Abstract
Molecular genetic testing for the 11p15-associated imprinting disorders Silver–Russell and Beckwith–Wiedemann syndrome (SRS, BWS) is challenging because of the molecular heterogeneity and complexity of the affected imprinted regions. With the growing knowledge on the molecular basis of these disorders and the demand for molecular testing, it turned out that there is an urgent need for a standardized molecular diagnostic testing and reporting strategy. Based on the results from the first external pilot quality assessment schemes organized by the European Molecular Quality Network (EMQN) in 2014 and in context with activities of the European Network of Imprinting Disorders (EUCID.net) towards a consensus in diagnostics and management of SRS and BWS, best practice guidelines have now been developed. Members of institutions working in the field of SRS and BWS diagnostics were invited to comment, and in the light of their feedback amendments were made. The final document was ratified in the course of an EMQN best practice guideline meeting and is in accordance with the general SRS and BWS consensus guidelines, which are in preparation. These guidelines are based on the knowledge acquired from peer-reviewed and published data, as well as observations of the authors in their practice. However, these guidelines can only provide a snapshot of current knowledge at the time of manuscript submission and readers are advised to keep up with the literature.
Clinical background
Silver–Russell syndrome
Silver–Russell syndrome (Russell–Silver syndrome; SRS; OMIM 180860) is mainly defined by severe intrauterine and postnatal growth restriction (<P3) associated with a variable spectrum of further features (for a review see Saal1). The typical SRS phenotype includes a relative macrocephaly, a triangular-shaped face with a prominent forehead, body and limb asymmetry, and feeding difficulties (for a review see Azzi et al2 and Wakeling et al3). Some patients show a mild motor and cognitive developmental delay, including learning difficulties and speech delay.3, 4 Furthermore, adult follow-up data indicate an increased risk for metabolic disorders in later life.5
The clinical presentation is variable and probably partly influenced by the mosaic distribution of molecular changes.6 Although several scoring systems have been suggested (for a review see Azzi et al7), the clinical diagnosis of SRS is difficult because of its molecular and clinical heterogeneity (Tables 1 and 2). Therefore, molecular testing is often applied to patients who do not fulfill the strict clinical criteria, resulting in a detection rate lower compared with that expected from studies based on clinically well-defined cohorts.8
Table 1. Molecular subtypes and their frequencies in the 11p15.5-associated imprinting disorders (frequencies of molecular subtypes are taken from refs 1, 6, 9).
| Chromosome | Type of mutations/epimutation | Frequency | MLID | Mosaicism possible | Recurrence risk |
|---|---|---|---|---|---|
| SRS (OMIM 180860)1,6 | |||||
| 11p15.5 | H19/IGF2:IG-DMR hypomethylation | 40–60% | 7–10% | Yes | Empirically low, only in rare cases increased due to genomic trans-acting mutations (eg, MLID) |
| Duplications/deletions | <1% | No | No | Up to 50%, depending on the gene content of the aberration and the sex of the parent contributing the affected allele. In case of duplication of the whole 11p15 region, SRS in case of a maternal and BWS in case of a paternal transmission. | |
| UPD | 1 case | — | Yes | Empirically low | |
| CDKN1C point mutation | 1 family | No | No | 0% or 50%, depending on the sex of the parent contributing the affected allele | |
| IGF2 point mutation | 1 family | No | No | 0 or 50%, depending on the sex of the parent contributing the affected allele | |
| 7 | upd(7)mat | 4–10% | No | No | Empirically low, but some may be high because of familial translocations |
| upd(7q)mat | Single cases | No | No | Empirically low | |
| Duplications/deletions/translocations affecting 7p13 and 7q32 | Single cases | No | No | Up to 50% | |
| 14q32 | upd(14)mat, epimutation, duplications | Single cases | Unknown | yes | Refer to TS14 literature |
| Whole genome | (Mosaic) maternal unidiploidy | Single cases | Yes | No | |
| (submicroscopic) chromosomal imbalances | ~1% | No | No | Up 50%, depending on the chromosome and type of rearrangement | |
| BWS (OMIM130650)9 | |||||
| 11p15.5 | KCNQ1OT1:TSS-DMR hypomethylation | 50–60% | 25% | Yes | Empirically low, only in some cases increased because of genomic trans-acting mutations (eg, MLID) |
| H19/IGF2:IG-DMR hypermethylation | 5–10% | No | Yes | 20% of patients carry OCT4/SOX2 binding site mutations or deletions within the ICR1. In general, the recurrence risk is empirically low, but if an underlying genetic defect is present it may be up to 50%, depending on the gender of the contributing carrier | |
| upd(11)pat | 20–25% | No | Yes | Empirically low, but some may be high due to translocations | |
| Duplications/deletions/(translocations) | 1–2% | No | No | Up to 50%, depending on the gene content of the aberration and the sex of the parent contributing the affected allele. In case of duplication of the whole 11p15 region, SRS in case of a maternal and BWS in case of a paternal transmission | |
| CDKN1C mutations | 5% (sporadic) 50% (familial) | No | No | 0% or 50%, depending on the sex of the parent contributing the affected allele | |
| Whole genome | (Mosaic) paternal unidiploidy | ~1%? | Yes | No | |
Abbreviations: BWS, Beckwith–Wiedemann syndrome; MLID, multilocus imprinting disturbance; SRS, Silver–Russell syndrome.
Table 2. Overview on the currently applied methods for 11p15.5 testing and their limitations.
| Suitability to detect | ||||||||
|---|---|---|---|---|---|---|---|---|
| Method | Loci per test | UPD | Epimutation | CNV | SNP | Suitable for prenatal testing | Average amount of DNA needed | Limitations |
| MS methods based on methylation-sensitive enzymes | ||||||||
| MS Southern blot | 1 | Y | Y | Y | N | (Y) | 5 μg | Large amounts of DNA, time-consuming. Main limitation is restriction site interrogation of epimutation at single CpGs within the DMRs Sensitivity is relatively low in comparison with the other techniques. No commercially available kit Only indirect discrimination of CNVs. In case of a positive result, further testing is required to discriminate the underlying change |
| MS-MLPA | Up to ~46a | Y | Y | Y | N | Y | 50–100 ng | Commercial kits. May have reduced sensitivity for detection of mosaic epimutation and UPD. SNVs can interfere. Dependent on the probe composition of the kit, a discrimination between the types of (epi)mutations and UPD can be possible |
| MS methods based on bisulfite treatment: these methods are not MS in sensu stricto, as they are based on primers equally amplifying unmethylated or methylated alleles | ||||||||
| MS PCR | 1 | Y | Y | Y | N | Y | 1 μg | No commercially available kit. Only indirect discrimination of CNVs. In case of a positive result, further testing is required to discriminate the underlying change |
| ASMM RTQ-PCR | 1 | Y | Y | N | N | Y | 150 ng | No commercially available kit. Only indirect discrimination of CNVs. In case of a positive result, further testing is required to discriminate the underlying change |
| MS-HRMA | 1 | Y | Y | Y | Y | Y | 1 μg | Does not examine discrete CpG sites but looks at regional profile. No commercially available kit Only indirect discrimination of CNVs. In case of a positive result, further testing is required to discriminate the underlying change |
| MS pyrosequencing | 1 | N | Y | Y | (Y) | Y | >100 ng | May have reduced sensitivity for detection of mosaic epimutation. Read-length limits analyzed CpGs per read. Only indirect discrimination of CNVs. In case of a positive result, further testing is required to discriminate the underlying change |
| MS-SNuPE | Up to 10 | Y | Y | Y | N | Y | 100 ng | No commercially available kit. Only indirect discrimination of CNVs. In case of a positive result, further testing is required to discriminate the underlying change |
| Non-MS tests for CNV and/or UPD detection | ||||||||
| Microsatellite analysis (STR) | 1 | Y | N | Y | N | Y | <20 ng per locus | DNA of at least one parent required; restricted to the detection of CNVs and UPD |
| qPCR | 1 | N | N | Y | N | Y | 200 ng | Only unbalanced alterations are detected. Flexible design targeting the region of interest; small CNVs can be detected |
| Molecular karyotyping (arrayCGH, SNP array) | Whole genome: depends on resolution | Y (SNP) N (CGH) | N | Y | N | Y | 250 ng | Only unbalanced alterations are detected Does not detect epimutations. No UPD detection by arrayCGH. Uneven probe coverage can leave CNVs undetected |
| DNA sequencing | Locus-specific | N | N | N | Y | Y | Depending on the size of the gene: ~60–200 ng | Only sequencing variants can be detected, other genomic variants including CNVs escape detection. In case of homozygosity of SNP large deletions should be considered |
Abbreviations: ASMM RTQ-PCR, allele-specific methylated multiplex real-time quantitative PCR; CHG, comparative genomic hybridization; CNV, copy number variation (deletions/duplications); MLPA, multiplex ligation probe-dependent amplification; MS, methylation-specific; MS-HRMA, MS-high-resolution melting analysis; MS-SNuPE, methylation-specific single-nucleotide primer extension; N, no; qPCR, quantitative PCR; SNP, single nucleotide variation – monogenic disease-causing variant; UPD, uniparental disomy; Y, yes.
