Abstract
Objective:
To determine the prognostic value of selected biomarkers in clinically isolated syndromes (CIS) for conversion to multiple sclerosis (MS) and disability accrual.
Methods:
Data were acquired from 2 CIS cohorts. The screening phase evaluated patients developing clinically definite MS (CIS-CDMS) and patients who remained as CIS during a 2-year minimum follow-up (CIS-CIS). We determined levels of neurofascin, semaphorin 3A, fetuin A, glial fibrillary acidic protein, and neurofilament light (NfL) and heavy chains in CSF (estimated mean [95% confidence interval; CI]). We evaluated associations between biomarker levels, conversion, disability, and magnetic resonance parameters. In the replication phase, we determined NfL levels (n = 155) using a 900 ng/L cutoff. Primary endpoints in uni- and multivariate analyses were CDMS and 2010 McDonald MS.
Results:
The only biomarker showing significant differences in the screening was NfL (CIS-CDMS 1,553.1 [1,208.7–1,897.5] ng/L and CIS-CIS 499.0 [168.8–829.2] ng/L, p < 0.0001). The strongest associations were with brain parenchymal fraction change (rs = −0.892) and percentage brain volume change (rs = −0.842) at 5 years. NfL did not correlate with disability. In the replication phase, more NfL-positive patients, according to the cutoff, evolved to MS. Every 100-ng/L increase in NfL predicted CDMS (hazard ratio [HR] = 1.009, 95% CI 1.005–1.014) and McDonald MS (HR = 1.009, 95% CI 1.005–1.013), remaining significant for CDMS in the multivariate analysis (adjusted HR = 1.005, 95% CI 1.000–1.011). This risk was lower than the presence of oligoclonal bands or T2 lesions.
Conclusions:
NfL is a weak independent risk factor for MS. Its role as an axonal damage biomarker may be more relevant as suggested by its association with medium-term brain volume changes.
Identifying patients with clinically isolated syndromes (CIS) who will present a second attack and determining the degree of disability accrual over the medium- to long-term are crucial goals in multiple sclerosis (MS). Although MRI is the most reliable prognostic marker1,2 and oligoclonal bands (OCBs) also have an important role,3,4 the disease heterogeneity hinders a more individualized prognosis. Therefore, the search for biomarkers that capture the different aspects of MS is still necessary. Although many have been identified, few are currently useful in the clinical setting.5 Consequently, we aimed to determine the prognostic value of selected biomarkers for conversion to MS and disability accrual, and to assess their associations with MRI inflammatory activity and neurodegenerative parameters.
METHODS
Patients.
We acquired longitudinal data from CIS cohorts at the Multiple Sclerosis Center in Vall d'Hebron Hospital, Barcelona,6 and the Neurology Department of Ramón y Cajal Hospital, Madrid. Inclusion criteria were age younger than 50 years and baseline clinical evaluation within the first 3 months of disease onset. We recorded baseline demographic and clinical characteristics and conducted follow-up visits every 3 to 6 months, assessing for relapses, clinical worsening, and excluding patients in whom we reached alternative diagnoses. A second attack established the diagnosis of clinically definite MS (CDMS).7 Definitions of disability accrual were a sustained increase in the Expanded Disability Status Scale (EDSS) of 1.0 point over 1 year or confirmed EDSS score ≥3.0.
Biological samples.
We collected venous blood and CSF within 3 months of disease onset for OCB determination in the 2 hospitals. Remnant samples were aliquoted and stored at −80°C until further use.
MRI acquisition and analysis.
At Vall d'Hebron Hospital, brain MRI acquisition on 1.5- or 3.0-tesla superconductive magnets included the following sequences, obtained with a contiguous 3- to 5-mm slice thickness: transverse proton density and T2-weighted conventional or fast spin-echo, transverse and sagittal T2 fluid-attenuated inversion recovery, and unenhanced and contrast-enhanced T1-weighted spin-echo. We included routine baseline spinal cord MRIs in November 2007. Baseline scans were done within 5 months of disease onset and follow-up MRIs at 1 year and every 5 years thereafter.6 The Ramón y Cajal cohort followed a similar acquisition protocol, performing baseline and follow-up MRIs within 3 months after the CIS and at 1 year, including spinal cord MRI in case of myelitis.
Neuroradiologists scored T2 lesion number and location, and gadolinium (Gd)-enhancing and new T2 lesion number. T2 lesion volume was calculated using the semiautomated Jim medical image display package (Xinapse Systems, Ltd., Colchester, UK). Brain volume parameters were obtained using the Structural Image Evaluation, using Normalization, of Atrophy (SIENA) software, part of FSL (FMRIB Software Library).8 Single-time-point analysis to obtain brain parenchymal fraction (BPF) estimates was performed with SIENAx. Two-time-point analysis was performed with SIENA, estimating the percentage brain volume change (PBVC) between 2 input images of the same subject at different time points.
Biological markers.
We selected fetuin A (FA),9 semaphorin 3A (sema3A),10,11 glial fibrillary acidic protein (GFAP),12–15 neurofilament heavy (NfH) and light (NfL) chains,13–20 and neurofascin21,22 after a PubMed search in 2008, which we updated regularly throughout the study duration, including the terms “biological markers” or “biomarkers” and “multiple sclerosis” or “clinically isolated syndromes.”
