Abstract
Introduction:
Bloodstream infection (BSI) is a leading cause of mortality in critically ill patients. The mortality directly attributable to BSI has been estimated to be around 16% and 40% in general hospital population and Intensive Care Unit (ICU) population, respectively. The detection rate of these infections increases with the number of blood samples obtained for culture. The newer continuous monitoring automated blood culture systems with enhanced culture media show increased yield and sensitivity. Hence, we aimed at studying the role of single and multiple blood specimens from different sites at the same time in the outcome of automated blood culture system.
Materials and Methods and Results:
A total of 1054 blood culture sets were analyzed over 1 year, the sensitivity of one, two, and three samples in a set was found to be 85.67%, 96.59%, and 100%, respectively, which showed a statistically significant difference (P < 0.0001). Similar findings were seen in few more studies, however, among individual organisms in contrast to other studies, the isolation rates of Gram-positive bacteria were less than that of Gram-negative Bacilli with one (or first) sample in a blood culture set. In our study, despite using BacT/ALERT three-dimensional continuous culture monitoring system with FAN plus culture bottles, 15% of positive cultures would have been missed if only a single sample was collected in a blood culture set.
Conclusion:
The variables like the volume of blood and number of samples collected from different sites still play a major role in the outcome of these automated blood culture systems.
Keywords: BacT/ALERT 3D, bacteremia, blood stream infections, FA plus, number of samples
Introduction
Bloodstream infection (BSI) is a leading cause of mortality in critically ill patients. The mortality directly attributable to BSI has been estimated to be around 16% and 40% in general hospital population and Intensive Care Unit population, respectively. Timely diagnosis and appropriate antimicrobial therapy have a major role in predicting the outcome of these infections.[1]
The prompt detection of bacteremia and fungemia is a critical function of the clinical microbiology laboratory, and blood culture is a critical tool for the detection of BSIs.[2] Blood cultures are considered to be one of the most significant specimen types that a microbiology laboratory handles and every laboratory has a strict notification policy to ensure that positive blood cultures are promptly reported to the physician.
The detection rate of these infections increases with the number of blood samples obtained for culture. The sensitivity of 80% with single blood cultures has been found to increase to more than 90% with two culture samples. However, more than three cultures do not increase the diagnostic yield significantly. The usage of single blood culture, as well as more than three blood cultures, is highly discouraged.[1] Despite the fact that newer blood culture media and improved continuous monitoring blood culture systems detect organisms faster and more frequently, the volume of blood and a number of samples remain as major variables predicting the outcome of these culture systems.
Currently, there are no recommendations for an optimal time difference between multiple blood culture samples, and recent studies suggest obtaining multiple samples for blood culture at the same time but from different venipuncture sites.[1,3]
Here, in this study, we intended to compare the outcome of blood cultures from a single specimen and multiple specimens collected at the same time from different venipuncture sites, using BacT/ALERT three-dimensional (3D) automated blood culture system.
Materials and Methods
This hospital-based, prospective study was conducted from July 2015 to June 2016 (1 year) in our department. Approval from the institutional ethics committee was duly obtained.
Sample collection: Blood specimens were obtained at the bedside by nursing staff from wards, critical care units, or by trained phlebotomist. The skin was disinfected with 2% chlorhexidine. The antecubital, median cubital fossa were the preferred sampling sites using a needle and syringe. Whenever, multiple specimens were taken, peripheral veins from different sides and extremities were used. Maximum of three samples was collected from each patient at the same time. The samples collected from a single patient at the same time (from different sites) were included under single blood culture set. In our study, a set can have a maximum of three samples
Volume standards: A volume of 5–10 ml and 2–4 ml of blood was collected from adult and pediatric patients, respectively, for every single specimen
The BacT/ALERT® 3D Microbial Detection System (BioMerieux, France) was used for culture and BacT/ALERT® FA plus and BacT/ALERT® PF plus culture bottles were employed for adult and pediatric patients, respectively
All the samples were processed according to standard guidelines recommended by the manufacturer.
Data analysis
All the relevant data were entered in Microsoft Excel (v16.0.4266), and demographic parameters were analyzed using the same. Comparison of results between single, paired, and three blood culture specimens was done. Statistical analysis was done using the IBM SPSS Statistics for Windows, v22.0. (IBM Corp. Armonk, NY; 2013). Individual samples Kruskal–Wallis test (nonparametric) was used to compare the difference in isolation rates between three groups. Stratifications were applied for individual organisms and organism groups based on clinical importance.
