Abstract
Humans are continuously exposed to hazardous chemicals in the environment. These chemicals or their electrophilic metabolites can form adducts with genomic DNA, which can lead to mutations and the initiation of cancer. The identification of DNA adducts is required for understanding exposure and the etiological role of a genotoxic chemical in cancer risk. The analytical chemist is confronted with a great challenge because the levels of DNA adducts generally occur at less than one adduct per 107 nucleotides, and the amount of tissue available for measurement is limited. Ion trap mass spectrometry has emerged as an important technique to screen for DNA adducts because of the high level sensitivity and selectivity, particularly, when employing multi-stage scanning (MSn). The product ion spectra provide rich structural information and corroborate the adduct identities even at trace levels in human tissues. Ion trap technology represents a significant advance in measuring DNA adducts in humans.
Keywords: Biomonitoring, DNA adducts, Ion Trap Mass spectrometry
INTRODUCTION
Humans are continuously exposed to genotoxicants through environment, tobacco, diet, medication, and reactive electrophiles are formed endogenously through normal cellular processes. Some known and possible human carcinogens are polycyclic aromatic hydrocarbons (PAHs), N-nitroso compounds (NOCs), aromatic amines, and heterocyclic aromatic amines (HAAs). These chemicals originate from air pollution, automobile exhaust, tobacco smoke, or cured and cooked red meat (IARC Monograph, 1983; IARC Monograph, 1986; IARC Monograph, 2012). Representative structures of some of these carcinogens and their major DNA adducts are shown in Figure 1. All of these procarcinogens undergo metabolism to form reactive electrophiles that covalently adduct to macromolecules, such as DNA and proteins (Loeb and Harris, 2008; Miller, 1970). Biomonitoring of protein adducts of carcinogens has been used to assess exposures to hazardous chemicals, and adduction products of blood proteins, such as serum albumin and hemoglobin, have been employed as biomarkers in human populations (Liebler, 2002; Rappaport et al., 2012; Skipper et al., 1994). However, protein adduct formation may not accurately represent the extent of DNA damage that occurs in distant target organs. DNA adducts, if not repaired, can induce mutations in critical genes, leading to the disruption of critical cellular functions and the initiation of cancer (Loeb and Harris, 2008). DNA adducts are a measure of exposure and also provide mechanistic information about metabolism and chemical carcinogenesis. Moreover, the patterns of mutation spectra in tumor related genes can reveal a causal role of chemicals in carcinogenesis (Grollman, 2013; Loeb and Harris, 2008; Pfeifer et al., 2002).
Figure 1.
Representative structures of known and probable human carcinogens and their DNA adducts.
The molecular and epidemiological data that have linked chemical exposures to aflatoxin B1 (AFB1) and aristolochic acids (AAs), and their DNA adducts and characteristic mutations in tumor related genes are the “gold standards” in epidemiological studies that have examined the role of chemical exposures and cancer risk (Figure 1). AFB1, a mycotoxin produced by the stored grain stuffs contaminated by fungi Aspergillus flavus, is widely found in areas of Southeast Asia and Sub-Saharan African and one of the most potent natural product carcinogens (McGlynn and London, 2011). Epidemiology studies have shown a strong relation exists among dietary AFB1 exposure, human primary hepatocellular carcinoma (HCC) and specific mutations at codon 249 of the TP53 tumor suppressor gene (Aguilar et al., 1994). The reactive AFB1 epoxide metabolite binds to DNA to form 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 (AFB1-N7-Gua), an unstable adduct that rearranges to the ring-opened formamidopyrimidine (FAPy) derivative AFB1-FAPy. It is AFB1-FAPy which is responsible for the G–T transversion mutation at codon 249 of the TP53 (Smela et al., 2002; Stone et al., 2011).
AAs are a structurally related family of nephrotoxic and carcinogenic nitrophenanthrene compounds found in Aristolochia herbaceous plants. The exposure to AAs in the rural villages of the Balkan countries has been shown to occur through the ingestion of home-baked bread contaminated with Aristolochia seeds. The ingestion of traditional Chinese medicines contaminated with Aristolochia sp represents another major source of exposure to AAs and occurs world-wide (Grollman, 2013). AAs are responsible for the clinical syndromes known formerly as Balkan endemic nephropathy (BEN) (Grollman, 2013; Jelakovic et al., 2012) and Chinese herbs nephropathy (Chen et al., 2012; Vanherweghem et al., 2003). Both diseases are associated with a high incidence of urothelial carcinomas of the upper urinary tract (UUC), (Chen et al., 2012; Grollman, 2013; Jelakovic et al., 2012) constituting a disease entity now termed aristolochic acid nephropathy (AAN) (De Broe, 2012). One major DNA adduct of AA is 7-(deoxyadenosin-N6-yl)-aristolactam (dA-AL-I). This adduct is responsible for the characteristic A-T transversion mutation in the TP53 tumor suppressor gene, as well as mutations throughout the tumor genome in patients with AA-induced urothelial carcinomas of the upper urinary tract (Grollman, 2013; Hoang et al., 2013). dA-AL-I has been detected by ion trap mass spectrometry in kidney tissue of patients from Croatia, Taiwan, China, Romania, and the United States (Yun et al., 2015).
METHODS TO MEASURE DNA ADDUCTS
The major methods to measure DNA adducts include 32P-postlabeling, (Phillips, 2012; Randerath et al., 1981) immunochemistry, (Poirier et al., 2000; Santella, 1999) gas chromatography mass spectrometry (GC/MS) (primarily for oxidized DNA bases following chemical derivatization (Dizdaroglu et al., 2015)), liquid chromatography (LC) coupled with electrochemical, fluorescence or MS (Brown, 2012; Klaene et al., 2013; Phillips et al., 2000; Singh and Farmer, 2006). The 32P-postlabeling method was developed more than 30 years ago by Randerath, Gupta and colleagues, (Randerath et al., 1981) and it is still a commonly used method for in vivo studies because it only requires 10 μg DNA per sample. Depending upon the adduct, a level of detection 1 adduct per 1010 nucleotides can be achieved (Phillips, 2012). Regardless of the excellent level of sensitivity, the use of highly radioactive phosphorus is a health hazard, and the lack of internal standards precludes quantitation, and the absence of spectral data for adduct identification greatly hinders the usefulness of 32P-postlabeling in human studies.
Immunodetection is another sensitive method to detect DNA adduct (Poirier et al., 2000; Santella, 1999). Once an antibody is developed, it can be employed in an enzyme-linked immunosorbent assays (ELISA) with DNA isolated from tissues, or used in an immunohistochemistry assay to probe for DNA adducts in specific cell types of intact tissues (Chen et al., 2002; Gammon et al., 2002; Hsu et al., 1997; Motykiewicz et al., 1995; Pratt et al., 2011). However, an important drawback of immunoassays is that the specificity of many antibodies, even monoclonal antibodies, for DNA adducts is uncertain, and the antibodies may cross-react with other DNA lesions, leading to errors in identification and quantification. The uncertainty in specificity and the semi-quantitative nature of the assay confines the usage of this screening method in human population studies.
The central role of MS in measuring the DNA adduct formation and their biological consequences has been recently reviewed by Liu et. al.(Liu and Wang, 2015) LC-tandem MS (MS/MS) with stable isotope dilution is the primary method for quantitation of DNA adducts (Liu and Wang, 2015; Singh and Farmer, 2006; Tretyakova et al., 2012). The adducts are usually detected at the nucleoside level after digestion of the DNA with nucleases, although in some instances adducts can be measured as the modified bases. Over the past 20 years, the triple quadrupole (QqQ) MS has been the principal MS instrument employed to measure DNA adducts due to its relatively low cost, excellent sensitivity and robustness, and ease of operation. The DNA adducts are routinely measured in the selected reaction monitoring (SRM) mode. However, the need to confirm adduct structure by a higher level of fragmentation of the adducted base or higher resolving power have turned researchers to other mass analyzers such as Ion Trap (IT) or Time-of-Flight (TOF).(Tretyakova et al., 2012) Our laboratory has extensively used IT-MS employing the multi-stage scanning (MSn) feature to screen for DNA adducts present at trace levels in humans (Goodenough et al., 2007; Gu et al., 2012; Yun et al., 2015).
FUNDAMENTAL OF THE ION TRAP
The IT is derived from the quadrupole mass analyzer. Ion motions are subjected to the quadrupolar field in both IT and quadrupole. The IT applies additional electric potential along the axial axis to stop ions exiting the device, achieving a “trapping” effect. In tandem MS, IT permits ion storage, isolation, fragmentation and sequential ejection to occur in a time-dependent approach (“tandem-in-time”), in contrast to the QqQ-MS, where three quadruples are connected sequentially to perform a single MS2 scan in a “tandem-in-space” manner.
Quadrupole and quadrupolar field
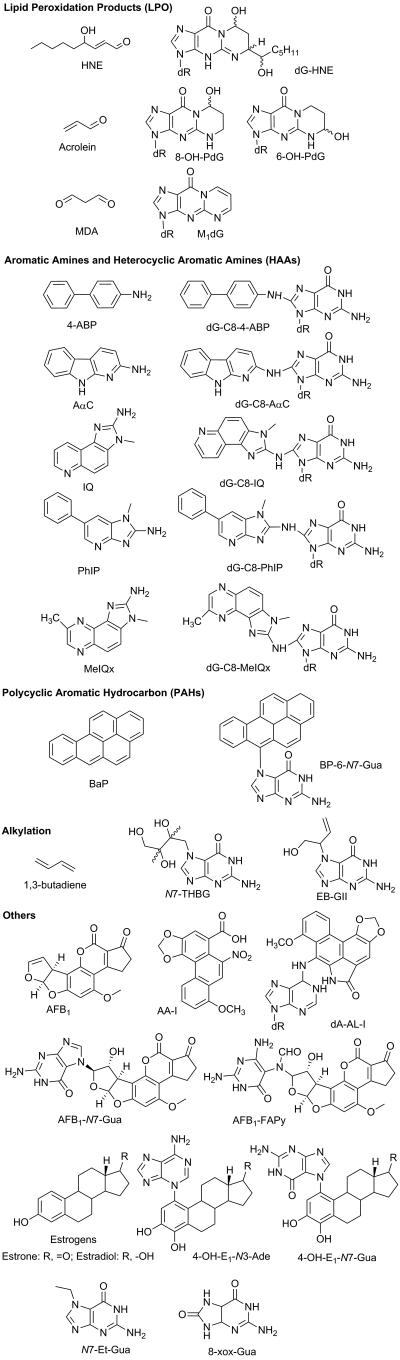
Knowledge on the principle of the quadrupole is required to understand the operation of the ion trap. For the convenience, all mass analyzer principles are explained in the positive ionization mode. In the negative mode, the ion behavior can be extrapolated in analogous fashion. Figure 2 (A) shows the scheme of a single-quadrupole instrument. A quadrupole mass analyzer is composed of four parallel rods of hyperbolic shape. Rods opposite each other are connected electrically, where a combination of Direct Current (DC) and Alternating Current (AC) voltages are applied to the two pairs of rods with the same amplitude but opposite in polarity. The AC is operated at a constant frequency while both voltages of AC and DC vary. The AC voltage oscillates in the range of radio frequency (RF), thus it is also referred to as RF voltage. Upon entering a quadrupole, ions maintain the original speeds (a trajectory) along the axial direction (z-axis). On the radial direction (x-y plane), ions are accelerated by the applied voltage to move towards a rod, followed by deceleration once the RF voltage changes phase. Ions that fail to change the direction of movement by the time the electric field switch the polarity will collide on the rod. Thus, one pair of rods performs as a high mass filter, while the other set of rods serves as a low mass filter. Collectively, ions traverse the quadrupole in a movement resembling a spiral motion (de Hoffmann and Stroobant, 2007).
Figure 2.
Scheme of (A) a single-quadrupole mass analyzer and (B) stability diagram.