Except for NfH, we determined biomarker levels using commercial sandwich ELISA kits according to the manufacturers' recommendations (FA: BioVendor, Brno, Czech Republic; NfL: UmanDiagnostics AB, Umeå, Sweden; GFAP: Abnova, Taipei City, Taiwan; neurofascin: USCN Life Science Inc., Wuhan, China; and sema3A: USCN Life Science Inc.). We optimized the assays using varying sample dilutions measured in duplicates. NfH levels were determined using an electrochemiluminescence-based solid-phase sandwich immunoassay17 at the University Hospital Basel, Switzerland, selected among different assays for its higher sensitivity.18,23,24 All samples were tested blinded to clinical data and outcome measures. Whenever possible according to availability, we tested samples from the same patients using a different, unthawed aliquot.
Experimental design.
Screening phase.
We selected 2 opposite groups with a 2-year minimum follow-up from the Vall d'Hebron cohort: CIS-CDMS (n = from 33 to 38), including patients with 3 to 4 Barkhof-Tintore criteria on baseline MRI and presence of OCBs who converted to CDMS; and CIS-CIS (n = from 33 to 39), comprising patients with 0 Barkhof-Tintore criteria and absence of OCBs who remained as CIS (figure e-1A at Neurology.org).
Replication phase.
Only NfL levels showed significant differences in the screening and were thus evaluated in a replication phase. We selected consecutive patients with available CSF samples from the Vall d'Hebron and Ramón y Cajal cohorts (n = 155). Patients from Vall d'Hebron were selected from a different time range (2009–2011) than the NfL screening phase because of sample availability. NfL determination was performed in each hospital separately using the above-mentioned assay (figure e-1B).
Statistical analysis.
We performed parametric and nonparametric descriptive statistics depending on the normality of the distributions of the continuous variables.
Screening phase.
For each biomarker, we calculated sample size for 80% power at the 0.05 level of significance based on previous data. We used generalized linear models to compare CIS groups in terms of conversion to CDMS and assess biomarker levels and disability accrual, with the Bonferroni correction for multiple comparisons when applicable. Data are reported as the estimated mean (95% confidence interval [CI]). Potential covariates were age, sex, CIS topography, time from CIS to lumbar puncture, sample storage time, and disease-modifying treatment (DMT) before CDMS. We calculated Spearman correlations for MRI inflammatory activity parameters at baseline, 1, and 5 years (number of T2, Gd-enhancing, and new T2 lesions; T2 lesion volume), and partial correlations for neurodegenerative parameters at 1 and 5 years of follow-up (BPF change [BPFΔ] adjusted for age and baseline Gd-enhancing lesions, and PBVC). We calculated BPFΔ by subtracting baseline from 1- and 5-year follow-up SIENAx estimates and dividing by the baseline values, then multiplying by 100. We assessed the PBVC estimated by SIENA for changes from baseline to 1 and 5 years. Finally, we assessed Spearman correlations between NfL and NfH levels in a subgroup of 42 patients with determination of both biomarkers.
Replication phase.
Primary endpoints were conversion to CDMS and 2010 McDonald MS according to NfL status (positive/negative) based on a 900 ng/L cutoff value, established as the mean ± 3 SD of NfL levels in a control group of patients with noninflammatory neurologic diseases.25 We performed uni- and multivariate Cox proportional hazard regression analyses for NfL levels as a continuous or dichotomic variable. Covariates were OCBs, baseline T2 lesion number, DMT before MS, and hospital where NfL levels were determined. We used generalized linear models to compare NfL levels in terms of disability accrual (EDSS score ≥3.0), and Spearman correlations or generalized linear models to determine associations between NfL and MRI inflammatory parameters at baseline and 1 year. We could not assess brain volume changes because of the few estimated measures at the time of the analysis.
