Abstract
Background
Colon cancer is one of the most prevalent and deadly cancers worldwide. It is still necessary to further define the mechanisms and explore therapeutic targets of colon cancer. Dysregulation of long noncoding RNAs (lncRNAs) has been shown to be correlated with diverse biological processes, including tumorigenesis. This study aimed to characterize the biological mechanism of taurine-upregulated gene 1 (TUG1) in colon cancer.
Material/Methods
qRT-PCR was used to analyze the expression level of TUG1 and p63 in 75 colon cancer tissues and the matched adjacent non-tumor tissue. In vitro, cultured colon cancer cell lines HCT-116 and LoVo were used as cell models. TUG1 and p63 were silenced via transferring siRNA into HCT-116 or LoVo. The effects of TUG1 were investigated by examining cell proliferation, apoptosis, and migration.
Results
Among the 75 colon cancer cases, the expression of TUG1 was significantly higher in colon cancer tissues compared with the matched adjacent non-tumor tissue, while p63 expression was lower in the tumor tissue. In HCT-116 and LoVo, the expression of TUG1 was significantly increased by p63 siRNA transfection. Furthermore, down-regulation of TUG1 by siRNA significantly inhibited the cell proliferation and promoted colon cancer cell apoptosis. In addition, inhibition of TUG1 expression significantly blocked the cell migration ability of colon cancer cells.
Conclusions
LncRNA TUG1 may serve as a potential oncogene for colon cancer. Overexpressed TUG1 may contribute to promoting cell proliferation and migration in colon cancer cells.
MeSH Keywords: Colonic Neoplasms; RNA, Long Noncoding; Transcellular Cell Migration
Background
Colon cancer is the third most common type of cancer, with approximately 1 million new cases each year world-wide, and is the second most frequent cause of cancer-related deaths worldwide [1]. Radio- and chemotherapy are the primary treatment in both resectable and advanced colon cancer [2,3]. Colon cancer transformation from the normal colonic mucosa arises through a progressive accumulation of genetic changes [4]. Therefore, the mechanisms of genetic changes and signaling pathways related to formation and development of colon cancer have been further studied.
Long noncoding RNAs (LncRNAs) is a transcribed RNA molecule greater than 200 nt in length and show complex overlapping patterns of expression and regulation. LncRNAs regulate gene expression at the epigenetic level, transcriptional level, and post-transcriptional level and are widely involved in the physiological and pathological process of human disease [5]. LncRNAs and their roles in colon cancer development have attracted much attention. Some of them, including ATB, CCAT1, and HOTAIR, have been reported to be involved in the initiation and progression of colon cancer and to modulate the biological properties of cancer cells [6–9].
Recently, LncRNA TUG1 has been reported to be up-regulated in osteosarcoma via regulation of transcription variants n37730 [10]. Another study demonstrated that TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth by epigenetically silencing KLF2 [11]. These studies show that TUG1 may be an important regulatory gene in cancer. However, the role of TUG1 in colon cancer remains unknown.
In this study, we investigated the function of TUG1 in colon cancer tumorigenesis and found that the expression of TUG1 was significantly higher in colon cancer tissues compared with the matched adjacent non-tumor tissue, and p63 expression was lower in the tumor tissue. Silenced p63 in HCT-116 or LoVo significantly increased the expression of TUG1. Furthermore, down-regulation of TUG1 by siRNA significantly inhibited the cell proliferation and promoted colon cancer cell apoptosis. In addition, inhibition of TUG1 expression significantly blocked cell migration ability of colon cancer cells. Overall, the results presented here indicate that LncRNA TUG1 may serve as a potential oncogene for colon cancer.
Material and Methods
Patients and tissue samples
The original research was approved by the Medical Ethics Committee of our hospital and every patient provided written informed consent. A total of 75 colon cancer patients undergoing surgery for colon cancer were included from Yantai Yuhuangding Hospital (China) since January 2011 in this study. The study protocol was approved by the Institutional Ethics Committee [no. (2010)152]. Tumor specimens and corresponding adjacent normal tissues were collected and stored in liquid nitrogen until use. The characteristics of subjects enrolled in the study are shown in Table 1.
