Abstract
Background:
Immune cell infiltration in tumor is an emerging prognostic biomarker in breast cancer. The gold standard for quantification of immune cells in tissue sections is visual assessment through a microscope, which is subjective and semi-quantitative. In this study, we propose and evaluate an approach based on antibody-guided annotation and deep learning to quantify immune cell-rich areas in hematoxylin and eosin (H&E) stained samples.
Methods:
Consecutive sections of formalin-fixed parafin-embedded samples obtained from the primary tumor of twenty breast cancer patients were cut and stained with H&E and the pan-leukocyte CD45 antibody. The stained slides were digitally scanned, and a training set of immune cell-rich and cell-poor tissue regions was annotated in H&E whole-slide images using the CD45-expression as a guide. In analysis, the images were divided into small homogenous regions, superpixels, from which features were extracted using a pretrained convolutional neural network (CNN) and classified with a support of vector machine. The CNN approach was compared to texture-based classification and to visual assessments performed by two pathologists.
Results:
In a set of 123,442 labeled superpixels, the CNN approach achieved an F-score of 0.94 (range: 0.92–0.94) in discrimination of immune cell-rich and cell-poor regions, as compared to an F-score of 0.88 (range: 0.87–0.89) obtained with the texture-based classification. When compared to visual assessment of 200 images, an agreement of 90% (κ = 0.79) to quantify immune infiltration with the CNN approach was achieved while the inter-observer agreement between pathologists was 90% (κ = 0.78).
Conclusions:
Our findings indicate that deep learning can be applied to quantify immune cell infiltration in breast cancer samples using a basic morphology staining only. A good discrimination of immune cell-rich areas was achieved, well in concordance with both leukocyte antigen expression and pathologists’ visual assessment.
Keywords: breast cancer, convolutional neural network, digital pathology, tumor-infiltrating immune cells, tumor microenvironment
INTRODUCTION
Abundance of tumor-infiltrating lymphocytes (TILs) is associated with a favorable outcome in breast cancer.[1] Several retrospective studies report a prognostic value of TILs and a potential role in prediction of response to treatment.[2,3] Especially in patients with triple-negative and human epidermal growth factor receptor-2-positive disease, high level of TILs in the primary tumor is reported to correlate with better prognosis.[4,5]
TILs are commonly quantified through microscopy of tissue sections stained for basic morphology with H&E and reported as the proportion of the stroma and tumor compartments that the immune cell infiltrates cover.[6] Challenges in the visual evaluation relate to subjectivity; evaluations suffer from intra- and inter-observer variability, and detailed manual evaluation of large series of whole-slide samples is time-consuming and semi-quantitative.[7] Methodologies enabling robust and high throughput TIL assessment are required to better understand the clinical significance of TILs in breast cancer.
Computer vision methods have potential to improve reproducibility, reduce the evaluation time, and make the readouts more quantitative. Automated and semi-automated computer vision methods have shown promising results in analysis of histological samples, such as detection of mitotic cells,[8] classification of tissue morphologies,[9] and quantification of immunohistochemical (IHC) markers.[10] Recently, convolutional neural networks (CNNs) have achieved state-of-the-art performance in computer vision tasks from object classification and detection to segmentation,[11] and a number of studies have demonstrated the applicability of CNNs in histological samples.[12,13,14]
Here, we study whether TILs can be quantified in digitized H&E-stained whole-slides obtained from patients with primary breast cancer. First, we stained consecutive tissue sections using H&E and the pan-leukocyte marker CD45. The CD45 staining was used to guide the annotation of training set of leukocyte-rich and leukocyte-poor tissue regions in the consecutive, digitized H&E-stained tissue section. Guiding the annotation with a specific antibody staining decreases subjectivity in defining the ground-truth and accelerates the otherwise laborious process. Second, we applied a CNN model pretrained on a large dataset of natural images as a feature extractor in quantification of TIL-rich regions (infiltrations) from other tissue entities. Deep neural networks trained on large and diverse image data form convolutional filters that are highly generalizable.[15] The concept of applying a pretrained deep learning model on another data domain is known as transfer learning, and therefore, we designate the proposed approach as antibody-supervised deep learning. To evaluate the approach, we compared it first to a classification with texture features and then to visual evaluations by two pathologists.
