Abstract
Colorectal cancer (CRC) is one of the most common neoplasms in the world. Fanconi anemia (FA) is a very rare genetic disease causing bone marrow failure, congenital growth abnormalities and cancer predisposition. The comprehensive FA DNA damage repair pathway requires the collaboration of 53 proteins and it is necessary to restore genome integrity by efficiently repairing damaged DNA. A link between FA genes in breast and ovarian cancer germline predisposition has been previously suggested. We selected 74 CRC patients from 40 unrelated Spanish families with strong CRC aggregation compatible with an autosomal dominant pattern of inheritance and without mutations in known hereditary CRC genes and performed germline DNA whole-exome sequencing with the aim of finding new candidate germline predisposition variants. After sequencing and data analysis, variant prioritization selected only those very rare alterations, producing a putative loss of function and located in genes with a role compatible with cancer. We detected an enrichment for variants in FA DNA damage repair pathway genes in our familial CRC cohort as 6 families carried heterozygous, rare, potentially pathogenic variants located in BRCA2/FANCD1, BRIP1/FANCJ, FANCC, FANCE and REV3L/POLZ. In conclusion, the FA DNA damage repair pathway may play an important role in the inherited predisposition to CRC.
Introduction
Colorectal cancer (CRC) is the third most frequent neoplasm in the world and its average lifetime risk in the general population is ∼5%.1 There is some degree of familial aggregation in up to 35% of CRC patients, but the majority of the underlying germline predisposition factors remain still unidentified. The Mendelian CRC syndromes, with Lynch syndrome and familial adenomatous polyposis being the most common, correspond to 5% of total CRC cases and are mainly due to germline mutations in APC, MUTYH and the mismatch repair genes (ie, MLH1, MSH2, MSH6, PMS2). Recently, next-generation sequencing efforts in familial CRC have identified additional causative germline mutations in genes such as POLE, POLD1 and NTHL1.2, 3, 4
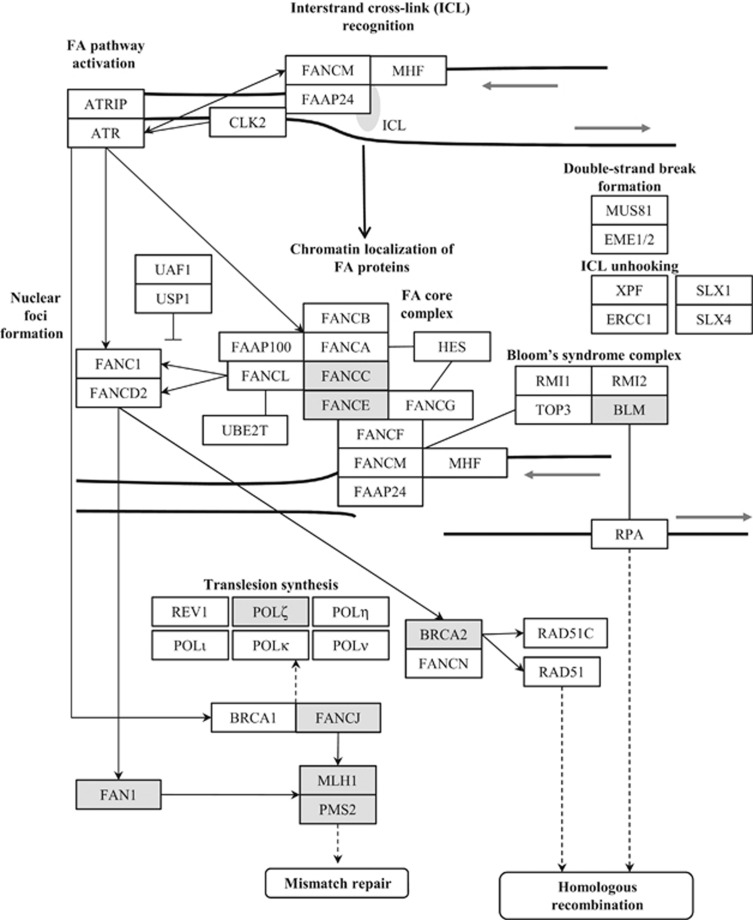
Fanconi anemia (FA) is a very rare genetic disease with an incidence of 1–3 per 500 000 births and it causes bone marrow failure, congenital growth abnormalities and cancer predisposition. FA patients have chromosome fragility and hypersensitivity to drugs that induce DNA interstrand crosslinks (ICLs).5 It corresponds to an autosomal recessive condition and it has been associated with germline mutations in 18 FA genes. Among them, monoallelic mutations in FANCD1/BRCA2, FANCJ/BRIP1, FANCN/PALB2 and FANCC have also been linked to breast and ovarian cancer genetic predisposition.6, 7 The comprehensive FA DNA damage repair pathway requires the collaboration of 53 proteins and it is necessary to restore genome integrity by efficiently repairing damaged DNA, especially ICLs (Figure 1). ICLs affect both DNA strands impeding transcription and replication-fork progression and also complicating correct DNA repair as there is no unaffected template available.8 Besides the link between FA genes and breast and ovarian cancer, some other genes not contributing to FA but part of the FA DNA damage repair pathway have additionally been involved in the same cancer predisposition and include BRCA1, RAD51C and FANCM.9 Very recently, mutations in some FA DNA damage repair pathway genes have also been postulated to be the germline triggers in familial CRC cases, including BRCA2,10, 11 FAN112 and BLM.13