The electric force subjected to the ions inside a quadrupolar field can be described using a second-order linear differential equation, and integrated by the Mathieu equation.(de Hoffmann and Stroobant, 2007) Simplification of the equation yields two parameters, a and q, which can be plotted to predict the ion motions. Such a plot is known as a “stability diagram” (Figure 2 (B)). Mathematically, only ions located inside the triangle have stable trajectories. Equations 1 and 2 show the expressions of a, q and the amplitudes of DC (U) and AC (V) after rearrangement. The parameters ω and r0 are constant for a given quadrupole instrument. Thus, the resolution of the quadrupole depends on the ratio of U/V: the bigger the slope, the higher the resolution.
| (Eq. 1) |
| (Eq. 2) |
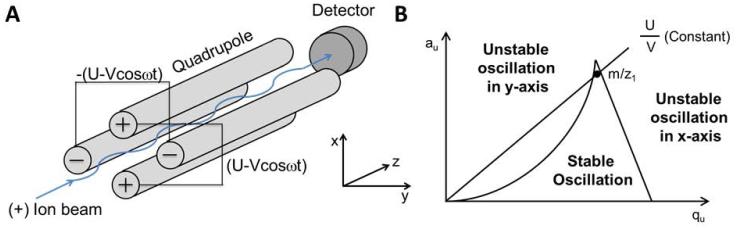
If DC is switched off (U=0, referred to as “RF-only”), the scan line falls to the qu axis, which transforms the device from ion selection to ion transfer. A collision gas can be introduced into such RF-only quadrupole (depicted as “q”, differing from the q in the stability diagram), and such device serves as the centerpiece for a QqQ instrument. Figure 3 shows the tandem MS scan modes that are possible for a QqQ instrument. Since the fragmentation occurs as a space-dependent mode in q, only MS2 fragmentation can be performed. The QqQ operating under selected reaction monitoring (SRM) is one of the most widely used MS methods in small molecule detection and quantitation (see below) (Tretyakova et al., 2012).
Figure 3.
Tandem MS scan modes of the QqQ-MS. Ions of different m/z are depicted by the size of circles. Ions measured by Q1 are indicated by dash lines in the mass spectra at the far right and those by Q3, solid lines.
Ion Trap
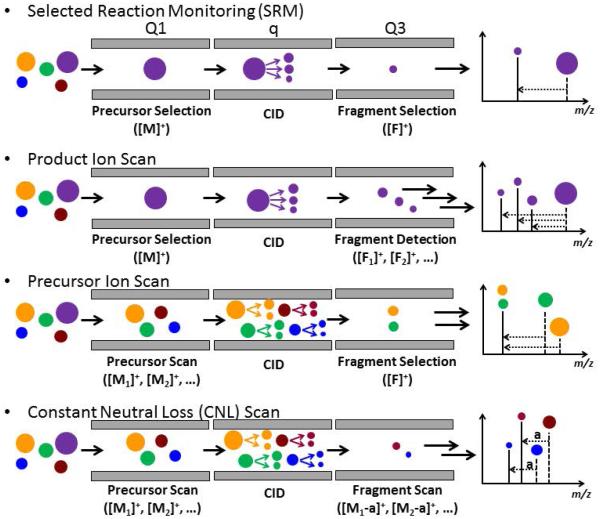
An IT stores ions in three or two dimensions through a combination of RF oscillation at the radial direction and an electric potential at the axial direction. IT can be classified into two categories: 3-Dimension (3D) and 2-Dimension (2D) ion traps. The 3D ion trap (Figure 4 (A)), which is also referred to as a quadrupole ion trap (QIT), was invented in the 1960s.(Paul, 1990) It is made up of a circular electrode (ring electrode) with two ellipsoid caps on the top and the bottom (endcap electrodes). Ions are injected into the trap through the entrance electrode, oscillating in a boomerang type circular motion, under the influence of the quadrupolar field by the ring-electrode and the DC voltage by the endcap electrode. The trap is filled with an inert gas such as helium (damping gas), which removes excess energy from the ions by collisional cooling and allowing ions to migrate to the center of the ion trap. The damping gas results in a more uniform ejection process of the ions leading to improved mass spectral performance.(Stafford, 2002) QIT-MS has been used extensively in proteomics and sequencing of peptides.(Olsen et al., 2009; Schwartz et al., 2002) The 2D ion trap is also called a linear ion trap (LIT) (Figure 4 (B)). Similarly, ions are circulating in an LIT under a quadrupolar field generated by the four-rod quadrupole and the electric field applied by the end lens that repel the ions in the direction opposite to the direction of entry.
Figure 4.
Scheme of (A) Quadrupole Ion Trap and (B) Linear Ion Trap mass analyzers with radial ejection.
The LIT has a higher ion storage capacity than QIT, (Douglas et al., 2005; Schwartz et al., 2002) however, sample matrix effects can impact the performance of all IT MS instruments. An IT has limited storage capacity, and only a finite number of ions can be collected in the trap. Overfilling the trap will result in an effect known as “space charging”, due to columbic repulsions of the like charges, which results in the spreading of the ion packet into a diffuse cloud, reducing the mass resolution and ion signal (Payne and Glish, 2005). Thus, the number of ions which enter the ion trap is controlled by setting a maximum total ion count and a dynamic ion injection time setting. The injection time is set by applying a short prescan to measure the ion influx, which is used to calculate the actual time the gate lens needs to remain open to fill the trap to the maximum capacity.
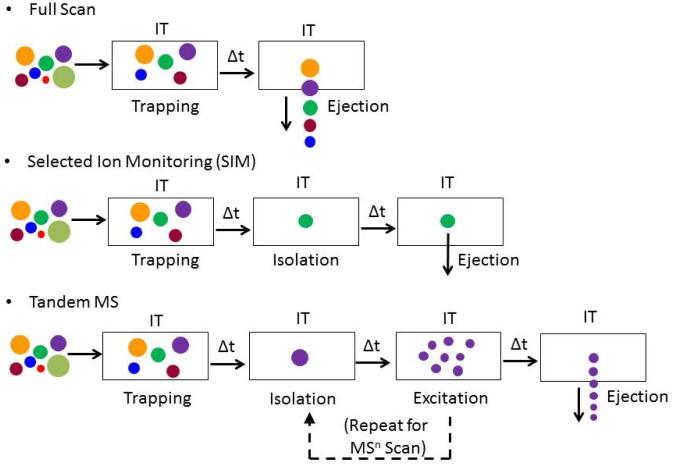
Four basic functions are performed by all ITs: trapping, isolation, excitation and ejection. Figure 5 depicts the scan modes available for IT. In the full scan mode, all ions are collected and ejected from low to high mass, resulting in a spectrum with an ion count for each mass. In the selected ion monitoring (SIM) mode, once all of the ions are collected in the trap, an isolation waveform is applied so that all ions are ejected but a single ion or a collection of ions over narrow m/z range, which are sequentially ejected and detected. In the MS2 scan stage, all ions are collected and a single ion (precursor ion) is isolated. Then the voltages are increased to excite the precursor ion and cause it to fragment by colliding with the damping gas, and the product ions are scanned out for detection. Since the fragmentation is performed in a time-dependent manner, the process of isolation and excitation can be repeated continuously, resulting in multistage MSn scanning. The MSn data provide additional information about the structure of the adduct. The multistage scanning of the analyte, also termed consecutive reaction monitoring, permits extensive mass spectral characterization of DNA adduct structures (Goodenough et al., 2007; Yun et al., 2012).
Figure 5.
Scan modes of the IT-MS. Ions of different m/z are depicted by the size of circles.
Quadrupole Based Mass Analyzers: QqQ versus Ion Trap Analyses of DNA Adducts
The QqQ instruments generally employ the SRM mode for DNA adduct measurements (Liu and Wang, 2015; Singh and Farmer, 2006; Tretyakova et al., 2012). In this mode, the protonated adducts ([M+H]+) are selectively transmitted by the first mass analyzer (Q1) and subjected to collision induced dissociation (CID), typically with argon gas, in the second quadrupole (q). The glycosidic linkage of the DNA adduct is normally the most facile bond to undergo cleavage in the gas phase.(Wolf and Vouros, 1994) The CID conditions are optimized to cleave the deoxyribose moiety [M+H-116]+ to form the protonated base adduct, also called the aglycone [BH2]+. The aglycone is then selectively transmitted through the third quadrupole (Q3). The SRM mode is particularly effective for precise quantification because of the rapid duty cycle of QqQ. The operation of the QqQ in the Product Ion Scan mode with elevated collision energy results in a dissociation cascade or a multiple-step fragmentation of the aglycone and additional structural information can be obtained on the aglycone (Turesky and Vouros, 2004). However, the product ion scan mode is rarely employed for the characterization of DNA adducts in vivo because the rate of scanning is slow and not sufficiently sensitive to detect adducts at trace levels.(Paehler et al., 2002) Thus, the analyst must rely on the retention time (tR) and a single SRM transition ([M+H-116]+) as criteria for analyte identification.
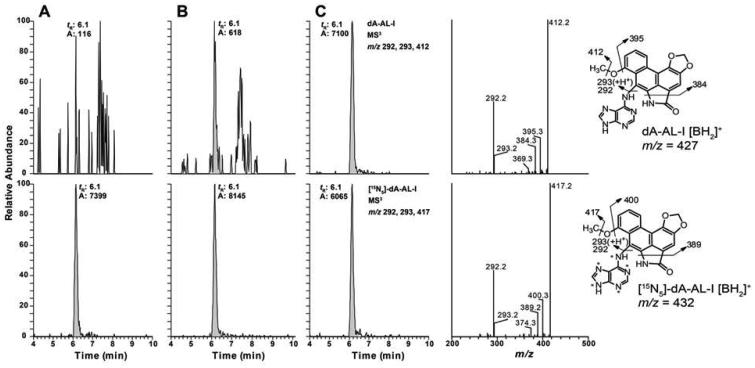
In contrast, both QIT and LIT can acquire full product ion MSn spectra with excellent sensitivity. The product ion spectra provide characteristic fragment ions of the adduct, and the method selectivity can be improved by analyzing the adduct at MS3 or even MS4 level if necessary (Goodenough et al., 2007; Kozekov et al., 2010; Sangaraju et al., 2014). The CID occurs in the quadrupolar field of the ion trap filled with helium gas by applying resonance excitation frequency matched to m/z of the precursor ion, such that only a single-step fragmentation occurs because the product ions are no longer in resonance with the excitation frequency. Thus, fragment ions with higher relative abundance are often generated by the IT mass spectrometer compared to those generated by CID in the QqQ. Because of the design of the IT and its rapid scanning rate, full product ion and MSn spectra are readily acquired with low amounts of analyte. Thus, both spectral characterization and quantification of DNA adducts can be achieved at the MS2 or MS3 scan stages by the LIT-MS. Specific product ions are used to reconstruct the mass chromatograms for quantitative measurements. An example of the power of the LIT-MS to measure DNA adducts at the MS3 scan stage is shown in Figure 6. The chromatograms of the aristolactam DNA adduct, dA-AL-l, were acquired on DNA from the renal cortex of patients with upper urothelial cancer.(Yun et al., 2015) The identity of the adduct was confirmed by its characteristic fragment ions shown in the product ion spectra at the MS3 scan stage for both unlabeled and its stable isotope labeled internal standard.
Figure 6.
UPLC-LIT-MS3 reconstructed ion chromatograms of dA-Al-I from human kidney cortex of patients with UUC from Taiwan at levels (A) below LOQ, (B) 0.4 adducts, and (C) 5.9 adducts per 108 nucleotides. The product ion spectra of dA-AL-I and its internal standard obtained from subject C, the 15N (*) labeled internal standard was spiked into DNA at a level of 5 adducts per 108 nucleotides. Reprinted from Yun (2012) with permission from American Chemical Society.
LITERATURE REVIEW
Studies investigating DNA adduct formation in experimental laboratory animals commenced in the 1940s (Price et al., 1948). In the ensuing decades, the methods of examining DNA adduct formation in experimental laboratory animals or in cell culture generally required radioactive carcinogens of high specific activity. The chemically modified DNA underwent digestion with nucleases to form the mononucleoside radiolabeled adducts, which were analyzed by HPLC with a radioactive detector, and synthetic DNA adduct standards served as ultraviolet (UV) markers (Beland et al., 1983; Beland and Kadlubar, 1985). Thereafter, many animal model studies employed 32P-postlabeling (Phillips, 2012; Randerath et al., 1981). During the past two decades, soft ionization techniques such as matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) have emerged as techniques to detect non-volatile and thermally labile compounds (Fenn et al., 1989; Nordhoff et al., 1994). The on-line coupling of HPLC combined, in particular, with ESI-MSn has been employed to measure DNA adducts of many classes of carcinogens over the past 20 years (Liu and Wang, 2015; Singh and Farmer, 2006; Tretyakova et al., 2012). Unlike studies conducted with laboratory animals treated with large doses of chemicals where ample tissue is available, the measurement of DNA adducts in human tissues is a far greater analytical challenge because chemical exposures are much lower than those employed in animal studies. Furthermore, the levels of DNA adducts usually occur only in the range of 0.01 – 10 per 108 nucleotides, and the amount of tissue available for assay is often limiting in human studies (Liu and Wang, 2015; Singh and Farmer, 2006; Tretyakova et al., 2012).