METHODS
Patient Material
Formalin-fixed paraffin-embedded (FFPE) tumor samples of twenty patients operated for primary breast cancer within the Hospital District of Helsinki and Uusimaa, Finland, were used in the study. The samples were stored in archives of the Helsinki University Hospital Laboratory (HUSLAB, Helsinki, Finland) and the Head of the Division of Pathology and Genetics approved use of the samples. The samples were anonymized and all patient-related data and unique identifiers were removed, and therefore, the study did not require ethical approval in compliance with Finnish legislation regulating human tissues obtained for diagnostic purposes (act on the use of human organs and tissue for medical purposes 2.2.2001/101). Samples represented different histological types: Ductal carcinoma (n = 13, 65%), lobular carcinoma (n = 3, 15%), medullary carcinoma (n = 2, 10%), adenosquamous carcinoma (n = 1, 5%), and cribriform carcinoma (n = 1, 5%) and different histological grades: Grade-I (n = 3, 15%), Grade-II (n = 3, 15%), and Grade-III (n = 14, 70%).
Staining Protocols
From each FFPE block, we cut two consecutive sections (3.5 μm): One for H&E staining and one for staining with the pan-leukocyte CD45 antibody. The fresh sections were mounted on electrically charged glass slides (SuperFrost Plus, Thermo Scientific, Waltham, MA, USA) and dewaxed using alcohol-xylene series. For H&E staining, we used undiluted Mayer's hematoxylin (Merck, Darmstadt, Germany) and 0.5% eosin (Merck). For IHC, we used a CD45 antibody (Agilent Technologies, Santa Clara, CA, USA) diluted to 1:500, 3,3’-diaminobenzidine as chromogen, and Mayer's hematoxylin (Agilent Technologies) as a counterstain with a 1:10 dilution.
Sample Digitization
Samples were digitized with a whole-slide scanner (Pannoramic 250 FLASH, 3DHISTECH Ltd., Budapest, Hungary) equipped with a plan-apochromat 20× objective (numerical aperture 0.8), a VCC-F52U25CL camera (CIS, Tokyo, Japan) with three image sensors (1,224 × 1,624; 4.4 × 4.4 μm/pixels), and a 1.0 adapter. The scanned images (0.22 μm/pixel) were compressed into a wavelet format (Enhanced Compressed Wavelet, ECW, ER Mapper, Intergraph, Atlanta, GA) with a compression ratio of 1:9 and stored on a whole-slide image management server (WebMicroscope, Fimmic Oy, Helsinki, Finland). The average size of the digital samples was 8.5 × 109 pixels (range: 2.3 × 109–12.4 × 109).
Annotation of the Training Set
Based on the CD45-expression, we annotated a training set of image regions (n = 1,116) in the twenty H&E-stained whole-slide images [Figure 1]. While viewing the consecutively cut H&E and CD45 sections side-by-side, we labeled the regions with a raster graphic editor (Adobe Photoshop, Adobe Systems, Mountain View, CA, USA) in downscaled H&E-stained image (1:10, 2.2 μm/pixel). Five entities, four representing different tissue categories and one representing background (BG), were labeled: (1) leukocyte-rich (LR) regions – tissue regions in epithelium and stroma densely populated with TILs. (2) Epithelial (EP) tissue – regions of normal and malignant epithelium with none or few TILs. (3) Stroma predominant regions (SR) – regions of stromal tissue including tissue folds and other tissue types not separately defined with none or few TILs. (4) Adipose tissue (AD) and (5) BG. The TIL-rich and TIL-poor regions were confirmed and selected based on the CD45 expression in the consecutive section.
Figure 1.
