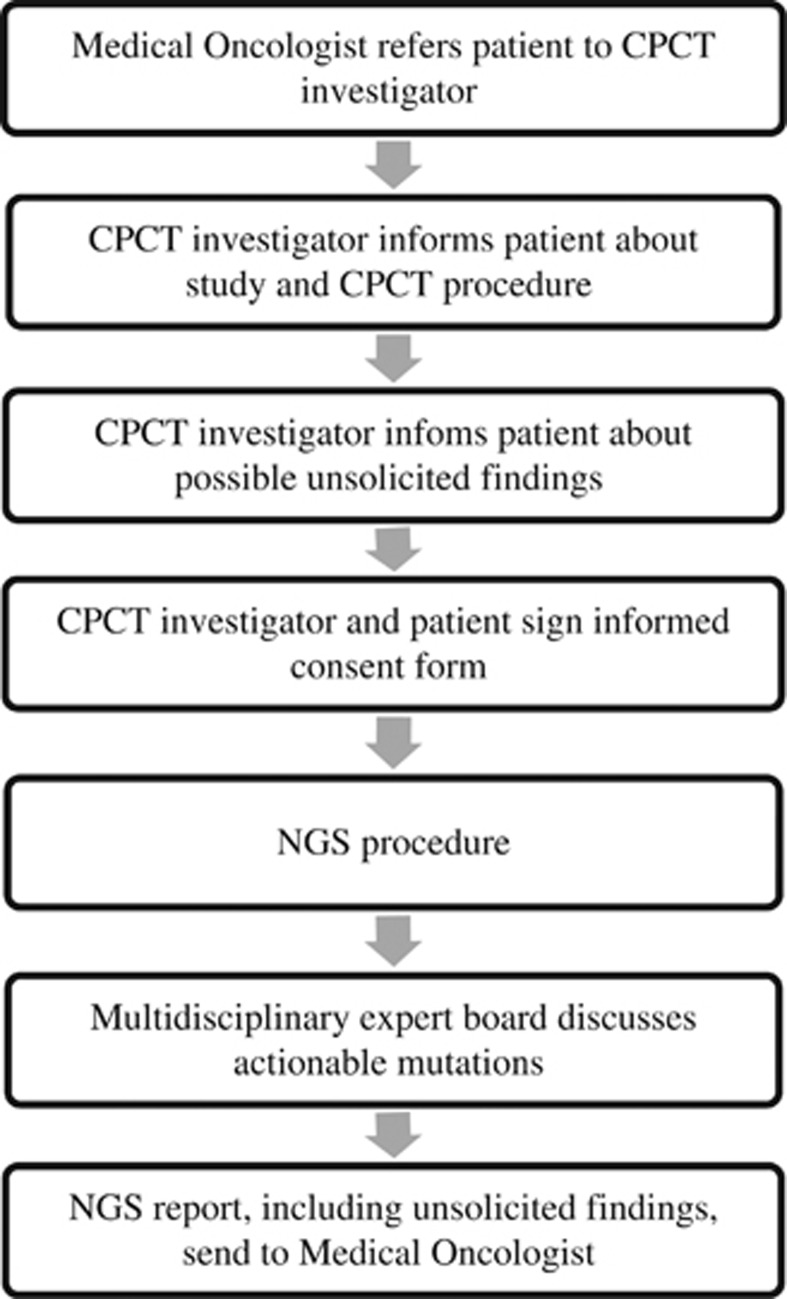
Patients who undergo tumor biopsies for NGS within the CPCT consortium, all have advanced staged cancer. Our procedure is visualized in Figure 1. Main selection criteria are: age ≥18 year, locally advanced (irresectable) or metastatic cancer from a solid tumor, indication for systemic treatment with anticancer agents, evaluable disease (by for instance radiological imaging, physical examination and/or blood tumor marker), safe biopsy of a metastatic or locally advanced lesion possible, expected adequacy to follow-up and a written informed consent.
Figure 1.
