Abstract
Antibody class-switch recombination and somatic hypermutation critically depend on the function of activation-induced cytidine deaminase (AID). Rare variants in its gene AICDA have been reported to cause autosomal recessive AID deficiency (autosomal recessive hyper-IgM syndrome type 2 (HIGM2)). Exome sequencing of a multicase Finnish family with an HIGM2 phenotype identified a rare, homozygous, variant (c.416T>C, p.(Met139Thr)) in the AICDA gene, found to be significantly enriched in the Finnish population compared with other populations of European origin (38.56-fold, P<0.001). The population history of Finland, characterized by a restricted number of founders, isolation and several population bottlenecks, has caused enrichment of certain rare disease-causing variants and losses of others, as part of a phenomenon called the Finnish Disease Heritage. Accordingly, rare founder mutations cause the majority of observed Finnish cases in these mostly autosomal recessive disorders that consequently are more frequent in Finland than elsewhere. Screening of all currently known Finnish patients with an HIGM2 phenotype showed them to be homozygous for p.(Met139Thr). All the Finnish p.(Met139Thr) carriers with available data on their geographic descent originated from the eastern and northeastern parts of Finland. They were observed to share more of their genome identity by descent (IBD) than Finns in general (P<0.001), and they all carried a 207.5-kb ancestral haplotype containing the variant. In conclusion, the identified p.(Met139Thr) variant is significantly enriched in Finns and explains all thus far found AID deficiencies in Finland.
Introduction
Primary immunodeficiency disorders (PIDDs) encompass a wide range of genetically determined inborn errors of immunity. Presently, >5000 variants affecting the function have been reported in over 250 genes as causative of 265 PIDDs, and novel defects continue to be discovered.1
Hyper-IgM syndromes (HIGMs) include a genetically heterogeneous group of PIDDs defined by early-onset recurrent infections and autoimmunity, absence or very low levels of IgG, IgA and IgE, but elevated or normal serum IgM levels.2 This phenotype typically results from inherited defects in proteins involved in class-switch recombination (CSR) and somatic hypermutation (SHM). Classical HIGM-causing genes include CD40LG, AICDA, CD40 and UNG.3 CSR, SHM and central B-cell tolerance critically depend on normal activation-induced cytidine deaminase (AID) function.4, 5, 6 AID also participates in removal of epigenetic memory by active demethylation.7 The immunologic HIGM phenotypes of AID and uracil DNA glycosylase (UNG) deficiencies closely resemble each other and are relatively easy to screen. In these, no CD19+CD27+IgD-IgM- switched memory B (smB) cells can be found in blood, whereas marginal zone CD19+CD27+IgD+IgM+ B (MZB) cells are normal or high. AID deficiency is estimated to affect <2/107 individuals.8
The population history of Finland is characterized by a restricted number of founders, isolation, several population bottlenecks and recent expansion of the population. This has led to the enrichment of some deleterious variants and loss of others, creating a phenomenon called the Finnish Disease Heritage (FDH). By definition, FDH disorders are more frequent in Finland than elsewhere, and a majority of the Finnish patients share the same founder mutation.9
Here, we have identified the Finnish founder allele causing HIGM2 and assessed its prevalence in Finns compared with other populations.
Materials and methods
This study was conducted in accordance to the principles of the Helsinki Declaration and was approved by the Coordinating Ethics Committee of Helsinki University Hospital. Written informed consent was obtained from all subjects.
Patients
The index case of the family I (I-I) was immunologically characterized in Helsinki University Hospital. She has undetectable level of smB cells and a typical clinical picture of HIGM2 (Table 1). Next, patient cohorts of the pediatric and adult immunodeficiency units of all five Finnish university hospitals were screened for patients with a phenotype compatible with either AID or UNG deficiency. All subsequent patients (families II–IV) with (1) low or absent IgA, IgG and IgE levels but normal or high IgM levels according to the laboratory reference values, together with (2) missing smB cells but normal or high levels of MZB in B-cell phenotyping (for methods see Haapaniemi et al10) were included in the study. Study subjects underwent clinical and immunological evaluations at Helsinki, Kuopio, Oulu and Tampere University Hospitals. All available patient records since June 1959 were reviewed and patients interviewed. Altogether, four families were identified (Table 1 and Figure 1). Patient histories are described in detail in the Supplementary Information.