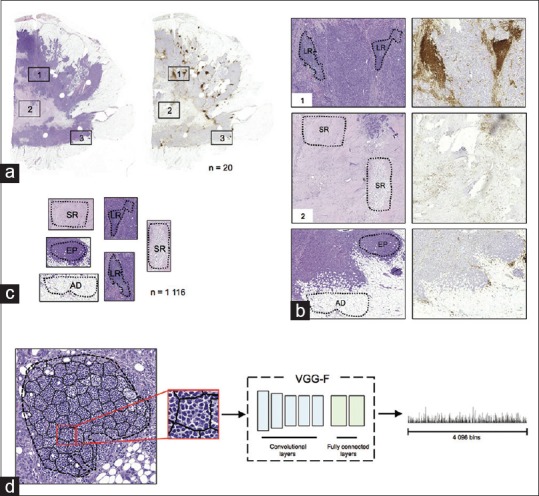
Antibody-supervised deep learning. (a) Each pair (n = 20) of consecutively cut tumor sections were stained with H&E (left) and the pan-leukocyte CD45 antibody (right). (b) Guiding annotation by the CD45 expression, regions representing different tissue categories were marked in the H&E section (LR: leukocyte-rich, EP: epithelium, SR: stroma predominant, and AD: adipose). (c) Marked tissue regions (n = 1,116) were extracted from the H and E sections. (d) An example of superpixel segmentation and feature extraction
Annotation of the Test Set
To compare our approach to pathologists’ visual assessment at the patient level, we randomly selected 10 images (1,000 × 1,000 pixels, 440 × 440 μm2) excluding areas containing BG from each of the 20 whole-slide image. Two pathologists (P.E.K. and M.M.) visually estimated relative proportions of the different tissue categories of interest (LR, EP, SR, and AD) in this test set of 200 images. The experts were blinded from the results of the automated quantification, and they were asked to estimate the proportions of tissue categories on a continuous scale and independently of each other.
Computerized Quantification of Tumor-infiltrating Lymphocytes
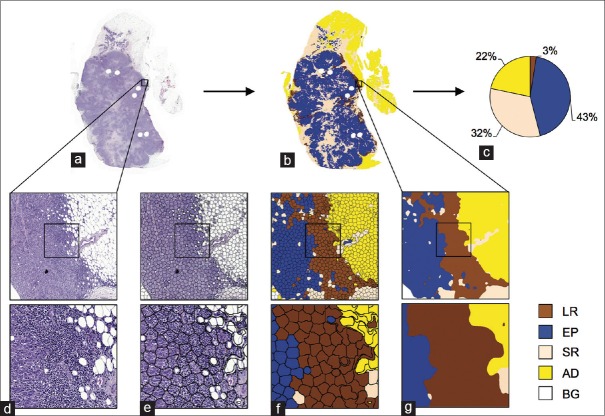
The whole-slide images were downscaled (0.44 μm/pixel) and divided into nonoverlapping tiles (3,000 × 3,000 pixels) for the analysis. The proposed approach is composed of three main components: (i) regional segmentation into superpixels, (ii) classification of superpixels with CNN activations, and (iii) postprocessing [Figure 2].
Figure 2.
Computerized quantification of tumor infiltrating immune cells in whole-slides. (a) An example of a whole-slide image subject to analysis and (b) corresponding analysis result with (c) a pie chart visualizing proportional ratios of different tissue categories excluding background. (d) An example of an image tile that is divided into (e) homogeneous tissue areas, superpixels, which are (f) classified into different tissue categories (LR: Leukocyte-rich, EP: Epithelium, SR: Stroma, AD: Adipose, and BG: Background). (g) Classification results were smoothed with spatial filtering
Superpixel segmentation is a low-level segmentation method that divides an image into locally similar segments, i.e., superpixels. The motivation to use superpixel segmentation is to first over-segment tissue structures into homogeneous regions [Figure 2e], which are subsequently classified into the tissue categories or to BG [Figure 2f]. Prior to the superpixel segmentation, we downscaled the image tiles (0.88 μm/pixel), and median filtered the color channels with ten pixels’ radius. Then, we further filtered the image tiles with an average filter with radius of 15 pixels and converted the images into Lab color-space for simple linear iterative clustering (SLIC)[16] superpixel segmentation algorithm. Filtering images before SLIC smoothen the fine image structures and guide superpixel formation to global changes in color and intensity. The regions’ size and shape parameters of SLIC were set to 50 and 150, respectively.
Inspired by work on transfer learning,[17,18,19,20] we used a pretrained CNN model (the VGG-F network)[21] as feature extractor. From each superpixel, we extracted activations of the penultimate layer of the VGG-F network, a model trained with the ImageNet image dataset.[22] The superpixels were scaled to fit the input of the pretrained network (224 × 224 pixels). A linear multiclass support vector machine (SVM)[23] (one vs. rest, L2 regularized L2-loss) classifier was used in the classification of the superpixels. The classifier's cost parameter (C) was optimized with a 3-fold cross-validation grid search over C = [2-10, 2-7,… 27, 210]. In training, we weighted the C-parameter according to the relative proportion of training samples in the different categories.