We performed statistical tests on the 0.05 level of significance, using the IBM SPSS Statistics (version 20.0; IBM Corp., Armonk, NY). We also prepared figures using GraphPad Prism 5.02 for Windows (GraphPad Software, La Jolla, CA).
Standard protocol approvals, registrations, and patient consents.
This study received approval from the corresponding local ethical committees in both hospitals and all patients signed a written informed consent.
RESULTS
Screening phase.
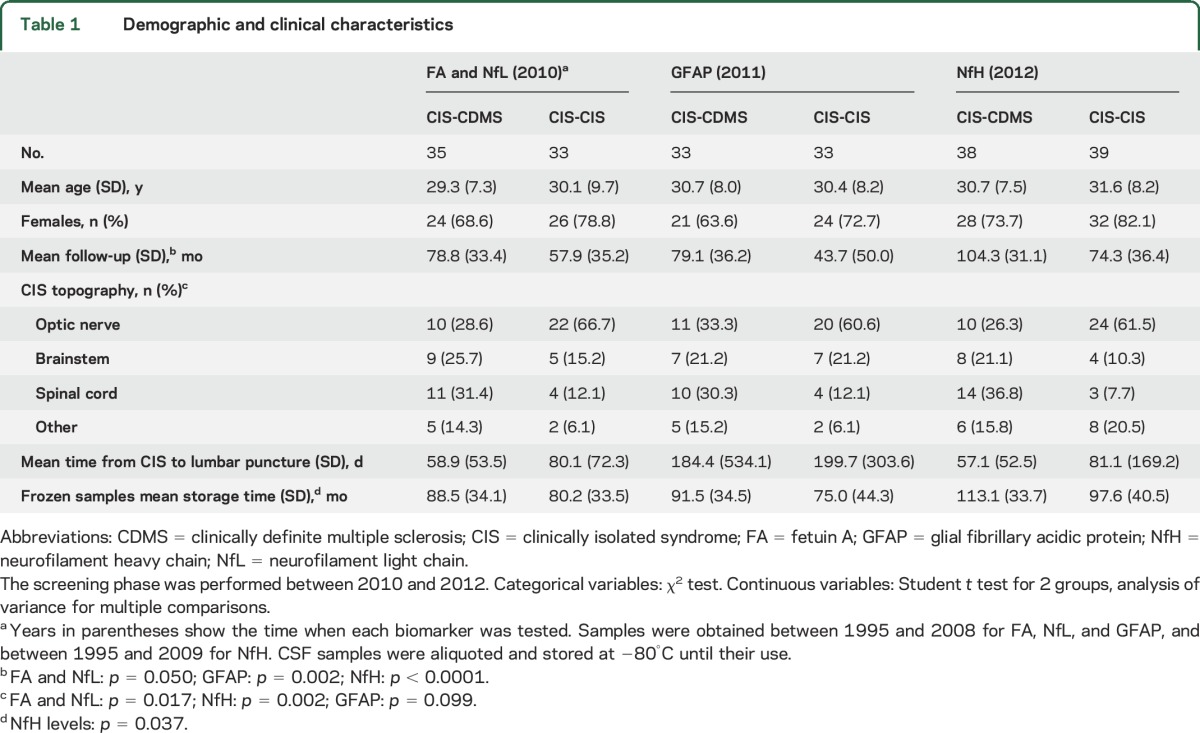
Appendix e-1A describes preliminary analyses. Baseline demographic and clinical characteristics were comparable between CIS groups (table 1) except for the higher frequency of optic neuritis in CIS-CIS regarding FA, NfL, and NfH. The proportion of patients on DMT before CDMS varied between 8.8% and 17.7% for each biomarker. Mean follow-up was longer for CIS-CDMS than CIS-CIS tested for GFAP and NfH. The differences in storage time between CIS-CDMS and CIS-CIS samples for NfH did not correlate with this biomarker's levels (rs = −0.131, p = 0.256). NfH levels correlated with age (rs = 0.230, p = 0.044). We found no differences in time from CIS to lumbar puncture. Therefore, for each protein, we adjusted the analyses for age, topography, and/or DMT.
Table 1.
Demographic and clinical characteristics
FA, GFAP, and NfH results were negative (figure e-2, A–C). We could not detect neurofascin and sema3A.
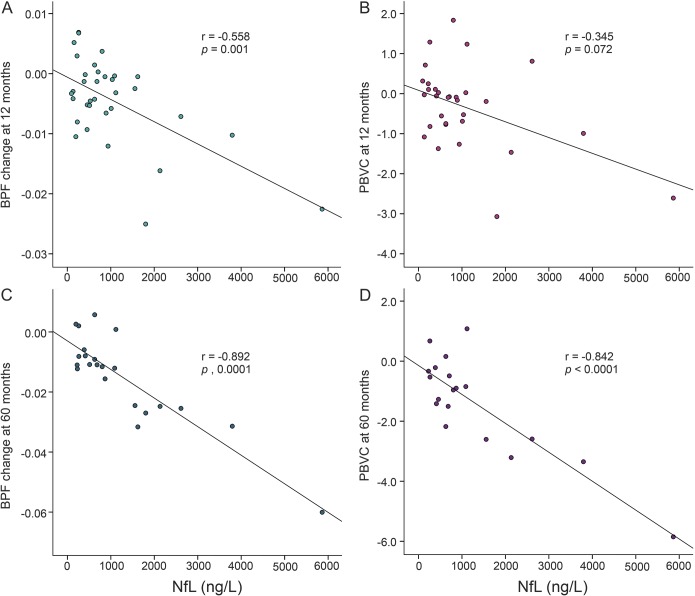
As for NfL levels, we found a significant difference between CIS-CDMS (1,553.1 [1,208.7–1,897.5] ng/L) and CIS-CIS (499.0 [168.8–829.2] ng/L) (p < 0.0001) (figure 1). The average intra- and interplate coefficients of variation (CVs) were 9.7% and 0.5%, respectively. We found no significant differences for EDSS progression of 1 point or ≥3.0, even though in the latter case, NfL levels were 1,506.1 (645.2–2,367.1) ng/L in the 15 patients with EDSS score ≥3.0 and 906.6 (693.2–1,120.1) ng/L if <3.0 (p = 0.185). We found significant correlations between NfL levels and inflammatory activity parameters on MRI, except for Gd-enhancing lesions at 5 years (table 2). As for neurodegenerative parameters, NfL and both BPFΔ and PBVC at 5 years showed the strongest correlations (table 2, figure 2).
Figure 1. NfL levels in the 2 CIS groups.

Results adjusted for CIS topography and disease-modifying treatment. CDMS = clinically definite multiple sclerosis; CIS = clinically isolated syndrome; NfL = neurofilament light chain.
Table 2.
Correlations between baseline neurofilament light chain levels and MRI measures at baseline, 1 year, and 5 years: Screening phase

Figure 2. Scatterplots showing the correlations between NfL levels and brain volume changes.
The graphs represent the raw data. The correlation coefficients and p values correspond to the partial correlations adjusted for age and baseline gadolinium-enhancing lesions. (A) BPF change at 1 year. (B) PBVC at 1 year. (C) BPF change at 5 years. (D) PBVC at 5 years. BPF = brain parenchymal fraction; NfL = neurofilament light chain; PBVC = percentage brain volume change.
Finally, we found a correlation between NfL and NfH levels (rs = 0.466, p = 0.002).
Replication phase.
Appendix e-1B describes preliminary analyses. Baseline demographic and clinical characteristics are shown in table e-1. Median (interquartile range) NfL levels were 1,238.3 (1,782.1) ng/L for patients converting to CDMS and 555.8 (825.5) ng/L for CIS-CIS. Average intra- and interplate CV were 4.9% and 7.0% at Vall d'Hebron (n = 93) and 5.1% and 8.8% at Ramón y Cajal (n = 62).
