Abstract
Structural genetic variants like copy number variants (CNVs) comprise a large part of human genetic variation and may be inherited as well as somatically acquired. Recent studies have reported the presence of somatically acquired structural variants in the human genome and it has been suggested that they may accumulate in elderly individuals. To further explore the presence and the age-related acquisition of somatic structural variants in the human genome, we investigated CNVs acquired over a period of 10 years in 86 elderly Danish twins as well as CNV discordances between co-twins of 18 monozygotic twin pairs. Furthermore, the presence of mosaic structural variants was explored. We identified four mosaic acquired uniparental disomy events on chromosome 4q and 14q in the follow-up samples from four individuals, and our study thereby supports the increasing prevalence of somatic mosaic variants with age.
Introduction
Over the last 10 years, it has become increasingly evident that a major part of the genetic variation between individuals is caused by structural variants, for example, copy number variants (CNVs), which are deletions and duplications of DNA segments.1 CNVs arise during mitotic and meiotic cell division and can be inherited as well as somatically acquired.2
One way to distinguish the inherited CNVs from the somatically acquired is by comparing longitudinal samples from the same individual. Generally, it seems that aging may be accompanied by an accumulation of both smaller and larger mosaic variants, that is, variants present in only a subset of cells.3, 4, 5 Furthermore, longitudinal studies of specific structural variants suggest that the mosaic cell proportion may increase as well as decrease over time.3, 5
Another way to identify somatic variants is by studying the genomes of monozygotic (MZ) twins. MZ twins are genetically identical at conception, and therefore, any genetic differences between them must be somatic.6 So far, this strategy has been applied in a number of studies of phenotypically discordant as well as healthy or unselected MZ twins, and both mosaic and non-mosaic differences have been found.6, 7, 8
In this study, we attempt to further explore the plasticity of the aging genome by searching for somatic structural variants acquired over a 10-year period in longitudinal samples from 86 elderly Danish MZ and dizygotic (DZ) twins. Furthermore, in a subset analysis, the genomes of the MZ twin pairs are compared to identify post-zygotically acquired somatic structural variants.
Materials and Methods
Study population
The study population included 18 MZ and 25 DZ twin pairs selected from the Longitudinal Study of Aging Danish Twins (LSADT). LSADT includes twins aged 70 years or older and was initiated in 1995.9 The 86 individuals included in the present study were sampled twice: in 1997 (intake) and in 2007 (follow-up). They had a mean age of 75.7 years at intake (age range: 73.0–81.3 years) and were phenotypically unselected and generally healthy enough to engage in a 2 h interview including several physical and cognitive tests and blood drawing at each sampling.
Permission to collect blood samples and the use of register-based information was granted by The Regional Scientific Ethical Committees for Southern Denmark.
Genotyping
DNA was extracted from whole blood using standard methods,10 and genotyping was performed according to the manufacturer's instructions using the Illumina HumanOmni2.5 or HumanOmniExpress BeadChips (Illumina Inc., San Diego, CA, USA). The data have been deposited to the NCBI Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) under series accession number GSE76390.
Only the 716 299 single nucleotide polymorphisms (SNPs) overlapping between the two arrays were included in the subsequent CNV detection.
In the quality control, sample sex and twin zygosity were genotypically confirmed, and samples with genotyping call rates <95% were excluded.
CNV detection
PennCNV11 was used as the primary CNV detection algorithm. Sample estimates of signal intensity values (log R ratio (LRR) and B allele frequency (BAF)) for each SNP were exported from GenomeStudio (Illumina Inc.), and the population frequency of the B allele of included SNPs was supplied from another study.12 GC correction13 was applied to correct signal intensity waves associated with the GC content in the 500 kb on each side of the SNPs as specified by the UCSC GC annotation file (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/gc5Base.txt.gz).
QuantiSNP14 was used as the secondary CNV detection algorithm using default settings and the GC correction option.
The CNVs detected by PennCNV and QuantiSNP were detected on autosomes only and were based on at least three consecutive SNPs. Here, these CNVs are referred to as non-mosaic somatic CNVs, that is, acquired somatic CNVs that are present in a sufficiently high proportion of cells to be detected by the applied algorithms.
The evaluation of the non-mosaic structural variants was based on predefined and structured criteria and consisted of the steps shown below. The unique CNVs remaining after each step and their prevalence are listed in Supplementary Table S1.
CNV detection by PennCNV. After detection, CNVs of the same type (deletion or duplication) were merged if they overlapped with at least 50% of the length of the smaller CNV.
Comparison of paired samples, that is, samples from MZ twins or longitudinal samples from the same individuals. Only CNVs found to be discordant between paired samples were included in the downstream analysis.
