Abstract
The RNA binding proteins Zfp36l1 and Zfp36l2 act redundantly to enforce the β-selection checkpoint during thymopoiesis, yet their molecular targets remain largely unknown. Here, we identify these targets on a genome wide scale in primary mouse thymocytes and show that Zfp36l1/l2 regulate DNA damage response and cell cycle transcripts to ensure proper β-selection. DN3 thymocytes lacking Zfp36l1/l2 share a gene expression profile with post-selected DN3b cells despite the absence of intracellular TCRβ and reduced IL-7 signaling. Our findings show that in addition to controlling the timing of proliferation at β-selection post-transcriptional control by Zfp36l1/l2 limits DNA damage responses which are known to promote thymocyte differentiation. Zfp36l1/l2 therefore act as post-transcriptional safeguards against chromosomal instability and replication stress by integrating pre-TCR and IL-7 signaling with DNA damage and cell cycle control.
Keywords: Zfp36l1, Zfp36l2, Cell cycle, DNA damage, Tis11b, Tis11d, lymphocyte development, beta-selection, T-ALL, leukemia
Introduction
Early thymocyte development occurs in multiple quasi-discrete stages during which cells are required to pass through stringent developmental checkpoints. These checkpoints ensure T cell receptor (TCR) reactivity and prevent transformation into highly proliferative states. During the first stages of T cell differentiation in mice CD4- CD8-double negative (DN) thymocytes can be subdivided into DN1-4 populations based on surface expression of CD44 and CD25 (1–3). The DN3 stage is further divided into DN3a and DN3b cells, the latter of which have successfully recombined the variable (V), diversity (D), and joining (J) gene segments of the Trb locus and express intracellular (ic) TCRβ. They are selected by a process known as the β-selection checkpoint at which icTCRβpositive DN3b cells undergo a proliferative burst and have an increased metabolic state as shown by CD98 expression (3, 4). This dramatically expands the pool of thymocytes with successful Trb rearrangments which can progress to the double positive (DP) stage of development (2). During VDJ recombination double strand DNA breaks (DSBs) are formed by the Recombinase Activating Gene (RAG) complex and activate the DNA damage response (DDR) pathway. These lead to activation of Atm (ataxia-telangiectasia-mutated), DNA-PKcs (DNA-dependent kinase catalytic subunit), and Atr (Atm- and Rad3-related) (5, 6). A critical target of these kinases is histone variant H2AFX, which is phosphorylated (P-H2AFX) at the site of DNA damage (7). P-H2AFX then recruits other DDR factors to the break site, and stabilizes cleaved DNA ends prior to joining (8–11). Atm and DNA-PKcs are also responsible for the activation of the Chk1 and Chk2 protein kinases which phosphorylate multiple downstream effectors, including Cdc25a and p53, leading to cell cycle arrest and DSB resolution/repair (12, 13). Remarkably, the activation of these pathways have been linked to the promotion of thymocyte differentiation (14, 15) as well as transformation.
The ZFP36 family of RNA binding proteins (RBP) comprises three gene family members in humans and four in mice. These RBPs bind to A/U rich elements (ARE) in the 3’ untranslated region (3’UTR) of messenger RNA (mRNA), and promote RNA decay (16). As such, many mRNAs have been proposed as targets of ZFP36 family proteins, although few have been shown to be physiologically relevant (16). Constitutive knock out (KO) of Zfp36 leads to viable animals which develop an autoimmune disease caused by the overexpression of the pro-inflammatory cytokine TNF (17–19), while Zfp36l1- or Zfp36l2-null mice die in utero or shortly after birth due to disorganized vasculature or anemia respectively (20–22). During early B cell development Zfp36l1/l2 act redundantly to enforce quiescence and enable recombination of the immunoglobulin genes (23). Although the development of B cells lacking both Zfp36l1 and Zfp36l2 is impaired, these mice do not develop B cell malignancy. By contrast, the conditional deletion of both Zfp36l1 and Zfp36l2 (DCKO) in thymocytes results in the bypass of the β-selection checkpoint and development of T cell acute lymphoblastic leukemia (T-ALL) (24). These tumors are dependent on Notch1 whose expression is increased following the release of its mRNA from post-transcriptional repression by Zfp36l1/l2. However the details of how the beta-selection checkpoint is circumvented remain unknown. A better understanding of the spectrum of mRNAs bound by Zfp36l1/l2 in thymocytes is necessary to elucidate the molecular mechanisms through which they regulate the development and proliferative properties of thymocytes.