The CPCT NGS procedure.
First, the treating medical oncologist will refer his or her patient to a CPCT investigator at the local institute. This CPCT investigator is a physician and preferably someone else than the patient's own medical doctor to reduce the therapeutic misconception, which occurs when someone misunderstands the distinction between the aims of a scientific study and clinical care.7 Then, the CPCT investigator informs the patient about the aims of the intended study, the study-related procedures (including the biopsy procedure and blood draw to obtain tumor DNA, as well as germline DNA) and the possibility of discovering unsolicited genetic findings. After the patient has had a reasonable time to consider participation in a NGS procedure, patients willing to participate sign informed consent. After written informed consent is obtained, the CPCT investigator subsequently initiates baseline screening to determine CPCT study enrollment. After definite trial inclusion, a blood sample and a biopsy from a metastatic lesion are taken, both as part of the CPCT study-related procedures. Snap-frozen biopsy material and blood samples are transported to the central core facility at the department of Pathology at the University Medical Center Utrecht for centralized histological assessment and DNA analysis. DNA sequencing and variant reporting is performed in the ISO15189-certified genome diagnostics lab of the Medical Genetics Department. Reliable genetic variants are reviewed in a multidisciplinary team involving CPCT investigators, bioinformaticians, pathologists, medical oncologists, molecular geneticists and clinical geneticists to discuss possible actionable somatic mutations. This information is then reported back to the treating medical oncologist in order to inform their patients about potential treatment options. Patients with subsequent identified unsolicited findings are offered a referral to a clinical geneticist for further counseling and validation of the genetic variant in a second blood sample. This second blood sample is then analyzed in the DNA diagnostic laboratory of a Clinical Genetic Center.
Before 2014, if the patient consented to be informed about unsolicited findings which could lead to an increased risk of the development of cancer in their relatives, these (germline) findings were to be disclosed by their treating medical oncologist during a consultation. The policy of the research ethics committee (REC) regarding the return of research results, however, has been to return clinically relevant and actionable unsolicited findings from studies, genetic findings included. Participants who do not want to receive these results were excluded from participation in order to prevent the researcher facing a serious dilemma when confronted with imaging findings or genetic risk information that may be of interest to the participant. Therefore, we amended the study protocol and informed consent forms in 2014 in order to align to the REC policy.
Hence, an opt out option is no longer available. Patients who do not want their genetic risk information returned, currently have to decide either not to participate in the trial or to consent with receiving unsolicited findings. Currently, the suitability of this disclosure policy is under discussion, as the emergence of NGS has changed the circumstances under which many disclosure policies were designed.8
Results
From January 2011 until August 2015, 3 out of 376 patients participating in CPCT trials were confronted with unsolicited findings derived from a NGS procedure of their tumor biopsies with matching germline blood samples. They participated in the NGS procedure with the hope that possible genetic information could be identified as a target for anticancer drugs, which would allow specific treatment options. All three gave informed consent when an opt out on the return of unsolicited findings was available. Until August 2015, a total of 376 patients signed informed consent and underwent tumor biopsy. Table 1 shows the characteristics of 185 patients whose tumor biopsies are already sequenced.
Table 1. Patient characteristics.
| Total patients sequenced in August 2015 | n=185 |
|---|---|
| Age | 63 (33–89) years |
| Gender | |
| Male | 103 (55.7%) |
| Female | 82 (44.3%) |
| Ten most common cancer diagnosisa | |
| Melanoma | 31 (16.8%) |
| Colorectal | 29 (15.7%) |
| Breast | 23 (12.4%) |
| Sarcoma | 9 (4.9%) |
| Liver | 8 (4.3%) |
| Kidney | 8 (4.3%) |
| Esophageal | 8 (4.3%) |
| Pancreatic | 7 (3.8%) |
| CUP | 7 (3.8%) |
| Lung | 5 (2.7%) |
| Other cancer diagnosis | 50 (27.0%) |
All patients are advanced staged cancer patients.