Results
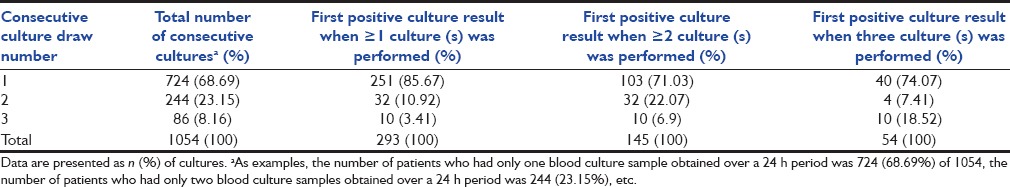
A total of 1054 blood culture sets have been collected from 758 patients. The mean age of patients was 35.56 (±23.23) with a gender ratio (M: F) of 1.61. Among the 1054 sets, 86 sets (8.16%) had three blood cultures collected at the same time from three different collection sites of the same patient; 244 sets (23.15%) had two blood cultures collected at the same time from two different collection sites of the same patient and rest 724 (68.69%) were single blood culture samples.
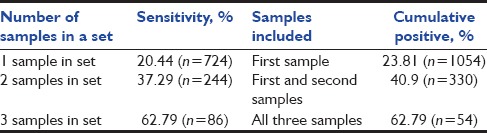
Among the 1054 blood culture sets, 293 showed growth of clinically significant organisms in at least one sample from a set. The sensitivity of the blood cultures increased from 85.67% to 96.59% when the number of samples was increased from one to two, and a rise of another 3.41% was seen when three samples were taken. The Kruskal–Wallis ANOVA test revealed a significant difference between all the three groups (P < 0.0001). Among the blood culture sets which had three samples (n = 86), the increase in sensitivity was from 74.07% to 81.48% when two samples were considered, and an increase of another 18.52% was seen with the third sample. The sensitivity of blood cultures with one, two, and three samples in a set were 20.44% (n = 724), 37.29% (n = 244), and 62.79% (n = 86), respectively. The complete split-up of the positive sets versus total sets with the consecutive culture draw number has been shown in Table 1.
Table 1.
Relationship between the number of consecutive blood cultures (blood culture sets) with samples obtained over a 24 h
The sensitivity of the blood culture sets with one, two, and three samples and the cumulative positive percentage have been shown in Table 2.
Table 2.
Sensitivity of blood culture sets and cumulative positive percentage
The isolation rates of different bacteria in different samples of a blood culture set have been shown in Table 3 and the isolation rates based on different organism groups have been shown in Table 4.
Table 3.
The isolation rates of different bacteria in different samples a blood culture set
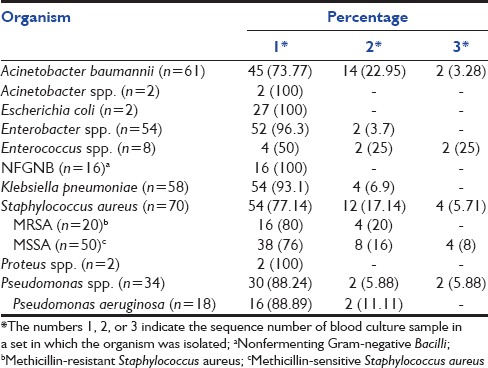
Table 4.
The isolation rates of different groups of bacteria in different samples of a blood culture set
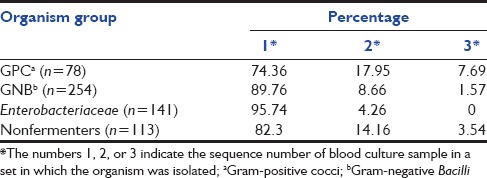
Among the different bacterial groups, the isolation rates of Gram-positive cocci were significantly less than that of Gram-negative Bacilli when only one (or the first) sample is considered in a blood culture set (P = 0.0011). Among Gram-negative Bacilli, the isolation rates of nonfermenters were significantly less when only one (or the first) sample is considered in a blood culture set (P = 0.001).
With the number of blood samples increased to two in a culture set, though the total isolation rates increased significantly (P < 0.0001), the difference in isolation rates between Gram-positive cocci and Gram-negative Bacilli remained significantly high (P = 0.0172); however, the difference between Enterobacteriaceae and nonfermenters among Gram-negative Bacilli had reduced to an extent where there is no significant difference between them (P = 0.0810).