Table 1.
Clinicopathological features of 75 colon cancer patients.
| Characteristics | Total case N (N%) | |
|---|---|---|
| Gender, N(%) | Male | 39 (52) |
| Female | 36 (48) | |
| Age | Media | 62.34±10.56 |
| Range | 33–85 | |
| Smoker, N(%) | Non-smoker | 40 (53.33) |
| Smoker | 35 (46.67) | |
| TNM stage, N(%) | I | 12 (16) |
| II | 28 (37.33) | |
| III | 17 (22.67) | |
| IV | 18 (24) | |
| Grade, N(%) | 1 | 20 (26.67) |
| 2 | 41 (54.67) | |
| 3 | 14 (18.66) | |
| 4 | 0 (0) | |
| Tumor invasion depth, N(%) | T1+T2 | 16 (21.33) |
| T3+T4 | 59 (78.67) | |
| Lymph node metastasis, N(%) | N0 | 36 (48) |
| N1 | 20 (26.67) | |
| N2 | 19 (25.33) | |
| Distant matastasis, N(%) | M0 | 54 (72) |
| M1 | 21 (28) | |
Cell culture
The human colon cancer cell lines HCT-116 and LoVo were purchased from the Cell Bank of the Institute of Biochemistry and Cell Biology, China Academy of Sciences (Shanghai, China). The cells were maintained in DMEM/F-12 (1:1) medium (HyClone, USA) containing 10% fetal bovine serum (FBS, Gibco, Carlsbad, USA) at 37°C in humidified air containing 5% CO2.
SiRNA transfection
Small interfering RNA that targeted TUG1-RNA (TUG1-siRNA) and p63-RNA (p63-siRNA) and a scrambled negative control (Scrambled-siRNA) were generously provided by the RiboBio Company (Guangzhou, China). Human colon cancer cells HCT-116 and LoVo were transfected with either 50 nM siRNA or Scrambled-siRNA for 24 h using Lipofectamine 2000 transfection reagent according to the manufacturer’s protocol (Life Technologies).
RNA extraction and quantitative real-time PCR assay
Total RNA was isolated from tissues using TRIZOL reagent (Invitrogen) according to the manufacturer’s protocol. RNA was reverse transcribed into cDNA using a reverse transcriptase kit (Takara, Dalian, China). The PCR amplification was performed for 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, on a ABI 7500 fast realtime PCR system (Applied Biosystems) with SYBR Green Real-time PCR Master Mix (Takara, Dalian, China). The expression level of each sample was internally normalized against that of the GAPDH. The relative quantitative value was expressed by the 2−ΔΔCt method. Each experiment was performed in triplicate.
Cell proliferation assay
The CCK-8 assay kit (Dojindo; Japan) was used to determine the proliferation of HCT-116 and LoVo cells. In each 96-well plate, the cells (1×103/well) were seeded and cultured for 12 h, then transfected with Scrambled-siRNA or TUG1-siRNA for 24 h, and further incubated for 24, 48, and 72 h. At each time point, 10 μL CCK-8 reagents were added to each well, the plate was re-incubated for 2 h, and the absorbance at 450 nm was subsequently detected. Each sample was tested in triplicate and all experiments were performed 3 times.
Cell apoptosis assay
Cells were cultured and transfected with TUG1-siRNA or Scrambled-siRNA for 48 h, as described above. Cells were harvested and washed twice with ice-cold phosphate-buffered saline (PBS) and then stained using the Annexin V-FITC Apoptosis Detection Kit (BD, San Jose, USA) according to the manufacturer’s instructions. Cells were then analyzed by using a flow cytometer (BD FACS Calibur, Becton, Dickinson and Company Biosciences, San Jose, USA).