Table 1. Characteristics of Finnish AID deficiency patients.
| N | Origin | Genotype AICDA p.(Met139Thr) | Sex | Clinically affected | Age at onset (y) | Serum or plasma levels at diagnosis (g/l) | IgM+ memory B cells (% of B cells) | IgG+ memory B cells (% of B cells) | Lymphatic hyperplasiaa | Infectionsa | Autoimmunity and immunodysregulationa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgM | IgG | IgA | IgE | |||||||||||
| (0.47–2.84 g/l) | (6.8–15 g/l) | (0.52–4.02 g/l) | (0–110 IU/l) | (7.2–30.8%) | (6.5–29.2%) | |||||||||
| I-I | Kuopio | Homozygote | F | Yes | <1 | 9.8 | ND | 0.04 | ND | Normal | ND | Yes | Otitis media, mastoiditis, pansinusitis, purulent pleuritis, recurrent pneumonia and bronchiectasis, during immunosuppressive treatment CMV viremia, Pneumocystis jirovecii pneumonia and fatal enterococcal sepsis | Sjögren's syndrome, interstitial nephritis and renal tubular acidosis, membranous glomerulonephritis, bronchial asthma, severe leukocytoclastic vasculitis of lower limbs |
| I-III | Kuopio | NA | M | Yes | <1 | NA | NA | NA | NA | NA | NA | Yes | Otitis media, fatal bacterial meningitis at age 2 | No |
| I-IV | Kuopio | Homozygote | M | Yes | <1 | 28.4 | 0.07 | 0.06 | ND | NA | NA | Yes | Otitis media, mastoiditis, sinusitis, purulent pleuritis, recurrent pneumonia and bronchiectasis, pulmonary and suspected cervical and inguinal lymph node tuberculosis | Recurrent idiopathic nondestructive monoarthritis of knees, mild asymptomatic duodenal villous atrophy and intraepithelial lymphocytosis, bronchial asthma and primary emphysema |
| I-X | Kuopio | Homozygote | F | Yes | <10 | 13.5 | 1.98 | 0.04 | ND | NA | NA | Yes | Otitis media, mastoiditis, sinusitis, recurrent pneumonia, massive cervical granulomatous lymphadenopathy with cutaneous fistula | No |
| II-I | Kuusamo | Homozygote | F | Yes | <2 | 54 | 2.1 | ND | ND | 18.40% | ND | No | Otitis media, sinusitis | No |
| II-III | Kuusamo | Homozygote | M | Yes | <1 | 16.5 | ND | ND | ND | 38.90% | ND | No | Otitis media, neck abscess, recurrent pneumonia | No |
| III-III | Ii/Vehmersalmi | Homozygote | F | Yes | <3 | 16.8 | 0.1 | 0.06 | ND | 24.8% | ND | No | High number of upper respiratory infections | Mild autoimmune cholangitis |
| IV-I | Kiuruvesi/ Ikaalinen | Homozygote | M | Yes | <3 | 4 | 0.1 | 0.04 | ND | 53.70% | ND | No | Otitis media, sinusitis, neck abscess | No |
Abbreviations: F, female; M, male; Ig, immunoglobulin; NA, not assessed; ND, not detectable (values under the detection threshold of the analysis method). The p.(Met139Thr) variant in the AICDA gene refers to the following reference sequences: RefSeq NG_011588.1 and NM_020661.2 (GRCh37.p13).
For more detailed history see Supplementary Information.