DNA adducts have been measured in human tissues, but also fluids such as blood (leukocytes), (Balbo et al., 2011; Ma et al., 2014; Sangaraju et al., 2013; Sangaraju et al., 2014) saliva (buccal cells) (Balbo et al., 2012; Bessette et al., 2010) and urine (exfoliated epithelial cells) (Yun et al., 2015). The collection of these biofluids is a relatively non-invasive procedure and more facile to obtain than biopsy samples. Human DNA adduct analysis by IT-MS are grouped by DNA sources and summarized in Table 1. DNA adducts can be analyzed as the nucleobases, following acid/neutral hydrolysis of the DNA. These hydrolysis methods are most suitable for adducts with labile glycosidic linkages such as alkylated N7-Gua or N3-Ade adducts, which undergo facile depurination (Boysen et al., 2009). However, most adducts are generally analyzed at the deoxynucleoside level after digestion of DNA with an enzyme cocktail containing several nucleases such as DNase, nuclease P1, phosphodiesterase or alkaline phosphatase. The advantage of analyzing adducts at the deoxynucleoside level is that the near universal loss of the deoxyribose (dR) moiety ([M+H]+ > [M+H-116]+) upon MS2 fragmentation can be monitored for targeted adduct analysis or non-targeted screening in DNA adductomics (Balbo et al., 2014b; Kanaly et al., 2007). By employing LIT, the aglycone ion [BH2]+ can undergo further fragmentation at the MS3 scan stage to confirm the adduct’s structure and by comparing the product ion spectrum to that of the stable isotope labeled internal standard.(Goodenough et al., 2007; Yun et al., 2012) Thus, multistage scanning can be used to characterize the structure of putative adducts, especially at quantities that are far too low for characterization by NMR (Bessette et al., 2009).
Table 1.
Human DNA adduct measured by IT-MS in the literature.
| Adduct | DNA source | DNA Isolation Method | μg DNA assayed |
Sample cleanup | MS | LOD/LOQ | Adduct level | reference |
|---|---|---|---|---|---|---|---|---|
| BP-6-N7-Gua | Urine | IA | -- | HPLC fractionation |
QIT-MS2 | 5 to 10 fmol/mL urine |
20 – 50 fmol/mg urine creatinine |
(Bhattacharya et al., 2003) |
| Acro-dG | Hippocampus/parahippocam pal gyrus from brains |
N. R. | 10 μg | SPE | QIT-MS2 | 5 adducts per 108
nucleotides |
2.8 – 5.1 adducts per 106 nucleotides |
(Liu et al., 2005) |
| HNE-dG | Hippocampus/parahippocam pal gyrus and ingerior parietal lobule regions from brains |
N. R. | 10 μg | SPE | QIT-MS2 | 4 adducts per 108
nucleotides |
2.4 – 5.6 adducts per 107 nucleotides |
(Liu et al., 2006) |
| 4-OH-E1-1-N3-Ade | Breast tissues | LLE | -- | SPE, HPLC fractionation |
QIT-MS2 | 5-20 fmol/g tissue | 20-70 fmol/g tissue | (Zhang et al., 2008) |
| dG-4-ABP | 4-ABP treated human hepatocyte |
Phenol/chloroform extraction |
2-10 μg | SPE | LIT-CNL-MS3 | -- | -- | (Bessette et al., 2009) |
| dG-C8-4-ABP | Exfoliated urothelial cells | Qiagen™ Blood & Cell Culture Mini kit |
1 μg | Microfluidic chip |
QIT-MS2 | 5 adducts per 109
nucleotides |
N. D. | (Randall et al., 2010) |
| dG-C8 adducts of 4- ABP, AαC, IQ, MeIQx and PhIP |
Saliva buccal cells | Phenol/chloroform extraction |
9-51 μg | SPE | LIT-MS3 | 3 adducts per 109
nucleotides |
1-9 adducts per 108
nucleotides |
(Bessette et al., 2010) |
| N7-Et-Gua | Leukocytes | Qiagen™ buffy coat kit | 180 μg | SPE | LIT-HRMS2 | 8 fmol/μmol guanine |
10 – 180 fmol/umol guanine |
(Balbo et al., 2011) |
| dG-C8 adducts of 4- ABP, AαC, IQ, MeIQx and PhIP |
Carcinogen treated human hepatocyte |
Phenol/chloroform extraction |
2-10 μg | Online trapping | LIT-MS3 | 3 adducts per 109
nucleotides |
3.4 – 140/107
nucleotides |
(Nauwelaers et al., 2011) |
| dG-C8-PhIP and 4- ABP |
tumor-adjacent normal mammary tissues |
Phenol/chloroform extraction |
2-10 ug | Online trapping | LIT-MS3 | 3 adducts per 109
nucleotides |
3 adducts per 109
nucleotides |
(Gu et al., 2012) |
| dA-AL-I | Normal renal cortex and upper urinary tract tumor tissues |
Phenol/chloroform extraction |
10-20 μg | Online trapping | LIT-MS3 | 3 adducts per 109
nucleotides |
9-338 adducts per 108
nucleotides |
(Yun et al., 2013; Yun et al., 2012) |
| 8-oxo-dG | Urine | -- | -- | RP and SCX column coupling |
QIT-MS2 | 0.5 ng/mL urine | 1.16 ± 0.46 nmol/mmol creatine |
(Rota et al., 2013) |
| N7-THBG | Leukocytes | Qiagen™ buffy coat kit | 35-150 μg | SPE | LIT-HRMS2 | 2 adducts per 109
nucleotides |
0.6-19 adducts per 109
nucleotides |
(Sangaraju et al., 2013) |
| EB-GII | Leukocytes | Qiagen™ buffy coat kit | 3-76 μg | HPLC fractionation |
LIT-HRMS3 | 0.4 adducts per 109
nucleotides |
< LOQ | (Sangaraju et al., 2014) |
| M1dG | Leukocytes | Qiagen™ buffy coat kit | 39-244 μg | SPE, HPLC fractionation |
LIT-HRMS2 | 0.125 fmol/mg DNA | 0.004 – 9.15 adducts per 108 nucleotides |
(Ma et al., 2014) |
| dA-AL-I | Exfoliated urinary cells | Qiagen™ kit | 11 – 33 μg | Online trapping | LIT-MS3 | 3 adducts per 109
nucleotides |
3 to 581 adducts per 108
nucleotides |
(Yun et al., 2015) |
N. R. Not reported.
LLE. Liquid-liquid extraction.
N. D. Not detected.
RP. Reversed-phase.
SCX. Strong cation exchange.
DNA Adducts from Different Human Organs
Acrolein, a highly reactive α,β-unsaturated aldehyde, is an ubiquitous environmental contaminant (Izard and Libermann, 1978) and lipid peroxidation product produced endogenously (Uchida, 1999). Acrolein reacts with deoxyguanosine (dG) to form the cyclic 1,N2-propanodeoxyguanosine adducts (Acro-dG), 3H-8-hydroxy-3-(β-D-2′deoxyribofuranosyl)-5,6,7,8-tetrahydropyrido[3,2-α]purin-9-one (8-OH-PdG) and 3H-6-hydroxy-3-(β-D-2′deoxyribofuranosyl)-5,6,7,8-tetrahydropyrido[3,2-α]purin-9-one (6-OH-PdG). These adducts are major exocyclic DNA adducts detected in animal tissues assayed by 32P-postlabeling (Nath and Chung, 1994; Nath et al., 1996; Nath et al., 1998). Liu et. al. developed an HPLC-QIT-MS2 method to quantify 8-OH-PdG and 6-OH-PdG in human autopsy brain tissues using a high capacity QIT in 2005 (Liu et al., 2005). They reported a statistically significant 2-fold increase in the levels of Acro-dG in the hippocampus/parahippocampal gyrus of Alzheimer’s disease (AD) subjects compared to age-matched controls. Later on, Liu et. al.(Liu et al., 2006) employed a QIT to screen for DNA adducts formed with another major α,β-unsaturated aldehyde, trans-4-hydoxy-2-nonenal (HNE), which forms 4 isomeric cyclic 1,N2 adducts with dG (Kozekov et al., 2010). The HNE-dG adducts were identified in hippocampus/parahippocampal gyrus (HPG) and inferior parietal lobule (IPL) regions of postmortem brains by HPLC-QIT-MS2. However, there was no statistically significant difference between adduct levels in AD subjects and age-matched control of both regions of the brain.
Prolonged exposure to estrogens has been linked to the risk of developing breast cancer (Clemons and Goss, 2001). The hydroxylated estrogen metabolite, 3,4-catechol-estrogen quinone, can react with DNA, to form 4-hydroxyestrogen-1-N7-guanine (4-OH-E1-1-N7-Gua) and 4-hydroxyestrogen-1-N3-adenine (4-OH-E1-1-N3-Ade), which are labile and undergo depurination. Rogan and Cavalieri are pioneers in studying the role of unbalanced metabolism of estrogen in the etiology of hormonal associated cancers, such as breast cancer, ovarian cancer and thyroid cancer. They developed an HPLC-QqQ-MS2 method employing SRM mode to quantify metabolites and DNA adducts of estrogens in human urine (Gaikwad et al., 2009; Zahid et al., 2014; Zahid et al., 2013). Zhang et. al. identified and quantitated one of the depurinated estrogen-metabolite-modified DNA adducts, 4-OH-E1-1-N3-Ade, by nanoHPLC-QIT-MS2, in tumor-adjacent mammary tissue (Zhang et al., 2008). The adduct was detected in breast tissues from five out of six subjects, including one non-cancer patient. Genotoxic chemicals formed in well-done cooked meats and tobacco smoke also may contribute to breast cancer. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is a potential human carcinogen formed in well-done cooked meat, (Sugimura et al., 2004) and 4-aminobiphenyl (4-ABP) is a human carcinogen present in tobacco smoke and the environment (IARC Monograph, 1986; Luceri et al., 1993; Patrianakos and Hoffmann, 1979). Putative DNA adducts of PhIP and 4-ABP have been detected by IHC or 32P-postlabeling in mammary tissue or exfoliated epithelial cells in milk of lactating mothers (Faraglia et al., 2003; Gorlewska-Roberts et al., 2002; Zhu et al., 2003). However, Gu et. al. employed a very selective and sensitive HPLC-LIT-MS3 method (Goodenough et al., 2007) to measure the dG-C8 adducts of PhIP and 4-ABP in DNA of tumor-adjacent normal mammary tissues from newly diagnosed breast cancer subjects.(Gu et al., 2012) They did not find dG-C8-4-ABP adduct, and dG-C8-PhIP was only detected in one out of 70 biopsy samples even though the level of sensitivity of the LIT-MS based method was 100-fold lower than the average level of PhIP adducts reported by IHC (Zhu et al., 2003). Thus, the IHC assay may have detected DNA adducts other than dG-C8-PhIP or dG-C8-4-ABP. Further studies are warranted to determine the potential of PhIP and 4-ABP to form DNA adducts in mammary tissue and contribute to breast cancer risk.
In 2012, Yun et. al. developed a highly sensitive and robust HPLC-LIT-MS3 method and measured DNA adducts of AAs from renal cortex of patients from Taiwan with upper urothelial cancer (Yun et al., 2012). The aristoloactam DNA adduct, dA-AL-I was quantified at levels ranging from 9 to 338 adducts per 108 nucleotides. This method built the foundation for a series of publications on the measurement of aristolactam DNA adducts in fresh frozen but also in formalin-fixed paraffin-embedded (FFPE) renal tissues, and exfoliated urothelial cells. Employing HPLC-LIT-MS3, dA-AL-I was quantified in renal cortex and upper urothelial tumor tissues from nearly 200 patients with upper urothelial cancer from Taiwan, the Balkan endemic regions, and Romania (Yun et al., 2013; Yun et al., 2012; Yun et al., 2015; Yun et al., 2014).