In this report we combine the detailed phenotypic analyses of early thymocytes from DCKO mice with genome-wide approaches to identify the molecular mechanisms regulated by the RBPs. We integrate RNAseq gene expression data with Individual-nucleotide resolution Cross-Linking and ImmunoPrecipitation (iCLIP) (25) to identify RBP binding positions within their mRNA targets. Our results show that DN3 thymocytes lacking Zfp36l1/l2 closely share gene expression profiles with post-selection DN3b wild-type thymocytes, despite having reduced VDJ recombination of Trbv gene segments and being icTCRβ-neg. Furthermore DCKO thymocytes have elevated expression of positive cell cycle regulators, and show increased cycling and DDR pathway activation in vivo. Conversely overexpression of a GFPZFP36L1 transgene reduces cell cycle entry. Inhibition of the cell cycle in DCKO mice by treatment with a Cdk4/6 inhibitor partially rescues icTCRβ expression in DN3 thymocytes. Thus Zfp36l1/l2 limit the cell cycle in developing thymocytes and the persistence of DSBs in cycling cells.
Materials and Methods
Mouse strains
C57BL/6 mice were from Jackson Laboratories and bred at the Babraham Institute. Zfp36l1fl/fl; Zfp36l2fl/fl; CD2cre double conditional knockout (DCKO) mice were previously described (24). GFPZFP36L1 transgenic mice were generated by targeting the ROSA26 locus using standard methods (23). For cell type specific Cre expression CD2cre (Tg(CD2-cre)4Kio) mice were used (26) and for assessing Myc expression GFP-myc knock-in mice (27) were crossed to DCKO mice. All animal procedures were approved by the Animal Welfare and Experimentation Committee of the Babraham Institute and the UK Home Office.
Flow cytometry
Single cell suspensions of thymocytes were preincubated with Fc-block (anti-mouse CD16/CD31, clone 2.4G2; Bio X Cell) in staining buffer (PBS, 2% FBS, 2 mM EDTA) for 10 min at 4°C and stained with surface antibodies for 20 min at 4°C. For intracellular staining of CD3ε and TCRβ, the BD Cytofix/Cytoperm™ kit was used. For detection of phosphoproteins (Akt, Erk, Zap70/Syk, Stat5, H2afx) and Ccnd3/Ccne2, cells were fixed with BD Lyse/Fix Buffer and permeabilized with BD Phosflow™ Perm Buffer III. Afterwards, surface and intracellular proteins were stained in staining buffer for 1 h at room temperature (RT). For detection of phosphorylated ATM substrates, cells were fixed with 4% PFA and permeabilized with 90% methanol. Fixable Viability Dye eFluor 780 or DAPI (4,6-diamidino-2-phenylindole, 0.1µg/ml) was used for assessing cell viability. For activated caspases CaspGLOW pan caspases kit (Biovision) was used according to the manufacturer’s instructions. Samples were acquired on an LSRFortessa (BD Biosciences) and analyzed with FlowJo software (Tree Star). Antibodies were purchased from the following companies:
Abcam: Cyclin E2 (E142).
BD: B220 (RA3-6B2), CD3ε (2C11), CD4 (RM4-5), CD25 (PC61), CD25 (7D4), Cyclin D3 (1/Cyclin D3), Kit (ACK45), Ly6G (RB6-8C5), NK1.1 (PK136), P-Akt (M89-61), P-Stat5 (Y694, BD-TL), P-Zap70/Syk (17A/P-ZAP70), TCRγδ (GL3), rat anti-mouse IgG2b (R9-91), BrdU (3D4).