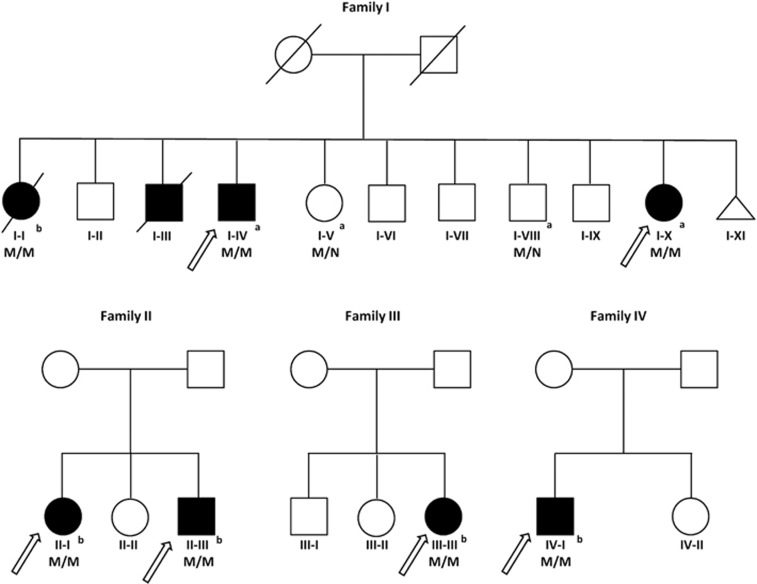
Figure 1.
AICDA variants in four families with HIGM2. Solid symbols indicate affected patients and open symbols unaffected family members. Triangles represent stillborn individuals. Slashes indicate deceased persons (reported cause of death is sepsis (65 y.o.) for I-I, and meningitis (2 y.o.) for I–III). The original familial probands (index cases) are pointed by arrows. The AICDA p.(Met139Thr) variant is indicated by M, wild-type alleles by N. aIndividuals evaluated by whole-exome sequencing. bTargeted analysis of the p.(Met139Thr) variant by Sanger sequencing.
Molecular genetics
Genomic DNA of the studied individuals was isolated using standard salt precipitation protocols.
Exome sequencing was performed in the two index patients of family I and in two of their healthy relatives to investigate the genetic basis of their familial disease presentation. A NexteraRapid Capture Exome kit (Illumina, San Diego, CA, USA) was used for library preparation and exome enrichment and sequencing was performed on a HiSeq 1500 platform (Illumina). The data were analyzed using a version 2.7 of the in-house developed analysis pipeline for quality control and variant identification (VCP).11 Detailed sequencing statistics and procedures for read alignment and variant calling are provided in the Supplementary Information. Additional patients were screened for AIDCA allelic variants by Sanger sequencing. The analysis of the AICDA gene (GRCh37.p13:12:8754762-8765463) and the primer design were performed using the following genomic and transcript sequences: RefSeq NG_011588.1 and NM_020661.2. The identified variants and other patient data were deposited in the LOVD database (http://databases.lovd.nl/shared/genes/AICDA) (variant ID: AICDA_000004; individual IDs: 00058568, 00058569, 00058570, 00058572, 00058573, 00058574 and 00058575).
DNA variants were verified by restriction endonuclease digestion. See Supplementary Information for the sequence of primers and further description of the procedures.
Population analysis
We performed a population-based analysis of the identified sequence variant frequency by using data of 60 786 individuals from Exome Aggregation Consortium including 34 699 individuals of European origin, of whom 3013 were Finnish.
The geographic distribution in Finland of the p.(Met139Thr) alleles (RefSeq NM_020661.2; c.416T>C; rs200858797) was illustrated based on the information obtained from the study subjects and from the carriers included in the SISu project (http://sisu.fimm.fi/)12 for which such data were available and in three Finnish sample collections (the Finnish Twin Cohort study, the National Finrisk Study and the Migraine Family Study; Supplementary Information,Supplementary Table 1 and Figure 2).