DNA Adducts in Human Biofluids
Benzo[a]pyrene (B[a]P) is a prototypical PAH and classified as a Group 1 human carcinogen by the IARC (IARC Monograph, 1983). Penning has extensively studied the role of aldo-keto reductases (AKRs) in the metabolic activation of PAHs(Penning, 2014). Using B[a]P as a prototypical PAH, an HPLC-LIT-MS3 method was developed to identify stable DNA adducts of benzo[a]pyrene-7,8-dione, the bioactivated form of B[a]P catalyzed by AKRs, in human lung cell lines (Huang et al., 2013). Alternatively, B[a]P can be bioactivated to the radical cation and form DNA adducts at sites such as C8-, N7-Gua, and N3- and N7-Ade (Cavalieri and Rogan, 1998). In 2003, Bhattacharya et. al. measured DNA base adducts of B[a]P, 7-(benzo[a]pyren-6-yl)-adenine (BP-6-N7-Ade) and 7-(benzo[a]pyren-6-yl)-guanine (BP-6-N7-Gua) in urine (Bhattacharya et al., 2003). Urine samples were processed by an immunoaffinity (IA) column with high specificity towards BP-6-N7-Gua and BP-6-N7-Ade. The adducts were isolated by HPLC with fluorescence detection and the fractions containing adducts were analyzed by HPLC-QIT-MS. BP-6-N7-Gua was detected from coal smoke-exposed women urine while the unexposed controls were negative. Such a method may be employed to screen individuals at high risk of PAH exposure from cigarette or other forms of smoke.
The exposure to oxidizing agents, such as tobacco smoke, as well as normal cellular processes, can lead to the formation of reactive oxygen species (ROS). Due to its low redox potential, guanine is a susceptible target for ROS-induced DNA damage,(Ravanat et al., 2012) and oxidative DNA damage can be studied by monitoring the levels of 8-oxo-7,8-dihydroguanine (8-oxo-Gua) or 8-oxo-dG in tissues. However, an overestimation of levels of 8-oxo-dG can occur due to artefactual oxidation of dG during the isolation of DNA, or the subsequent digestion of DNA, or derivatization procedures employed for GC/MS.(Ravanat et al., 1995) The Standard Committee on Oxidative DNA Damage has published recommended protocols to minimize the artefactual oxidation of 8-oxo-dG (Collins et al., 2004; Gedik et al., 2005). 8-Oxo-dG in urine, on the other hand, has been reported to be stable and free of artefactual DNA oxidation due to the minimum sample manipulation (Lin et al., 2004; Matsumoto et al., 2008). Rota et. al. employed serial coupling LC columns and measured 8-oxo-dG in human urine samples by HPLC-QIT-MS2 (Rota et al., 2013). However, after normalizing the concentration of 8-oxo-dG in urine with the creatinine content, there was no statistically significant difference in 8-oxo-dG levels among the healthy male and female volunteers.
Exfoliated urothelial cells in urine are a feasible source of DNA for biomonitoring DNA-damaging agents of the urinary system. Randall et. al. developed a highly sensitive method by operating at a nano-flow rate and online enrichment with a microchip to quantify dG-C8-4-ABP adduct from only 1 μg of DNA employing an ion trap MS from Agilent.(Randall et al., 2010) Spiking studies showed that the limit of quantification was 5 adducts per 109 DNA bases. The authors did not detect dG-C8-4-ABP in urine of non-smokers; urine of smokers were not screened for adducts. Yun et al. (Yun et al., 2015) assayed for dA-Al-I adduct in exfoliated urinary cells from patients who had ingested traditional Chinese herbal medicines containing Aristolochia sp. Four out of five subjects contained dA-Al-I in urinary cells at a level above LOQ (3 adducts per 109 bases), when assayed by QIT/MS. The adducts were still detected in exfoliated cells up to 3 months after cessation of drug usage. The one subject without detectable dA-Al-I had no recorded history of Aristolochia sp. usage.
The oral and nasal cavities are the first portals encountering carcinogens in the diet and tobacco smoke. In 2010, Bessette et. al. characterized and quantified the dG-C8 adducts of tobacco smoke and cooked meat carcinogens PhIP, 2-amino-9H-pyrido[2,3-b]indole (AαC), 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) and 4-ABP in salivary DNA by HPLC-LIT-MS3.(Bessette et al., 2010) Adducts levels ranged from 1 to 9 per 108 nucleotides with dG-C8-PhIP being found most frequently and present in 15 out of 37 subjects, whereas dG-C8-AαC and dG-C8-MeIQx were detected in 3 current smokers and dG-C8-4-ABP, from 2 current smokers. The successful measurement of DNA adducts suggests that human saliva may be a promising noninvasive source for biomonitoring the exposure of dietary and tobacco specific carcinogens.
Leukocytes are another accessible source of DNA for biomonitoring DNA adducts. Balbo et. al. quantified N7-ethyl-guanine (N7-Et-Gua), an adduct derived from ethylating agents of unknown structures, from leukocytes of smokers and non-smokers.(Balbo et al., 2011) A nanoHPLC-LIT equipped with high resolution (HR) Orbitrap MS was established with an LOQ of 8 fmol/μmol Gua (3 adducts per 108 nucleotides) with 180 μg DNA used for assay. N7-Et-Gua was detected in the majority of subjects in the smoker and non-smoker groups; however, no statistically significant difference was observed between the adduct levels from the two groups. Sangaraju et. al. employed nanoHPLC-LIT-HRMSn for the quantitation of DNA adducts formed with 1,3-butadiene (BD) (Sangaraju et al., 2013; Sangaraju et al., 2014). BD is a known human carcinogen present in automobile exhaust and cigarette smoke.(Lewtas, 1993) The epoxide metabolites of BD can alkylate DNA to form adducts such as N7-(2,3,4-trihydroxybut-1-yl) guanine (N7-THBG) and N7-(1-hydroxy-3-buten-2-yl) guanine (EB-GII). These adducts may contribute to the carcinogenicity of BD in animal models (Goggin et al., 2011). Both adducts are released from DNA backbone by thermal hydrolysis and serve as biomarkers for monitoring BD exposure. Utilizing the HR feature of LIT-Orbitrap, N7-THBG was quantified in DNA from human leukocyte at MS2 scan stage with an LOQ of 2 adducts per 109 nucleotides.(Sangaraju et al., 2013) However, their attempts to quantify EB-GII, by QqQ-MS in SRM mode and nanoHPLC-LIT-HRMS2, were unsuccessful due to co-eluting isobaric interferences. The authors showed that only HRMS3 could provide sufficient selectivity and sensitivity, but the amount of EB-GII in human blood samples was below the LOQ (0.4 adducts per 109 nucleotides). Ma et. al. employed nanoHPLC-LIT-HRMS2 method to measure the cyclic dG adduct of malondialdehyde (MDA) in human leukocytes.(Ma et al., 2014) MDA is a principal product of lipid peroxidation and an endogenous genotoxicant (Marnett, 1999). DNA adducts of MDA may contribute to the inflammation-mediated diseases such as cancer and cardiovascular diseases. The predominant DNA adduct of MDA is 3-(2-deoxy-β-D-erythro-pentafuranosyl)pyrimido-[1,2-α]purin-10(3H)-one deoxyguanosine (M1dG), a mutagenic lesion that induces T > A mutations in E. coli (Fink et al., 1997). The method to measure M1dG reached an ultra-low LOQ value of 0.125 fmol/mg DNA (4 adducts per 1011 nucleotides), when 200 μg of leukocyte DNA was assayed.
DNA extracted from Human cells treated with carcinogens
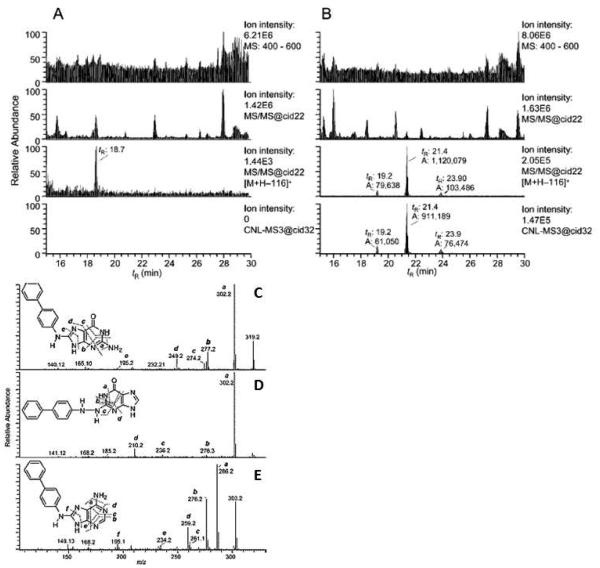
Freshly isolated human hepatocytes maintained in primary culture are excellent sources to study the metabolism and bioactivation of carcinogens that occurs in humans because the xenobiotic metabolism enzymes and cofactors are present at physiological concentrations. Human hepatocytes have been successfully employed to study the metabolism of AFB1 and HAAs.(Langouet et al., 1995; Langouet et al., 2002; Langouet et al., 2001) In 2009, Bessette et. al. utilized HPLC-LIT-MS3 to screen for multiple DNA adducts formed in primary human hepatocyte cultures by an untargeted Data-Dependent Constant Neutral Loss (DD-CNL) method, where the loss of dR (m/z 116) triggered the MS3 fragmentation of the aglycone adducts (Bessette et al., 2009). Four adducts were detected, by the DD-CNL-MS3 method, in DNA of hepatocytes treated with 4-ABP. The major adduct was dG-C8-4-ABP, followed by dA-C8-4-ABP, and the proposed adducts dG-N2,N4-4-ABP, dG-N2-4-ABP (Figure 7). The product ion spectra at the MS3 scan stage supported the proposed structures of the 4-ABP-DNA adducts. This was the first report of utilizing DD-CNL with the LIT as a tool to screen for multiple DNA adducts. Nauwelaers et. al. incubated 4-ABP, AαC, 2-amino-3-methylimidazo[4,5-f]quinoline (IQ), MeIQx or PhIP in freshly cultured rodent hepatocytes or human hepatocytes from eight different donors and quantified the major dG-C8 adducts of these carcinogens by HPLC-LIT-MS3.(Nauwelaers et al., 2011) Interestingly, their findings showed that human hepatocytes were more efficient than the rodent hepatocytes at forming DNA adducts of these chemicals, suggesting that rat model commonly used in carcinogenesis bioassays may underestimate the potential genotoxicity of HAAs in humans.
Figure 7.
CNL-MS3 data-dependent scanning of 4-ABP adducts in human hepatocytes, either (A) untreated or (B) treated with 4-ABP (10 μM). The chromatograms from top to bottom represents: the full scan mode from m/z 400-600; total ion chromatograms of all MS2 scan events; peaks filtered by the CNL of m/z 116, and peaks detected by data-dependent CNL-MS3 scan mode. Spectra in the bottom panel shows the CNL-MS3 products of (C) dG-C8-4-ABP, (D) proposed dG-N2,N4-4-ABP and (E) dA-C8-4-ABP adducts. Reprinted from Bessette (2009) with permission from American Chemical Society.
HOW TO ACHIEVE SUCCESSFUL MEASUREMENT
Sample Preparation
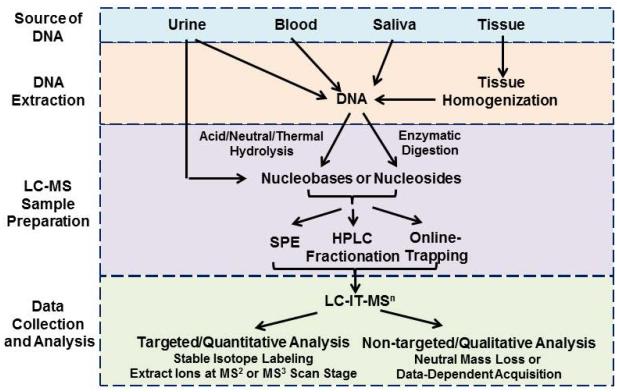
The general steps in DNA sample preparation and adduct analysis by LC-IT-MSn are depicted in Figure 8. The success in analyzing DNA adducts at trace levels roots from consistent daily good laboratory practice. The quality of sample preparation, beginning from the collection of tissues, the isolation DNA, followed by the enzymatic or thermal hydrolysis of DNA, and the enrichment of DNA adducts prior to LC-MS analysis, determines the success of the assay. Contamination can easily occur but is often difficult to trace and eliminate. Designated working area and dedicated instruments such as a dedicated auto-sampler and LC-MS system, speed vacuum concentrators for solvent evaporation, and dedicated pipettes using only tips with filters are required for trace-analysis manipulations and adduct measurements.