The number of 46 probes refers to the conventional MLPA kits.
Beckwith–Wiedemann syndrome
Beckwith–Wiedemann syndrome (BWS; OMIM1306509) was initially called EMG syndrome from its three main features exomphalos/umbilical hernia, macroglossia and (neonatal) gigantism. Additional signs include neonatal hypoglycemia, hemihypertrophy, organomegaly (essentially kidney, liver, spleen, pancreas), earlobe creases, polyhydramnios, hemangioma and cardiomyopathy. In 5–7% of children, embryonal tumors (most commonly Wilm's tumor (WT)) are diagnosed (for a review see Brioude et al10). The clinical diagnosis of BWS is often difficult because of its variable presentation and overlap with other overgrowth syndromes (for a review see Shuman et al11), and hence several scoring systems have been suggested12, 13 (reviewed by Ibrahim et al14). Robust genotype/epigenotype–phenotype correlations have been established10, 14, 15, 16 (Table 1): hemihypertrophy is strongly associated with upd(11)pat, exomphalos with H19/IGF2:IG-DMR hypermethylation and CDKN1C mutations, and, most importantly, the risk for WT is predominant in H19/IGF2:IG-DMR hypermethylation and upd(11)pat. By contrast, other embryonal tumors such as hepatoblastoma, neuroblastoma and adrenal tumors are observed in patients with KCNQ1OT1:TSS-DMR or upd(11)pat, although at a much lower incidence. In patients with a mosaic paternal uniparental diploidy (also named 'genome-wide UPD'), further tumors can occur, with a later age of onset than in case of the other molecular subtypes. Hence, the determination of the molecular subtype is important for an individual prognosis and management. Nevertheless, phenotypic transitions are fluid, ranging from some features that include ‘incomplete BWS' to the full clinical picture. Thus, molecular testing for BWS is also suggested in case of isolated hemihypertrophy as molecular changes in 11p15.5 have also been identified in patients with isolated body asymmetry. It should furthermore be considered in bilateral WT, familial WT cases and WT patients with at least one feature of BWS.
The chromosomal region 11p15.5
One of the major imprinted regions in humans is localized to the short arm of chromosome 11 (11p15.5) (Figure 1). Several of the 11p15 genes are involved in human growth and development, as well as in tumorigenesis. As a result, SRS and BWS, as the two imprinting disorders (IDs) associated with 11p15.5 alterations, are clinically characterized by disturbed growth and, in the case of BWS, associated with an increased risk for tumors.
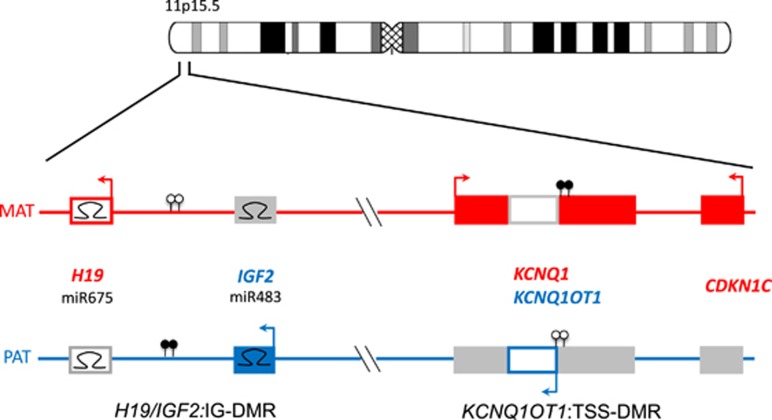
Figure 1.
The imprinted gene cluster in 11p15.5. It is divided in two functional domains whose imprinting is dependent on distinct ICRs (H19/IGF2:IG-DMR, KCNQ1OT1:TSS-DMR). (Filled boxes, protein coding genes; empty boxes, noncoding genes; Ω miRNAs; filled lollipops, methylated regions; empty lollipops, unmethylated regions; red, genes expressed from the maternal (mat) chromosome; blue, genes expressed from the paternal (pat) chromosome. Arrows above the genes, transcription direction of sense genes; arrows below the genes, transcription direction of anti-sense genes)
The chromosomal region 11p15.5 spans ~1 Mb and harbors two separate imprinting control regions (ICRs): the chromosomal region 11p15.5 spans ~1 Mb and harbors two separate ICRs (Supplementary Table 1): the telomeric one, ICR1, includes the H19/IGF2:IG-DMR, which is methylated on the paternal allele, whereas the centromeric one, ICR2, includes the KCNQ1OT1:TSS-DMR, which is maternally methylated.
In the telomeric ICR1 a different chromatin architecture of the parental alleles leads to a reciprocal expression of H19 and IGF2 (insulin-like growth factor 2). The ICR1 contains seven CTCF (CCCTC-binding factor) target sites in the DMR, which is located 2 kb upstream of H19. The zinc-finger binding factor CTCF binds to the maternally unmethylated ICR1 copy and thereby forms a chromatin boundary that blocks IGF2 and promotes H19 transcription of the maternal 11p15 genomic region. In contrast, the paternal methylation pattern forms alternative bounds and therefore expresses IGF2, which is involved in fetal development and growth.17 While the contribution of IGF2 to the pathoetiology of SRS has recently been shown,18 the function of H19 remains unknown, although it encodes several noncoding RNAs and binds factors that are implicates in epigenetic marks maintenance.19, 20, 21
The centromeric ICR2 in 11p15 regulates the expression of CDKN1C, KCNQ1 (potassium channel KQT-family member 1) and further genes and is methylated only on the maternal allele. Loss-of-function mutations in the maternally expressed CDKN1C gene account for 5% of sporadic and up to 50% of familial BWS cases (Table 1) (for mutations see: http://databases.lovd.nl/shared/genes/CDKN1C). Gain-of-function mutations are associated with growth retardation syndromes (IMAGe syndrome, SRS). The gene encodes a cyclin-dependent kinase inhibitor (p57KIP2) and is part of the p21CIP2Cdk inhibitor family. Another noncoding RNA gene in 11p15, KCNQ1OT1 (also known as LIT1), starts in intron 10 of the KCNQ1 gene. KCNQ1OT1 is expressed by the paternal allele and probably represses the expression of the CDKN1C gene on the same allele. Loss of methylation of the maternal ICR2 allele correlates with an increased expression of KCNQ1OT1. In BWS, one central physiological change caused by ICR2 (epi)mutations (hypomethylation at ICR2 as well as CDKN1C point mutations) is the reduced expression of (functional) CDKN1C.