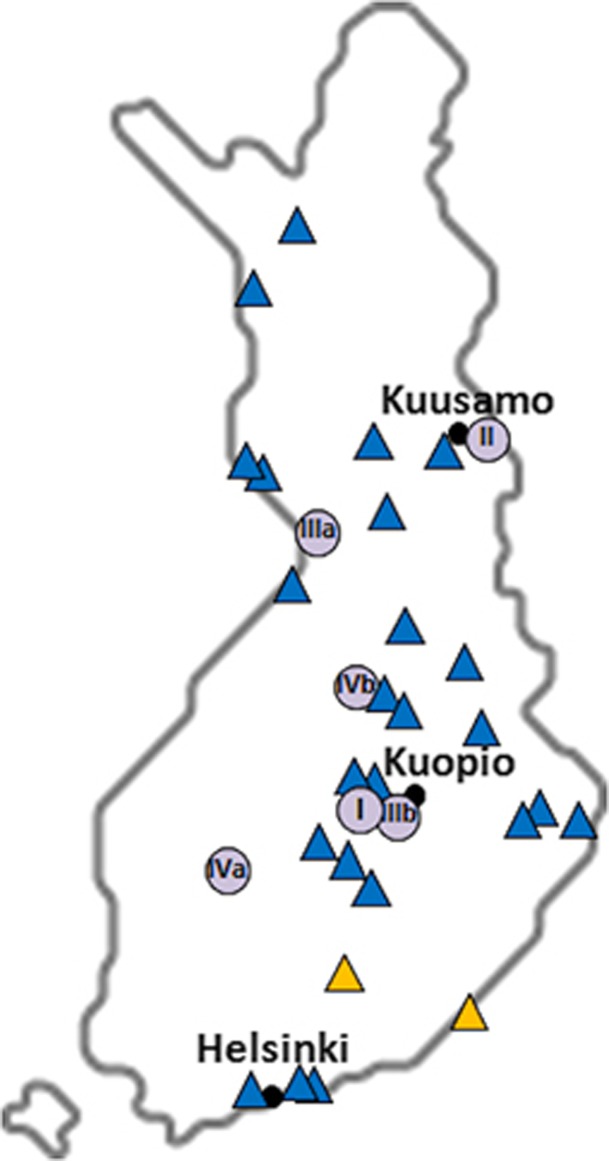
Figure 2.
Distribution of the AICDA p.(Met139Thr) carriers in Finland. Blue triangles point to the geographical origin of the Finnish carriers (n=27) of the p.(Met139Thr) variant included in SISu and in epidemiological and clinical Finnish sample collections (the Finnish Twin Cohort study, the National Finrisk Study and the Migraine Family Study) (Supplementary Table 1). Yellow symbols indicate the birthplaces of carriers' parents, if discordant. The birthplaces of the patients identified in this study are indicated by a purple spot, listing the number of the family (from I to IV). For families III and IV, the mother corresponds to ‘a' and the father to ‘b'. The black dots mark the main municipal areas.
The analysis of pairwise segmental sharing was conducted on a set of 6755 Finns included in epidemiological and clinical Finnish sample collections, of whom 20 were p.(Met139Thr) carriers, using 113 common markers genotyped using HumanCoreExome-24 BeadChips (Illumina; 1000 Genomes; www.1000genomes.org; accessed 18 April 2014). A cryptic relatedness analysis was performed by using the identity by descent (IBD) estimation on the above-mentioned set of unaffected individuals (Supplementary Information).
In parallel, a segregation analysis of polymorphisms in a 2-Mb region encompassing the p.(Met139Thr) was executed in the 32 carriers included in the above mentioned clinical and epidemiological sample collections in addition to the SISu data set (Supplementary Table 2) by utilizing PLINK (v. 1.07, http://pngu.mgh.harvard.edu/purcell/plink/).13 Next, according to the observed haplotype blocks, a total of 80 markers located in a 1.1-Mb region surrounding the p.(Met139Thr) were screened for a putative shared allele, carried on in the comprehensive group of 32 carriers (Supplementary Table 3).
Statistical analysis
Pearson's χ2 test (108 simulations) was used to evaluate the different p.(Met139Thr) allelic distributions among Finns and the more heterogeneous European populations (Supplementary Table 4), and the load of hidden relatedness among the carriers and the general population was weighed using Welch's two-sample t-test (108 simulations).
Results
Genetic analysis
We first studied a family with four affected individuals originating from Eastern Finland (Figure 1). The index of the family had previously been tested in a reference laboratory to have wild-type AICDA and UNG. However, exome sequencing revealed a known biallelic AIDCA variant in the living affected members (p.(Met139Thr)) that has previously been shown to cause HIGM2.14 The two healthy relatives carried one copy of the variant. Targeted Sanger sequencing of an archived sample from the index verified the presence of the same biallelic sequence change (I-I, Figure 1). Thereafter, all remaining Finnish patients with a compatible phenotype (n=4) were screened and found homozygotes for the p.(Met139Thr) variant (Figure 1 and Table 1).