To fine-tune the final segmentation, we smoothed the classification results with spatial filtering [Figure 2g]. First, the decision value channel of the BG was dilated with a circular structuring element (radius of 50 pixels) to minimize possible classification errors on the tissue borders. Then, we filtered each decision value channel with a disk-shaped average filter bank: Radii of {20, 25, 25, 25, 50} pixels for corresponding channels {LR, EP, SR, AD, BG}. Finally, we formed the final segmentation result by pixel-wise majority voting through the decision channels and labeled all AD regions smaller than 40,000 pixels as stroma (SR).
The analysis methods were implemented in a numerical computing environment (MATLAB, MathWorks, Natick, MA, USA), with libraries for computer vision and classification; LIBLINEAR,[24] VLFeat,[25] and MatConvNet.[26] On a 3.1 GHz quad-core processor with 16GB memory, the average time to analyze an image tile (3,000 × 3,000 pixels) was 1 min, including download from a remote server, superpixel segmentation, and feature extraction.
Texture Features
A joint distribution of local binary patterns (LBPs)[27] and local variance (VAR) was used to capture the texture content of the superpixels. We extracted rotation uniform 2 LBP/VAR features without (LBP/VAR) and with (LBP/VAR-KCHI2) an explicit kernel mapping.[28] The LBP/VAR features were computed in a neighborhood of 16 sampling points (n) on radius (r) of 4 pixels. The quantification limits for VAR were set based on 10 random images of the different tissue categories by dividing the VAR distribution into quartiles.
Statistical Methods
The classification performance was evaluated based on F-score, area under receiver operating characteristics curve (AUC), and with accuracy, sensitivity, specificity, and precision. F-score is defined as a harmonic mean of precision and sensitivity: 2 × (sensitivity × precision)/(sensitivity + precision). The inter-rater agreement was assessed with Cohen's kappa coefficient (k) with quantization to: {0%, 20%, 40%, 60%, 80%, 100%}. The same quantization was used in evaluation of percent-agreement. For evaluating correlation, we computed pairwise Pearson's linear (two-tailed) correlation coefficient (r). For statistical evaluation, we used 3-fold cross-validation and leave-one-out cross-validation. In 3-fold cross-validation, a dataset is divided into three random partitions from which one at the time is used in evaluation and the rest are used for training whereas in leave-one-out cross-validation only one sample at the time is used in evaluation and the rest are used for training.
RESULTS
Classification Accuracy According to Feature Extraction Method
First, we evaluated how features extracted with the deep learning (VGG-F) network compare to texture features (LBP/VAR) in discrimination of TILs and different tissue categories. Details of the features are listed in Table 1. The training samples (n = 1,116) were segmented into superpixels with SLIC, resulting in a set of 123,442 superpixels representing different categories (LR n = 9,995, EP n = 25,749, SR n = 28,784, AD n = 31,269, and BG n = 27,645). For statistical evaluation, we ran a 3-fold cross-validation 10 times.
Table 1.
Image feature details
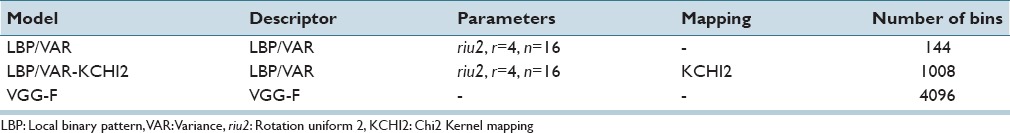
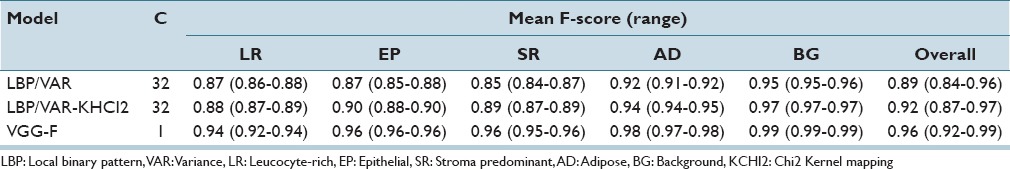
Overall F-scores for VGG-F, LBP/VAR, and LBP/VAR-KCHI2 were 0.96, 0.89, and 0.92, respectively, while corresponding AUCs were 0.996, 0.983, and 0.984, respectively. The mean sensitivity in the discrimination of TIL-rich regions with the VGG-F based model was 91% (range: 88%–92%), specificity 100% (range: 100%–100%), and precision 96% (range: 96%–97%), respectively.