Molecular disturbances in SRS and BWS
The types of mutations and epimutations in SRS and BWS affect the same genomic regions and genes, but in an opposite manner (Table 1). Four different types of molecular changes have been reported: epimutations (i.e. aberrant methylation at a DMR without a genomic alteration in the DMR itself), UPD, deletions/duplications of the DMRs and genomic point mutations in a gene the expression of which is regulated by the two germline DMRs. In particular, the identification of microduplications and microdeletions within 11p15.5 helped to define regulatory elements (i.e. DMRs, OCT4/SOX2 binding sites, enhancer motifs) responsible for a fine-tuned methylation and/or expression of 11p15.5 factors.22, 23 For molecular diagnostic testing, it is important to keep in mind that the predominant alterations in both disorders, the H19/IGF2:IG-DMR hypomethylation in SRS and the upd(11)pat and 11p15 epimutations in BWS, can occur in a mosaic state.
Silver–Russell syndrome
The majority of patients with the classical SRS phenotype7 carry molecular changes in 11p15: hypomethylation of the H19/IGF2:IG-DMR accounts for 40–60% of patients.7 About 1% of patients carry duplications or deletions affecting the region 11p15.5 (Table 1): the phenotype of these carriers is influenced by the size and gene content of the aberrations (for a review refs. 24, 25, 26). Upd(11p)mat and point mutations in the 11p15.5-encoded genes CDKN1C and IGF2 have only been described once each.18, 27, 28
Between 4 and 10% of SRS patients carry maternal UPD of chromosome 7 (upd(7)mat) or segmental upd(7q)mat (for a review see Eggermann et al29), in single cases chromosomal imbalances affecting either the GRB10 gene in 7p13 or the MEST gene in 7q32 have been reported (an LOVD entry for SRS-specific structural variations is currently in the submission process). Additionally, in a considerable number of patients (submicroscopic) alterations of chromosomes other than 7 and 11 may be detected.7, 30, 31 A number of SRS patients exhibiting (epi)mutations in 14q32 has recently been reported, and these molecular alterations correspond to findings in patients with Temple syndrome (TS14, OMIM616222). TS14 is an ID with changes affecting the IG-DMR and/or MEG3-DMR in 14q32, and its phenotype32 overlaps with SRS (for a review see Kagami et al33). In single cases, maternal uniparental disomy of chromosomes 16 and 20 (upd(16)mat, upd(20)mat) have been reported7, 34 (for a review see Eggermann et al35).
With the exception of patients with H19/IGF2:IG-DMR hypomethylation, the clinical findings in carriers of the other molecular changes do not always fit the clinical scoring suggested by different groups (for a review see Azzi et al7). In particular, in neonates and adults the decision on molecular testing cannot always be based on a convincing phenotype, but testing might also be applied to patients with less obvious growth parameters and dysmorphic signs.
Beckwith–Wiedemann syndrome
In more than 80% of BWS patients chromosome 11p15.5 epimutations or mutations can be detected affecting the ICR1 and/or ICR2 DMRs (for a review see Brioude et al10and Mussa et al16) (Table 1). Most BWS cases are sporadic but familial inheritance is observed in up to 15% of all cases.11
The most frequent change is the hypomethylation of the KCNQ1OT1:TSS-DMR, accounting for up to 60% of patients. The second most frequent alteration in BWS is upd(11)pat, detectable in nearly 20%. The H19/IGF2:IG-DMR is affected by hypermethylation in 5–10% of BWS cases. For this subgroup, it has recently been shown that OCT4/SOX2 binding site mutations or deletions encompassing these binding sites within the ICR1 cause the aberrant methylation36 by preventing de novo methylation of the maternal allele. Deletions, duplications and even balanced translocations in 11p15.537, 38 also contribute to the mutational spectrum, with the size and parental origin of the affected region influencing the phenotype. CDKN1C mutations are not only frequent in familial cases but are also of importance in sporadic BWS with a frequency of 5%.39 In the latter group, a significant number of patients exhibit a cleft palate.40 Familial cases mainly present with CDKN1C mutations (50%), chromosomal duplications/deletions or mutations in other genes/regulative elements (eg, OCT4/SOX2, CTCF binding sites).
Multilocus imprinting disturbances
The association between aberrations at specific imprinted genes and distinct congenital disorders is well known, but the number of reports on patients with generally disturbed methylation patterns (MLID) has increased recently (for a review see Mackay et al41). While the patients often exhibit a specific ID phenotype, for example, BWS, molecular testing reveals that aberrant methylation does not only affect the disease-specific imprinted loci (eg, 11p15 in BWS) but also other imprinted regions. In BWS, MLID is detected in ~25% of patients with hypomethylation of the KCNQ1OT1:TSS-DMRs, and in SRS in 7–10% of H19/IGF2:IG-DMR patients (Table 1). A common cause of MLID has not yet been reported, but mutations in trans-acting genes/factors have been identified in some cases.42, 43, 44 Thus, whole-exome/genome sequencing strategies or candidate gene analyses might be considered in these patients on a research basis and in cooperation with reference centers. Assisted reproductive technologies have also been associated with an increased risk of MLID.45
Molecular genetic testing strategies
Several diagnostic assays for the molecular diagnosis of SRS and BWS have been developed (Table 2), but the decision on the tests to be applied in a laboratory depends on many factors, including the equipment of the laboratory, the methodological experience, the pattern of referral, the in-house guidelines for diagnostic workup and the national strategy of diagnostic workup.
In any case, the laboratory has to be aware of the advantages and limitations of a test, and the diagnostic workup should follow the diagnostic algorithms shown in Figure 2. This diagnostic algorithm refers to samples received to confirm the clinical diagnosis of SRS or BWS. It does not account for cases referred for UPD analyses based on chromosomal findings or a precedent (family) history with an already known molecular disturbance.
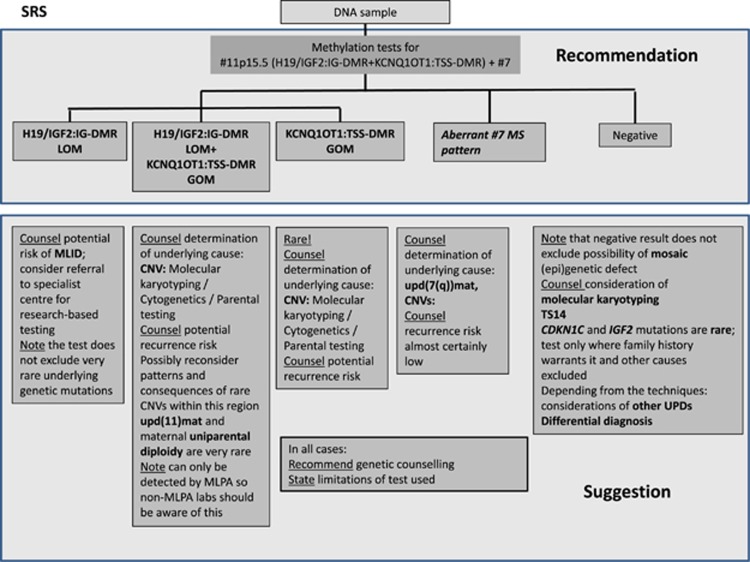
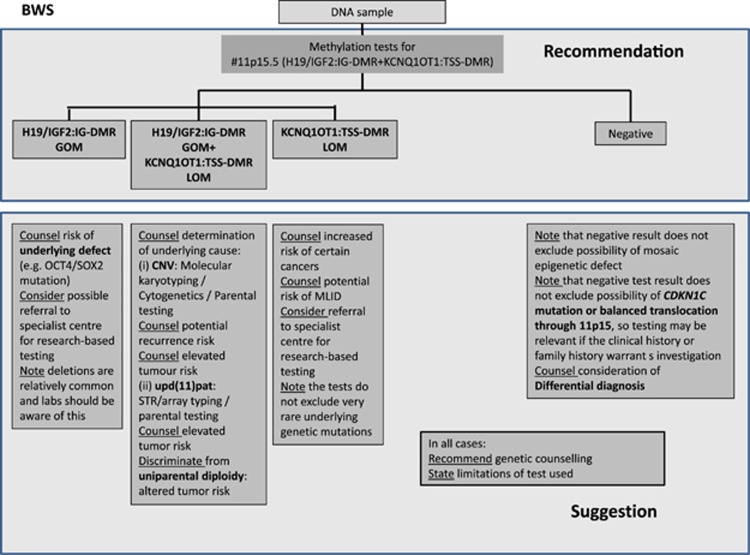
Figure 2.