Population analysis
Overall, we found the HIGM2 causing p.(Met139Thr) alteration to have a frequency of 0.012% in a total of 57 391 exomes provided by the Exome Aggregation Consortium (ExAC). More detailed analysis of the data revealed an allelic frequency of 0.0047% in 31 686 individuals of European ancestry (non-Finns) and the absence of the variant in non-European populations (22 692 individuals). Compared with other populations of European origin, a statistically significant 38.56-fold allelic frequency was observed in Finns with 11 uniallelic carriers in 3013 exomes (0.18%, P<0.001; Supplementary Table 4), resulting in the calculated theoretical frequency of AID deficiency of 0.81/106 in those of Finnish ancestry. Other AICDA variants showed no substantial differences in frequencies between the populations (data not shown).
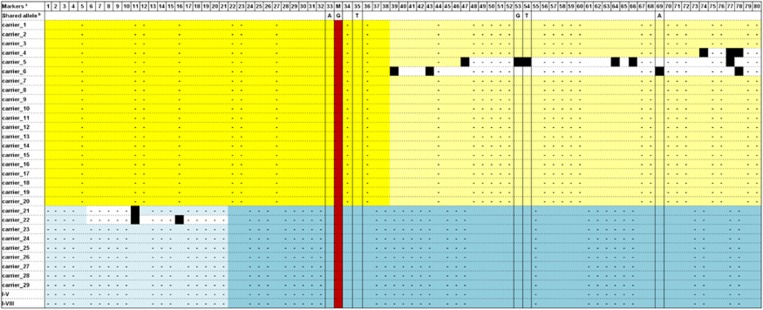
Because of the enrichment of the p.(Met139Thr) variant in Finland, we studied its geographical distribution based on the information on birthplace retrieved from the studied subjects, and from those 27 out of 31 carriers within the SiSu cohort and other Finnish sample collections with such data available. Interestingly, all of the AID deficiency patients and 24 of the 27 carriers originated from the late settlement regions of Eastern and Northeastern Finland, suggesting shared origin for the p.(Met139Thr) alleles in all these individuals (Figure 2). The remaining three carriers were born in Helsinki area that has experienced substantial immigration from the rest of the country during recent centuries. Thus, we searched for possible shared haplotype in the region surrounding AICDA by utilizing the exome data for 3325 individuals of the SiSu cohort, including 11 p.(Met139Thr) carriers. We first retrieved the haplotype structure of the 2 Mb genomic region encompassing the p.(Met139Thr) and observed clear haplotype blocks 90 kb upstream and 51 kb downstream of the variant (Supplementary Table 5). Further examination of the genomic region flanking AICDA using the UCSC Genome Browser15 revealed the presence of a 10-kb recombination hot spot encompassing the gene that likely weakens the possibility of tracking a conserved ancestral allele. Nonetheless, by combining the genetic data of all the 31 carriers of the two different population-based data sets (exome data of the SiSu cohort and genotyping data of the Finnish epidemiological and clinical cohorts) and the two exome sequenced familial carriers, and by monitoring the alleles seen in each haplotype block, we identified a 207.5-kb core haplotype including the p.(Met139Thr) variant shared by all the carriers (Figure 3). The minimal shared region was restricted by recombination in five individuals, whereas the core haplotype extended significantly further in the others (Figure 3). Further comparison of the pairwise genome-wide IBD showed higher values in the group of p.(Met139Thr) carriers (average piHat=0.007±0.0027) than in the general population (piHat=0.003±0.005), displaying significant increased relatedness within the carriers (P=1.59E−12).
Figure 3.