The first patient with ovarian cancer participated in a CPCT study and underwent a tissue biopsy to retrieve tumor material and a blood draw for germline NGS testing. A germline BRCA2 1-bp deletion was reported. Patient had consented to be informed about possible unsolicited findings arising from the NGS procedure and was informed about the results by her medical oncologist. An appointment with a genetic counselor was made. The germline BRCA2 1-bp deletion was confirmed with a second specific and validated test in DNA extracted from a new blood sample. Patient 1 was relieved to hear the cause of her illness and immediately informed her family members and encouraged further investigations. She did so because she felt the urge to warn her relatives for the risks of breast and ovarian cancer. Recently, her daughter, who opted for predictive DNA testing and was diagnosed a mutation carrier too, has undergone prophylactic mastectomy to reduce her risk of breast cancer. At the time patient 1 was sequenced, in the Netherlands, screening for BRCA mutation was not routine for ovarian cancer patients. This patient was the first person in her family to be diagnosed with ovarian cancer; no family members with breast cancer were known and because of her age at diagnosis (above 60) she was not routinely referred for a diagnostic BRCA testing. As this BRCA mutation would not have been detected if she had not participated in this study, we consider this an unsolicited finding.
At this moment, patient 1 has stable disease after her first-line chemotherapy. Once disease progression occurs, she will be eligible for poly (ADP-ribose) polymerase inhibition.
The second patient with metastatic breast cancer participated in screening for the CPCT phase 1 trials. A blood draw and tumor biopsy for NGS were subsequently performed. The NGS results showed a p16-Leiden mutation associated with Familial Atypical Multiple Mole Melanoma (FAMMM) syndrome. The germline mutation was confirmed and patient was informed. We do not know if this patient has informed her relatives about the FAMMM syndrome, which is unrelated to the patient's breast cancer. This patient did not attend the appointment we arranged with our clinical geneticist. No further treatment options were available for this patient with an expected survival of just a few months. She interpreted the results as just another cancer-related ‘symptom' without feeling the necessity of further investigations or counseling. However, FAMMM syndrome may have serious consequences for family members, as this syndrome presents with life-threatening diseases, such as melanoma and pancreatic cancer. Patients carrying the p16-Leiden mutation qualify for regular surveillance to detect melanoma or pancreatic cancer at an early stage.
The third patient was diagnosed with a melanoma. He participated in a CPCT study to reveal whether he would be a candidate for future anticancer treatment with selective inhibitors of mutant BRAF V600E. A BRCA 2 missense mutation was discovered by NGS. Unfortunately, the patient died before genetic results were available. After validating this missense mutation, we could re-classify this missense mutation as a variant of unknown significance.
Discussion of current recommendations in the literature
Consensus now emerges that genetic risk information should be returned to patients9 but disagreement exists what results should be communicated, how and by whom and to who. With the rapidly expanding use of NGS procedures generating large amounts of genetic data, informed consent procedures become increasingly important, but at the same time very challenging. Our recent experiences show that the return policy of unsolicited findings is of utmost importance to integrate in the NGS procedure. The question how, to who, by whom and which genetic risk information should be returned to patients is a very real one, which is expected to become more important. Our own experiences and discussions are reflected in ongoing international debates.
First, there has been an ongoing debate regarding the appropriate type of informed consent for NGS, including both the content and the procedure. Although the majority of experts state that the option to refuse genetic results should be addressed at the time of the informed consent, recently there are suggestions that patients should be able to reconsider their choices.10 This means patients do not have to follow through on their earlier decisions. There is, in other words, a growing plea for facilitating the ongoing changing mind of participants during the study and after signing the informed consent form. This so-called ‘dynamic consent' provides additional functionality to allow ongoing engagement and maintenance of research participants' consent preferences.10, 11
Overall, the informed consent process and informed consent form should clarify the circumstances in which a patient may be re-contacted in the future. A topic for further debate is whether professionals should actively contact participants when new findings are found.