Ranking of discordant CNVs based on length and the PennCNV confidence score followed by evaluation of CNVs in the upper 10% for each sample by visual inspection of LRR and BAF plots. If less than five discordant CNVs remained, the top five were used.
CNV detection by QuantiSNP. After detection, CNVs of the same type (deletion or duplication) were merged if they overlapped with at least 50% of the length of the smaller CNV.
Disposal of CNVs remaining after step 3 if the CNV calls made by QuantiSNP did not confirm the discordance between paired samples.
Visual inspection of LRR and BAF plots of remaining CNVs to verify the discordance and select the best candidates for qPCR validation.
qPCR validation
qPCR validation of selected CNVs (Supplementary Table S2) was performed with pre-designed TaqMan Copy Number Assays (Life Technologies, Carlsbad, CA, USA (Supplementary Table S3)) according to the manufacturer's instructions using the TaqMan Copy Number Reference Assay RNase P as internal control and a sample of pooled DNA from 10 unrelated individuals as a calibrator sample.
PCR cycling and fluorescence detection was carried out using the StepOnePlus Real-Time PCR System (Life Technologies). Fluorescence intensity data were exported to the CopyCaller Software (Life Technologies) for calculation of copy numbers using the ΔΔCT method.
Detection of mosaic variants
Mosaic variants were detected using the Mosaic Alteration Detection tool,15 which is incorporated in the R-GADA software package.16 We applied the same parameters as described by Jacobs et al4 and only autosomal structural variants ≥1 Mb in size were investigated. Prior to the detection, LRR estimates were GC-corrected as described above, and samples with BAF SD>0.05 and LRR SD>0.33 were excluded.
The detected mosaic variants were filtered against PennCNV calls, and LRR and BAF plots were visually inspected. Estimation of the mosaic proportion of cells was performed as in the study by Rodriguez-Santiago et al.17
Results
A total of 83 individuals were available for longitudinal analysis of non-mosaic somatic CNVs after quality control. The identified putative CNVs acquired within the 10-year period (Supplementary Table S1) were evaluated based on length, the PennCNV confidence score and LRR and BAF plots, and were filtered against QuantiSNP calls. The remaining variants were verified by visual inspection of LRR and BAF plots, and the three variants showing the strongest discordance were selected for qPCR validation (Supplementary Table S2). However, none of them could be validated.
Detection of non-mosaic somatic CNV differences between MZ co-twins was performed for 18 twin pairs. For 17 of these, the comparison was possible at intake as well as at follow-up. Using the same approach as in the longitudinal analysis, only one CNV was eligible for validation with qPCR (Supplementary Table S2), but it could not be validated.