In all pairwise/inter-category comparisons, features derived with the VGG-F CNN outperformed the LBP/VAR texture features [Table 2]. Kernel mapping improved performance of the texture features in all categories although the difference in classification of TIL-rich regions was marginal (F-score: 0.88 vs. 0.87). AD tissue and BG are more homogeneous in comparison to other tissue categories, and all features discriminated AD and BG superpixels better than others. By definition, the TIL-rich category is composed of the immune cells mixed with different tissue morphologies, and overall it obtained the lowest F-scores.
Table 2.
Classification accuracies for different image features
Comparison to Visual Evaluations
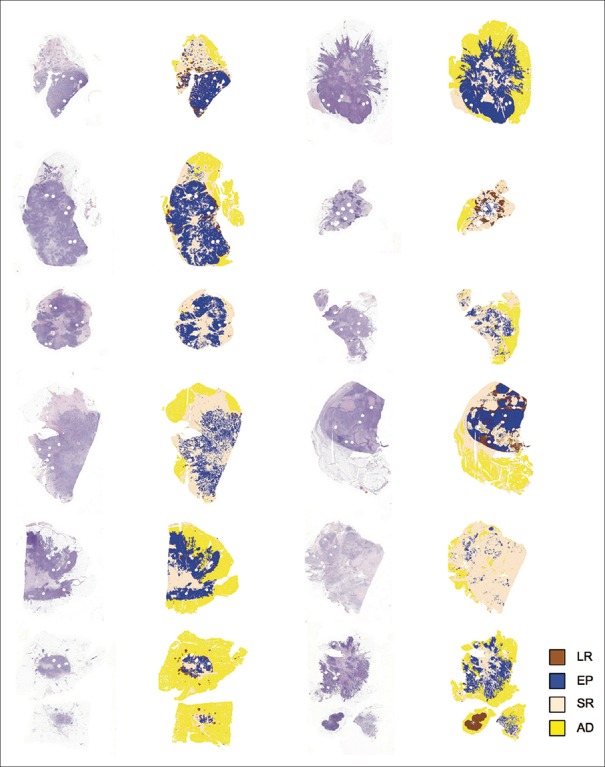
To further evaluate the proposed approach, we compared the automated quantification to visual assessment of TILs and other tissue entities performed by two pathologists in the test set (n = 200). First, we analyzed the whole-slides (n = 20) using a leave-one-out cross-validation, processing all the samples independently [Figure 3]. Then, we extracted the regions corresponding to the test images from the segmentation result images for evaluation [Figure 4].
Figure 3.
Analysis of whole-slide images. H&E sections were segmented into four tissue categories and background (LR: Leukocyte-rich, EP: Epithelium, SR: Stroma, and AD: Adipose)
Figure 4.
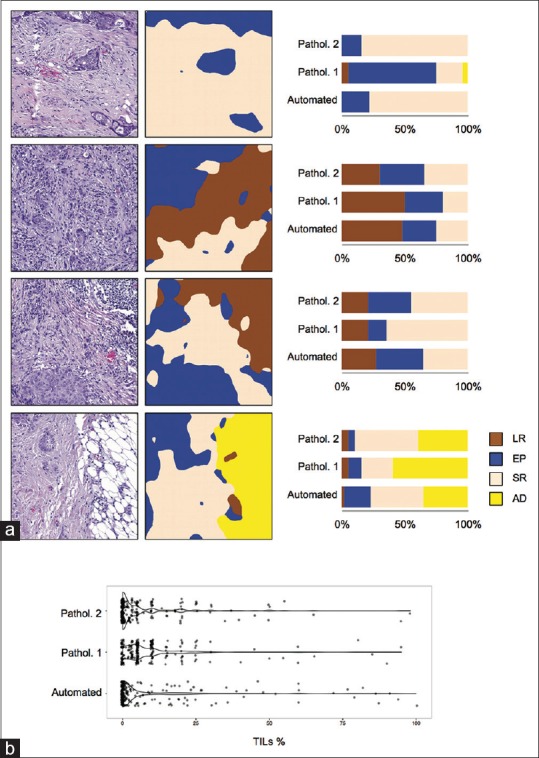
Comparison to pathologists’ assessments. Test images (n = 200) were randomly selected from the whole-slides, and visually evaluated for immune cell-rich regions (LR), epithelium (EP), stroma (SR), and adipose (AD) by two pathologists, and the results were compared to automated quantifications. (a) Four example images: H&E images (left), automated quantification (middle), and a bar-plot illustrating the visual assessments and automated quantification (right). (b) Distributions of automated quantification and visual assessments of immune infiltration in test images (n = 200)
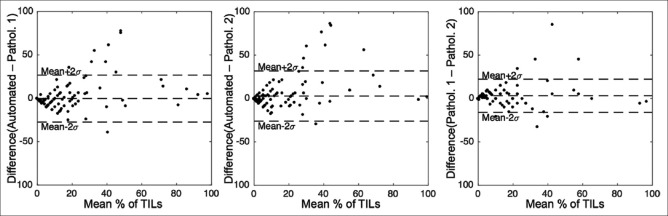
On a patient level, the average agreement between the automated CNN-based and the pathologists’ visual TIL quantification was 90% (κ = 0.79) while the TIL quantification agreement between the pathologists was 90% (κ = 0.78). The largest differences in TIL quantification were seen in the middle range, between values 25% and 75%, where the automated quantification slightly overestimated values in comparison to visual evaluations as seen in the Bland–Altman plots [Figure 5].