Second, there has been debate how patients should be informed. Several authors proposed to experiment with novel types of consent, among which tiered consent.2, 12, 13, 14, 15 A tiered consent will give the participant a set of choices or well-defined packages (see for example, Table 2) and allows the participant to choose, so it gives the patient greater control over the potentially available information.12, 13
Table 2. Four categories of possible NGS test results (Bredenoord et al, 2011).
| Category 1 | Category 2 | Category 3 | Category 4 |
|---|---|---|---|
| A gene variant that predisposes you to a disease that can be prevented or treated. | A gene variant that predisposes you to a disease that cannot be prevented or treated. | A gene variant that does not affect your own health, but that may be important to the health of your other relatives, such as your children or future offspring. | Uncertain gene variants, meaning they may or may not be important to your health or the health of your relatives. |
| Example: you have a gene variant, which means you are much more likely to develop breast cancer. In this case, we may recommend that you more closely monitor your breasts or have prophylactic surgery. | Example: you have a gene variant, which implies that you are more likely to develop Alzheimer's disease. Alzheimer's disease cannot be treated or prevented. | Example: you could learn that you have a variant in the gene that may cause cystic fibrosis (CF) in future offspring, if the father would have this variant in his gene too. | Example: you have an so-called unclassified variant, which implies you do have a variant, for example, for an increased risk of breast cancer but the significance is unknown. |
These predefined options could consist, for example, of a default package and several optional packages.2, 16 The default package contains actionable information that is highly relevant for the patient, like directly life-saving information or information indicating serious health problems. The optional packages may include data of moderate clinical validity, or a package with reproductive information or data of ‘personal or recreational' interest. In our consortium, optional packages are not yet offered, but we currently perform an empirical ethics study to test the suitability of such a disclosure policy.
Third, there has been debate what genetic information should be returned and how the family should be involved. The American Society of Clinical Oncology (ASCO) and the American Society of Human Genetics (ASHG) stimulate health-care professionals to inform patients about the potential for genetic risks to their relatives. Also the CPCT, as well as other research groups are concerned with the question how this can be addressed appropriately. Our first patient encouraged her oncologist to further investigate her incidental finding, as she felt a responsibility for her family members.
Another family matter, postmortem disclosure of NGS results, should be taken into account as well, particularly in the context of cancer,17 as is sadly illustrated by our third case who died before genetic results were available.
Fourth, there has been debate when patients should be informed about the possibility of unsolicited findings. Within our consortium, patients are informed beforehand about the possibility of unsolicited genetic risk information. Initially, they are briefly informed by their own medical doctor, and later by the CPCT investigator involved in the biopsy procedures. In other institutes, for example at the University of Michigan, all patients undergoing NGS procedure of their tumor had to meet with a genetic counselor before consenting to genomic analysis.18 This may have been preferable in our second patient. She did not feel the necessity of further investigations or counseling concerning her unsolicited finding. Counseling by a genetic counselor in advance of her NGS procedure could possibly have altered her attitude toward receiving genetic information, so she could have opted out for results not directly associated with her current threatening diagnosis before the NGS procedure was performed. However, doing so in all patients eligible for NGS testing would be a large burden for both patients and professionals, as the vast majority of cases will have no unsolicited findings at all.
Fifth, there has been debate whether patients should have the option to opt out from receiving genetic results. The American College of Medical Genetics and Genomics (ACMG) recommends a broader obligation to returning genetic information: they suggest a minimum list of 56 genes that should be routinely reported to the ordering clinician. In 2013, the ACMG recommended that these findings should be reported without asking upfront preferences from the patient and family and without considering the limitations associated with patient's age.19 This ACMG policy statement sparked intensive discussions and was considered controversial because it could affect patients' autonomy and their potential interest in not knowing this genetic information. As a result, this policy statement is now withdrawn and the option to opt out is added. To opt out means that patients should have the option to refuse the return of genomic test results, both those related to the study purpose and those that are unsolicited findings, unless the study aims are related to the return of these data. Although we assume that the majority of patients are willing to receive not only the default package but also additional packages, which is confirmed in studies as well,20 patients do not have an obligation to learn genetic information. Patients who are contacted regarding such results should have the right to decline receiving those results.21 We earlier recommended to always allow an opt out for patients who participate in genome studies for receiving genetic information, also in case of unsolicited findings arising from the default package.8 If an opt out was offered, our second patient had had the option to opt out for return of results except those relevant for her current breast cancer treatment.