Diagnostic algorithm for SRS (a) and BWS (b). For both disorders, methylation-specific tests for both imprinted regions in 11p15.5, and in case of SRS for the DMRs on chromosome 7 are recommended. The lab has to be aware of the possible outcomes and their interpretation. In case of a positive testing result, the subsequent discrimination is mandatory and has at least to be suggested in the report. Ditto, the further proceeding after a negative result has to be advised by the reporting geneticists.
First step: Methylation test for the 11p15.5 DMRs in SRS and BWS and for chromosome 7 in SRS
In both SRS and BWS, a methylation-sensitive (methylation-specific (MS)) approach targeting the two DMRs in 11p15.5 is recommended (Figure 2). In case of SRS, first-step analysis also has to include chromosome 7 methylation testing. Indeed, the majority of patients with chromosome 7 alterations carry an UPD of the whole chromosome, but there is a growing number on reports of patients with a segmental UPD for 7q (upd(7q)mat) including MEST.29 Furthermore, several patients with copy number variations affecting either the GRB10 locus in 7p13 or the MEST imprinted region in 7q32 have been reported, thus both imprinted regions have to be analyzed.
Further procedure for SRS and BWS after a positive testing result
With a positive result of methylation testing for 11p15.5 (or chromosome 7), the clinical diagnosis is confirmed. However, the discrimination between the different molecular subtypes is required (epimutations, UPDs, copy number variations (CNVs), translocations) as the basis for a personalized clinical management in BWS, and a directed genetic counseling in both disorders.16 Depending on the applied method, this discrimination may be possible in a single analysis or multiple tests have to be carried out (Figure 2). It depends on the diagnostic repertoire of the lab whether it offers this workup or whether it forwards the samples to reference centers. In case only the first step of diagnosis is performed, the required subsequent diagnostic steps and putative molecular results must be included in the report, including the associated recurrence risks (Figure 2 and Table 1).
Further procedure for SRS after a negative testing result
In case of a negative 11p15.5 and chromosome 7 testing result, testing for TS14-associated alterations and molecular karyotyping can be considered (Figure 2).
In contrast, mutations in CDKN1C and IGF2 have only been described in a single family each,18, 27 and screening studies in larger cohorts of sporadic SRS patients have so far been negative, and therefore these genes should only be analyzed after a careful re-evaluation of clinical findings and family history. The sequencing should not be included in routine diagnostic analysis for SRS. Additionally, differential diagnoses have to be considered (for a review see Saal1).
Indeed, further molecular changes have been identified in single cases (eg, upd(16)mat, upd(20)mat); thus, if warranted by the clinical features and/or family history of the patient, testing for these very rare (epi)mutations may be considered.
The complex molecular workup in SRS, including differential diagnosis (see below) can, in part, be circumvented by replacing single-step analyses for each imprinted locus by tests targeting different DMRs simultaneously in the same run ('multilocus testing'), thereby allowing the one-step identification of aberrant methylation patterns at different DMRs caused by other imprinting defects (eg, TS14, MLID) or uniparental diploidy.
Further procedure for BWS after a negative testing result
In case of a negative result of the methylation test for the 11p15.5 DMRs in a BWS patient, sequencing of the coding exons and the exon–intron boundaries of the CDKN1C gene should be considered in case of a positive family history, cleft palate and abdominal wall defect (umbilical hernia or exomphalos).
Nevertheless, nearly 30% of patients with a characteristic BWS phenotype still remain without a molecular diagnosis, and differential diagnoses should also be discussed after clinical re-evaluation (for a review see Shuman et al11).
Mosaicism in SRS and BWS
Mosaicism affects the majority of molecular disturbances in SRS and BWS,7, 41 that is, in a given sample the (epi)genetic defect is present only in a fraction of the total number of cells. Low-level mosaicism within a tissue may not be detected because of the detection limit of the methods used, leading to false-negative results. The fraction of aberrant cells may vary among different tissues. After clinical re-evaluation testing other tissues might be considered to exclude tissue-specific mosaicism.
Molecular genetic testing methods
Several techniques have been implemented in the molecular diagnostic testing algorithm of both SRS and BWS, and as the first EMQN pilot schemes for SRS and BWS in 2014 showed (http://www.imprinting-disorders.eu/wp-content/uploads/2015/07/EMQN-SRS-Summary-Report.pdf; http://www.imprinting-disorders.eu/wp-content/uploads/2015/07/EMQN-BWS-Summary-Report.pdf), different methods (Table 2) are suitable for diagnostic analysis. However, the choice of a technique to identify 11p15.5 and chromosome 7 disturbances should consider the loci to be analyzed and their precise genomic localization, the investigated tissue and the spectrum of occurring mutations and epimutations. Furthermore, none of these tests are certified for diagnostic use. Therefore, they have to be fully validated in the laboratory, and the normal reference ranges may be challenged by the possible occurrence of mosaicism. Furthermore, MS-specific tests should contain controls for complete digestion or bisulfite treatment.
Several methods do not allow the discrimination between the different types of molecular changes (upd, CNVs, epimutations) (Table 2), in this situation a combination of techniques has to be applied.
MS-specific techniques
Assays targeting altered methylation patterns can be roughly subdivided into two groups, those based on bisulfite-converted DNA and those using methylation-sensitive restriction enzymes (Table 2). With the exception of Southern blotting, all MS assays are based on the hybridization of short probes or primers; therefore, these tests are prone to be affected by single-nucleotide polymorphisms (SNPs) influencing the hybridization efficacy of complementary probe–primer sequences. Thus, false-positive results might occur, and positive results should therefore be confirmed – if available – by a second technique or other probes–primers, or at least by a second independent run.
The EMQN pilot scheme revealed that MS-specific multiplex ligation probe-dependent amplification (MLPA) is by far the most widely used test (>80%) in routine diagnostics for SRS and BWS and will be described below in more detail; for the other tests see Table 2.
MS-MLPA (Supplementary Figure 1) as an adaption to normal MLPA provides a tool to simultaneously detect changes of copy numbers and methylation. DNA is digested with a methylation-sensitive restriction enzyme after hybridization of the MLPA probes. Comparison of peak heights of undigested and digested DNA indicates the methylation level of a particular probe. Monitoring complete digestion is an essential quality control of the experiment. MS-MLPA just requires general laboratory equipment (thermocycler, capillary electrophoresis platform), and the bioinformatics analysis can be performed by freely available or commercially available software packages (eg, Coffalyser.Net (JSI, Freiburg, Germany); GeneMarker software (SoftGenetics, State College, PA, USA)). For data normalization, the parallel analysis of samples with normal genotype/epigenotype is recommended by the manufacturer. Quality and purity of DNA is an essential prerequisite for definitive MLPA data interpretation, and the labs should therefore be aware of it. In principle, DNA samples isolated with different methods and from different tissues might be used in parallel in the same experiment, but the methylation pattern in the tissues tested must be known and identical to the tissue tested. According to general good practice laboratory guidelines, a non-template control and at least one positive control should be run in parallel. Several MS-MLPA Kits are commercially available from MRC Holland (Amsterdam, Netherlands; http://www.mrc-holland.com).