According to the cutoff, 63 patients (40.6%) were NfL positive. Significantly more NfL-positive patients converted to CDMS or fulfilled McDonald MS over time (data not shown).
When evaluating NfL levels as a continuous variable in the univariate analyses, we observed a 1-point increase in risk of both CDMS and McDonald MS for every 100-ng/L increment. The adjusted hazard ratio (aHR) for CDMS remained significant in the multivariate analysis (table 3).
Table 3.
Uni- and multivariate analyses for CDMS and 2010 McDonald MS with NfL as a continuous variable
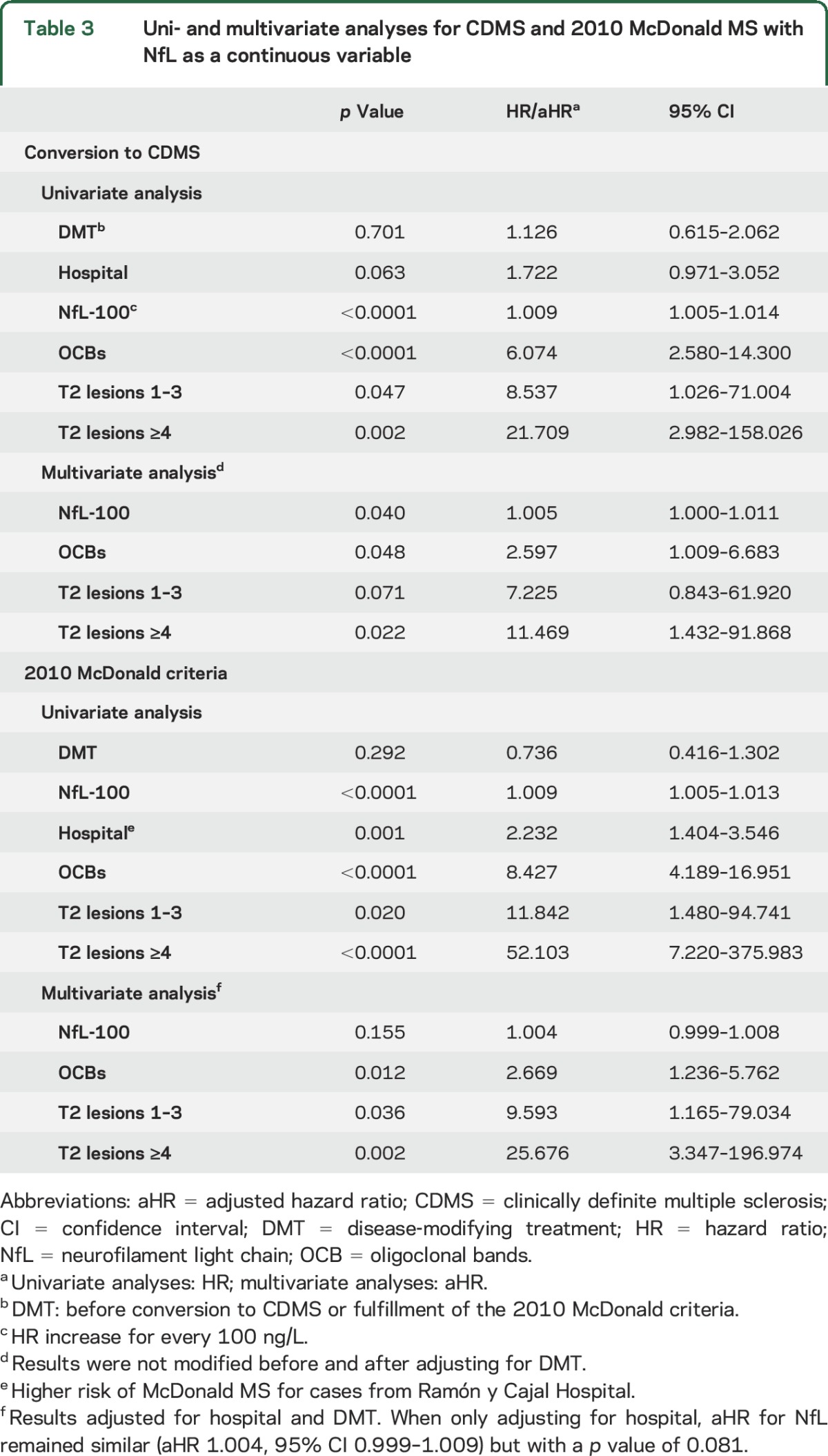
As for NfL status, NfL positivity was a predictor of CDMS (HR 2.279, 95% CI 1.283–4.049, p = 0.005) and McDonald MS (HR 2.801, 95% CI 1.761–4.455, p < 0.0001) in the univariate analyses; however, this significance was lost in the multivariate models (aHR 1.220, 95% CI 0.656–2.269, p = 0.530 and aHR 1.347, 95% CI 0.809–2.244, p = 0.252, respectively).
Regarding disability, although NfL levels were 2,635.6 (1,226.4–4,044.8) ng/L in the 11 patients with EDSS score ≥3.0 and 1,560.5 (934.8–2,186.2) ng/L if <3.0, this difference was not statistically significant (p = 0.172).
Finally, we found significant correlations between NfL levels and T2 lesion volume and Gd-enhancing lesion number on baseline MRIs (n = 63, rs = 0.517, p < 0.0001 and n = 146, rs = 0.469, p < 0.0001, respectively) and follow-up MRIs (n = 36, rs = 0.420, p = 0.011 for new T2 lesion volume and n = 94, rs = 0.231, p = 0.025 for Gd-enhancing lesions). Higher NfL levels were associated with a higher T2 lesion number at baseline and 1 year (data not shown).