Cell migration analysis
Cell migration ability assessment was performed using Transwell polycarbonate membrane inserts (Millipore, Schwalbach, Germany). The cells (1×105) were plated onto an insert and 20% FBS was added to the cell-free medium in the lower chamber. After incubation 24 h at 37°C, the inserts were washed in PBS, and the cells were fixed to the membranes with 4% paraformaldehyde, then stained with Hoechst (10 μg/ml). The migrated cells were counted per high-power field.
Statistical analysis
Data are presented as mean ±SD. Graphs were drawn using GraphPad Prism Software. Data were analyzed using a one-way analysis of variance (ANOVA) followed by the t test. For all statistical analyses, P<0.05 was considered statistically significant.
Results
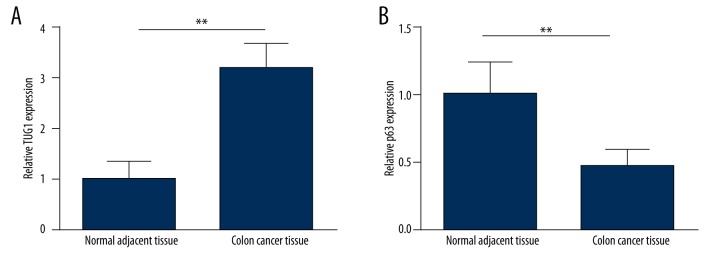
LncRNA TUG1 was significantly up-regulated in colon cancer tissues
To assess the changes of LncRNA TUG1 and p63 expression in cases of colon cancer, the expression of TUG1 and p63 was examined in 75 tissue specimens. As shown in Figure 1A, the expression level of TUG1 was significantly higher in the primary tumor tissue of patients than in matched non-tumor tissue, while p63 expression was lower in the tumor tissue (Figure 1B). The results suggest a connection between TUG1expression and colon cancer. Based on this expression pattern, we therefore used HCT-116 and LoVo cells for the next studies.
Figure 1.
Expression of LncRNA TUG1 and p63 in colon cancer tissue specimens. qRT-PCR analysis of LncRNA TUG1 (A) and p63 expression (B) in 75 pairs of primary colon cancer tumor tissues and their corresponding adjacent tissues. ** P<0.01.
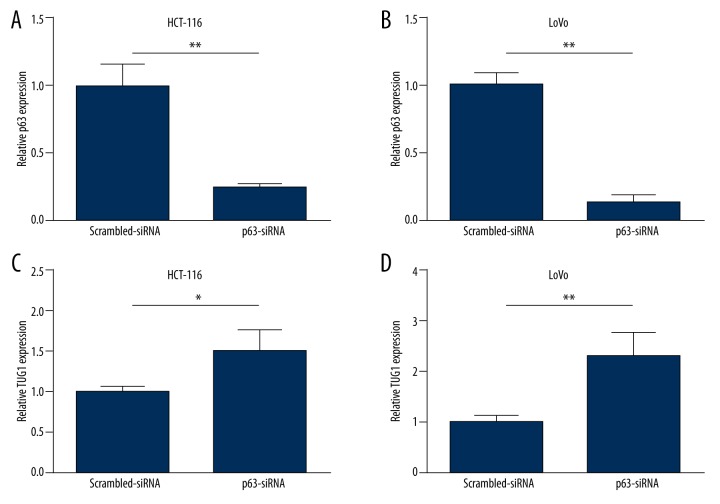
Interference of p63 up-regulated the expression of LncRNA TUG1 in colon cancer cells
Further investigation in cell lines was carried out. P63-siRNA was used to influence the expression of p63 in HCT-116 and LoVo. As Figure 2A, 2B show, p63-siRNA significantly decreased the mRNA level of p63 in HCT-116 and LoVo. The mRNA level of TUG1 was significantly increased in HCT-116 after transfection with p63-siRNA (Figure 2C). Similarly, after transfection with p63-siRNA, the expression of TUG1 was also remarkably increased in LoVo cells (Figure 2D).