Discussion
Blood cultures are regarded as the “gold standard” for diagnosis of bloodstream infections. Since the duration and magnitude of bacteremia vary significantly between patients, the site of collection, volume of blood, and number of samples play a major role in the outcome of these blood cultures.[4] Here, in this study, we have studied the role of a number of samples (taken from different sites at the same time) in the outcome of blood cultures using BacT/ALERT 3D automated culture system.
The higher isolation rates with more number of samples collected from different sites at the same time have been seen in many studies conducted with different culture systems [Table 5].
Table 5.
Sensitivity (isolation rates) of single and multiple specimens seen with different culture systems
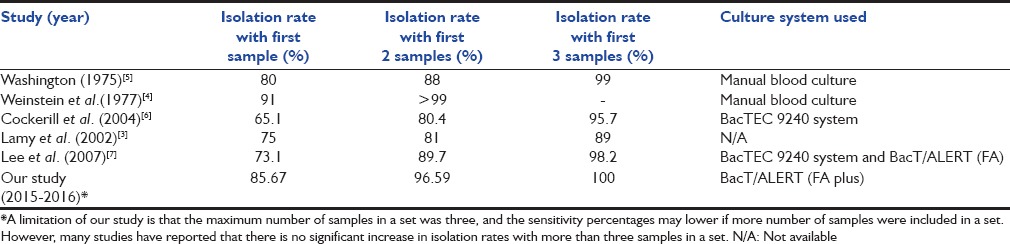
Most of the studies conducted in a similar setting recommend collecting at least 2–3 samples from independent sites at the same time. The recent continuous monitoring blood culturing systems incorporate special components in their media to increase the yield of blood cultures. However, it is evident that the need of more number of samples for blood culture has not reduced even with these systems.[1]
The isolation rates of different organisms with the different number of samples were analyzed. Few studies reported that among different organisms, the isolation rate of Staphylococcus aureus even with a single (or first) sample is higher than other organisms. Lee et al. reported 93% and 87% isolation rate of S. aureus only with the first sample.[5] However, in our study, the isolation rates of Gram-negative Bacilli, especially members of Enterobacteriaceae showed higher recovery with one (or first) sample. The recovery rates of Gram-positive cocci such as S. aureus (77.14%) and Enterococcus spp. (50%) were less compared to other organisms.
Conclusion
In our study, despite using BacT/ALERT 3D continuous culture monitoring system with FAN plus culture bottles, 15% of positive cultures would have been missed if only a single sample was collected in a blood culture set. Although improvements in culture techniques and media preparations can improve the sensitivity of blood cultures, the variables like the volume of blood and number of samples collected from different sites still play a major role in the outcome of these blood culture systems.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Seifert H, Wisplinghoff H. Bloodstream infection and endocarditis. In: Mahy BW, ter Meulen V, Borriello SP, Murray PR, Funke G, Kaufmann SH, et al., editors. Topley and Wilson's Microbiology and Microbial Infections. Chichester, UK: John Wiley and Sons, Ltd; 2010. [Google Scholar]
- 2.Kirn TJ, Mirrett S, Reller LB, Weinstein MP. Controlled clinical comparison of BacT/alert FA plus and FN plus blood culture media with BacT/alert FA and FN blood culture media. J Clin Microbiol. 2014;52:839–43. doi: 10.1128/JCM.03063-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamy B, Roy P, Carret G, Flandrois JP, Delignette-Muller ML. What is the relevance of obtaining multiple blood samples for culture? A comprehensive model to optimize the strategy for diagnosing bacteremia. Clin Infect Dis. 2002;35:842–50. doi: 10.1086/342383. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein MP, Murphy JR, Reller LB, Lichtenstein KA. The clinical significance of positive blood cultures: A comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. II. Clinical observations with special reference to factors influencing prognosis. Rev Infect Dis. 1983;5:54–70. doi: 10.1093/clinids/5.1.54. [DOI] [PubMed] [Google Scholar]
- 5.Washington JA., 2nd Blood cultures: Principles and techniques. Mayo Clin Proc. 1975;50:91–8. [PubMed] [Google Scholar]
- 6.Cockerill FR, 3rd, Wilson JW, Vetter EA, Goodman KM, Torgerson CA, Harmsen WS, et al. Optimal testing parameters for blood cultures. Clin Infect Dis. 2004;38:1724–30. doi: 10.1086/421087. [DOI] [PubMed] [Google Scholar]
- 7.Lee A, Mirrett S, Reller LB, Weinstein MP. Detection of bloodstream infections in adults: How many blood cultures are needed? J Clin Microbiol. 2007;45:3546–8. doi: 10.1128/JCM.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