Figure 5.
Bland–Altman plots for immune cell infiltration assessment in the test images (n = 200)
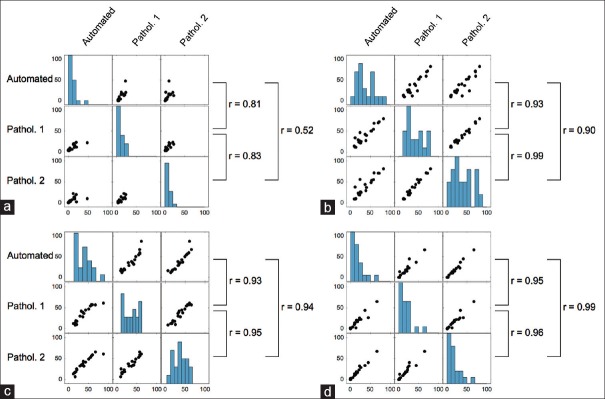
The average agreement between the CNN-based quantification and the visual assessments across all tissue entities was 83% (κ = 0.73), whereas the agreement between the pathologists was 84% (κ = 0.75). The average correlation between the automated quantification and visual assessments was r = 0.87, as compared to a correlation of r = 0.93 observed between pathologists’ evaluations [Figure 6].
Figure 6.
Correlation-plots of pathologists’ assessments and automated quantification of (a) immune cell-rich regions, (b) epithelial tissue, (c) stromal tissue, and (a) adipose tissue at patient level (n = 20)
In quantification of EP tissue, the correlation between automated quantification and the pathologists’ assessment (r = 0.92) was lower than the correlation between the two pathologists (r = 0.99). In quantification of stromal and AD tissue, the correlation between a computerized and human observer assessment was on par or higher (stroma: r = 0.94, AD: r = 0.99) as compared to correlations between the two human observers (stroma: r = 0.95, AD: r = 0.96).
CONCLUSIONS
Our results indicate that it is feasible to quantify tumor-infiltrating immune cells in H&E-stained breast cancer samples. The proposed deep learning immune cell quantification approach achieved an agreement with human observers (90%, κ = 0.79) that is comparable to the agreement between two human observers (90%, κ = 0.78) on a patient level. Furthermore, we report an F-score of 0.94 and an AUC of 0.99 in discrimination of TIL-rich and TIL-poor tissue regions and show that features extracted with the pretrained CNN-model outperform texture features in classification of TILs and other tissue categories. In addition, we present an application of antibody staining as a guide for image annotation and definition of ground-truth.
Automated quantification of TILs in H&E-stained breast cancer samples has not been studied extensively. A study using Markov random fields reported a cross-validation accuracy of 90% in classification of TIL-rich and TIL-poor images[29] in a set of 41 images (600–700 × 500–600 pixels; 0.33 μm/pixel). Another study evaluated a method based on expectation-maximization driven geodesic active contours[30] for detection of lymphocytes in 100 images (400 × 400 pixels), obtaining a sensitivity of 86% and a precision of 64%. Both active contours and Markov random fields are suitable algorithms for cell segmentation; however, because of high computational requirements, they might not scale for analysis of whole-slide images. In fact, detailed cell/nuclei segmentation might not be necessary in TIL assessment and could be replaced with an evaluation of proportional area covered by TIL-rich regions, as recommended by an international TIL-working group.[6]
The laborious nature of image annotation, especially when cell-level annotations are required, and lack of common databases are limiting the number of training and test samples available for researchers. An approach using Haralick features and an SVM classifier resulted in a cell level classification sensitivity of 58% in a test of 168 annotated cells, including 94 lymphocytes.[31] In another study, a method based on a transferable belief model obtained 94% sensitivity and 81% precision in lymphocyte detection on cell level when evaluated with 10 images (400 × 400 pixels).[32] Evaluation done with only a few images or cells might not reflect the true performance, making the comparison to other studies difficult.