Chromosome 11p15.5 testing (ME030 BWS/RSS Kit, MRC Holland)
The assay includes more than 20 probes specific for the region 11p15.5. Two probes serve as a control of the HhaI digestion in the methylation run. As controls for copy number quantification, probes outside 11p15.5 are included. Ten of the 11p15.5-specific probes are methylation-specific and target the germline H19/IGF2:IG-DMR (four probes) and KCNQ1OT1:TSS-DMR (four probes), additionally one probe each for the somatic DMRs of IGF2 and CDKN1C is included. The analysis of the same germline DMR by several probes reduces the risk of false-positive or false-negative results due to SNPs. Furthermore, if one probe fails and artificial hypomethylation of single probes occurs (eg, due to DNA extraction method or cell culture), there are three remaining probes to assess the methylation status.
The dosage analysis of the kit allows the identification of duplications and deletions in 11p15.5. In cases of a large duplication affecting the whole 11p15.5 region all probes are affected. Additionally, it also detects smaller CNVs restricted to parts of the region. However, the precise size and content of 11p15.5 CNVs should be determined by further suitable methods (eg, molecular karyotyping, qPCR).
Owing to the opposite imprinting patterns of the two 11p15.5 DMRs (H19/IGF2:IG-DMR is paternally methylated and KCNQ1OT1:TSS-DMR is maternally methylated), the methylation analysis of the ME030 assay allows both the identification and discrimination between the epimutations, and, in combination with the dosage analysis, between epimutations, UPDs and CNVs affecting both DMRs. Indeed, segmental UPDs of only one of the DMRs might be indistinguishable from isolated epimutations, but these have not yet been reported. In these situations, microsatellite typing might be applied.
Chromosome 7 testing (ME032 UPD7-UPD14 Kit, MRC Holland)
With this assay, upd(7)mat can be identified (Supplementary Figure 1). Similar considerations as for the interpretation of the ME030 Kit should be made, but in contrast to the 11p15.5 assay, and because of the methylation status of the targeted DMRs, a differentiation between UPD and epimutations is not possible and requires additional workup. However, with the exception of isolated hypermethylation of the GRB10 DMR,46 epimutations have not yet been reported. The advantage of this assay is the inclusion of imprinted loci on chromosomes 14 and 6, and thus molecular changes associated with TS14 as well as some MLID patterns can be detected in parallel.
Non-MS assays
Depending on the design and informativity of the initially applied MS test, an identification, confirmation and further characterization of genomic alterations (CNVs, SNPs, UPD) might be required. For these purposes, the following assays have been implemented with different advantages and limitations (Table 2):
Microsatellite typing (short tandem repeat typing, STR) is frequently applied for detection of UPDs, in particular in prenatal testing. In addition to the patient's DNA sample, a sample from at least one parent is required to delineate the parental origin of the two STR alleles. For whole arm UPD detection STRs are an efficient and inexpensive tool, but it has to be considered that a discrimination between (mosaic)UPD and deletions or duplications are not possible in every case (Supplementary figure 2). Furthermore, segmental UPDs of small fragments might escape detection. In case UPD has been determined solely by STR typing, the lab should be aware of the scenario of non-paternity. Microsatellite markers suitable for UPD analysis in context with SRS and BWS should be selected on the basis of their informativity (PIC>0.7) and their genomic localization (markers used in diagnostic testing are listed in Supplementary table 2).
Molecular karyotyping (Chromosomal Microarray) is a suitable method to determine the size and gene content of CNVs, but it has to be considered that the probe content of arrays is not standardized, and different assays have different coverages of the regions of interest. Therefore, very small imbalances might remain undetected and might rather be analyzed by qPCR. By SNP array analysis, uniparental isodisomic segments can in principal be detected and are therefore applicable in BWS testing,47 but small isodisomic segments cannot be discriminated from small homozygous stretches, and heterodisomy can only be identified by parallel analysis of parental samples and the use of additional bioinformatic tools. aCGH is not suitable for UPD detection.
Identification of single-nucleotide variants/point mutations should be performed by genomic sequencing, analyzing the genomic sequences defined as reference sequences (Supplementary Table 2). In case of sequencing genes (eg, CDKN1C), the appropriate guidelines (eg, of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (2015)48 or the Association for Clinical Genetic Sciences49 (http://www.acgs.uk.com/media/774853/evaluation_and_reporting_of_sequence_variants_bpgs_june_2013_-_finalpdf.pdf) should be followed. It must be noted that sequencing analysis of CDKN1C is complex as the gene is GC-rich and contains several non-pathogenic in-frame deletion variants.
Future methodological developments
Owing to the heterogeneous molecular findings in SRS and BWS, and the limitations of several of the currently applied tests, the diagnostic workup including discrimination of the molecular subtypes might become time-consuming and expensive. Therefore, the development of multilocus tests is in progress, either on the basis of conventional MS-MLPA by combining probes from different imprinted loci or by next-generation sequencing approaches. The latter harbors the potential to be more sensitive, as massive parallel sequencing might allow the detection of low-level mosaicism, and comprehensive inclusion of possible genomic and epigenetic changes.
Interpretation of diagnostic testing results
The precise identification and discrimination of the mutations or epimutations in patients referred with SRS or BWS features is the prerequisite for clinical management in BWS and a well-directed genetic counseling.
LOM at one of the 11p15.5 DMRs in SRS and BWS/MLID
LOM (Hypomethylation) at one of the two DMRs in 11p15.5 represents the most frequent diagnostic finding in the two 11p15.5-associated IDs (Table 1) and their identification confirms the clinical diagnosis of SRS and BWS, respectively. In general, the recurrence risk is low in these molecular subgroups,
However, there are reports on families with two BWS sibs,43, 50 with one sib showing a KCNQ1OT1:TSS-DMR hypomethylation and the second one showing the same epimutation but as part of an MLID. Although this situation seems to be rare, it illustrates that MLID also has to be considered in BWS testing. In general, MLID accounts for 7–10% of SRS with H19/IGF2:IG-DMR hypomethylation, and for 25% of patients with KCNQ1OT1:TSS-DMR hypomethylation in BWS. Assuming a general frequency for this molecular subtype of 50% in BWS cohorts, MLID is detectable in 12.5% of BWS patients. In case of MLID the recurrence risk is difficult to estimate because mutations in genes associated with a generally disturbed methylation have only been recently reported. The disease-causing variants may either have an autosomal recessive effect (eg, ZFP57 in transient neonatal diabetes mellitus42) or represent the recently defined maternal effect mutations, that is, the maternal genotype is responsible for the aberrant methylation in her offspring.43, 44
GOM at one of the DMRs in 11p15.5
Up to now, isolated GOM (hypermethylation) in 11p15.5 has been reported for BWS: hypermethylation of the H19/IGF2:IG-DMR might be difficult to detect in case of a low-level mosaicism. Although GOM mainly occurs sporadically, up to 20% of GOM patients carry genomic disease-causing variants within the OCT4/SOX2 binding site (BS) 5′ to the DMR36 and referral to specialist research centers can be considered. In case of detection of an OCT4/SOX2 BS pathogenic variants, it has to be evaluated individually. Another group of genomic variants affecting the H19/IGF2:IG-DMR are microdeletions that reduce the number of CTCF target sites and might be associated with the inactivation of the DMR: their size and spacing influence the degree of DNA methylation and the clinical phenotype.22 Careful characterization of these microdeletions is therefore needed to predict recurrence risks and phenotypical outcomes. In these cases, analysis of parental DNA should be considered to determine the mode of transmission, and the recurrence risk is 50% in the event of a maternally inherited disease-causing variant.