Finally, we have to consider how family members should be involved. In our study, this is highly relevant as we are dealing with patients with advanced malignancies with sometimes short life expectancies. This may result in difficult situations when one should decide whether and by whom the family members should be informed about discovered unsolicited findings, after the participant is deceased, which is particularly relevant for highly penetrant, dominant genetic mutations.17 As a default, Boers et al propose a passive disclosure under at least three conditions. First, before the NGS procedure, patients should be counseled on the familial importance of genomic information and about possible postmortem disclosure to relatives. Second, an appropriate procedure for informing and counseling relatives should be agreed upon before implementing NGS. Finally, there should be agreement on the selection of results, including those of immediate clinical significance, which are eligible for postmortem disclosure to relatives. Debate is necessary on whether and when and by active disclosure is more appropriate, and also by whom this should be done: family members or professionals?17 Ormondroyd et al22 also describe a role for genetic counseling services, they concluded that genetic counselors should be involved instead of family members of deceased persons and inform relatives about genetic risk information.
Empirical research recently observed that patients who participate in trials are highly motivated to learn results and that there are numerous medically actionable results that could be derived from whole-exome sequencing and whole-genome sequencing.23, 24, 25
Stimulating the development of educational materials that clearly communicate disease associations or the development of decision tools for patients and physician's is an open research field.14, 26 Further research is needed to compare different ways of disclosing results, also from patients' perspective and preferences in this field of rapid evolving NGS strategies in daily practice.
Conclusions and implications for clinical practice
As NGS has become part of the current diagnostic armamentarium, there is a need to explicitly inform patients about possible unsolicited findings. The question how, to who, by whom and which genetic risk information should be returned to patients is a very real one, which is expected to grow because of the rapid developments in NGS. In our experience, at least 1% of patients (3 out of 376) had unsolicited findings. These unsolicited findings have to be confirmed by a validated test and patients should be counseled by a genetic counselor. Informed consent procedures need to be more explicit in asking patients if they want to be informed about unsolicited findings and what genetic risk information exactly they want to be returned. For our CPCT consortium, and centers alike, a tiered informed consent, offering predefined packages can be used with options for patients to opt in and opt out for the return of unsolicited genetic results. More research, especially toward the needs and preferences of patients concerning the return of genetic risk information is needed.
Acknowledgments
We thank Edwin PJG Cuppen, PhD, for his careful and critical reading of our discussion paper. Furthermore, we would like to acknowledge Marianne Keessen, Rob B van der Luijt, PhD, Ies J Nijman, and all collaborators at the Center for Personalized Cancer Treatment for substantive discussion and feedback on the CPCT's informed consent procedure.
The authors declare no conflict of interest.