Figure 1.
The Fanconi anemia (FA) DNA damage repair pathway. Genes linked to colorectal cancer (CRC) predisposition by the present report and previous evidence are shaded in gray (adapted from KEGG database, http://www.genome.jp/kegg/pathway.html).
Materials and methods
We selected 74 CRC probands from 40 unrelated Spanish families (4 affected relatives from 1 family, 3 affected relatives from 8 families, 2 affected relatives from 15 families and 16 CRC unrelated patients) with strong CRC aggregation compatible with an autosomal dominant pattern of inheritance and without point mutations or large rearrangements in the most common hereditary CRC genes (APC, MUTYH and mismatch repair genes). Families were selected based on the following criteria: ≥3 relatives with CRC, ≥2 consecutive affected generations and at least one CRC diagnosed before the age of 60 years. This study was approved by the institutional ethics committee of each participating hospital and written informed consent was obtained at CRC diagnosis.
Sequencing, raw data analysis and variant filtering was performed as previously described for a subset of 42 patients.14 In this regard, it should be noted that this previous cohort was completed with 31 additional new CRC patients, corresponding to 11 new families and 5 new cases in previously analyzed families, totaling 74 CRC probands from 40 families. Briefly, germline DNA whole-exome sequencing (WES) used the HiSeq2000 platform (Illumina, San Diego, CA, USA) and SureSelectXT Human All Exon for exon enrichment V4 (Agilent, Santa Clara, CA, USA). Mean coverage was >95 × in all samples and 51 Megabases was the target size that required ∼4 Gigabytes of sequencing per sample. After sequencing and data analysis, variant prioritization selected only those very rare alterations (0–0.1%), shared by individuals sequenced from the same family, producing a putative loss of function and located in genes with a role compatible with cancer (Supplementary Table 1). Variants were validated by Sanger sequencing (GATC Biotech, Köln, Germany) and segregation analysis of the prioritized variants was performed in additional affected family members (CRC and advanced adenoma) when constitutive DNA was available. Genetic variants have been submitted to the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/; accession numbers SCV000262600, SCV000262601, SCV000262602, SCV000262603, SCV000262604 and SCV000262605). In addition, somatic loss of heterozygosity (LOH) was studied by Sanger sequencing or microsatellites in tumor DNA of patients (one per family) carrying the selected variants when possible. DNA was extracted from a percentage of tumor cells of 70–80% in most cases.
Results and discussion
The aim of our study was to find candidate germline predisposition variants by performing exome sequencing in a cohort of familial CRC patients compatible with an autosomal dominant inheritance and without an alteration in the previously known hereditary CRC genes in order to facilitate genetic counseling and correctly address prevention strategies. Our preliminary results for a subset of CRC patients were previously published14 but additional family members and new families were whole-exome sequenced more recently and the present report corresponds to the analysis of the complete cohort of 74 samples.