DISCUSSION
In the screening phase, we aimed to identify differences in biomarker levels under the rationale that, if none were found between 2 opposite CIS groups,26 none would be found in consecutive patients, a scenario that better resembles clinical practice. In addition, this phase allows a strict use of biological samples. Only NfL levels were significantly higher in patients who converted to MS compared to nonconverters. Neurofilaments are type IV intermediate filaments specific for neurons, released into the CSF when axonal damage occurs.16,27,28 A study with 38 patients reported higher NfL levels in CIS-CDMS compared to CIS-CIS,18 a finding unconfirmed in other investigations including 9 to 36 patients per group.20,29,30 Such discrepancies could be attributable to a small sample size in the converter group,20,30 homogeneity in baseline inflammatory parameters such as proportion of OCBs present in both CIS groups,29 or the method used to detect NfL.30 In addition, sample size calculations are not referred and some groups may be too small to properly demonstrate any differences. Our study was adequately powered and thus confirms the differences with a well-balanced distribution between groups.
We found no associations between NfL levels and disability by the EDSS. Except for one, most studies showed significant correlations, but the analyses included CIS and other MS phenotypes, observing the strongest correlations in progressive forms.13,14,18,31–33 Khalil et al.20 evaluated 67 patients with CIS exclusively and found a marginal correlation coefficient of rs = 0.324 (p < 0.05). When assessing NfL levels according to EDSS score ≥3.0 in both phases, they were higher in patients with more severe disability but the difference was not significant, a finding probably influenced by the few patients who reached such EDSS score.
We observed significant associations between NfL and MRI inflammatory parameters on baseline and follow-up MRIs, with similar findings during the replication phase. Regarding Gd-enhancing lesions, in the screening, we found correlations with NfL levels at baseline and 1 year, observed again in the replication phase, but not at 5 years. There are previous reports of correlations with T2 lesion number and Gd-enhancing lesions in CIS and/or MS groups,18,25 unconfirmed in other CIS-specific studies.20,34 The most striking findings concern MRI neurodegenerative parameters: our results suggest baseline NfL levels increase not only in association with lesion load, accrual, and activity, but also independently of these parameters.35,36 On the contrary, a previous study did not find any correlations between NfL levels and brain volume change in CIS.20 Although the authors did adjust for age and used the SIENA software, they assessed the follow-up only at 1 year and did not control by Gd-enhancing lesions. We additionally evaluated the association between baseline NfL levels and brain volume changes at 5 years. Our findings are partly supported by those of Burman et al.,15 in which NfL levels were elevated irrespective of Gd-enhancing lesions in progressive MS. Unfortunately, our appraisal is limited: our sample size was small and we were unable to repeat this evaluation in the replication phase. However, by estimating both BPFΔ and PBVC, the latter a robust method, we believe our results are reliable and should be assessed in future studies.
Of note, there was a significant correlation between NfL and NfH levels, in accordance with previous publications on CIS and relapsing-remitting MS,18,20,31 although contrary to our results, some of these studies demonstrated associations between NfH and EDSS or MRI parameters.18,20 Thus, the origin of this correlation in our study is a matter of debate and deserves further study.
In the replication phase, considering that a dichotomic biomarker could be more useful in the clinical practice, we explored a 900 ng/L cutoff,25 observing that more NfL-positive than -negative patients evolved to CDMS and McDonald MS. Two other studies explored cutoff values to either identify relapses or predict evolution to secondary progressive MS.13,37 Their cutoff values, however, are lower than ours, a finding possibly related not only to the different outcomes but also to the use of a different assay,13 or the calculation methods based on NfL levels in patients with MS,13,37 whereas we established our cutoff using a control group with noninflammatory neurologic diseases under the rationale that this value would be more stable across studies. To assess whether this is a better approach, the cutoff should be evaluated in other cohorts. One study explored NfL levels and fulfillment of the 2010 dissemination in space criteria in optic neuritis,34 whereas we used the cutoff value to investigate the risk of developing both CDMS and 2010 McDonald MS at the time of a CIS. Another contribution is the assessment of NfL as an independent risk factor for MS. In the univariate analyses, we found a higher risk of CDMS and McDonald MS for every 100-ng/L increase in NfL levels, maintained for CDMS in the multivariate analysis. The nonsignificant results for McDonald MS could be conditioned by the distribution of patients over the different categories in the multivariate analysis, the correlations between NfL levels and T2 lesions, and the strong predictive value of T2 lesions for dissemination in space fulfillment. When assessing NfL status, the increased risk of both CDMS and McDonald MS in the univariate analyses was lost in the multivariate models. Thus, although NfL status could be more practical in the clinical setting, in our study, only NfL as a continuous variable is an independent risk factor for CDMS. Nevertheless, the value of NfL should be considered in context with OCB and T2 lesion results and, according to a recent study, it could be considered a weak risk factor for MS.6
Despite conducting measures to minimize bias, a limitation in the replication phase concerns the NfL determination in 2 different centers. Although the corresponding intra- and interplate CVs were within the assay's standardized limits, we did not perform quality controls such as testing the same samples in both centers.38 Besides, the shorter follow-up of the Vall d'Hebron cohort in the replication phase may preclude excluding patients with potential alternative diagnoses; nevertheless, the specific inclusion of typical CIS presentations lowers this risk.6 Furthermore, a shortcoming of NfL levels is their determination in CSF because some centers do not perform lumbar punctures routinely. It is impossible to quantify serum NfL levels with the available ELISA29,30; however, a new electrochemiluminescence array showed promising results in detecting higher serum NfL levels in patients with CIS compared to healthy controls.39
Our biomarker list was probably not thoroughly comprehensive. We did, however, search for updates throughout the study duration. Besides, the properties of the selected assays could have influenced our findings. Another limitation is the difficulty in assessing all biomarkers in the same patients without compromising the sample size given the limited CSF aliquot availability. Nevertheless, the homogeneous data collection and systematic sample storage procedures could aid to minimize bias. Finally, we did not compare the NfH electrochemiluminescence assay to other methods.