Biolegend: CD4 (RM4-5), CD8 (53-6.7), CD25 (PC61), CD44 (IM7), CD98 (RL388), CD127 (A7R34), Kit (ACK2), NK1.1 (PK136), P-H2ax (2F3), Sca1 (E13-161.7), TCRβ (H57-597), Thy1.1 (OX-7), Streptavidin.
Cell Signaling: ATM substrates phospho-(Ser/Thr), P-Erk (D13.14.4E).
eBioscience: B220/CD45R (RA3-6B2), CD4 (GK1.5), CD11b (M1/70), CD24 (30F1), Ly6G (RB6-8C5), TCRγδ (eBioGL3).
mdBioproducts: ST2 (DJ8).
Jackson: Donkey anti-rabbit IgG.
Molecular Probes: Goat anti-rabbit IgG.
Gating strategies
Lineage-negative excludes CD4, CD8, CD11b, TCRγδ, NK1.1, B220 and GR1. DN thymocytes are CD4-, CD8- and DN1 (CD44+, CD25-), DN2 (CD44+, CD25+), DN3 (CD44-, CD25+), DN3a (CD44-, CD25+, CD98low), DN3b (CD44-, CD25intermediate, CD98+), DN4 (CD44-, CD25-), except for Figure 1E, F where DN2 is (Lineage-negative, CD44+, Kithigh) and DN3 is (Lineage-negative, CD44low, Kitlow, CD25+). Intermediate single positive cells (iSP) are (CD8+, CD24high) and mature CD8 (CD8+, CD24-). DP (CD4+, CD8+) cells are divided into early (CD98+, CD71+) and late (CD98+, CD71-) DPs. ILC2 cells are gated as (Lineage-negative, Thy1.1+, Sca1high, ST2+).
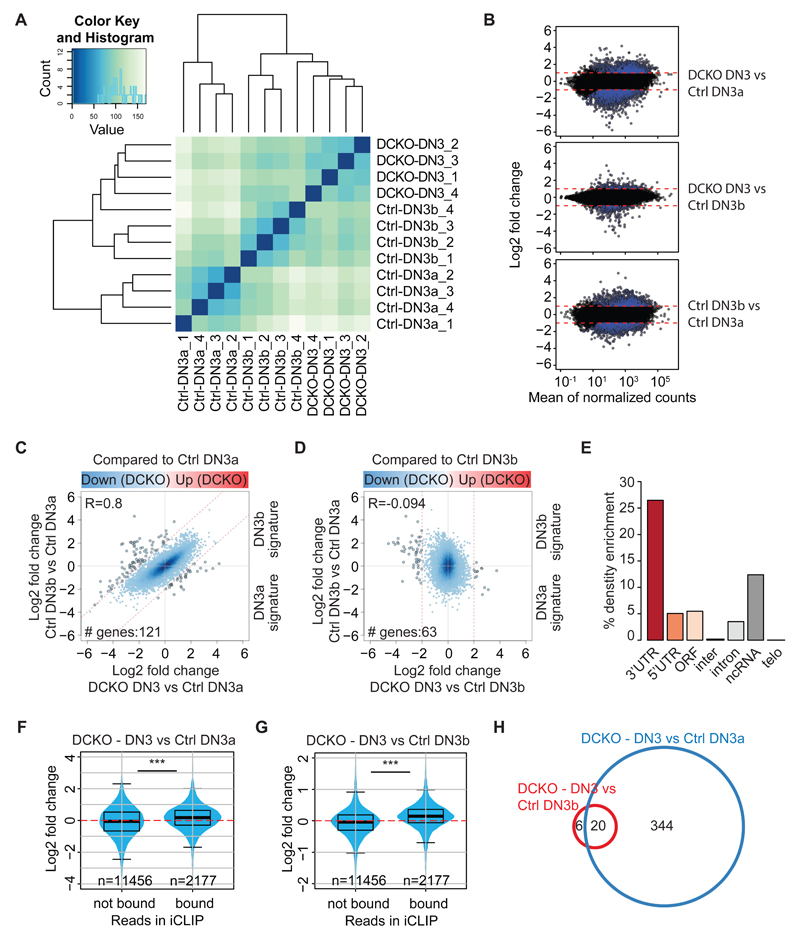
Figure 1. Zfp36l1/l2-deficient thymocytes are committed to the T cell lineage but fail beta-selection.