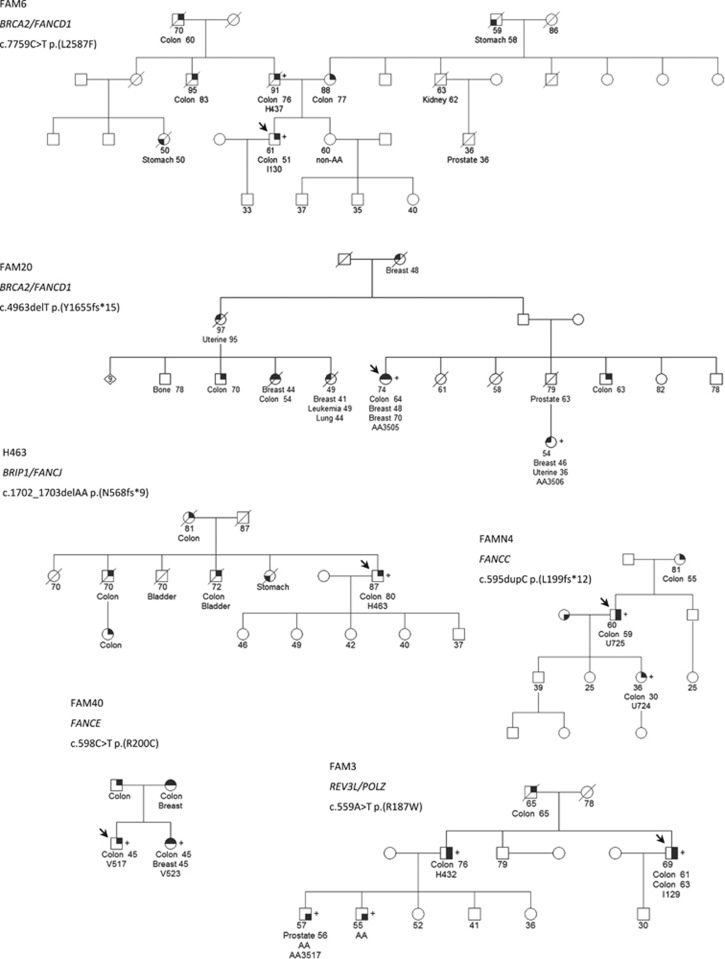
Interestingly, our data revealed heterozygous, rare, potentially pathogenic variants in six families located in genes belonging to the FA DNA damage repair pathway including BRCA2/FANCD1, BRIP1/FANCJ, FANCC, FANCE and REV3L/POLZ after data analysis and variant prioritization (Table 1 and Figure 2). All six prioritized variants were validated by Sanger sequencing (Supplementary Figure 1) and segregation analysis was correct in family members (CRC and advanced adenoma). It is also interesting to point out that among the 1–57 variants identified by WES that remained after filtering in each family, the genetic variants finally prioritized corresponded to the best candidate for genetic predisposition to CRC (not reported or with a very low frequency <0.01% in external control exome data sets including a Spanish database, potential loss-of-function variant affecting residues highly conserved in evolution, previous implication with cancer predisposition and correct segregation). It is important to highlight that three of the variants corresponded to frameshift alterations and three to missense variants. The former are expected to truncate the protein and, therefore, are likely pathogenic, whereas for the latter pathogenicity is plausible but has not been proven with functional studies. On the other hand, the identified missense variants comply with several criteria to be potentially deleterious and two of them (those in BRCA2 and FANCE) that fall in protein domains where pathogenic mutations have been previously reported in FA or familial breast and ovarian cancer patients in the ClinVar database or the FA mutation database (http://www.rockefeller.edu/fanconi/). In addition, when comparing with a publically available genetic variation control data set (Exome Aggregation Consortium), we detected a clear enrichment for putative loss-of-function variants in these FA DNA damage repair pathway genes in our familial CRC cohort, when considering nonsense, canonical splice site, frameshift and missense variants with a likely pathogenicity prediction (CADD >15, Combined Annotation Dependent Depletion, CADD, http://cadd.gs.washington.edu/) and a genotype frequency <0.01% (6/40, 15% vs 2546/60 706, 4.2% χ2 test, uncorrected for multiple testing P-value=0.0025), or only nonsense, canonical splice site and frameshift with a genotype frequency <0.01% (3/40, 7.5% vs 306/60 706, 0.5% χ2 test, uncorrected for multiple testing P-value <0.0001). It is also worth mentioning that in 5 of the 6 families carrying the reported variants, other neoplasms besides CRC were present with an age of onset <60 years, including breast cancer, endometrial cancer, prostate cancer, lung cancer, leukemia and gastric cancer. No relevant clinical or histopathological characteristics were detected among variant carriers, although a small sample size could be precluding the detection of such correlation.