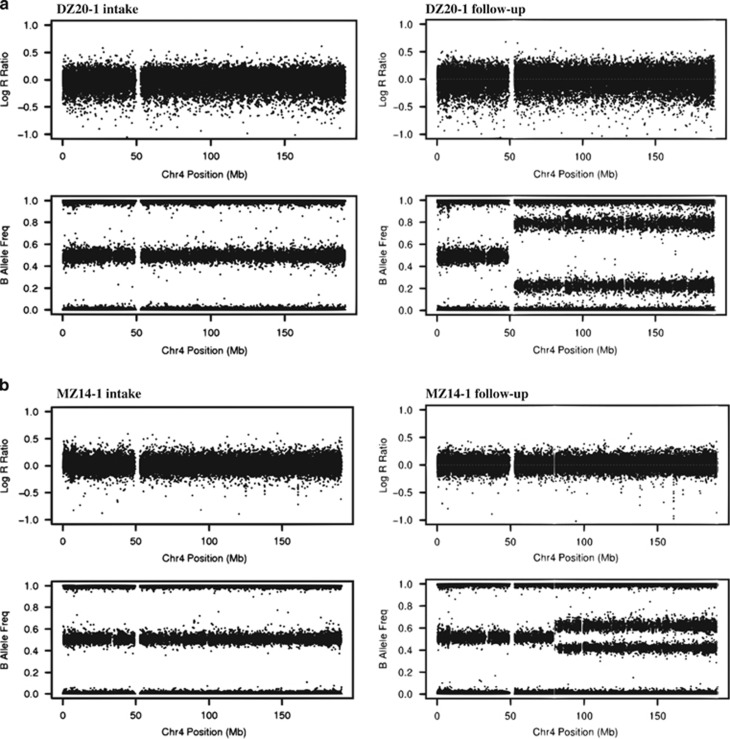
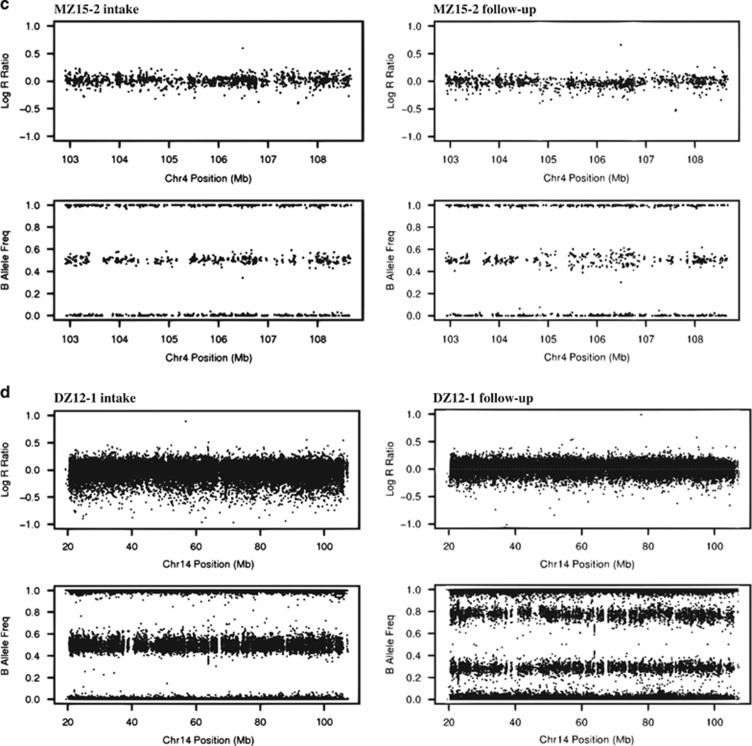
A total of 162 samples from 83 participants passed quality control prior to the detection of mosaic variants. Disposal of non-mosaic variants detected by PennCNV and visual re-evaluation left us with four acquired uniparental disomy (aUPD) events in four participants (Table 1 and Figure 1). All four events were detected in follow-up samples.
Table 1. Detected mosaic structural variants.
| Participant | Sample | Positiona | Length (Mb) | Type | Mosaic cell proportion (%) |
|---|---|---|---|---|---|
| DZ20-1 | Follow-up | Chr4:g.52697856_190915650 | 138.2 | aUPD | 46 |
| MZ14-1 | Follow-up | Chr4:g.79840956_190915650 | 111.1 | aUPD | 20 |
| MZ15-2 | Follow-up | Chr4:g.104830724_106753845 | 1.9 | aUPD | 9 |
| DZ12-1 | Follow-up | Chr14:g.19327823_107287663 | 88.0 | aUPD | 40 |
The position of the mosaic structural variants with information on chromosome and start and stop base pair positions based on the GRCh37/hg19 genome build.
Figure 1.
Plots of log R ratio and B allele frequency for the detected mosaic structural variants. aUPD was detected in (a) the entire chromosome 4q in ~46% of cells in the follow-up sample from DZ20-1, (b) a 111 Mb region on 4q in ~20% of cells in the follow-up sample from MZ14-1, (c) a 1.9 Mb region on 4q in ~9% of cells in the follow-up sample from MZ15-2, and (d) the entire chromosome 14q in ~40% of cells in the follow-up sample from DZ12-1.
Discussion
In this study, we searched for somatically acquired structural genetic differences in phenotypically unselected elderly Danish twins by comparing time-separated samples from the same individual as well as samples from MZ twins. In total, we identified four mosaic aUPD events in follow-up samples from 4 out of 83 participants.
The presence of non-mosaic somatic CNV discordances between phenotypically unselected MZ twins has been investigated in two recent studies,7, 8 and both studies indicate that such events are rare. This is supported by our study, where no reproducible non-mosaic CNV differences were found in the included MZ twin pairs. This may be a consequence of the sample size, but it may also reflect that somatic CNVs are more likely to be present as mosaics. This hypothesis is in line with the results detected in our study as well as in the studies by Bruder et al6 and Forsberg et al.3 In addition, our study confirms the previously reported trend of an accumulation of mosaic variants with age.3, 4, 5 Accordingly, the mosaic structural variants detected in our study are all found in follow-up samples, and although the included individuals have advanced ages already at intake, this could imply that the somatic variants primarily arise at even older ages. Alternatively, the mosaic cell proportion, as previously seen,3, 5 fluctuates with age and may thus have been below the detection limit of the applied method in the intake samples.