Figure 8.
Sample preparation steps for DNA adduct analysis by LC-IT-MS.
One must be aware of the cell types/tissue regions to be collected in order to avoid potential dilution of the DNA adducts from cells irrelevant to the assay. For example, cells exfoliated from the epithelial lining of the vagina of females can dilute the levels of DNA adducts in exfoliated urinary cells when studying DNA adduct formation of urothelial carcinogens,(Talaska et al., 1990) or muscle cells in bladder tissues (detrusor muscle), when adducts are formed in the urothelium layer (transitional epithelium), where cancers are thought to originate.
The retrieval of DNA with high purity from tissues is another crucial step in the successful measurement of DNA adducts. The most commonly used protocols include phenol-chloroform extraction or solid-phase extraction (SPE) of DNA employing silica-based resins. The phenol-chloroform extraction method removes denatured proteins, lipids and other cell components, which partition into the organic phase. The DNA in the aqueous phase is then collected by precipitation with ethanol in the presence of high salt.(Gupta, 1993) Freshly distilled phenol saturated with TE (10 mM Tris buffer, pH 8, and 1 mM EDTA) buffered between pH 7.0 - 8.0 is used to isolate DNA. The pH affects the partitioning of DNA between the organic and aqueous phases. At an acidic pH 5.0, the elevated H+ ions will neutralize the negative phosphate charges on DNA backbone, and the macromolecule will partition into the organic phase. The phenol must be of high quality as outdated, oxidized phenol can cause ring-opening at the imidazole moiety of arylamine and heteroarylamine substituted dG-C8 adducts (Shibutani et al., 1990; Tang et al., 2013).
Solid-phase DNA extraction (silica-based) methods are commonly used in commercial kits, which have higher throughput than the solution-phase extraction with phenol-chloroform. The silica-based technology binds DNA in the presence of high salt and chaotropic solutions. The chaotrope disrupts the shell of hydration around the biomolecules and silica, which allows cations from the solution to form a salt bridge between the negatively charged DNA backbone and the negatively charged silica. The DNA is then washed with high salt and ethanol, followed by elution of DNA with a low salt solution that disrupts the salt bridge. In the magnetic beads technology, the magnetic or paramagnetic core is encapsulated by a layer of silica, which will bind the DNA, followed by purification employing the same solvent conditions described above. Bench top automated instruments can reduce the time of sample preparation, and further increase the throughput of the method. Although time consuming, our experiences show that phenol/chloroform extraction usually surpasses solid-phase methods with respect to DNA yield and purity. In addition, issues such as lot-to-lot variation of silica-based kits, insufficient removal of the magnetic beads, and even the shelf-life and handling of kit solutions can affect the yield and purity of the DNA. It is critical to evaluate the stability and recovery of the DNA adducts by comparing the different methods of DNA extraction.
The enzymatic digestion of DNA is done with a cocktail of enzymes. Different laboratories employ different enzymes and conditions of digestion. Our laboratory routinely employs DNase I (Type IV, bovine pancreas), alkaline phosphatase (Escherichia coli), nuclease P1 (from Penicillium citrinum), and Phosphodiesterase I (from Crotalus adamanteus venom). This mixture of enzymes efficiently digests a variety of different classes of DNA adducts, including those formed with aromatic amines, heterocyclic aromatic amines, aristolochic acid, polycyclic aromatic hydrocarbons, tobacco specific N-nitroso compounds, and lipid peroxidation adducts (Bessette et al., 2010; Goodenough et al., 2007; Guo et al., 2016; Kozekov et al., 2010; Yun et al., 2012). The analysis of the DNA digest by HPLC with UV provides a good estimate of DNA purity and the efficacy of the nucleases to fully digest the non-modified DNA bases. In our laboratory, a portion of DNA digest, equivalent to 0.5 to 1 μg DNA, is analyzed by HPLC-UV by comparing the peak areas of the four dNs at A260 to that of dNs standards of equal amount assayed. Our HPLC conditions employ a C18 reversed phase column, 20 mM ammonium acetate, pH 4.5 as LC solvent A and acetonitrile as solvent B (Gu et al., 2012). A gradient is used to resolve the dNs. The detection of guanosine and uridine, which are baseline resolved from the dNs, signifies RNA contamination. Normally, the percent contamination of RNA is on the order of one percent or less. The efficacy of nuclease digestion of non-modified DNA should be quantitative and small molecular weight oligonucleotides should not be seen in the chromatogram. Ideally, oligonucleotides containing defined amounts of adducted carcinogens (Dong et al., 2006) or DNA modified with known levels of carcinogens that have been characterized by other means (Beland et al., 1999; Brown et al., 2001; Divi et al., 2002) should be employed as positive controls to confirm the recovery of bulky DNA adducts, which may inhibit the efficacy of nuclease digestion.
The level of DNA adducts can be reported by different means. For example, the adduct level can be expressed as pmol of adduct per μmol Gua; pmol adduct per mg DNA; or mole ratio of adduct per nucleotides. The first unit is often reported when adducts are measured as free base, and Gua is quantified by HPLC-UV (Hecht et al., 1986). The other two units are commonly reported in studies where adducts are analyzed as dNs, following enzymatic digestion. In these cases, DNA can be estimated before digestion by UV spectroscopy, using a value of 1 absorbance unit at A260 as an equivalent to 50 μg/mL double-stranded DNA (Maniatis et al., 1982). The ratio of A260/A280 is used to assess the purity of DNA (Glasel, 1995). A ratio value of 1.8 is generally accepted as “pure” DNA. Significant protein contamination is suggested when the A260/A280 ratio is below 1.6, and a ratio above 2.0 indicates the presence of RNA. However, these ratio values are only an estimation of the DNA purity, and conditions such as pH and ionic strength of the solution, the aromatic amino acid content of residual proteins, and the performance of the spectrophotometer can affect these ratio values.(Wilfinger et al., 1997) Alternatively, the dG content can be measured by HPLC, following enzymatic digestion. Under the assumption that 1 mg DNA contains 3 μmol nucleotides, and dG accounts for 22% of the total nucleosides, the estimates of DNA adducts can be determined (Lao et al., 2007).
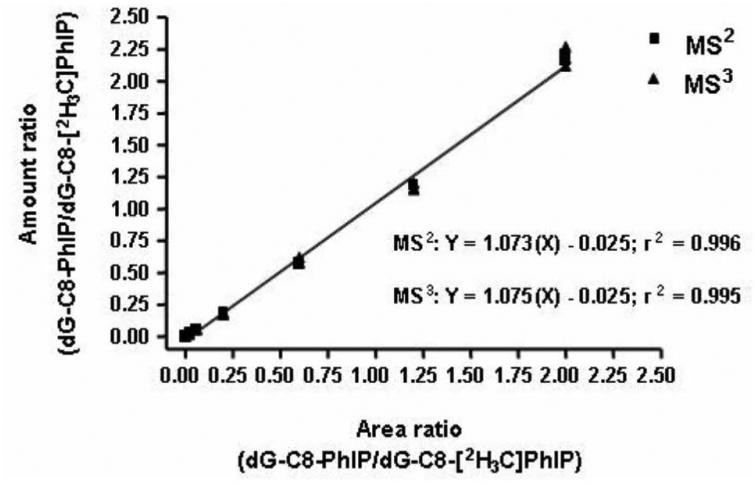
DNA adducts are quantified by the employment of calibration curves. The calibration curves are constructed by spiking variable amount of synthetic DNA adduct standards and constant levels of their stable isotope labeled internal standards into the DNA digest. The level of internal standard is usually chosen in the middle of the range of unlabeled adducts and a linearity of at least 50-fold can be achieved. A representative calibration curve of dG-C8-PhIP is shown in Figure 9. The calibration curves display excellent linearity (r2 > 0.99) when assayed at either the MS2 or MS3 scan stages with the HPLC-IT-MSn (Goodenough et al., 2007).
Figure 9.
Calibration curves of dG-C8-PhIP constructed at the MS2 and MS3 scan stages. Reprinted from Goodenough (2007) with permission from American Chemical Society.
After acid/neutral thermal hydrolysis or enzymatic digestion, the DNA adducts must be enriched from the non-modified nucleobases or dNs, which are present at one million-fold or greater levels than the targeted adduct. The enzymatic digestion conditions introduce salt, protein and potential contaminants into the sample. As a result, MS signal can be suppressed or components in the digest may introduce isobaric interferences that impact the reliability and sensitivity of the assay.(Klaene et al., 2015) Such chemical complexity has been noticed by several groups and possible solutions include optimizing the nuclease:DNA ratio, removal of the nucleases, or employing “cleaner” recombinant enzymes rather than enzymes obtained from animal or bacterial sources (Balbo et al., 2014a; Klaene et al., 2015). DNA adducts can be enriched by SPE, off-line HPLC fractionation, or on-line trapping prior to LC/MS. Isobaric interferences also can be introduced from impurities in the solvents and the SPE resin, or by chemicals co-extracted from the frits of the SPE cartridge, which can result in ion suppression (Goodenough et al., 2007). The employment of SPE cartridges without frits can circumvent some of these interferences. LC-MS grade solvents are recommended in all steps involved in the sample preparation. The aforementioned techniques have been extensively reviewed (Balbo et al., 2014b; Liu and Wang, 2015; Tretyakova et al., 2012). Our lab utilizes an online C18 trap column to enrich hydrophobic adducts prior to LC-MS analysis. This method dramatically reduces sample preparation time. The DNA digest is concentrated to dryness by vacuum centrifugation, resuspended in appropriate solvent, and injected directly on to the trap column (Yun et al., 2012).
HPLC-IT-MSn Analysis
Each DNA adduct possesses its own analytical challenge. There is no universal method for assaying all types of DNA adducts. A variety of HPLC stationary phases with different chemical selectivities are available and should be evaluated for optimal performance. The choice of LC solvent, the type and concentration of buffer (ion-pair agent) are all critical aspects for good chromatography, but also for improving MS sensitivity (Tretyakova et al., 2012; Yin et al., 2013). Our laboratory generally employs Magic C18 AQ (Michrom) resins for chromatography, and dilute formic acid (0.01 – 0.05% v/v) and acetonitrile as the organic solvent for adduct measurements.
Since the ionization source of online LC-MS, ESI, is a concentration-dependent detector device, (Davis et al., 1995; Hopfgartner et al., 1993) the sensitivity of the analysis can be improved by decreasing the flow rates from conventional HPLC (0.1 – 1 mL/min) to capillary- (1 – 10 μL/min) or nanoLC (0.1 – 0.3 μL/min) systems. Nanosources are commercially available from a number of vendors, and nanoLC flow should be considered for ultra-trace DNA adduct measurements. The decreased flow rate and the diameter of the emitter tip can greatly improve the MS sensitivity by improving the ionization and analyte transmission efficiency, and also decrease the degree of ion suppression (El-Faramawy et al., 2005; Shen et al., 2002). However, the spraying conditions, the inner diameter size of the emitter and its position in the source, the spray voltage, and solvent composition can greatly affect the sensitivity of nanoESI and must be carefully optimized (Schmidt et al., 2003).
IT-MS instruments are commercially available from several vendors. Each instrument has its own unique strengths and advantages. As was discussed in the Section on Fundamental of the Ion Trap, the most critical feature that affects the instrument performance in an IT is the control of the number of ions inside the trap at any point in time. If the ion count is too high, “space charging” will occur and cause a large decrease in mass accuracy and resolution. However, a low total ion count will result in poor ion statistics, resulting in poor quality mass spectra. Our laboratory is equipped with the Velos Pro LIT-MS from Thermo Fisher Scientific Inc. Several critical instrument parameters of the Thermo Fisher LIT-MS are discussed here. The control of the ion population in the trap is achieved by setting Automatic Gain Control (AGC) and Injection Time (Scigelova and Makarov, 2013). In this instrument, the gate lens will open for a short period of time to measure the ion influx, which determines the Injection Time required for the lens to open to fill the trap with target number of ions specified by AGC. Thus, the influx of ions will reach the AGC with an actual injection time smaller than the specified value, or arrive at the maximum Injection Time with fewer ions collected than the designated AGC value. Increasing the Injection Time improves the quality of spectra because the trap will be filled with more ions up to the predetermined AGC value, but the number of scans acquired across the peak will decrease. The injection time and AGC must be optimized to assure that a sufficient number of scans are acquired across the chromatographic peak. This is particularly important for narrow eluting peaks. We strive to acquire a minimum of ten scans across the peaks for quantitative measurements of DNA adducts and other analytes (Guo et al., 2015).