Haplotype structure of the flanking region of the AICDA gene in the 31 Finnish carriers of p.(Met139Thr) variant. The haplotypes of the carriers analyzed by genotyping chip (the Finnish Twin Cohort study, the National Finrisk Study and the Migraine Family Study) are shown on horizontal lines on yellow background in the top part of the panel. The haplotypes of the carriers analyzed by WES (SISu project and study subjects of family I) are presented on blue background. The red column shows the position of the p.(Met139Thr) variant. Missing genotypes are marked by ‘-'. The yellow/blue squares show the identified shared haplotype in each mutation carrier, white filling indicates noninformative genotypes and black squares label recombination event (ie, absence of the allele included in the above mentioned haplotype). The minimum regions shared by all mutation carriers in each data set are indicated by darker color. aThe markers used in the analysis are indicated with numbers in the top row (marker names listed in Supplementary Table 3). bThe columns framed by black lines highlight the markers shared by both data sets, and the alleles seen in the shared haplotype are shown above the column.
Discussion
In the current study, we identified a Finnish founder mutation for AID deficiency. The rare recessive p.(Met139Thr) allelic variant in the AICDA gene causes the disease in all known Finnish patients. The variant, previously confirmed to affect the AID function in a single HIGM2 patient of unknown origin,14 exhibits a significant (38.56-fold) enrichment in Finns compared with the data from other European populations.
There are at least 117 previously published cases of AID deficiency and, currently, at least 43 autosomal recessive or dominant negative causative AICDA variants have been reported.16, 17, 18, 19 The observed p.(Met139Thr) change affects an evolutionarily conserved amino acid residue in the APOBEC-like domain, and in silico analyses are consistent with a deleterious effect, resulting in severely impaired CSR.15 Interestingly, a different causative missense substitution affecting the same amino acid has been found in three Turkish patients with HIGM2 (RefSeq NM_020661.2; c.415A>G, p.(Met139Val); rs104894321).20, 21 This disrupts the AID activity in vitro.22
Given the known frequency of uniallelic p.(Met139Thr) in 3013 Finns, the predicted prevalence for homozygous individuals was ~0.81/106. However, the presently known prevalence of AID deficiency in Finland is 1.5/106, greatly exceeding the estimate in literature, although this was partly based on the incidence in French-Canadians, the other known population with an AICDA founder mutation.8 Currently, there are relatively few Northern and Northeastern Finns in the SISu cohort, potentially explaining the observed difference between theoretical and known prevalence. To evaluate the contribution of other variants causing HIGMs in the Finnish population, we performed a similar population-based comparison of allele frequencies for all the variants in genes affecting CSR (AICDA, UNG, CD40 and CD40LG). None of the other allelic variants were significantly enriched in the Finnish cohort compared with other European populations.
Finland's population history has led to an enrichment of some disease-causing variants and losses of others. In each FDH disorder, a causative Finmajor founder mutation accounts for most, if not all, affected individuals and is more frequent in Finland than elsewhere.23 FDH thus far has included three PIDDs: cartilage-hair hypoplasia (CHH), autoimmune polyendocrinopathy (APECED) and Cohen syndrome, with a prevalence of 50/106, 36/106 and 10/106, respectively.9, 24, 25, 26, 27, 28 The currently available exome data further confirmed the Finmajor mutations causing APECED (rs121434254, 6.25-fold) and Cohen syndrome (rs180177327, 47.11-fold) to be significantly enriched in Finns compared with other populations. No reliable data are available for the CHH-associated variants.
As almost all p.(Met139Thr) carriers originated from the late settlement areas of Eastern and Northeastern Finland, the geographic distribution of the variant fits well with the known inhabitation patterns of the country and suggest a single origin.29, 30 Consequently, we made an effort to validate this hypothesis by analyzing the genomic region encompassing the AICDA gene in the individuals of the SiSu cohort. We identified linkage disequilibrium blocks upstream and downstream the p.(Met139Thr), suggesting the actual architecture of the region as reflecting the remnants of a wider previous haplotype that has potentially included the variant. The small number of the carriers and, mostly, the presence of a recombination hot spot of 10 kb surrounding the AICDA gene31 could have limited our ability to identify the ancestral allele. In order to overcome these limitations, we further studied the haplotype structure of the surrounding areas in all the p.(Met139Thr) carriers included in the SiSu project and three epidemiological and clinical Finnish sample collections. A segregation analysis revealed a shared haplotype of 207.57 kb inclusive of the variant, with no apparent recombination events in the 31 carriers. This was surrounded by a partially conserved genomic region of 901 kb where a limited amount of recombination events had taken place. The outlined genetic structure comprises the likely ancestral founder allele. Our hypothesis of a single mutation event and shared ancestry was also further strengthened by the finding that all p.(Met139Thr) carriers shared more of their genome than the general population (2.25-fold increased IBD).