It has been suggested that the age-related accumulation of mosaic structural variants in blood seen here and in previous studies3, 4, 5 may be a consequence of an age-related reduction in the number of cell clones in the blood.3 With a less diverse clonal makeup of the blood, it could be speculated that the likelihood of certain cell clones to rise to a detectable level increases. Also, the accumulation could be due to increased rates of somatic mutation or a decrease in genome maintenance.4 The participants included in this study are somewhat selected in terms of age and probably also in terms of health. Hence, if a decrease in genome maintenance is associated with an increased propensity to accumulate mosaic structural variants, it could be speculated that their genomes would carry less somatic structural variations, compared with individuals of equal age or younger in a less good health, as a well-functioning genome maintenance system is positively correlated with longevity.18
All of the four mosaic structural variants detected in this study are aUPD events, which have been found to be the most prevalent type of mosaic events.4, 5 Generally, UPDs are the result of the inheritance of both copies of a pair of chromosomes from one parent only, and in the acquired form, they are thought to most commonly arise as a consequence of mitotic nondisjunction/anaphase lag or mitotic recombination.19, 20 Interestingly, three of the four detected mosaic variants are located on chromosome 4q, whereas the fourth spans the entire q-arm of chromosome 14. In a very recent meta-analysis5 based on data from >100 000 individuals, aUPD on chromosome 14q was one of the most frequently detected mosaic chromosome anomalies, and chromosome 4q aUPD was observed several times as well. It could be speculated that the chromosomal distribution of mosaic events seen in this study may reflect that some chromosomal regions are more prone to rearrangements than others, or that some chromosomal rearrangements are more likely to undergo clonal expansion than others.
The phenotypic consequences of mosaic variants are likely to be less severe than constitutive variants, for example, because the proportion of mutant cells may be too small to cause an effect, or because the mutant cells may only be present in a tissue where the variant has no effect.2 The most well-known potential consequence of somatic mutations is cancer, and although aUPD events are common features of several cancers,20 none of the carriers of mosaic variants in this study have been diagnosed with any kind of cancer.
In conclusion, our study confirms that mosaic structural variants can accumulate with age. The variants detected in the present study have no apparent phenotypic consequences and could thus represent examples of normal variation in the aging genome.
Acknowledgments
We thank the Institute of Clinical Molecular Biology, Christian-Albrechts-University, Kiel, Germany for technical assistance with genotyping. This study was financially supported by the European Union's Seventh Framework Programme (FP7/2007-2011) under grant agreement n° 259679, the VELUX Foundation, and The Danish National Program for Research Infrastructure 2007 (grant no. 09-063256).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Feuk L, Carson AR, Scherer SW: Structural variation in the human genome. Nat Rev Genet 2006; 7: 85–97. [DOI] [PubMed] [Google Scholar]
- Campbell IM, Shaw CA, Stankiewicz P, Lupski JR: Somatic mosaicism: implications for disease and transmission genetics. Trends Genet 2015; 31: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LA, Rasi C, Razzaghian HR et al: Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet 2012; 90: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs KB, Yeager M, Zhou W et al: Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012; 44: 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Zhou W, Sampson JN et al: Characterization of large structural genetic mosaicism in human autosomes. Am J Hum Genet 2015; 96: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder CE, Piotrowski A, Gijsbers AA et al: Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet 2008; 82: 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdellaoui A, Ehli EA, Hottenga JJ et al: CNV concordance in 1,097 MZ twin pairs. Twin Res Hum Genet 2015; 18: 1–12. [DOI] [PubMed] [Google Scholar]
- McRae AF, Visscher PM, Montgomery GW, Martin NG: Large autosomal copy-number differences within unselected monozygotic twin pairs are rare. Twin Res Hum Genet 2015; 18: 13–18. [DOI] [PubMed] [Google Scholar]
- Skytthe A, Kyvik K, Holm NV, Vaupel JW, Christensen K: The Danish Twin Registry: 127 birth cohorts of twins. Twin Res 2002; 5: 352–357. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D et al: PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007; 17: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard M, Debrabant B, Tan Q et al: Copy number variation associates with mortality in long-lived individuals: a genome-wide assessment. Aging Cell 2016; 15: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin SJ, Li M, Hou C et al: Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res 2008; 36: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM et al: QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res 2007; 35: 2013–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JR, Rodriguez-Santiago B, Caceres A et al: A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinformatics 2011; 12: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Caceres A, Gonzalez JR: R-Gada: a fast and flexible pipeline for copy number analysis in association studies. BMC Bioinformatics 2010; 11: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Santiago B, Malats N, Rothman N et al: Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am J Hum Genet 2010; 87: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Suh Y: Genome instability and aging. Annu Rev Physiol 2013; 75: 645–668. [DOI] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG et al: Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet 2010; 19: 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuna M, Knuutila S, Mills GB: Uniparental disomy in cancer. Trends Mol Med 2009; 15: 120–128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.