Other parameters, such as ion optics voltages, Isolation Width, Activation Q, Activation Time and Collision Energy also affect the sensitivity of the MS and the quality of the mass spectra. The influence of these parameters will vary for different adducts. The signal response of the adducts should be optimized by directly infusing synthetic DNA adduct standards into the MS and optimizing ion optics voltages and ensure high transmission efficiency of ion beams entering the trap. The Isolation Width is the baseline width of a window for selecting a mass peak (m/z). The width should be sufficiently narrow to maintain high precursor selectivity but too low a value results in loss of sensitivity because not all of the ions in a mass peak are isolated. In addition, McClellan and coworkers noticed a drop off of MS sensitivity and mass resolution when applying the same Isolation Width to some “fragile” compounds (McClellan et al., 2002). Their observations are confirmed by our experiences and show that such fragile molecules, including some DNA adducts, dissociate at lower collision energy and are prone to fragmentation during the applied isolation waveform especially at slower scan speed, and therefore, the time period of ion isolation must be reduced. Thus “fragile” DNA adducts require wider Isolation Width (m/z 3 – 5) than other adducts (m/z 1 - 2) (Bessette et al., 2010; Goodenough et al., 2007). The Activation Q is the RF frequency used to trap fragment ions. A lower activation Q results in lower energy deposition and allows detection of the lower m/z fragmentation ions. A higher activation Q results in greater energy deposition, however lower m/z fragmentation ions are not stable in the ion trap and are not scanned out. Decreasing the activation q value of CID in the ion trap will lower mass cutoff for MS/MS, thereby allowing the scanning of low m/z product ions. However, this approach also places the precursor ions into a shallower trapping potential, possibly leading to decreased ion stability and undesired ion losses during ion activation. We observed a need to increase the Activation Q from the default value of 0.25 to 0.35 for the fragmentation of PhIP in LIT-MS; the default value of 0.25 yielded a 10-fold lower ion intensity (Guo et al., 2015). In contrast, no difference signal was observed when employing Activation Q of 0.25 or 0.35 to dissociate the dG adducts of PhIP or other HAAs (Bessette et al., 2010; Goodenough et al., 2007). The Activation Time together with the Collision Energy can be chosen by the user to perform either a short, hard fragmentation (shorter Activation Time with larger Collision Energy), or a long, soft fragmentation (longer Activation Time with smaller Collision Energy).
CONCLUSIONS AND FUTURE DIRECTIONS
IT-MS is a powerful technique for measuring DNA adducts with high sensitivity and selectivity, and multi-stage scanning provides high quality spectral data to corroborate DNA adduct structure and improves the selectivity for quantitative measurements of DNA adducts. The paucity of freshly frozen tissues and biofluids, which are often not available for large population studies have impeded the use of DNA adducts as biomarkers in large scale molecular epidemiology studies. However, formalin-fixed paraffin-embedded (FFPE) tissues with clinical diagnosis of disease are routinely archived and are a valuable resource for DNA adduct biomarker research. Our laboratory has established methods to retrieve DNA adducts of AA from FFPE tissues,(Yun et al., 2015) and we are evaluating the potential of employing archived DNA for the measurement of multiple classes of DNA adducts (Guo et al., 2016). With the motivation to screen for multiple DNA adducts, the MSn scanning of the IT-MS can be employed in the data dependent-scan mode to screen for putative adducts based on the loss of dR (m/z 116) (Bessette et al., 2009). This application is being extended to HRMS instruments, such as the Orbitrap, with accurate mass measurement, which can improve the selectivity of screening by a precise mass difference of 116.0475 (Balbo et al., 2014a). Further improvements in software tools enabling the bioanalyst to make full use of the HR mass spectral data generated by the Orbitrap analyzers are required for adductomics. Comprehensive, publicly available data bases of libraries of fragmentation spectra must be curated to facilitate the identification of DNA adducts as no one single lab will have reference standards for all known DNA adducts. With the maturation of the methodology and instrumentation, we expect to see a fast growth in the number of publications reporting the measurement of DNA adducts by IT-MS. These data will advance our understanding of chemical exposures, DNA adducts, and risk factors for cancer.
ACKNOWLEDGEMENT
Data reported from the authors’ laboratory were supported by NIH grants R21ES014438, R01CA122320, R33CA186795, R01ES019564, and NCI Center Support Grant No. CA077598 (all to RJT), which supports the Mass Spectrometry core facility, a part of the Analytical Biochemistry shared resource at Masonic Cancer Center, University of Minnesota.
LITERATURE CITED
- Aguilar F, Harris CC, Sun T, Hollstein M, Cerutti P. Geographic variation of p53 mutational profile in nonmalignant human liver. Science. 1994;264:1317–1319. doi: 10.1126/science.8191284. [DOI] [PubMed] [Google Scholar]
- Balbo S, Hecht SS, Upadhyaya P, Villalta PW. Application of a high-resolution mass-spectrometry-based DNA adductomics approach for identification of DNA adducts in complex mixtures. Anal Chem. 2014a;86:1744–1752. doi: 10.1021/ac403565m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo S, Meng L, Bliss RL, Jensen JA, Hatsukami DK, Hecht SS. Kinetics of DNA adduct formation in the oral cavity after drinking alcohol. Cancer Epidemiol Biomarkers Prev. 2012;21:601–608. doi: 10.1158/1055-9965.EPI-11-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo S, Turesky RJ, Villalta PW. DNA Adductomics. Chem Res Toxicol. 2014b;27:356–366. doi: 10.1021/tx4004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo S, Villalta PW, Hecht SS. Quantitation of 7-ethylguanine in leukocyte DNA from smokers and nonsmokers by liquid chromatography-nanoelectrospray-high resolution tandem mass spectrometry. Chem Res Toxicol. 2011;24:1729–1734. doi: 10.1021/tx200262d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beland FA, Beranek DT, Dooley KL, Heflich RH, Kadlubar FF. Arylamine-DNA adducts in vitro and in vivo: their role in bacterial mutagenesis and urinary bladder carcinogenesis. Environ. Health Perspect. 1983;49:125–134. doi: 10.1289/ehp.8349125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beland FA, Doerge DR, Churchwell MI, Poirier MC, Schoket B, Marques MM. Synthesis, characterization, and quantitation of a 4-aminobiphenyl-DNA adduct standard. Chem Res Toxicol. 1999;12:68–77. doi: 10.1021/tx980172y. [DOI] [PubMed] [Google Scholar]
- Beland FA, Kadlubar FF. Formation and persistence of arylamine DNA adducts in vivo. Environ Health Perspect. 1985;62:19–30. doi: 10.1289/ehp.856219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, Turesky RJ. Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal Chem. 2009;81:809–819. doi: 10.1021/ac802096p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, Turesky RJ. Identification of carcinogen DNA adducts in human saliva by linear quadrupole ion trap/multistage tandem mass spectrometry. Chem Res Toxicol. 2010;23:1234–1244. doi: 10.1021/tx100098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Barbacci DC, Shen M, Liu JN, Casale GP. Extraction and purification of depurinated benzo[a]pyrene-adducted DNA bases from human urine by immunoaffinity chromatography coupled with HPLC and analysis by LC/quadrupole ion-trap MS. Chem Res Toxicol. 2003;16:479–486. doi: 10.1021/tx020060d. [DOI] [PubMed] [Google Scholar]
- Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. The formation and biological significance of N7-guanine adducts. Mutat Res. 2009;678:76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. Methods for the detection of DNA adducts. Methods Mol Biol. 2012;817:207–230. doi: 10.1007/978-1-61779-421-6_11. [DOI] [PubMed] [Google Scholar]
- Brown K, Guenther EA, Dingley KH, Cosman M, Harvey CA, Shields SJ, Turteltaub KW. Synthesis and spectroscopic characterization of site-specific 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine oligodeoxyribonucleotide adducts. Nucleic Acids Research. 2001;29:1951–1959. doi: 10.1093/nar/29.9.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri EL, Rogan EG. Role of aromatic hydrocarbons in disclosing how catecholestrogens initiate cancer. Adv Pharmacol. 1998;42:837–840. doi: 10.1016/s1054-3589(08)60876-5. [DOI] [PubMed] [Google Scholar]
- Chen CH, Dickman KG, Moriya M, Zavadil J, Sidorenko VS, Edwards KL, Gnatenko DV, Wu L, Turesky RJ, Wu XR, Pu YS, Grollman AP. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci U S A. 2012;109:8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Wang LY, Lunn RM, Tsai WY, Lee PH, Lee CS, Ahsan H, Zhang YJ, Chen CJ, Santella RM. Polycyclic aromatic hydrocarbon-DNA adducts in liver tissues of hepatocellular carcinoma patients and controls. Int J Cancer. 2002;99:14–21. doi: 10.1002/ijc.10291. [DOI] [PubMed] [Google Scholar]
- Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001;344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- Collins AR, Cadet J, Moller L, Poulsen HE, Vina J. Are we sure we know how to measure 8-oxo-7,8-dihydroguanine in DNA from human cells? Arch Biochem Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Davis MT, Stahl DC, Hefta SA, Lee TD. A microscale electrospray interface for on-line, capillary liquid chromatography/tandem mass spectrometry of complex peptide mixtures. Anal Chem. 1995;67:4549–4556. doi: 10.1021/ac00120a019. [DOI] [PubMed] [Google Scholar]
- De Broe ME. Chinese herbs nephropathy and Balkan endemic nephropathy: toward a single entity, aristolochic acid nephropathy. Kidney Int. 2012;81:513–515. doi: 10.1038/ki.2011.428. [DOI] [PubMed] [Google Scholar]
- de Hoffmann E, Stroobant V. Mass Spectrometry: Principles, and Applications. 3rd John Wiley $ Sons Ltd; Chichester, West Sussex, England: 2007. [Google Scholar]
- Divi RL, Beland FA, Fu PP, Von Tungeln LS, Schoket B, Camara JE, Ghei M, Rothman N, Sinha R, Poirier MC. Highly sensitive chemiluminescence immunoassay for benzo[a]pyrene-DNA adducts: validation by comparison with other methods, and use in human biomonitoring. Carcinogenesis. 2002;23:2043–2049. doi: 10.1093/carcin/23.12.2043. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M, Coskun E, Jaruga P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic Res. 2015;49:525–548. doi: 10.3109/10715762.2015.1014814. [DOI] [PubMed] [Google Scholar]
- Dong H, Suzuki N, Torres MC, Bonala RR, Johnson F, Grollman AP, Shibutani S. Quantitative determination of aristolochic acid-derived DNA adducts in rats using 32P-postlabeling/polyacrylamide gel electrophoresis analysis. Drug Metab Dispos. 2006;34:1122–1127. doi: 10.1124/dmd.105.008706. [DOI] [PubMed] [Google Scholar]
- Douglas DJ, Frank AJ, Mao D. Linear ion traps in mass spectrometry. Mass Spectrom Rev. 2005;24:1–29. doi: 10.1002/mas.20004. [DOI] [PubMed] [Google Scholar]