Figure 2.
Interference of p63 up-regulated the expression of LncRNA TUG1 in colon cancer cells. Relative p63 expression levels after transfection with p63-siRNA or Scrambled-siRNA for 24 h in HCT-116 (A) and LoVo (B); LncRNA TUG1 expression levels after transfection with p63-siRNA or Scrambled-siRNA for 24 h in HCT-116 (C) and LoVo (D). * P<0.05, ** P<0.01.
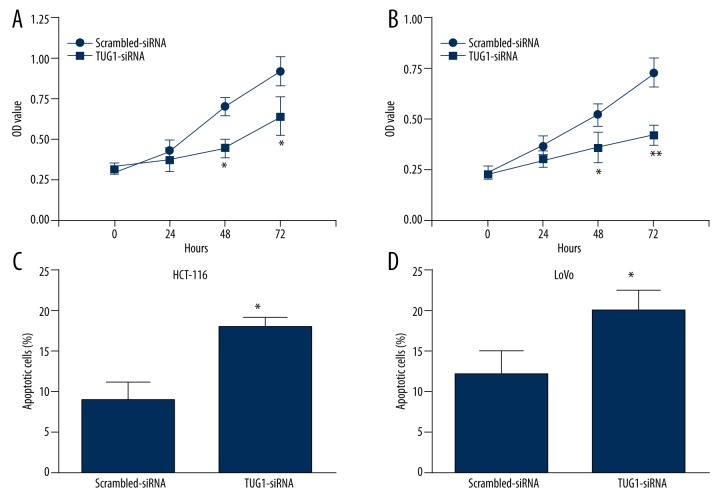
Down-regulation of TUG1 inhibited cell proliferation and promoted cell apoptosis of colon cancer cell lines
To further confirm the functional roles of TUG1, we then examined the effect of TUG1 on cell proliferation and cell apoptosis. CCK-8 assay showed that transfection with TUG1-siRNA significantly inhibited HCT-116 cell proliferation, as well as LoVo, at 48 h and 72h compared to Scrambled-siRNA (Figure 3A, 3B). Additionally, flow cytometry analysis showed that down-regulation of TUG1 in both HCT-116 and LoVo cells promoted cell apoptosis, but not in the control group (Figure 3C, 3D).
Figure 3.
Down-regulation of LncRNA TUG1 inhibited cell proliferation and promoted cell apoptosis of colon cancer cell lines. CCK-8 assay of cells transfected with TUG1-siRNA or Scrambled-siRNA for the indicated lengths of time (hours) in HCT-116 (A) and LoVo (B); Apoptosis assay of cells transfected with TUG1-siRNA or Scrambled-siRNA in HCT-116 (C) and LoVo (D); * P<0.05, ** P<0.01 vs. Scrambled-siRNA.
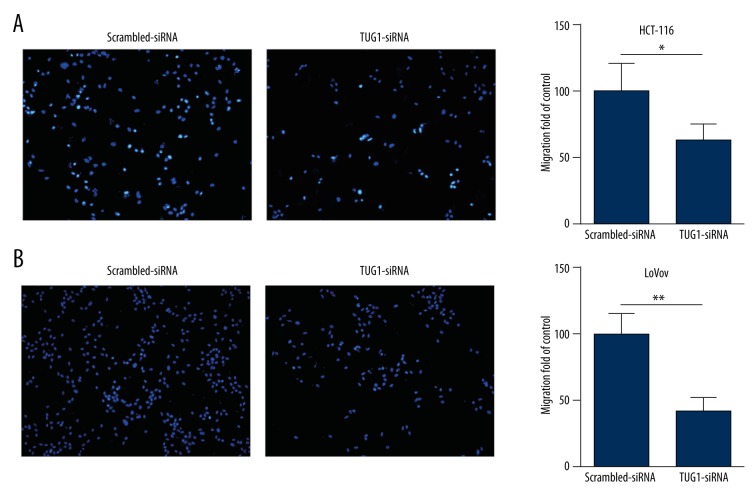
Down-regulation of TUG1 inhibited colon cancer cell lines migration ability
The number of migration HCT-116 cells in transwell assays was significantly decreased after transfection with TUG1-siRNA (Figure 4A). Similarly, after transfecting TUG1-siRNA, the number of migrating cells was also obviously reduced in LoVo cells (Figure 4B).