Therefore, elevated NfL levels indicate a higher risk of evolving to MS. However, the increase in risk for every 100 ng/L could be difficult to interpret in the daily clinical practice and, compared to OCBs and T2 lesions, NfL levels are a weak risk factor for MS. The search for a cutoff value should be assessed in other cohorts and, finally, the value of NfL as a marker of axonal damage may be more relevant as suggested by the correlations with medium-term brain volume loss.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Xavier Vidal for statistical analysis support (Department of Pharmacology, Vall d'Hebron University Hospital, Barcelona, Spain), the Red Española de Esclerosis Múltiple (REEM) (RD07/0060; RD12/0032) sponsored by the Fondo de Investigación Sanitaria (FIS), the Instituto de Salud Carlos III, the Ministry of Economy and Competitiveness in Spain, and the Ajuts per donar Suport als Grups de Recerca de Catalunya (2009 SGR 0793; 2014 SGR 1082) sponsored by the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) of the Generalitat de Catalunya in Spain.
GLOSSARY
- aHR
adjusted hazard ratio
- BPF
brain parenchymal fraction
- CDMS
clinically definite multiple sclerosis
- CI
confidence interval
- CIS
clinically isolated syndrome
- CV
coefficient of variation
- DMT
disease-modifying treatment
- EDSS
Expanded Disability Status Scale
- FA
fetuin A
- Gd
gadolinium
- GFAP
glial fibrillary acidic protein
- HR
hazard ratio
- MS
multiple sclerosis
- NfH
neurofilament heavy chain
- NfL
neurofilament light chain
- OCB
oligoclonal bands
- PBVC
percentage brain volume change
- sema3A
semaphorin 3A
- SIENA
Structural Image Evaluation, using Normalization, of Atrophy
Footnotes
Supplemental data at Neurology.org
Editorial, page 1068
AUTHOR CONTRIBUTIONS
Georgina Arrambide participated in the design of the work and on the acquisition, statistical analysis, and interpretation of data for the work, drafted and revised it, approved the present version to be published, and agreed to be accountable for all aspects of the work. Carmen Espejo participated in the design of the work and on the analysis and interpretation of data for the work, drafted and revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Herena Eixarch participated in the analysis and interpretation of data for the work, drafted and revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Luisa M. Villar participated in the acquisition, analysis, and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. José C. Alvarez-Cermeño participated in the acquisition, analysis, and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Carmen Picón participated in the acquisition and analysis of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Jens Kuhle participated in the acquisition, analysis, and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Giulio Disanto participated in the acquisition and analysis of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Ludwig Kappos participated in the analysis and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Jaume Sastre-Garriga participated in the design and acquisition, analysis, and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Deborah Pareto participated in the acquisition, analysis, and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Eva Simon participated in the acquisition of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Manuel Comabella participated in the acquisition and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Jordi Río participated in the acquisition and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Carlos Nos participated in the interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Carmen Tur participated in the acquisition and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Joaquín Castilló participated in the acquisition and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Angela Vidal-Jordana participated in the acquisition and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Ingrid Galán participated in the acquisition and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Maria J. Arévalo participated in the acquisition of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Cristina Auger participated in the analysis and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Alex Rovira participated in the analysis and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Xavier Montalban participated in the conception and design of the work and on the acquisition, analysis, and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work. Mar Tintore participated in the conception and design of the work and on the acquisition, analysis, and interpretation of data for the work, revised it for important intellectual content, approved the present version to be published, and agreed to be accountable for all aspects of the work.
STUDY FUNDING
This project was supported by FIS PI08/0788 and PI12/01313 from the FIS of the Ministry of Economy and Competitiveness of Spain, and by a McDonald Fellowship awarded to Georgina Arrambide by the MSIF. Carmen Espejo is partially supported by the Miguel Servet program (CP07/00146; CP13/00028), Herena Eixarch is supported by the Sara Borrell program (CD09/0363), and Angela Vidal-Jordana received support for research training contracts Rio Hortega (CM10/00032), all from the FIS, Instituto de Salud Carlos III, Ministry of Economy and Competitiveness of Spain. Jens Kuhle was supported by an ECTRIMS Research Fellowship Programme and by the Forschungsfonds of the University of Basel, Switzerland.