- El-Faramawy A, Siu KW, Thomson BA. Efficiency of nano-electrospray ionization. J Am Soc Mass Spectrom. 2005;16:1702–1707. doi: 10.1016/j.jasms.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Faraglia B, Chen SY, Gammon MD, Zhang Y, Teitelbaum SL, Neugut AI, Ahsan H, Garbowski GC, Hibshoosh H, Lin D, Kadlubar FF, Santella RM. Evaluation of 4-aminobiphenyl-DNA adducts in human breast cancer: the influence of tobacco smoke. Carcinogenesis. 2003;24:719–725. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- Fink SP, Reddy GR, Marnett LJ. Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc Natl Acad Sci U S A. 1997;94:8652–8657. doi: 10.1073/pnas.94.16.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaikwad NW, Yang L, Pruthi S, Ingle JN, Sandhu N, Rogan EG, Cavalieri EL. Urine biomarkers of risk in the molecular etiology of breast cancer. Breast Cancer (Auckl) 2009;3:1–8. doi: 10.4137/bcbcr.s2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon MD, Wolff MS, Neugut AI, Eng SM, Teitelbaum SL, Britton JA, Terry MB, Levin B, Stellman SD, Kabat GC, Hatch M, Senie R, Berkowitz G, Bradlow HL, Garbowski G, Maffeo C, Montalvan P, Kemeny M, Citron M, Schnabel F, Schuss A, Hajdu S, Vinceguerra V, Niguidula N, Ireland K, Santella RM. Environmental toxins and breast cancer on Long Island. II. Organochlorine compound levels in blood. Cancer Epidemiol Biomarkers Prev. 2002;11:686–697. [PubMed] [Google Scholar]
- Gedik CM, Collins A, Escodd Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- Glasel JA. Validity of nucleic acid purities monitored by 260nm/280nm absorbance ratios. Biotechniques. 1995;18:62–63. [PubMed] [Google Scholar]
- Goggin M, Sangaraju D, Walker VE, Wickliffe J, Swenberg JA, Tretyakova N. Persistence and repair of bifunctional DNA adducts in tissues of laboratory animals exposed to 1,3-butadiene by inhalation. Chem Res Toxicol. 2011;24:809–817. doi: 10.1021/tx200009b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MS n method for the characterization and quantification of 2'-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem Res Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlewska-Roberts K, Green B, Fares M, Ambrosone CB, Kadlubar FF. Carcinogen-DNA adducts in human breast epithelial cells. Environ Mol Mutagen. 2002;39:184–192. doi: 10.1002/em.10060. [DOI] [PubMed] [Google Scholar]
- Grollman AP. Aristolochic acid nephropathy: Harbinger of a global iatrogenic disease. Environ Mol Mutagen. 2013;54:1–7. doi: 10.1002/em.21756. [DOI] [PubMed] [Google Scholar]
- Gu D, Turesky RJ, Tao Y, Langouet SA, Nauwelaers GC, Yuan JM, Yee D, Yu MC. DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 4-aminobiphenyl are infrequently detected in human mammary tissue by liquid chromatography/tandem mass spectrometry. Carcinogenesis. 2012;33:124–130. doi: 10.1093/carcin/bgr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Yonemori K, Le Marchand L, Turesky RJ. Method to Biomonitor the Cooked Meat Carcinogen 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in Dyed Hair by Ultra-Performance Liquid Chromatography-Orbitrap High Resolution Multistage Mass Spectrometry. Anal Chem. 2015;87:5872–5877. doi: 10.1021/acs.analchem.5b01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Yun BH, Upadhyaya P, Yao L, Krishnamachari S, Rosenquist TA, Grollman AP, Turesky RJ. Multi-Class Carcinogenic DNA Adduct Quantification in Formalin-Fixed Paraffin-Embedded Tissues by Ultra-Performance Liquid Chromatography–Tandem Mass Spectrometry. Anal Chem. 2016 doi: 10.1021/acs.analchem.6b00124. DOI: 10.1021/acs.analchem.6b00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RC. 32P-postlabelling analysis of bulky aromatic adducts. IARC Sci Publ. 1993:11–23. [PubMed] [Google Scholar]
- Hecht SS, Trushin N, Castonguay A, Rivenson A. Comparative tumorigenicity and DNA methylation in F344 rats by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N-nitrosodimethylamine. Cancer Res. 1986;46:498–502. [PubMed] [Google Scholar]
- Hoang ML, Chen CH, Sidorenko VS, He J, Dickman KG, Yun BH, Moriya M, Niknafs N, Douville C, Karchin R, Turesky RJ, Pu YS, Vogelstein B, Papadopoulos N, Grollman AP, Kinzler KW, Rosenquist TA. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med. 2013;5:197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfgartner G, Bean K, Henion J, Henry R. Ion-Spray Mass-Spectrometric Detection for Liquid-Chromatography - a Concentration-Flow-Sensitive or a Mass-Flow-Sensitive Device. Journal of Chromatography. 1993;647:51–61. [Google Scholar]
- Hsu TM, Zhang YJ, Santella RM. Immunoperoxidase quantitation of 4-aminobiphenyl- and polycyclic aromatic hydrocarbon-DNA adducts in exfoliated oral and urothelial cells of smokers and nonsmokers. Cancer Epidemiol Biomarkers Prev. 1997;6:193–199. [PubMed] [Google Scholar]
- Huang M, Blair IA, Penning TM. Identification of stable benzo[a]pyrene-7,8-dione-DNA adducts in human lung cells. Chem Res Toxicol. 2013;26:685–692. doi: 10.1021/tx300476m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC Monograph . IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans. Polynuclear Aromatic Compounds , Part 1; Lyon, France: 1983. International Agency for Research on Cancer; pp. 1–477. [Google Scholar]
- IARC Monograph . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Tobacco smoking. International Agency for Research on Cancer; Lyon, France: 1986. International Agency for Research on Cancer. [Google Scholar]
- IARC Monograph . IARC Monograph on Chemical agents and related occupations. Lyon, France: 2012. International Agency for Research on Cancer. [Google Scholar]
- Izard C, Libermann C. Acrolein. Mutat Res. 1978;47:115–138. doi: 10.1016/0165-1110(78)90016-7. [DOI] [PubMed] [Google Scholar]
- Jelakovic B, Karanovic S, Vukovic-Lela I, Miller F, Edwards KL, Nikolic J, Tomic K, Slade N, Brdar B, Turesky RJ, Stipancic Z, Dittrich D, Grollman AP, Dickman KG. Aristolactam-DNA adducts are a biomarker of environmental exposure to aristolochic acid. Kidney Int. 2012;81:559–567. doi: 10.1038/ki.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly RA, Matsui S, Hanaoka T, Matsuda T. Application of the adductome approach to assess intertissue DNA damage variations in human lung and esophagus. Mutat Res. 2007;625:83–93. doi: 10.1016/j.mrfmmm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Klaene JJ, Flarakos C, Glick J, Barret JT, Zarbl H, Vouros P. Tracking matrix effects in the analysis of DNA adducts of polycyclic aromatic hydrocarbons. J Chromatogr A. 2015 doi: 10.1016/j.chroma.2015.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaene JJ, Sharma VK, Glick J, Vouros P. The analysis of DNA adducts: the transition from (32)P-postlabeling to mass spectrometry. Cancer Lett. 2013;334:10–19. doi: 10.1016/j.canlet.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozekov ID, Turesky RJ, Alas GR, Harris CM, Harris TM, Rizzo CJ. Formation of deoxyguanosine cross-links from calf thymus DNA treated with acrolein and 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 2010;23:1701–1713. doi: 10.1021/tx100179g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langouet S, Coles B, Morel F, Becquemont L, Beaune P, Guengerich FP, Ketterer B, Guillouzo A. Inhibition of Cyp1a2 and Cyp3a4 by Oltipraz Results in Reduction of Aflatoxin B-1 Metabolism in Human Hepatocytes in Primary Culture. Cancer Research. 1995;55:5574–5579. [PubMed] [Google Scholar]
- Langouet S, Paehler A, Welti DH, Kerriguy N, Guillouzo A, Turesky RJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in rat and human hepatocytes. Carcinogenesis. 2002;23:115–122. doi: 10.1093/carcin/23.1.115. [DOI] [PubMed] [Google Scholar]
- Langouet S, Welti DH, Kerriguy N, Fay LB, Huynh-Ba T, Markovic J, Guengerich FP, Guillouzo A, Turesky RJ. Metabolism of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in human hepatocytes: 2-amino-3-methylimidazo[4,5-f]quinoxaline-8-carboxylic acid is a major detoxification pathway catalyzed by cytochrome P450 1A2. Chem Res Toxicol. 2001;14:211–221. doi: 10.1021/tx000176e. [DOI] [PubMed] [Google Scholar]
- Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewtas J. Airborne carcinogens. Pharmacol Toxicol. 1993;72(Suppl 1):55–63. doi: 10.1111/j.1600-0773.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- Liebler DC. Proteomic approaches to characterize protein modifications: new tools to study the effects of environmental exposures. Environ. Health Perspect. 2002;110(Suppl 1):3–9. doi: 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HS, Jenner AM, Ong CN, Huang SH, Whiteman M, Halliwell B. A high-throughput and sensitive methodology for the quantification of urinary 8-hydroxy-2'-deoxyguanosine: measurement with gas chromatography-mass spectrometry after single solid-phase extraction. Biochem J. 2004;380:541–548. doi: 10.1042/BJ20040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang Y. Mass spectrometry for the assessment of the occurrence and biological consequences of DNA adducts. Chem Soc Rev. 2015;44:7829–7854. doi: 10.1039/c5cs00316d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lovell MA, Lynn BC. Development of a method for quantification of acrolein-deoxyguanosine adducts in DNA using isotope dilution-capillary LC/MS/MS and its application to human brain tissue. Anal Chem. 2005;77:5982–5989. doi: 10.1021/ac050624t. [DOI] [PubMed] [Google Scholar]
- Liu X, Lovell MA, Lynn BC. Detection and quantification of endogenous cyclic DNA adducts derived from trans-4-hydroxy-2-nonenal in human brain tissue by isotope dilution capillary liquid chromatography nanoelectrospray tandem mass spectrometry. Chem Res Toxicol. 2006;19:710–718. doi: 10.1021/tx0502903. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Harris CC. Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 2008;68:6863–6872. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luceri F, Pieraccini G, Moneti G, Dolara P. Primary aromatic amines from side-stream cigarette smoke are common contaminants of indoor air. Toxicol Ind Health. 1993;9:405–413. doi: 10.1177/074823379300900302. [DOI] [PubMed] [Google Scholar]
- Ma B, Villalta PW, Balbo S, Stepanov I. Analysis of a malondialdehyde-deoxyguanosine adduct in human leukocyte DNA by liquid chromatography nanoelectrospray-high-resolution tandem mass spectrometry. Chem Res Toxicol. 2014;27:1829–1836. doi: 10.1021/tx5002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning - A Laboratory Manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 1982. [Google Scholar]
- Marnett LJ. Lipid peroxidation-DNA damage by malondialdehyde. Mutat Res. 1999;424:83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Ogawa Y, Yoshida R, Shimamori A, Kasai H, Ohta H. The stability of the oxidative stress marker, urinary 8-hydroxy-2'- deoxyguanosine (8-OHdG), when stored at room temperature. J Occup Health. 2008;50:366–372. doi: 10.1539/joh.l7144. [DOI] [PubMed] [Google Scholar]
- McClellan JE, Murphy JP, 3rd, Mulholland JJ, Yost RA. Effects of fragile ions on mass resolution and on isolation for tandem mass spectrometry in the quadrupole ion trap mass spectrometer. Anal Chem. 2002;74:402–412. doi: 10.1021/ac015610b. [DOI] [PubMed] [Google Scholar]
- McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15:223–243. vii–x. doi: 10.1016/j.cld.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA. Carcinogenesis by chemicals: an overview--G. H. A. Clowes memorial lecture. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- Motykiewicz G, Malusecka E, Grzybowska E, Chorazy M, Zhang YJ, Perera FP, Santella RM. Immunohistochemical quantitation of polycyclic aromatic hydrocarbon-DNA adducts in human lymphocytes. Cancer Res. 1995;55:1417–1422. [PubMed] [Google Scholar]
- Nath RG, Chung FL. Detection of exocyclic 1,N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc Natl Acad Sci U S A. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath RG, Ocando JE, Chung FL. Detection of 1, N2-propanodeoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 1996;56:452–456. [PubMed] [Google Scholar]