Table 1. Description of the 6 genetic variants belonging to the Fanconi anemia DNA damage repair pathway detected in a cohort of 40 Spanish colorectal cancer families.
| Family | Gene | RefSeq | Genetic variant | Genotype frequency (ExAC, EVS, CSVS) | In silico | Family phenotype (age <60) | Cancer-AA carriers | LOH | Domain/region | OMIM |
|---|---|---|---|---|---|---|---|---|---|---|
| FAM6 | BRCA2/FANCD1 | NM_000059.3 | c.7759C>T p.(L2587F) | 5/60676 0/6503 0/572 | 5/5 | CRC, gastric | 2/2 | INCa | Interaction with DSS1 | Breast and ovarian cancer, FA |
| FAM20 | BRCA2/FANCD1 | NM_000059.3 | c.4963delT p.(Y1655fs*15) | 0/60706 0/6503 0/572 | FS | CRC, breast cancer, endometrial cancer, prostate cancer, lung cancer, leukemia | 2/2 | INCa | — | Breast and ovarian cancer, FA |
| H463 | BRIP1/FANCJ | NM_032043.2 | c.1702_1703delAA p.(N568fs*9)b | 0/60706 0/6503 0/572 | FS | CRC, gastric cancer | 1/1 | INCa | — | Breast and ovarian cancer, Spanish FA J familyc |
| FAMN4 | FANCC | NM_000136.2 | c.591_592dupC p.(L199fs*12) | 0/60706 0/6503 0/572 | FS | CRC, adenomas | 2/2 | N | — | Breast cancer, pancreatic cancer, FA |
| FAM40 | FANCE | NM_021922.2 | c.598C>T p.(R200C) | 0/60706 1/6503 0/572 | 5/5 | CRC, breast cancer | 2/2 | NA | Interaction with FANCC | FA, esophageal and gastric cancer |
| FAM3 | REV3L/POLZ | NM_002912.4 | c.559A>T p.(R187W) | 0/60706 0/6503 0/572 | 5/5 | CRC, prostate cancer, adenomas | 4/4 | Y | — | Lung cancer, chromosomal instability |
Abbreviations: AA, advanced adenoma; CRC, colorectal cancer; FA, Fanconi anemia; FS, frameshift; INC, inconclusive; LD, linkage disequilibrium; LOH, loss of heterozygosity; N, no; NA, not available; RefSeq, reference sequence; Seg, segregation; Y, yes.
Genotype frequency: presence or absence in external control exome data sets (ExAC (exome aggregation consortium), EVS (exome variant server) and CSVS (CIBERER Spanish variant server)).
In silico: number of deleterious predictions by bioinformatics tools used (CADD, PolyPhen, SIFT, PhyloP and LRT).
Cancer-AA carriers: number of cancer/advanced adenoma cases within the family that carry the variant.
LOH: depletion of the wild-type allele in tumor DNA in comparison with the germline.
Domain/region: the protein domain or region where the variant is located.
OMIM: OMIM database information including previous hereditary cancer involvement.
Minimal LOH was observed.
Previously reported in Esteban-Jurado et al.14
Same variant in our CRC cohort and in the Spanish FA J family.
Figure 2.
Pedigrees from families FAM3, FAMN4, FAM6, FAM20, FAM40 and H463 are shown. Filled symbol indicate affected for CRC (upper right quarter) adenoma/s (lower right quarter), stomach cancer (lower left quarter) and breast cancer (upper left quarter). Colon, breast, stomach, lung, prostate, kidney, uterine, leukemia and bladder refer to the type of cancer. AA/non-AA, advanced adenoma/nonadvanced adenoma; +, variant carrier. Index cases are indicated with an arrow.