(A) Representative FACS plots showing proportions of DN1, DN2, DN3 and DN4 thymocytes from control (Zfp36l1/2fl/fl) and Zfp36l1/2fl/fl; CD2Cre (DCKO) mice, gated on Lin- cells. Note that gating differs here from previous work (24) as we have identified that thymocytes previously gated as DN2 express icCD3ε, therefore are committed to the T cell lineage.
(B) Representative FACS plots showing proportions of Sca1+ cells from control and DCKO mice gated on CD4-, CD8- cells.
(C) Representative FACS plots showing proportions of Lin-, DN2 (cKithigh, CD44+) and DN3-4 (cKitlow, CD44low) cells from control and DCKO mice (left panel) and intracellular (ic) CD3ε expression in an overlay of DN2 and DN3-4 cells (right panel).
(D) Percentage of icTCRβ-positive cells from control and DCKO mice in DN2-4 as gated in A) and iSP (CD8+, CD24+), early DP (CD4+, CD8+, CD98+, CD71+), late DP (CD4+, CD8+, CD98+, CD71-) and CD8 (CD8+, CD24-). Data depicted as mean + SD.
(E) Representative FACS plot showing proportions of live CD4, CD8, DP and DN cells from control and DCKO mice.
(F) Percentages of DN1-4 cells from control and DCKO mice, gated on Lin- and: DN1 = CD44+, CD25-; DN2 = CD44+, cKithigh; DN3 = CD44-, cKitint-low, CD25+; DN4 = CD44-, CD25-.
(G) Absolute numbers of DN1, DN2, DN3 and DN4 cells from control and DCKO mice as described in (F).
(H) Total number of thymocytes from control or DCKO mice.
(I) Percentage DP cells derived from bone marrow progenitors of control and DCKO mice after 20 days of co-culture on OP or OP9-DL4 cells. Data depicted as mean + SD.
Lin panel = CD4, CD8, CD11b, TCRγδ, NK1.1, B220 and GR1. Horizontal lines indicate mean and statistical significances were calculated by 2way ANOVA with Sidak’s multiple comparisons test (D, F, G, I) or unpaired T test (H) (*p < 0.05, **p < 0.01, ***p < 0.001). Data are from several (D), 4 (F, G), more than 10 (H) and 2 pooled (I) independent experiments with numbers of mice indicated in the figure.
BrdU analysis
For cell cycle analysis, mice were injected intraperitoneally (i.p.) with 1mg BrdU (BD Biosciences) diluted in sterile PBS (200µl of a 5mg/ml solution). After 2.5 hrs, thymocytes were surface stained then fixed using BD Cytofix/Cytoperm™. Cells were treated with 30µg DNase I (provided with the BD BrdU kit) for 1 hour at 37°C then washed and stained intracellularly for BrdU and icTCRβ for 1 hour at RT. Cells were resuspended in BD perm/wash containing 1µg/ml DAPI.
OP9, OP9-DL1 and OP-DL4 culture
10,000 sorted DN3a (lineage-negative, CD44-, CD25+, CD98low) or DN3b (lineage-negative, CD44-, CD25intermediate, CD98+) thymocytes from GFPZFP36L1; CD2cre mice were seeded onto OP9-DL1 in 96wells, supplemented with 1ng/ml IL-7 and differentiation into CD4/CD8 double-positive cells was followed up to 3 days. Lineage markers were: CD11b, CD4, CD8, CD19, TCRγδ, GR1, Ter119. Cell sorting was done using a FACS Aria.