In SRS, GOM of the KCNQ1OT1:TSS-DMR is extremely rare and can be due to a small duplication of the maternal allele, thus requiring a careful workup by a specialized lab.
Detection of both LOM/GOM at the two 11p15.5 DMRs
Parallel occurrence of both LOM and GOM at the two 11p15.5 DMRs in the same patient either indicates UPD or CNVs. To discriminate these two alterations copy number analysis is required.
In BWS upd(11)pat contributes to up to 20% of cases, and it is mainly segmental (ie, restricted to the 11p15.5 region or the short arm of chromosome 11, and isodisomic),47 but some cases with upd(11)pat affecting the whole chromosome have been reported. STR determination of uniparental isodisomy can be hampered by the mosaic presence of the disturbance. Therefore, it might not only become a qualitative analysis but also a quantitative one. For upd(11)pat a postzygotic origin has been delineated and the recurrence risk is therefore not increased. Particular attention should be directed to (mosaic) uniparental diploidy (for a review see Kalish et al51). This genetic constitution is associated with further features additional to those of BWS, in particular with a severely increased risk for a broad-spectrum of (embryonal) tumors, and clinical surveillance and management is not covered by the conventional BWS tumor programs. A further problem in upd(11)pat is the variable degree of mosaicism, making the identification difficult or leading to a false interpretation: the increase of hybridization of the H19/IGF2:IG-DMR probes might be weaker than the decrease of the KCNQ1OT1:TSS-DMR probes, and might escape the detection.47 The application of a second method might be helpful to confirm the molecular suspicion of UPD, even in case the H19/IGF2:IG-DMR is in the upper control range.
Upd(11)mat is extremely rare in SRS,28 and only one case with mosaic maternal uniparental diploidy has been reported.
(Submicroscopic) chromosomal imbalances in 11p15.5 (CNV), that is, duplications, confirm the clinical diagnosis of SRS or BWS. After identification of a CNV in an SRS or a BWS patient, molecular karyotyping is indicated to confirm the abnormality and to determine its size as the content of genes and regulative elements of the aberration might explain additional clinical features of the patient, which might not belong to the SRS/BWS spectrum (an LOVD entry for SRS-specific CNVs is currently in the submission process). A cytogenetic/molecular-cytogenetic analysis of the patients and their parents might help to determine the nature of the imbalance and to estimate the recurrence risk in case of a familial rearrangement.
In case of a familial rearrangement, an autosomal-dominant inheritance with phenotypic expression depending on the sex of the contributing parent can be expected: in case the duplication affects the whole 11p15.5 region of the paternal allele, a BWS phenotype is probable, whereas an SRS-like clinical picture will occur in case the maternal chromosome carries the duplication. Thus, both phenotypes may occur in the same family.26
Smaller CNVs affecting only one of the 11p15.5 DMRs
Smaller duplications or deletions affecting only parts of the two 11p15.5 imprinting regions, may cause BWS, SRS or a normal phenotype, depending on the regulative and coding DNA sequences involved in the rearrangement and the sex of the contributing parent (for a review see Begemann et al25). Thus, a careful workup has to be applied in case of duplications or deletions of the imprinted regions in 11p15.5, and the geneticist has to estimate recurrence risks on the basis of the up-to-date literature.
Upd(7)mat in SRS patients
As the vast majority of upd(7)mat are products of a trisomy rescue, the recurrence risk is generally low and the only predisposing factor is advanced maternal age. However, two cases have been reported in which the upd(7)mat was the result of a familial chromosomal translocation, in this situation the recurrence risk is increased.52, 53 Thus, cytogenetic/molecular-cytogenetic analysis should be considered whenever there is a chance for a chromosomal aberration (eg, if a translocation or aberration in the family involving chromosome 7 or 11 is known), but need not be generally offered to UPD cases.
Pathogenic variants (in CDKN1C and IGF2)
In BWS, CDKN1C mutation analysis has to be considered owing to its frequency of up to 50% in familial cases. Indeed, in some labs it is performed as the first diagnostic step in families with BWS and in BWS patients with cleft palate. If a pathogenic variant in one of the genes is identified, genetic counseling should be offered and a segregation analysis in the families can be discussed. Comparable to chromosomal rearrangements, the recurrence risk is 50% for CDKN1C and IGF2 mutations, but it depends on the sex of the transmitting parent. A BWS phenotype will occur if a CDKN1C disease-causing variant is inherited from the mother, or if a de novo mutation affects the maternal allele, whereas there is no (or minimal) phenotypic effect in case the paternal gene copy carries the pathogenic variant. For SRS, maternal inheritance of a missense mutation located in the PCNA-binding domain of CDKN1C may lead to an SRS or IMAGe phenotype, whereas paternal inheritance of an IGF2 mutation results in SRS.
Normal chromosomes 11p15.5 and 7 methylation results in SRS
Normal chromosomes 11p15.5 and 7 methylation patterns do not rule out the clinical diagnosis of SRS, currently 30–40% of patients remain without a molecular confirmation of the clinical diagnosis.7 In routine screening without strict diagnostic inclusion criteria the detection rate is even lower.8 As mentioned before, molecular changes in the imprinted 14q32 region significantly contribute to the mutation spectrum7, 8, 33 and therefore overlap with TS14. TS14 testing is thus suggested as a further diagnostic step for patients with strong clinical suspicion of SRS, and it is already included in a commercially available MS-MLPA assay. Similar to that for SRS and BWS, TS14 testing should consist of appropriate assays to discriminate between the different molecular subgroups.
Furthermore, molecular karyotyping is indicated as a further molecular diagnostic step in patients with clear phenotypic features, and significantly contributes to the clarification of the cause of the phenotype and recurrence risks.
Owing to the mosaic tissue distribution of 11p15.5 epimutations,2 testing of other tissues (eg, fibroblasts) might also be considered after a careful clinical re-evaluation.
Normal 11p15.5 methylation result in BWS
The aforementioned (epi)mutations in the two germline DMRs in 11p15.5 account for 70–80% of patients with BWS, thus a negative result does not rule out BWS (Table 1). However, epimutations and upd(11)pat might escape detection owing to uneven mosaic distribution.54 Thus, the analysis of another tissue for epimutation might be considered if the routine 11p15.5 testing is negative.
For BWS no major further molecular subgroups have been identified, but differential diagnosis should also be considered.
Reporting
For different reasons it is not recommended to use the same templates for sample referral, reporting and interpretation of findings for BWS and SRS.
In general, the format of the reports has to correspond to general guidelines of reporting molecular genetic results according to International and the National Regulators (eg, from the Association for Clinical Genetic Science (Part of the British Society for Genetic Medicine; http://www.acgs.uk.com/media/949852/acgs_general_genetic_laboratory_reporting_recommendations_2015.pdf). Laboratories reporting (BWS/SRS) testing results should participate annually at least in external quality assessment. It is recommended that testing laboratories are accredited to international standards, for example, ISO 15189 or equivalent. In particular, it is essential that the report contains the following items:
unequivocal patient identifiers, numbering of pages,
the reason for referral,
source of DNA sample (eg, blood, buccal smear, amniocytes),
a consistent naming of the analyzed DMRs should be used in the same report, and recently a common nomenclature has been suggested the European Network of Congenital Imprinting Disorders EUCID.net (www.imprinting-disorders.eu)(Supplementary Table 1). The nomenclature of genomic mutations has to be in accordance with the suggestions of the HGVS (http://www.hgvs.org/mutnomen/),
applied methods and statement on the limitations and sensitivities including detection of mosaicism. Appropriate references should be given, if commercial kits are used, the kit version has to be mentioned (eg, ME030-C3; http://www.mrc-holland.com),
the precise result description,
interpretation of diagnostic testing results (see above),
statement whether the clinical diagnosis is confirmed or not,
statement on the significance of the molecular results for the clinical management: for SRS, the certainty of the molecular genetic diagnosis aids medical management and precludes further investigations for short stature. However, therapeutic measures such as growth hormone treatment, physiotherapy or managing of hypoglycemic are based on clinical symptoms rather than on the genetic diagnosis, and should therefore not be suggested in the report. In case of BWS, tumor surveillance should be advised in the report according to the national guidelines and as suggested by the Clinical Utility Gene card for BWS.9
in case the results of only single steps of the diagnostic algorithm (Figure 2) are reported, the consecutive steps must be suggested,
genetic counseling has to be recommended, and recurrence risk has to be mentioned.