- Nath RG, Ocando JE, Guttenplan JB, Chung FL. 1,N2-propanodeoxyguanosine adducts: potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res. 1998;58:581–584. [PubMed] [Google Scholar]
- Nauwelaers G, Bessette EE, Gu D, Tang Y, Rageul J, Fessard V, Yuan JM, Yu MC, Langouet S, Turesky RJ. DNA adduct formation of 4-aminobiphenyl and heterocyclic aromatic amines in human hepatocytes. Chem Res Toxicol. 2011;24:913–925. doi: 10.1021/tx200091y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhoff E, Kirpekar F, Karas M, Cramer R, Hahner S, Hillenkamp F, Kristiansen K, Roepstroff P, Lezius A. Comparison of IR- and UV-matrix-assisted laser desorption/ionization mass spectrometry of oligodeoxynucleotides. Nucleic Acids Res. 1994;22:2460–2465. doi: 10.1093/nar/22.13.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paehler A, Richoz J, Soglia J, Vouros P, Turesky RJ. Analysis and quantification of DNA adducts of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in liver of rats by liquid chromatography/electrospray tandem mass spectrometry. Chem Res Toxicol. 2002;15:551–561. doi: 10.1021/tx010178e. [DOI] [PubMed] [Google Scholar]
- Patrianakos C, Hoffmann D. Chemical studies on tobacco smoke LXIV. On the analysis of aromatic amines in cigarette smoke. J.Assoc.Off Anal.Chem. 1979;3:150–154. [Google Scholar]
- Paul W. Electromagnetic traps for charged and neutral particles (Nobel lecture) Agnew Chem Int Ed Engl. 1990;29:739–748. [Google Scholar]
- Payne AH, Glish GL. Tandem mass spectrometry in quadrupole ion trap and ion cyclotron resonance mass spectrometers. Methods Enzymol. 2005;402:109–148. doi: 10.1016/S0076-6879(05)02004-5. [DOI] [PubMed] [Google Scholar]
- Penning TM. Human aldo-keto reductases and the metabolic activation of polycyclic aromatic hydrocarbons. Chem Res Toxicol. 2014;27:1901–1917. doi: 10.1021/tx500298n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- Phillips DH. On the origins and development of the (32)P-postlabelling assay for carcinogen-DNA adducts. Cancer Lett. 2012;334:5–9. doi: 10.1016/j.canlet.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Phillips DH, Farmer PB, Beland FA, Nath RG, Poirier MC, Reddy MV, Turteltaub KW. Methods of DNA adduct determination and their application to testing compounds for genotoxicity. Environ Mol Mutagen. 2000;35:222–233. doi: 10.1002/(sici)1098-2280(2000)35:3<222::aid-em9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Poirier MC, Santella RM, Weston A. Carcinogen macromolecular adducts and their measurement. Carcinogenesis. 2000;21:353–359. doi: 10.1093/carcin/21.3.353. [DOI] [PubMed] [Google Scholar]
- Pratt MM, John K, MacLean AB, Afework S, Phillips DH, Poirier MC. Polycyclic aromatic hydrocarbon (PAH) exposure and DNA adduct semi-quantitation in archived human tissues. Int J Environ Res Public Health. 2011;8:2675–2691. doi: 10.3390/ijerph8072675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JM, Miller EC, Miller JA. The intracellular distribution of protein, nucleic acids, riboflavin, and protein-bound aminoazo dye in the livers of rats fed p-dimethylaminoazobenzene. J Biol Chem. 1948;173:345–353. [PubMed] [Google Scholar]
- Randall KL, Argoti D, Paonessa JD, Ding Y, Oaks Z, Zhang Y, Vouros P. An improved liquid chromatography-tandem mass spectrometry method for the quantification of 4-aminobiphenyl DNA adducts in urinary bladder cells and tissues. J Chromatogr A. 2010;1217:4135–4143. doi: 10.1016/j.chroma.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerath K, Reddy MV, Gupta RC. 32 P-labeling test for DNA damage. Proc Natl Acad Sci U.S.A. 1981;78:6162–6129. doi: 10.1073/pnas.78.10.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol Lett. 2012;213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanat JL, Cadet J, Douki T. Oxidatively generated DNA lesions as potential biomarkers of in vivo oxidative stress. Curr Mol Med. 2012;12:655–671. doi: 10.2174/156652412800792651. [DOI] [PubMed] [Google Scholar]
- Ravanat JL, Turesky RJ, Gremaud E, Trudel LJ, Stadler RH. Determination of 8-oxoguanine in DNA by gas chromatography--mass spectrometry and HPLC--electrochemical detection: overestimation of the background level of the oxidized base by the gas chromatography--mass spectrometry assay. Chem. Res. Toxicol. 1995;8:1039–1045. doi: 10.1021/tx00050a007. [DOI] [PubMed] [Google Scholar]
- Rota C, Cristoni S, Trenti T, Cariani E. A serially coupled stationary phase method for the determination of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine by liquid chromatography ion trap tandem mass spectrometry. Redox Biol. 2013;1:492–497. doi: 10.1016/j.redox.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaraju D, Villalta P, Goggin M, Agunsoye MO, Campbell C, Tretyakova N. Capillary HPLC-accurate mass MS/MS quantitation of N7-(2,3,4-trihydroxybut-1-yl)-guanine adducts of 1,3-butadiene in human leukocyte DNA. Chem Res Toxicol. 2013;26:1486–1497. doi: 10.1021/tx400213m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaraju D, Villalta PW, Wickramaratne S, Swenberg J, Tretyakova N. NanoLC/ESI+ HRMS3 quantitation of DNA adducts induced by 1,3-butadiene. J Am Soc Mass Spectrom. 2014;25:1124–1135. doi: 10.1007/s13361-014-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella RM. Immunological methods for detection of carcinogen-DNA damage in humans. Cancer Epidemiol. Biomarkers Prev. 1999;8:733–739. [PubMed] [Google Scholar]
- Schmidt A, Karas M, Dulcks T. Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: when does ESI turn into nano-ESI? J Am Soc Mass Spectrom. 2003;14:492–500. doi: 10.1016/S1044-0305(03)00128-4. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Senko MW, Syka JE. A two-dimensional quadrupole ion trap mass spectrometer. J Am Soc Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- Scigelova M, Makarov A. Fundamentals and Advances of Orbitrap Mass Spectrometry. John Wiley & Sons Ltd.; Chichester, West Sussex, England: 2013. [Google Scholar]
- Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74:4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- Shibutani S, Gentles RG, Iden CR, Johnson F. Facile Aerial Oxidation of the DNA-Base Adduct N-(2'-Deoxyguanosin-8-Yl)-2-Aminofluorene [Dg(C8)Af] Journal of the American Chemical Society. 1990;112:5667–5668. [Google Scholar]
- Singh R, Farmer PB. Liquid chromatography-electrospray ionization-mass spectrometry: the future of DNA adduct detection. Carcinogenesis. 2006;27:178–196. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- Skipper PL, Peng X, SooHoo CK, Tannenbaum SR. Protein adducts as biomarkers of human carcinogen exposure. Drug Metab.Rev. 1994;26:111–124. doi: 10.3109/03602539409029787. [DOI] [PubMed] [Google Scholar]
- Smela ME, Hamm ML, Henderson PT, Harris CM, Harris TM, Essigmann JM. The aflatoxin B(1) formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2002;99:6655–6660. doi: 10.1073/pnas.102167699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford G., Jr. Ion trap mass spectrometry: a personal perspective. J Am Soc Mass Spectrom. 2002;13:589–596. doi: 10.1016/S1044-0305(02)00385-9. [DOI] [PubMed] [Google Scholar]
- Stone MP, Banerjee S, Brown KL, Egli M. Chemistry and Biology of Aflatoxin-DNA Adducts. Frontiers in Nucleic Acids. 2011;1082:147–166. [Google Scholar]
- Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaska G, Dooley KL, Kadlubar FF. Detection and characterization of carcinogen-DNA adducts in exfoliated urothelial cells from 4-aminobiphenyl-treated dogs by 32P-postlabelling and subsequent thin-layer and high-pressure liquid chromatography. Carcinogenesis. 1990;11:639–646. doi: 10.1093/carcin/11.4.639. [DOI] [PubMed] [Google Scholar]
- Tang Y, Kassie F, Qian X, Ansha B, Turesky RJ. DNA Adduct Formation of 2-Amino-9H-pyrido[2,3-b]indole and 2-Amino-3,4-dimethylimidazo[4,5-f]quinoline in Mouse Liver and Extrahepatic Tissues During a Subchronic Feeding Study. Toxicol.Sci. 2013 doi: 10.1093/toxsci/kft077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova N, Goggin M, Sangaraju D, Janis G. Quantitation of DNA adducts by stable isotope dilution mass spectrometry. Chem Res Toxicol. 2012;25:2007–2035. doi: 10.1021/tx3002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turesky RJ, Vouros P. Formation and analysis of heterocyclic aromatic amine-DNA adducts in vitro and in vivo. J. Chromatogr.B Analyt.Technol.Biomed.Life Sci. 2004;802:155–166. doi: 10.1016/j.jchromb.2003.10.053. [DOI] [PubMed] [Google Scholar]
- Uchida K. Current status of acrolein as a lipid peroxidation product. Trends Cardiovasc Med. 1999;9:109–113. doi: 10.1016/s1050-1738(99)00016-x. [DOI] [PubMed] [Google Scholar]
- Vanherweghem JL, Debelle FD, Muniz Martinez MC, Nortier JL. Aristolochic acid nephropathy after Chinese herb remedies. In: De Broe ME, Porter GA, Bennett WM, Verpooten GA, editors. Clinical Nephrotoxins. 2nd Kluwer, Dordrecht: 2003. pp. 679–603. [Google Scholar]
- Wilfinger WW, Mackey K, Chomczynski P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques. 1997;22:474–476. 478–481. doi: 10.2144/97223st01. [DOI] [PubMed] [Google Scholar]
- Wolf SM, Vouros P. Application of capillary liquid chromatography coupled with tandem mass spectrometric methods to the rapid screening of adducts formed by the reaction of N-acetoxy-N-acetyl-2-aminofluorene with calf thymus DNA. Chem Res Toxicol. 1994;7:82–88. doi: 10.1021/tx00037a013. [DOI] [PubMed] [Google Scholar]
- Yin RC, Liu SQ, Zhao C, Lu ML, Tang MS, Wang HL. An Ammonium Bicarbonate-Enhanced Stable Isotope Dilution UHPLC-MS/MS Method for Sensitive and Accurate Quantification of Acrolein-DNA Adducts in Human Leukocytes. Analytical Chemistry. 2013;85:3190–3197. doi: 10.1021/ac3034695. [DOI] [PubMed] [Google Scholar]
- Yun BH, Rosenquist TA, Nikolic J, Dragicevic D, Tomic K, Jelakovic B, Dickman KG, Grollman AP, Turesky RJ. Human formalin-fixed paraffin-embedded tissues: an untapped specimen for biomonitoring of carcinogen DNA adducts by mass spectrometry. Anal.Chem. 2013;85:4251–4258. doi: 10.1021/ac400612x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun BH, Rosenquist TA, Sidorenko V, Iden CR, Chen CH, Pu YS, Bonala R, Johnson F, Dickman KG, Grollman AP, Turesky RJ. Biomonitoring of aristolactam-DNA adducts in human tissues using ultra-performance liquid chromatography/ion-trap mass spectrometry. Chem.Res.Toxicol. 2012;25:1119–1131. doi: 10.1021/tx3000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun BH, Sidorenko VS, Rosenquist TA, Dickman KG, Grollman AP, Turesky RJ. New Approaches for Biomonitoring Exposure to the Human Carcinogen Aristolochic Acid. Toxicol Res (Camb) 2015;4:763–776. doi: 10.1039/C5TX00052A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun BH, Yao L, Jelakovic B, Nikolic J, Dickman KG, Grollman AP, Rosenquist TA, Turesky RJ. Formalin-fixed paraffin-embedded tissue as a source for quantitation of carcinogen DNA adducts: aristolochic acid as a prototype carcinogen. Carcinogenesis. 2014;35:2055–2061. doi: 10.1093/carcin/bgu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahid M, Beseler CL, Hall JB, LeVan T, Cavalieri EL, Rogan EG. Unbalanced estrogen metabolism in ovarian cancer. Int J Cancer. 2014;134:2414–2423. doi: 10.1002/ijc.28565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahid M, Goldner W, Beseler CL, Rogan EG, Cavalieri EL. Unbalanced estrogen metabolism in thyroid cancer. Int J Cancer. 2013;133:2642–2649. doi: 10.1002/ijc.28275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Aft RL, Gross ML. Estrogen carcinogenesis: specific identification of estrogen-modified nucleobase in breast tissue from women. Chem Res Toxicol. 2008;21:1509–1513. doi: 10.1021/tx8001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine-DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:830–837. [PubMed] [Google Scholar]