DISCLOSURE
G. Arrambide has received compensation for consulting services from Biogen Idec and research support from Novartis. C. Espejo and H. Eixarch report no disclosures relevant to the manuscript. L. Villar has received speaking honoraria or research support from Bayer Schering Pharma, Merck Serono, Biogen Idec, Teva Pharmaceuticals, Sanofi-Aventis, Genzyme, and Novartis. J. Alvarez-Cermeño has received compensation for consulting services and speaking honoraria from Bayer Schering Pharma, Merck Serono, Biogen Idec, Teva Pharmaceuticals, Sanofi-Aventis, and Novartis. C. Picón reports no disclosures relevant to the manuscript. J. Kuhle has received research support from the Swiss MS Society, Swiss ALS Society, Protagen AG, Roche, Genzyme, and Novartis and served on scientific advisory boards for Genzyme/Sanofi-Aventis, Merck Serono, and Novartis Pharma. G. Disanto reports no disclosures relevant to the manuscript. L. Kappos has received and dedicated to research support fees for board membership, consultancy or speaking, or grants in the last 3 years from Actelion, Advancells, Allozyne, Bayer, Bayhill, Biogen Idec, BioMarin, CSL Behring, Eli Lilly, European Union, GeNeuro, Genmab, Gianni Rubatto Foundation, Glenmark, Merck Serono, MediciNova, Mitsubishi Pharma, Novartis, Novartis Research Foundation, Novo Nordisk, Peptimmune, Roche, Roche Research Foundation, Sanofi-Aventis, Santhera, Swiss MS Society, Swiss National Research Foundation, Teva, UCB, and Wyeth. J. Sastre-Garriga has received compensation for participating on advisory boards, speaking honoraria and travel expenses for scientific meetings, consulting services or research support from Novartis, Biogen Idec, Sanofi-Aventis, Teva, Serono Symposia International Foundation, Merck Serono, Almirall, and Genzyme. D. Pareto and E. Simon report no disclosures relevant to the manuscript. M. Comabella has received compensation for consulting services and speaking honoraria from Bayer Schering Pharma, Merck Serono, Biogen Idec, Teva Pharmaceuticals, Sanofi-Aventis, and Novartis. J. Río has received speaking honoraria and personal compensation for participating on advisory boards from Almirall, Bayer Schering Healthcare, Biogen Idec, Genzyme, Merck Serono, Novartis, Teva, and Sanofi-Aventis. C. Nos, C. Tur, J. Castilló, A. Vidal-Jordana, I. Galán, M. Arévalo, and C. Auger report no disclosures relevant to the manuscript. A. Rovira serves on scientific advisory boards for Neuro-Tec and on the editorial board of the American Journal of Neuroradiology and Neuroradiology, has received speaker honoraria from Bayer Schering Pharma, Sanofi-Aventis, Bracco, Merck Serono, Teva Pharmaceutical Industries Ltd., and Biogen Idec, receives research support from Bayer Schering Pharma, and serves as a consultant for Novartis. X. Montalban has received speaking honoraria and travel expenses for scientific meetings, has been a steering committee member of clinical trials or participated in advisory boards of clinical trials in the past with Almirall, Bayer, Biogen, Genzyme, Merck, Novartis, Receptos, Roche, Sanofi Genzyme, and Teva Pharmaceuticals. M. Tintore has received compensation for consulting services and speaking honoraria from Bayer Schering Pharma, Merck Serono, Biogen Idec, Teva Pharmaceuticals, Sanofi-Aventis, Novartis, Almirall, Genzyme, and Roche. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Fisniku LK, Brex PA, Altmann DR, et al. Disability and T2 MRI lesions: a 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain 2008;131:808–817. [DOI] [PubMed] [Google Scholar]
- 2.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 2011;69:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tintore M, Rovira A, Rio J, et al. Do oligoclonal bands add information to MRI in first attacks of multiple sclerosis? Neurology 2008;70:1079–1083. [DOI] [PubMed] [Google Scholar]
- 4.Masjuan J, Alvarez-Cermeno JC, Garcia-Barragan N, et al. Clinically isolated syndromes: a new oligoclonal band test accurately predicts conversion to MS. Neurology 2006;66:576–578. [DOI] [PubMed] [Google Scholar]
- 5.Comabella M, Montalban X. Body fluid biomarkers in multiple sclerosis. Lancet Neurol 2014;13:113–126. [DOI] [PubMed] [Google Scholar]
- 6.Tintore M, Rovira A, Rio J, et al. Defining high, medium and low impact prognostic factors for developing multiple sclerosis. Brain 2015;138:1863–1874. [DOI] [PubMed] [Google Scholar]
- 7.Poser CM, Paty DW, Scheinberg L, et al. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol 1983;13:227–231. [DOI] [PubMed] [Google Scholar]
- 8.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23(suppl 1):S208–S219. [DOI] [PubMed] [Google Scholar]
- 9.Tumani H, Lehmensiek V, Rau D, et al. CSF proteome analysis in clinically isolated syndrome (CIS): candidate markers for conversion to definite multiple sclerosis. Neurosci Lett 2009;452:214–217. [DOI] [PubMed] [Google Scholar]
- 10.Williams A, Piaton G, Aigrot MS, et al. Semaphorin 3A and 3F: key players in myelin repair in multiple sclerosis? Brain 2007;130:2554–2565. [DOI] [PubMed] [Google Scholar]
- 11.Eixarch H, Gutierrez-Franco A, Montalban X, Espejo C. Semaphorins 3A and 7A: potential immune and neuroregenerative targets in multiple sclerosis. Trends Mol Med 2013;19:157–164. [DOI] [PubMed] [Google Scholar]
- 12.Petzold A, Eikelenboom MJ, Gveric D, et al. Markers for different glial cell responses in multiple sclerosis: clinical and pathological correlations. Brain 2002;125:1462–1473. [DOI] [PubMed] [Google Scholar]