Prenatal testing
The growing knowledge on the molecular basis of SRS and BWS and the increasing number of positively tested patients results in an increasing demand for prenatal testing for these diseases. Owing to the complexity of molecular testing and the numerous questions that raised the context of prenatal testing, and ranging from methodological questions to ethical topics, the authors have addressed this topic separately.55, 56 In principal, prenatal testing might be offered in specific situations, but the decision on prenatal testing for SRS or BWS requires careful and comprehensive discussions (including the limitations of methods and information value, ethical issues) between the families, the genetic counselors, the obstetrics and laboratories.
Differential diagnosis
In both 11p15.5-associated IDs, the clinical diagnosis is often difficult because of its variable presentation during childhood and adolescence (eg, with age the facial characteristics tend to become attenuated in SRS, and normalized in BWS); furthermore, the molecular and phenotypic findings overlap with other IDs and syndromes. Thus, after exclusion of the major molecular changes detectable in SRS or BWS (Figure 2), a clinical re-evaluation should be performed and differential clinical diagnoses should be considered (eg, IMAGe syndrome, 3M syndrome for SRS; Sotos, Weaver, Perlman, Simpson–Golabi–Behmel syndrome for BWS).1, 11 Recent reports indicate the contribution of new, so far neglected loci and chromosomes to SRS: patients with upd(16)mat or upd(20)mat show symptoms overlapping with SRS.7, 34 However, it is still too early to include them in routine diagnostic workups, and they should rather be included in research strategies.
Concluding remarks
These guidelines are based on the knowledge acquired from peer-reviewed and published data, as well as observations of the authors in their practice. Current practice of molecular testing and reporting of SRS and BWS was assessed by a first external pilot quality assessment scheme organized by the EMQN in 2014. In the two-pilot schemes for SRS and BWS, a total of 42 laboratories from 19 European and non-European countries for BWS and 31 diagnostic centers from 14 countries for SRS participated. Based on the results from these schemes and in context with activities of the European Network of Imprinting Disorders (EUCID.net) towards a consensus in diagnostics and management of SRS and BWS, best practice guidelines have been developed. Members of institutions working in the field of SRS and BWS diagnostics were invited to comment, and in the light of feedback amendments were made. The final document was ratified in the course of an EMQN best practice guideline meeting for 11p15.5 IDs on 3 October 2015 and in accordance with the general SRS and BWS consensus guidelines, which are in preparation.
These guidelines can only provide a snapshot of current knowledge at the time of manuscript submission. Readers are advised to keep up with the literature.
Acknowledgments
The authors KE, JB, FB, KB, SR, ZT, DM, GM, IN, PL, LS, MB, DP, ERM, MM, AR, PL, KG, DJM and TE are members of the European Network of Congenital Imprinting Disorders (EUCID.net), supported by COST (BM1208). TE, KB and DP are supported by the BMBF (grant number 01GM115A/B).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Saal HM: Russell-Silver Syndrome; in Pagon RA, Adam MP, Ardinger HH et al (eds): GeneReviews(R) Seattle, WA, USA: University of Washington, 1993. [Google Scholar]
- Azzi S, Blaise A, Steunou V et al: Complex tissue-specific epigenotypes in Russell–Silver syndrome associated with 11p15 ICR1 hypomethylation. Hum Mutat 2014; 35: 1211–1220. [DOI] [PubMed] [Google Scholar]
- Wakeling EL, Amero SA, Alders M et al: Epigenotype–phenotype correlations in Silver–Russell syndrome. J Med Genet 2010; 47: 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder G, Liebl M, Woelfle J, Eggermann T, Blumenstock G, Schweizer R: Adult height and epigenotype in children with Silver–Russell syndrome treated with GH. Horm Res Paediatr 2013; 80: 193–200. [DOI] [PubMed] [Google Scholar]
- Takenouchi T, Awazu M, Eggermann T, Kosaki K: Adult phenotype of Russell–Silver syndrome: a molecular support for Barker–Brenner's Theory. Congenit Anom (Kyoto) 2015; 55: 167–169. [DOI] [PubMed] [Google Scholar]
- Bliek J, Terhal P, van den Bogaard MJ et al: Hypomethylation of the H19 gene causes not only Silver–Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am J Hum Genet 2006; 78: 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi S, Salem J, Thibaud N et al: A prospective study validating a clinical scoring system and demonstrating phenotypical-genotypical correlations in Silver–Russell syndrome. J Med Genet 2015; 52: 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T, Heilsberg AK, Bens S et al: Additional molecular findings in 11p15-associated imprinting disorders: an urgent need for multi-locus testing. J Mol Med (Berl) 2014; 92: 769–777. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Algar E, Lapunzina P et al: Clinical utility gene card for: Beckwith–Wiedemann syndrome. Eur J Hum Genet 2014, e-pub ahead of print 3 July 2013 doi:10.1038/ejhg.2013.132. [DOI] [PMC free article] [PubMed]
- Brioude F, Lacoste A, Netchine I et al: Beckwith–Wiedemann syndrome: growth pattern and tumor risk according to molecular mechanism, and guidelines for tumor surveillance. Horm Res Paediatr 2013; 80: 457–465. [DOI] [PubMed] [Google Scholar]
- Shuman C, Beckwith JB, Smith AC, Weksberg R: Beckwith–Wiedemann syndrome; in Pagon RA, Adam MP, Ardinger HH et al (eds): GeneReviews (R) Seattle, WA, USA: University of Washington, 1993. [Google Scholar]
- Elliott M, Bayly R, Cole T, Temple IK, Maher ER: Clinical features and natural history of Beckwith–Wiedemann syndrome: presentation of 74 new cases. Clin Genet 1994; 46: 168–174. [DOI] [PubMed] [Google Scholar]
- DeBaun MR, Tucker MA: Risk of cancer during the first four years of life in children from the Beckwith–Wiedemann Syndrome Registry. J Pediatr 1998; 132: 398–400. [DOI] [PubMed] [Google Scholar]
- Ibrahim A, Kirby G, Hardy C et al: Methylation analysis and diagnostics of Beckwith–Wiedemann syndrome in 1,000 subjects. Clin Epigenet 2014; 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparago A, Cerrato F, Vernucci M, Ferrero GB, Silengo MC, Riccio A: Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith–Wiedemann syndrome. Nat Genet 2004; 36: 958–960. [DOI] [PubMed] [Google Scholar]
- Mussa A, Russo S, De Crescenzo A et al: (Epi)genotype–phenotype correlations in Beckwith–Wiedemann syndrome. Eur J Hum Genet 2016; 24: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M, Hemberger M, Hughes J et al: Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 2002; 417: 945–948. [DOI] [PubMed] [Google Scholar]
- Begemann M, Zirn B, Santen G et al: Paternally inherited IGF2 mutation and growth restriction. N Engl J Med 2015; 373: 349–356. [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR: The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna 2007; 13: 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L: H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci USA 2013; 110: 20693–20698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Smits G, Fraser P, Reik W, Paro R: Non-coding transcripts in the H19 imprinting control region mediate gene silencing in transgenic Drosophila. EMBO Rep 2007; 8: 1068–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beygo J, Citro V, Sparago A et al: The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum Mol Genet 2013; 22: 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Crescenzo A, Sparago A, Cerrato F et al: Paternal deletion of the 11p15.5 centromeric-imprinting control region is associated with alteration of imprinted gene expression and recurrent severe intrauterine growth restriction. J Med Genet 2013; 50: 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt D, Enklaar T, Gartner-Rupprecht B et al: Microdeletion and IGF2 loss of imprinting in a cascade causing Beckwith–Wiedemann syndrome with Wilms' tumor. Nat Genet 2005; 37: 785–786 786–787. [DOI] [PubMed] [Google Scholar]