With 91% sensitivity, 96% precision, and 100% specificity in classification of TIL-rich and TIL-poor regions (superpixels), our results are well on par with the literature. By guiding the annotation with the antibody staining, we create a dataset of nearly 125,000 superpixels of which approximately 10,000 represents TIL-rich regions. Because of a superpixel can contain tens of cells, particularly when taken from a dense infiltration region, our dataset is considerably larger when compared to those reported in the literature. To the best of our knowledge, similar approach using antibody staining for annotation has not been presented earlier. Antibody-based supervision is potential for applications outside the current work, such as in more detailed classification of immune cell subtypes, detection of blood vessels or mitosis by use of corresponding antibodies and staining methods.
In addition to the evaluation of the TIL quantification on regional level, we compared the approach to human observers, which is the current gold standard in evaluation of H&E-stained tissue samples as well as IHC. The observed mean agreement between the automated quantification and pathologists’ assessments (90%, κ = 0.79) was in line with pathologists’ inter-observer agreement (90%, κ = 0.78). In concordance with the cross-validation in test set, the largest disagreements between the automated quantification and pathologists’ assessments were observed in the TIL-rich tissue category. This was also true for the agreement between the pathologists.
In addition to the analysis of FFPE samples, an image analysis application for detecting TILs in frozen tissue sections has been reported.[33] Morphological and contextual multiscale features together with an SVM classifier reached a cross-validation accuracy of 90% in the discrimination between cancer cells, lymphocytes, and stromal cells when tested with 871 annotated cells. Nevertheless, because of insufficient evidence outside the research settings, frozen sections are not presently recommended for TIL assessment[6] in clinical settings. In addition, a study based on local morphological scale[34] proposed a combination of epithelium-stroma-classifier and an IHC staining to assess TILs in ovarian cancer samples.
Deep learning algorithms have so far not been widely adapted for analysis of histological tumor samples. In general, the best performing deep learning models are trained with a vast number (million to 100 million) of labeled images. However, annotation of such amounts of training samples from histological material is challenging. Transfer learning is an attractive alternative to end-to-end training, first because of the availability of the best performing models, such as the VGG-networks,[21] AlexNet,[35] and GoogLeNet,[36] and second because of the good generalization that can be achieved already with a low number of training images.
Studies with LBP-based features have shown good agreement with human experts in classification of tissue morphologies, Linder et al. 2014 and Turkki et al. 2015[9,37] and they are broadly applied in medical image analysis.[38] Nevertheless, the features we extracted using the VGG-F model clearly outperformed the LBP-based features. This suggests that a similar approach could benefit applications that currently rely on texture features or other handcrafted image descriptors.