AID deficiency is clinically characterized by severe antibody deficiency, lymphatic nongranulomatous hyperplasia with hyperplastic germinal centers and inflammatory complications like hematologic autoimmunity, chronic hepatitis, diarrhea and aseptic arthritis.16, 17 Our patients display a uniform matching phenotype. The patients of family I have a longer follow-up than most patients in the current literature, lending insight into the long-term consequences of the disease. To the best of our knowledge, the patients have developed several previously unreported systemic, renal and gastrointestinal autoimmune complications (Table 1 and Supplementary Information). However, aggressively substituted younger patients in families II–IV seem to have few autoimmune problems. Unlike in common variable immunodeficiency, granulomatous lymphadenitis is not a previously described feature of AID deficiency. A pronounced and difficult to treat granulomatous lymphadenopathy was noted in family I and confirmed by biopsies. Unfortunately, no archived tissue samples were available. As this occurred during a familial tuberculous mini-epidemic, it suggests that infectious causes of granulomas should always be excluded in AID deficiency. Opportunistic lethal infections in I-I were likely caused by secondary immunosuppression and are also not a feature of AID deficiency. Whether AID deficiency is able to cause spontaneously terminated pregnancies should be further studied (cf. Supplementary Information).
In summary, we identified a single variant affecting the function of the protein accounting for all diagnosed AID deficiencies in Finns. In all likelihood, p.(Met139Thr) is a Finmajor founder mutation and AID deficiency belongs to the FDH. This phenomenon closely resembles the known p.Arg112Cys founder allele in French Canadians, but 3p.(Met139Thr) is even more prevalent in Finns.8 Taken together, these findings underline the correlation between the genetic structure of the population and the distribution of genetic disorders, and emphasize the benefits of researching population isolates with systematic health records available.
Acknowledgments
We are very grateful to Dr Jaana Hyytiäinen for providing additional data on our patients. We thank the Exome Aggregation Consortium, and a full list of contributing groups can be found at http://exac.broadinstitute.org/about. Teemu Kuulasmaa and Markku Laakso are thanked for kindly sharing the genetic data and geographic origin of p.(Met139Thr) carriers included in the METSIM study (http://www.nationalbiobanks.fi/index.php/studies2/10-metsim). We thank THL National FINRISK Study participants and its scientific team for their contribution to this study. Jaakko Kaprio is thanked for providing access to the Finnish Twin Cohort data, with the support of the Academy of Finland (Grants 265240 and 263278). The exome sequencing and primary data analysis was provided by the Sequencing and Bioinformatics Units of the FIMM Technology Centre.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Al-Herz W, Bousfiha A, Casanova JL et al: Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front Immunol 2014; 5: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulino AV, Notarangelo LD: Hyper IgM syndromes. Curr Opin Rheumatol 2003; 15: 422–429. [DOI] [PubMed] [Google Scholar]
- Durandy A, Taubenheim N, Peron S, Fischer A: Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv Immunol. 2007; 94: 275–306. [DOI] [PubMed] [Google Scholar]
- Kuraoka M, Holl TM, Liao D et al: Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc Natl Acad Sci USA 2011; 108: 11560–11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G, Ng YS, Bannock JM et al: Activation-induced cytidine deaminase (AID) is required for B-cell tolerance in humans. Proc Natl Acad Sci USA 2011; 108: 11554–11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogne M: Activation-induced deaminase in B lymphocyte maturation and beyond. Biomed J 2013; 36: 259–268. [DOI] [PubMed] [Google Scholar]