Figure 4.
Down-regulation of LncRNA TUG1 inhibited colon cancer cell lines migration ability. Transwell assays of cells transfected with TUG1-siRNA or Scrambled-siRNA showing the effects on migration ability of HCT-116 (A) and LoVo (B); Original magnification, 100×; * P<0.05, ** P<0.01.
Discussion
Colon cancer is the most commonly diagnosed cancer in males and females, with millions new cancer cases and deaths estimated to have occurred [12–14]. Despite improvements in cancer diagnosis and therapy, many patients are still diagnosed at the late stages of the disease. Research on candidate differentially expressed genes may help to find biological markers for the evaluation of cancer diagnosis and offer novel molecular targets for anticancer therapy [15].
A large number of LncRNAs are confirmed to play critical roles in carcinogenesis and participate in many diseases [16,17]. Recently, some LncRNAs were reported to be involved in colon cancer, such as ZFAS1 [18], UCA1 [19], and CCAT1 [20]. In this study, we found that the expression of LncRNA TUG1 was significantly higher in colon cancer tissues compared with the matched adjacent non-tumor tissue. This outcome indicates that TUG1 might be involved in the progression of colon cancer.
A great number of lncRNAs work via binding to polycomb repressive complex 2 (PRC2), including TUG1 [21]. TUG1 has multiple transcription factors, among which, p63 has drawn our attention. P63 is the oldest member of the p53 gene family. P63 regulate many p53 target genes due to their common structural features [22]. However, p63 has its own role, and several studies have reported that p63 is an independent predictor of poor outcome in gastric cancer patients [23]. The involvement of P63 in the differential diagnosis of acinic cell carcinoma versus mucoepidermoid carcinoma is also being researched currently [24]. Here, we found the expression of p63 was lower in tumor tissue. Silencing of p63 in HCT-116 or LoVo significantly increased the expression of TUG1. Therefore, we concluded that p63 is a regulation gene of TUG1.
LncRNAs can act as oncogenes or tumor suppressors and are involved in numerous cellular processes, playing roles in tumor progression by regulating cell differentiation, cell proliferation, and cell cycle [25–27]. TUG1 was also shown to have a cancerous function in several human tumors, such as urothelial carcinoma [28], osteosarcoma [10], and melanoma [29]. However, the role of TUG1 in colon cancer development and metastasis remains unknown. In our study, we found that down-regulation of TUG1 by siRNA significantly inhibited the cell proliferation in both HCT-116 and LoVo. In addition, down-regulation of TUG1 markedly increased apoptosis in HCT-116 and LoVo. These data suggest that interference in the expression of TUG1 may contribute to the growth and apoptosis of colon cancer cells and consequently promote the development of colon cancer. We also tested the migration capability of HCT-116 and LoVo after being transfected with TUG1-siRNA. We found that inhibition of TUG1 expression significantly blocked cell migration ability of colon cancer cells. This result indicates that the aberrant increasing level of TUG1 may promote cell migration. Thus, we confirmed that TUG1 may be a tumor oncogene in colon cancer.
Conclusions
Our findings further the understanding of colon cancer pathogenesis, and facilitate the development of lncRNA TUG1-directed diagnostics and therapeutics against colon cancer.
Footnotes
Conflict of interest statement
The authors do not have any possible conflicts of interest.