Taking into account our results and previous evidence,10, 11, 12, 13 our report draws the attention to the fact that the FA DNA damage repair pathway may play an important role in the inherited predisposition to CRC. It is also important to highlight that pleiotropy is becoming important in germline predisposition to cancer as a higher number of genes may be involved in the genetic predisposition to a broader spectrum of neoplasms, as evidenced by our results and others.11 Finally, it could be hypothesized that defects in the FA DNA damage repair pathway would affect correct homologous recombination and contribute to genome instability. However, the contribution to CRC predisposition of genetic variants in this pathway needs further investigation and collaborative efforts should be made in order to fully characterize it. If this involvement is further confirmed in additional familial CRC cohorts, it would become very relevant regarding the molecular genetic diagnosis for the hereditary forms of this neoplasm.
Acknowledgments
We are sincerely grateful to the patients and their families for their participation. CE-J and JM are supported by a contract from CIBERehd. MV-C is supported by Ministerio de Educación, Cultura y Deporte (FPU12/05138). PG is supported by a contract from the Fondo de Investigación Sanitaria (JR13/00013). CIBERehd and CIBERER are funded by the Instituto de Salud Carlos III. This work was supported by grants from the Fondo de Investigación Sanitaria/FEDER (13/02588, 14/00173, 14/00230), the Ministerio de Economía y Competitividad (SAF2014-54453-R), Fundación Científica de la Asociación Española contra el Cáncer (GCB13131592CAST), COST Action BM1206, Beca Grupo de Trabajo ‘Oncología' AEG (Asociación Española de Gastroenterología) and Agència de Gestió d'Ajuts Universitaris i de Recerca (Generalitat de Catalunya, 2014SGR255, 2014SGR135). We thank the Centre Nacional d'Anàlisi Genòmica and the Biobank of Hospital Clínic–IDIBAPS, Barcelona, for technical help. The work was carried out (in part) at the Esther Koplowitz Centre, Barcelona.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM: Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- Short E, Thomas LE, Hurley J, Jose S, Sampson JR: Inherited predisposition to colorectal cancer: towards a more complete picture. J Med Genet 2015; 52: 791–796. [DOI] [PubMed] [Google Scholar]
- Palles C, Cazier JB, Howarth KM et al: Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 2013; 45: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weren RD, Ligtenberg MJ, Kets CM et al: A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet 2015; 47: 668–671. [DOI] [PubMed] [Google Scholar]
- Kitao H, Takata M: Fanconi anemia: a disorder defective in the DNA damage response. Int J Hematol 2011; 93: 417–424. [DOI] [PubMed] [Google Scholar]
- Thompson ER, Doyle MA, Ryland GL et al: Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet 2012; 8: e1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Surrallés J: Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev 2015; 33: 32–40. [DOI] [PubMed] [Google Scholar]
- Walden H, Deans AJ: The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys 2014; 43: 257–278. [DOI] [PubMed] [Google Scholar]
- Economopoulou P, Dimitriadis G, Psyrri A: Beyond BRCA: new hereditary breast cancer susceptibility genes. Cancer Treat Rev 2015; 41: 1–8. [DOI] [PubMed] [Google Scholar]
- Garre P, Martín L, Sanz J et al: BRCA2 gene: a candidate for clinical testing in familial colorectal cancer type X. Clin Genet 2015; 87: 582–587. [DOI] [PubMed] [Google Scholar]
- Yurgelun MB, Allen B, Kaldate RR et al: Identification of a variety of mutations in cancer predisposition genes in patients with suspected Lynch syndrome. Gastroenterology 2015; 149: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguí N, Mina LB, Lázaro C et al: Germline mutations in FAN1 cause hereditary colorectal cancer by impairing DNA repair. Gastroenterology 2015; 149: 563–566. [DOI] [PubMed] [Google Scholar]
- de Voer RM, Hahn MM, Mensenkamp AR et al: Deleterious germline BLM mutations and the risk for early-onset colorectal cancer. Sci Rep 2015; 5: 14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban-Jurado C, Vila-Casadesús M, Garre P et al: Whole-exome sequencing identifies rare pathogenic variants in new predisposition genes for familial colorectal cancer. Genet Med 2015; 17: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.