- Kumar R, DiMenna L, Schrode N et al: AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature 2013; 500: 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y, Lavoie A, Cunningham-Rundles C et al: Mutations in activation-induced cytidine deaminase in patients with hyper IgM syndrome. Clin Immunol 2000; 97: 203–210. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T: Molecular genetics of the Finnish disease heritage. Hum Mol Genet 1999; 8: 1913–1923. [DOI] [PubMed] [Google Scholar]
- Haapaniemi EM, Kaustio M, Rajala HL et al: Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood 2015; 125: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulonen AM, Ellonen P, Almusa H et al: Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011; 12: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Wurtz P, Havulinna AS et al: Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 2014; 10: e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandy A, Peron S, Taubenheim N, Fischer A: Activation-induced cytidine deaminase: structure-function relationship as based on the study of mutants. Hum Mutat 2006; 27: 1185–1191. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS et al: The human genome browser at UCSC. Genome Res 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdaviani SA, Hirbod-Mobarakeh A, Wang N et al: Novel mutation of the activation-induced cytidine deaminase gene in a Tajik family: special review on hyper-immunoglobulin M syndrome. Expert Rev Clin Immunol 2012; 8: 539–546. [DOI] [PubMed] [Google Scholar]
- Caratao N, Cortesao CS, Reis PH et al: A novel activation-induced cytidine deaminase (AID) mutation in Brazilian patients with hyper-IgM type 2 syndrome. Clin Immunol 2013; 148: 279–286. [DOI] [PubMed] [Google Scholar]
- Lee WI, Huang JL, Yeh KW et al: Clinical features and genetic analysis of Taiwanese patients with the hyper IgM syndrome phenotype. Pediatr Infect Dis J 2013; 32: 1010–1016. [DOI] [PubMed] [Google Scholar]
- Cabral-Marques O, Klaver S, Schimke LF et al: First report of the Hyper-IgM syndrome Registry of the Latin American Society for Immunodeficiencies: novel mutations, unique infections, and outcomes. J Clin Immunol 2014; 34: 146–156. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y et al: Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 2000; 102: 565–575. [DOI] [PubMed] [Google Scholar]
- Quartier P, Bustamante J, Sanal O et al: Clinical, immunologic and genetic analysis of 29 patients with autosomal recessive hyper-IgM syndrome due to Activation-Induced Cytidine Deaminase deficiency. Clin Immunol 2004; 110: 22–29. [DOI] [PubMed] [Google Scholar]
- Ta VT, Nagaoka H, Catalan N et al: AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol 2003; 4: 843–848. [DOI] [PubMed] [Google Scholar]
- Norio R: The Finnish Disease Heritage III: the individual diseases. Hum Genet 2003; 112: 470–526. [DOI] [PubMed] [Google Scholar]
- Makitie O, Perheentupa J, Kaitila I: Growth in cartilage-hair hypoplasia. Pediatr Res 1992; 31: 176–180. [DOI] [PubMed] [Google Scholar]
- Neufeld M, Maclaren NK, Blizzard RM: Two types of autoimmune Addison's disease associated with different polyglandular autoimmune (PGA) syndromes. Medicine (Baltimore) 1981; 60: 355–362. [DOI] [PubMed] [Google Scholar]
- Norio R: Finnish Disease Heritage I: characteristics, causes, background. Hum Genet 2003; 112: 441–456. [DOI] [PubMed] [Google Scholar]
- Ridanpaa M, van Eenennaam H, Pelin K et al: Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 2001; 104: 195–203. [DOI] [PubMed] [Google Scholar]
- Polvi A, Linturi H, Varilo T et al: The Finnish disease heritage database (FinDis) update-a database for the genes mutated in the Finnish disease heritage brought to the next-generation sequencing era. Hum Mutat 2013; 34: 1458–1466. [DOI] [PubMed] [Google Scholar]
- Kittles RA, Perola M, Peltonen L et al: Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 1998; 62: 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palo JU, Ulmanen I, Lukka M, Ellonen P, Sajantila A: Genetic markers and population history: Finland revisited. Eur J Hum Genet 2009; 17: 1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J et al: A high-resolution recombination map of the human genome. Nat Genet 2002; 31: 241–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.