- Begemann M, Spengler S, Gogiel M et al: Clinical significance of copy number variations in the 11p15.5 imprinting control regions: new cases and review of the literature. J Med Genet 2012; 49: 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli L, Sparago A, De Crescenzo A et al: Silver–Russell syndrome and Beckwith–Wiedemann syndrome phenotypes associated with 11p duplication in a single family. Pediatr Dev Pathol 2010; 13: 326–330. [DOI] [PubMed] [Google Scholar]
- Brioude F, Oliver-Petit I, Blaise A et al: CDKN1C mutation affecting the PCNA-binding domain as a cause of familial Russell–Silver syndrome. J Med Genet 2013; 50: 823–830. [DOI] [PubMed] [Google Scholar]
- Bullman H, Lever M, Robinson DO, Mackay DJ, Holder SE, Wakeling EL: Mosaic maternal uniparental disomy of chromosome 11 in a patient with Silver–Russell syndrome. J Med Genet 2008; 45: 396–399. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Schonherr N, Jager S et al: Segmental maternal UPD(7q) in Silver–Russell syndrome. Clin Genet 2008; 74: 486–489. [DOI] [PubMed] [Google Scholar]
- Bruce S, Hannula-Jouppi K, Puoskari M et al: Submicroscopic genomic alterations in Silver–Russell syndrome and Silver–Russell-like patients. J Med Genet 2010; 47: 816–822. [DOI] [PubMed] [Google Scholar]
- Spengler S, Begemann M, Ortiz Bruchle N et al: Molecular karyotyping as a relevant diagnostic tool in children with growth retardation with Silver–Russell features. J Pediatr 2012; 161: 933–942. [DOI] [PubMed] [Google Scholar]
- Ioannides Y, Lokulo-Sodipe K, Mackay DJ, Davies JH, Temple IK: Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J Med Genet 2014; 51: 495–501. [DOI] [PubMed] [Google Scholar]
- Kagami M, Mizuno S, Matsubara K et al: Epimutations of the IG-DMR and the MEG3-DMR at the 14q32.2 imprinted region in two patients with Silver–Russell syndrome-compatible phenotype. Eur J Hum Genet 2015; 23: 1062–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulchandani S, Bhoj EJ, Luo M et al: Maternal uniparental disomy of chromosome 20: a novel imprinting disorder of growth failure. Genet Med 2015; 18: 309–315. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Perez de Nanclares G, Maher ER et al: Imprinting disorders: a group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin Epigenet 2015; 7: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi Habib W, Azzi S, Brioude F et al: Extensive investigation of the IGF2/H19 imprinting control region reveals novel OCT4/SOX2 binding site defects associated with specific methylation patterns in Beckwith–Wiedemann syndrome. Hum Mol Genet 2014; 23: 5763–5773. [DOI] [PubMed] [Google Scholar]
- Smith AC, Suzuki M, Thompson R et al: Maternal gametic transmission of translocations or inversions of human chromosome 11p15.5 results in regional DNA hypermethylation and downregulation of CDKN1C expression. Genomics 2012; 99: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach S, Capri Y, Rossignol S et al: Beckwith–Wiedemann syndrome and long QT syndrome due to familial-balanced translocation t(11;17)(p15.5;q21.3) involving the KCNQ1 gene. Clin Genet 2013; 84: 78–81. [DOI] [PubMed] [Google Scholar]
- Lam WW, Hatada I, Ohishi S et al: Analysis of germline CDKN1C (p57KIP2) mutations in familial and sporadic Beckwith–Wiedemann syndrome (BWS) provides a novel genotype-phenotype correlation. J Med Genet 1999; 36: 518–523. [PMC free article] [PubMed] [Google Scholar]
- Brioude F, Netchine I, Praz F et al: Mutations of the imprinted CDKN1C gene as a cause of the overgrowth Beckwith–Wiedemann syndrome: clinical spectrum and functional characterization. Hum Mutat 2015; 36: 894–902. [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Eggermann T, Buiting K et al: Multilocus methylation defects in imprinting disorders. Biomol Concepts 2015; 6: 47–57. [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM et al: Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 2008; 40: 949–951. [DOI] [PubMed] [Google Scholar]
- Meyer E, Lim D, Pasha S et al: Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith–Wiedemann Syndrome). PLoS Genet 2009; 5: e1000423–e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty LE, Rezwan FI, Poole RL et al: Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun 2015; 6: 8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee L, Lim DH, Dias RP et al: Epimutation profiling in Beckwith–Wiedemann syndrome: relationship with assisted reproductive technology. Clin Epigenet 2013; 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T, Schneider-Rätzke B, Begemann M, Spengler S: Isolated hypermethylation of GRB10 (7p12.2) in a Silver–Russell syndrome patient carrying a 20p13 microdeletion. Clin Genet 2014; 85: 399–400. [DOI] [PubMed] [Google Scholar]
- Keren B, Chantot-Bastaraud S, Brioude F et al: SNP arrays in Beckwith–Wiedemann syndrome: an improved diagnostic strategy. Eur J Med Genet 2013; 56: 546–550. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis Y, Payne S, McAnulty C et al: Practice Guidelines for the Evaluation of Pathogenicity and the Reporting of Sequence Variants in Clinical Molecular Genetics2013, Available from http://www.acgs.uk.com/media/774853/evaluation_and_reporting_of_sequence_variants_bpgs_june_2013_-_finalpdf.pdf.
- Niederhoffer KY, Penaherrera M, Pugash D et al: Beckwith–Wiedemann syndrome in sibs discordant for IC2 methylation. Am J Med Genet A 2012; 158a: 1662–1669. [DOI] [PubMed] [Google Scholar]
- Kalish JM, Conlin LK, Bhatti TR et al: Clinical features of three girls with mosaic genome-wide paternal uniparental isodisomy. Am J Med Genet A 2013; 161a: 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnecke A, Hinderhofer K, Jauch A, Janssen JW, Moog U: Silver–Russell syndrome due to maternal uniparental disomy 7 and a familial reciprocal translocation t(7;13). Clin Genet 2012; 82: 494–498. [DOI] [PubMed] [Google Scholar]
- Dupont JM, Cuisset L, Cartigny M et al: Familial reciprocal translocation t(7;16) associated with maternal uniparental disomy 7 in a Silver–Russell patient. Am J Med Genet 2002; 111: 405–408. [DOI] [PubMed] [Google Scholar]
- Alders M, Maas SM, Kadouch DJ et al: Methylation analysis in tongue tissue of BWS patients identifies the (EPI)genetic cause in 3 patients with normal methylation levels in blood. Eur J Med Genet 2014; 57: 293–297. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Brioude F, Russo S et al: Prenatal molecular testing for Beckwith–Wiedemann and Silver–Russell syndromes: a challenge for molecular analysis and genetic counseling. Eur J Hum Genet 2015, e-pub ahead of print 28 October 2015 doi:10.1038/ejhg.2015.224. [DOI] [PMC free article] [PubMed]
- Eggermann T, Buiting K, Temple IK: Clinical utility gene card for: Silver–Russell syndrome. Eur J Hum Genet 2011, e-pub ahead of print 8 December 2010; doi:10.1038/ejhg.2010.202. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.