- 13.Malmestrom C, Haghighi S, Rosengren L, Andersen O, Lycke J. Neurofilament light protein and glial fibrillary acidic protein as biological markers in MS. Neurology 2003;61:1720–1725. [DOI] [PubMed] [Google Scholar]
- 14.Norgren N, Sundstrom P, Svenningsson A, Rosengren L, Stigbrand T, Gunnarsson M. Neurofilament and glial fibrillary acidic protein in multiple sclerosis. Neurology 2004;63:1586–1590. [DOI] [PubMed] [Google Scholar]
- 15.Burman J, Zetterberg H, Fransson M, Loskog AS, Raininko R, Fagius J. Assessing tissue damage in multiple sclerosis: a biomarker approach. Acta Neurol Scand 2014;130:81–89. [DOI] [PubMed] [Google Scholar]
- 16.Petzold A. Neurofilament phosphoforms: surrogate markers for axonal injury, degeneration and loss. J Neurol Sci 2005;233:183–198. [DOI] [PubMed] [Google Scholar]
- 17.Kuhle J, Regeniter A, Leppert D, et al. A highly sensitive electrochemiluminescence immunoassay for the neurofilament heavy chain protein. J Neuroimmunol 2010;220:114–119. [DOI] [PubMed] [Google Scholar]
- 18.Teunissen CE, Iacobaeus E, Khademi M, et al. Combination of CSF N-acetylaspartate and neurofilaments in multiple sclerosis. Neurology 2009;72:1322–1329. [DOI] [PubMed] [Google Scholar]
- 19.Kuhle J, Leppert D, Petzold A, et al. Neurofilament heavy chain in CSF correlates with relapses and disability in multiple sclerosis. Neurology 2011;76:1206–1213. [DOI] [PubMed] [Google Scholar]
- 20.Khalil M, Enzinger C, Langkammer C, et al. CSF neurofilament and N-acetylaspartate related brain changes in clinically isolated syndrome. Mult Scler 2013;19:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathey EK, Derfuss T, Storch MK, et al. Neurofascin as a novel target for autoantibody-mediated axonal injury. J Exp Med 2007;204:2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawamura N, Yamasaki R, Yonekawa T, et al. Anti-neurofascin antibody in patients with combined central and peripheral demyelination. Neurology 2013;81:714–722. [DOI] [PubMed] [Google Scholar]
- 23.Shaw G, Yang C, Ellis R, et al. Hyperphosphorylated neurofilament NF-H is a serum biomarker of axonal injury. Biochem Biophys Res Commun 2005;336:1268–1277. [DOI] [PubMed] [Google Scholar]
- 24.Petzold A, Keir G, Green AJ, Giovannoni G, Thompson EJ. A specific ELISA for measuring neurofilament heavy chain phosphoforms. J Immunol Methods 2003;278:179–190. [DOI] [PubMed] [Google Scholar]
- 25.Villar LM, Picon C, Costa-Frossard L, et al. Cerebrospinal fluid immunological biomarkers associated with axonal damage in multiple sclerosis. Eur J Neurol 2015;22:1169–1175. [DOI] [PubMed] [Google Scholar]
- 26.Comabella M, Fernandez M, Martin R, et al. Cerebrospinal fluid chitinase 3-like 1 levels are associated with conversion to multiple sclerosis. Brain 2010;133:1082–1093. [DOI] [PubMed] [Google Scholar]
- 27.Nylen K, Csajbok LZ, Ost M, et al. CSF–neurofilament correlates with outcome after aneurysmal subarachnoid hemorrhage. Neurosci Lett 2006;404:132–136. [DOI] [PubMed] [Google Scholar]
- 28.Petzold A, Keir G, Warren J, Fox N, Rossor MN. A systematic review and meta-analysis of CSF neurofilament protein levels as biomarkers in dementia. Neurodegener Dis 2007;4:185–194. [DOI] [PubMed] [Google Scholar]
- 29.Fialova L, Bartos A, Svarcova J, Zimova D, Kotoucova J, Malbohan I. Serum and cerebrospinal fluid light neurofilaments and antibodies against them in clinically isolated syndrome and multiple sclerosis. J Neuroimmunol 2013;262:113–120. [DOI] [PubMed] [Google Scholar]
- 30.Avsar T, Korkmaz D, Tutuncu M, et al. Protein biomarkers for multiple sclerosis: semi-quantitative analysis of cerebrospinal fluid candidate protein biomarkers in different forms of multiple sclerosis. Mult Scler 2012;18:1081–1091. [DOI] [PubMed] [Google Scholar]
- 31.Kuhle J, Plattner K, Bestwick JP, et al. A comparative study of CSF neurofilament light and heavy chain protein in MS. Mult Scler 2013;19:1597–1603. [DOI] [PubMed] [Google Scholar]
- 32.Semra YK, Seidi OA, Sharief MK. Heightened intrathecal release of axonal cytoskeletal proteins in multiple sclerosis is associated with progressive disease and clinical disability. J Neuroimmunol 2002;122:132–139. [DOI] [PubMed] [Google Scholar]
- 33.Trentini A, Comabella M, Tintore M, et al. N-acetylaspartate and neurofilaments as biomarkers of axonal damage in patients with progressive forms of multiple sclerosis. J Neurol 2014;261:2338–2343. [DOI] [PubMed] [Google Scholar]
- 34.Modvig S, Degn M, Horwitz H, et al. Relationship between cerebrospinal fluid biomarkers for inflammation, demyelination and neurodegeneration in acute optic neuritis. PLoS One 2013;8:e77163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Miralles F, Sastre-Garriga J, Tintore M, et al. Clinical impact of early brain atrophy in clinically isolated syndromes. Mult Scler 2013;19:1878–1886. [DOI] [PubMed] [Google Scholar]
- 36.Bielekova B, McDermott MP. Will CSF biomarkers guide future therapeutic decisions in multiple sclerosis? Neurology 2015;84:1620–1621. [DOI] [PubMed] [Google Scholar]
- 37.Salzer J, Svenningsson A, Sundstrom P. Neurofilament light as a prognostic marker in multiple sclerosis. Mult Scler 2010;16:287–292. [DOI] [PubMed] [Google Scholar]
- 38.Petzold A, Altintas A, Andreoni L, et al. Neurofilament ELISA validation. J Immunol Methods 2010;352:23–31. [DOI] [PubMed] [Google Scholar]
- 39.Disanto G, Adiutori R, Dobson R, et al. Serum neurofilament light chain levels are increased in patients with a clinically isolated syndrome. J Neurol Neurosurg Psychiatry 2016;87:126–129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.