References
- Melford HC: Diagnostic exome sequencing: are we there yet? N Engl J Med 2012; 367: 1951–1953. [DOI] [PubMed] [Google Scholar]
- Bredenoord AL, Onland-Moret NC, van Delden JJM: Feedback of individual genetic results to research participants: in favor of a qualified disclosure policy. Hum Mutat 2011; 32: 861–867. [DOI] [PubMed] [Google Scholar]
- ACMG Board of Directors: ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 2015; 17: 68–69. [DOI] [PubMed] [Google Scholar]
- McGuire AL, Caulfield T, Cho MK: Research ethics and the challenge of whole-genome sequencing. Nat Rev Genet 2008; 9: 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J, Boddington P, De Vries J, Hawkins N, Melham K: Ethical implications of the use of whole genome methods in medical research. Eur J Hum Genet 2009; 18: 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El CG, Cornel MC, Borry P et al: Whole-genome sequencing in health care. Recommendations of the European Society of Human Genetics. Eur J Hum Genet 2013; 21: 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidz CW, Appelbaum PS: The therapeutic misconception: problems and solutions. Med Care 2002; 40: V55–V63. [DOI] [PubMed] [Google Scholar]
- Bredenoord AL, Bijlsma RM, van Delden JJM: Always allow an opt out. Am J Bioethics 2015; 15: 28–29. [DOI] [PubMed] [Google Scholar]
- Bredenoord AL, Kroes HY, Cuppen E, Parker M, van Delden JJM: Disclosure of individual genetic data to research participants: the debate reconsidered. Trends Genet 2011; 27: 41–47. [DOI] [PubMed] [Google Scholar]
- Williams H, Spencer K, Snaders C et al: Dynamic consent: a possible solution to improve patient confidence and trust in how electronic patient records are used in medical research. JMIR Med Inform 2015; 3: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machini K, Douglas J, Braxton A, Tsipis J, Kramer K: Genetic counselors' views and experiences with the clinical integration of genome sequencing. J Genet Counsel 2014; 23: 496–505. [DOI] [PubMed] [Google Scholar]
- Lolkema MP, Gadellaa-van Hooijdonk CG, Bredenoord AL, Kapitein P, Roach N, Cuppen E: Ethical, legal, and counseling challenges surrounding the return of genetic results in oncology. J Clin Oncol 2013; 31: 1842–1848. [DOI] [PubMed] [Google Scholar]
- McGuire AL, Hamilton JA, Lunstroth R, McCullough LB, Goldman A: DNA data sharing: research participants' perspectives. Genet Med 2008; 10: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt J, Cox R, Enns GM: Points to consider in the clinical use of NGS panels for mitochondrial disease: an analysis of gene inclusion and consent forms. J Genet Counsel 2014; 23: 594–603. [DOI] [PubMed] [Google Scholar]
- Stadler ZK, Schrader KA, Vijai J, Robson ME, Offit K: Cancer genomics and inherited risk. J Clin Oncol 2014; 32: 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Khoury MJ, Evans JP: Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med 2011; 13: 499–504. [DOI] [PubMed] [Google Scholar]
- Boers SN, van Delden JJ, Knoers NV, Bredenoord AL: Postmortem disclosure of genetic information to family members: active or passive? Trends Mol Med 2015; 21: 148–153. [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR et al: Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med 2011; 3: 111ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW et al: American College of Medical Genetics and Genomics (ACMG) recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitch K, Joseph G, Guiltinan J, Kianmahd J, Youngblom J, Blanco A: Lynch syndrome patients' views of and preferences for return of results following whole exome sequencing. J Genet Couns 2014; 23: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik GP, Amendola LM, Berg JS et al: Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Gen 2014; 94: 818–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormondroyd E, Moynihan C, Watson M et al: Disclosure of genetics research results after the death of the patient participant: a qualitative study of the impact on relatives. J Genet Couns 2007; 16: 527–538. [DOI] [PubMed] [Google Scholar]
- Biesecker LG: Opportunities and challenges for the integration of massively parallel genomic sequencing into clinical practice: lessons from the ClinSeq project. Genet Med 2012; 14: 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Burke W, Kohane I, Plon SE, Zimmern R: Next generation sequencing in the clinic: are we ready? Nat Rev Genet 2012; 13: 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmirzadi L, Chao EC, Palmaer E et al: Patient decisions for disclosure of secondary findings among the first 200 individuals undergoing clinical diagnostic exome sequencing. Genet Med 2014; 16: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FA, Hayeems RZ, Bytautas JP et al: Testing personalized medicine: patient and physician expectations of next-generation genomic sequencing in late-stage cancer care. Eur J Hum Genet 2014; 22: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]