Source of support: This study was supported by grants from the Yantai Scientific Development Project (2013WS216)
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Pathak S, Meng WJ, Nandy SK, et al. Radiation and SN38 treatments modulate the expression of microRNAs, cytokines and chemokines in colon cancer cells in a p53-directed manner. Oncotarget. 2015 doi: 10.18632/oncotarget.5815. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Improved survival with preoperative radiotherapy in resectable rectal cancer. Swedish Rectal Cancer Trial. N Engl J Med. 1997;336:980–87. doi: 10.1056/NEJM199704033361402. [DOI] [PubMed] [Google Scholar]
- 4.Wong HS, Chang WC. Correlation of clinical features and genetic profiles of stromal interaction molecule 1 (STIM1) in colorectal cancers. Oncotarget. 2015;6(39):42169–82. doi: 10.18632/oncotarget.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Yue B, Qiu S, Zhao S, et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2015 doi: 10.1111/jgh.13206. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 7.He X, Tan X, Wang X, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–88. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 8.Wu ZH, Wang XL, Tang HM, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 9.Sun QL, Zhao CP, Wang TY, et al. Expression profile analysis of long non-coding RNA associated with vincristine resistance in colon cancer cells by next-generation sequencing. Gene. 2015;572:79–86. doi: 10.1016/j.gene.2015.06.087. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Geng PL, Yin P, et al. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–15. doi: 10.7314/apjcp.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- 11.Huang MD, Chen WM, Qi FZ, et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 13.Gavriilidis P, Nikolaidou A. Colon adenocarcinoma associated with synchronous extramural gastrointestinal stromal tumor (GIST) of the ileum. Am J Case Rep. 2015;16:837–39. doi: 10.12659/AJCR.894845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai CC, Wu MN. Frequent ischemic stroke as first manifestation of occult colon cancer: A rare case. Am J Case Rep. 2015;16:723–27. doi: 10.12659/AJCR.895130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, Pan C, Hu J, Zhang S. The role of Reg IV in colorectal cancer, as a potential therapeutic target. Contemp Oncol. 2015;19:261–64. doi: 10.5114/wo.2015.54385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Z, Xiao Z, Liu F, et al. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1) Oncotarget. 2015;7(1):241–54. doi: 10.18632/oncotarget.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Ni H, Zhao Y, et al. Potential role of lncRNAs in contributing to pathogenesis of intervertebral disc degeneration based on microarray data. Med Sci Monit. 2015;21:3449–58. doi: 10.12659/MSM.894638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorenoor N, Faltejskova-Vychytilova P, Hombach S, et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7(1):622–37. doi: 10.18632/oncotarget.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med. 2015;8:11824–30. [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Z, Zhou M, Tian B, et al. Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal cancer and its clinical significance. Int J Clin Exp Med. 2015;8:3707–15. [PMC free article] [PubMed] [Google Scholar]
- 21.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonfloni S, Caputo V, Iannizzotto V. P63 in health and cancer. Int J Dev Biol. 2015;59:87–93. doi: 10.1387/ijdb.150045sg. [DOI] [PubMed] [Google Scholar]
- 23.Song Y, Liu D, He G. TKTL1 and p63 are biomarkers for the poor prognosis of gastric cancer patients. Cancer Biomark. 2015;15:591–97. doi: 10.3233/CBM-150499. [DOI] [PubMed] [Google Scholar]
- 24.Abd Raboh NM, Hakim SA. Diagnostic role of DOG1 and p63 immunohistochemistry in salivary gland carcinomas. Int J Clin Exp Pathol. 2015;8:9214–22. [PMC free article] [PubMed] [Google Scholar]
- 25.Pei J, Wang B. Notch-1 promotes breast cancer cells proliferation by regulating LncRNA GAS5. Int J Clin Exp Med. 2015;8:14464–71. [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng C, Yu X, Lai J, et al. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8:126. doi: 10.1186/s13045-015-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Zhi X, Wang L, et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135. doi: 10.1186/s13046-015-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555–59. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 29.van Heesch S, van Iterson M, Jacobi J, et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]