In summary, our findings show that automated quantification of TILs in digitized H&E-stained tissue samples from patients with breast cancer is feasible with an accuracy comparable to human experts. As biomarkers to support decision-making on immunological therapies are needed, automated methods would be beneficial in further exploration of the potentially predictive role of TILs in breast cancer. Enabling analysis of large cohorts of whole-slides, our work has potential implications in cancer research and clinical trials, as well as in clinical work by offering an automated report of the immunological characteristics of the tumor microenvironment based on a simple morphological stain only.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2016/7/1/38/189703
REFERENCES
- 1.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–41. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 2.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31:860–7. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 3.Denkert C, Loibl S, Noske A, Roller M, Müller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–13. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 4.Salgado R, Denkert C, Campbell C, Savas P, Nuciforo P, Aura C, et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: A secondary analysis of the neoALTTO trial. JAMA Oncol. 2015;1:448–54. doi: 10.1001/jamaoncol.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959–66. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–71. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson AN, et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA. 2015;313:1122–32. doi: 10.1001/jama.2015.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20:237–48. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Linder N, Konsti J, Turkki R, Rahtu E, Lundin M, Nordling S, et al. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn Pathol. 2012;7:22. doi: 10.1186/1746-1596-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: A publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12:R56. doi: 10.1186/bcr2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 12.Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):411–8. doi: 10.1007/978-3-642-40763-5_51. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Cruz-Roa A, Basavanhally A, Gilmore H, Shih N, Feldman M, et al. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging (Bellingham) 2014;1:034003. doi: 10.1117/1.JMI.1.3.034003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:26286. doi: 10.1038/srep26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? Advances in neural information processing systems. 2014:3320–8. [Google Scholar]
- 16.Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell. 2012;34:2274–82. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 17.Girshick R, Donahue J, Darrell T, Malik J. Region-based convolutional networks for accurate object detection and segmentation. IEEE Trans Pattern Anal Mach Intell. 2016;38:142–58. [Google Scholar]
- 18.Gong Y, Wang L, Guo R, Lazebnik S. Multi-scale orderless pooling of deep convolutional activation features. Springer International Publishing. 2014 [Google Scholar]
- 19.Liu F, Lin G, Shen C. CRF learning with CNN features for image segmentation. Pattern Recognition. 2015 [Google Scholar]
- 20.Cimpoi M, Maji S, Vedaldi A. Deep convolutional filter banks for texture recognition and segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2014 [Google Scholar]
- 21.Chatfield K, Simonyan K, Vedaldi A, Zisserman A. Return of the devil in the details: Delving deep into convolutional nets. arXiv preprint arXiv. 1405; 2014. [Google Scholar]
- 22.Socher R. ImageNet: A Large-scale Hierarchical Image Database. 2009 IEEE Conference on Computer Vision Pattern Recognition, IEEE. 2009:248–55. [Google Scholar]
- 23.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97. [Google Scholar]
- 24.Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A library for large linear classification. J Mach Learn Res. 2008;9:1871–4. [Google Scholar]
- 25.Vedaldi A, Fulkerson B. VLFeat: An Open and Portable Library of Computer Vision Algorithms. Proceedings of the 18th ACM international conference on Multimedia. ACM. 2010 [Google Scholar]
- 26.Vedaldi A, Lenc K. MatConvNet - Convolutional neural networks for MATLAB. Proceedings of the 23rd ACM international conference on Multimedia. ACM. 2015 [Google Scholar]
- 27.Ojala T, Pietikäinen M, Mäenpää T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell. 2002;24:971–87. [Google Scholar]
- 28.Vedaldi A, Zisserman A. Sparse kernel approximations for efficient classification and detection. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE. 2012 [Google Scholar]
- 29.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, et al. Computerized image-based detection and grading of lymphocytic infiltration in HER2 breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:642–53. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 30.Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, et al. Expectation-maximization-driven geodesic active contour with overlap resolution (EMaGACOR): Application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:1676–89. doi: 10.1109/TBME.2010.2041232. [DOI] [PubMed] [Google Scholar]
- 31.Kuse M, Sharma T, Gupta S. A classification scheme for lymphocyte segmentation in H and E stained histology images. Heidelberg: Springer Berlin; 2010. pp. 235–43. [Google Scholar]
- 32.Panagiotakis C, Ramasso E, Tziritas G. Lymphocyte segmentation using the transferable belief model. Heidelberg: Springer Berlin; 2010. pp. 253–62. [Google Scholar]
- 33.Yuan Y, Failmezger H, Rueda OM, Ali HR, Gräf S, Chin SF, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4:157ra143. doi: 10.1126/scitranslmed.3004330. [DOI] [PubMed] [Google Scholar]
- 34.Janowczyk A, Chandran S, Feldman M, Madabhushi A. Local morphologic scale: Application to segmenting tumor infiltrating lymphocytes in ovarian cancer TMAs. SPIE Medical Imaging. International Society for Optics and Photonics. 2011 [Google Scholar]
- 35.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Advantages in Neural Information Processing System. Advances in neural information processing systems. 2012:1097–105. [Google Scholar]
- 36.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015 [Google Scholar]
- 37.Turkki R, Linder N, Holopainen T, Wang Y, Grote A, Lundin M, et al. Assessment of tumour viability in human lung cancer xenografts with texture-based image analysis. J Clin Pathol. 2015;68:614–21. doi: 10.1136/jclinpath-2015-202888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanni L, Lumini A, Brahnam S. Local binary patterns variants as texture descriptors for medical image analysis. Artif Intell Med. 2010;49:117–25. doi: 10.1016/j.artmed.2010.02.006. [DOI] [PubMed] [Google Scholar]