Co-culture of bone-marrow derived hematopoietic stem cells (HSC) was performed as in (28). Briefly, Sca1-enriched HSC were obtained by magnetic cell sorting of lineage-negative cells and Sca1-enrichment with kits from Miltenyi. 40,000-75,000 HSCs were seeded per 6well onto OP9-DL4 or OP9 cells with 1ng/ml IL-7 and 5ng/ml Flt3L. Cells were passaged on day 7, 12 and 15-16 with Flt3L and in some cases IL-7 being withdrawn from day 12 to allow differentiation. There was no difference in the performance between OP9-DL1 or OP9-DL4 cells in our experiments.
IL-7 response and TCR signaling
For measuring IL-7 response, 10 mio thymocytes were rested for 2hrs at 37°C, then stimulated with 25ng/ml IL-7 for 5, 30, 60 and 120 min and immediately fixed with pre-warmed BD Lyse/Fix Buffer. For assessing TCR signaling, cells were rested as above and stimulated for 2, 5 and 10 min with 50ng/ml PdBU. After fixation, cells were permeabilized and stained for phosphoproteins as described in section Flowcytometry.
RNAseq
RNA was isolated from sorted Zfp36l1fl/fl; Zfp36l2fl/fl DN3a (Lineage-negative, CD44-, Kitlow, CD25+, CD98low) and DN3b (Lineage-negative, CD44-, Kitlow, CD25intermediate, CD98+) cells as well as Zfp36l1fl/fl; Zfp36l2fl/fl; CD2cre DN3 (Lineage-negative, CD44-, Kitlow, CD25+) cells with the RNeasy Micro Kit (Qiagen). RNAseq libraries were prepared from 20-200ng RNA using the TruSeq Stranded Total RNA and rRNA Removal Mix – Gold from Illumina. Libraries were sequenced by Hiseq 2000 in 100bp single-end reads. Sequencing data quality control was carried out with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were trimmed from adapter sequences using Trim Galore then mapped using Tophat (version 2.0.12) (29) to the genome assembly GRCm38. Multiply mapping reads were discarded. Reads aligning to genes were counted using htseq-count (30). DESeq2 (R/bioconductor package) was used for analysis of differentially expressed genes (31). Data was generated from four biological replicates. Genes regulated >1 log2FoldChange (2-fold up or down regulated) and padj<0.01 were considered as significantly regulated. Results tables from differential expression and pathway analyses can be obtained upon request.
Immunoprecipitation and Western blotting
Lysates (without crosslinking) and beads were prepared as described in the iCLIP procedure. Immunoprecipitation was performed with 500mg protein as described in the iCLIP procedure. Immunoblotting was performed by standard procedures. 50µg protein of crude lysate was loaded and 10% of the unbound fraction or IP, respectively. Antibodies for immunoblotting were: anti-BRF1/2 (Cell Signaling), anti-GFP (clone B2, Santa Cruz), anti-Tubulin (DM1A, Sigma).
Individual-nucleotide resolution Cross-Linking and ImmunoPrecipitation (iCLIP)
iCLIP experiments were preformed similar to previously described (25) and (23). Briefly, cell lysates were prepared from total thymocytes from C57BL/6 or DCKO mice which were irradiated with 300mJ/cm2 UV-ligh, then lysed in lysis buffer (50mM Tris HCL, pH 8.0, 100mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1:200 protease inhibitor cocktail (Sigma), 500U/ul RNasin (NEB)). 2.5mg protein supernatant was used per sample. Rabbit anti-ZFP36L1 antibody (BRF1/2, Cell Signaling) was conjugated to Protein A-Dynabeads (Invitrogen) over night.
Samples were treated with TURBO DNase and RNase I for partial RNA digest, and Zpf36l1/RNA complexes were precipitated using beads-coupled rabbit anti-ZFP36L1 antibody (BRF1/2, Cell Signaling). Washes were performed in high-stringency salt buffers (First iCLIP: 50 mM Tris-HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.2% SDS, 0.5% Sodium deoxycholate; second iCLIP: 50 mM Tris-HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.4% SDS, 0.5% Sodium deoxycholate) and PNK buffer (20 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 0.2% Tween-20). The RNA was then dephosphorylated, ligated to an RNA adaptor (L34: P-AUAGAUCGGAAGAGCGGUUCAG-Puromycin) and radioactively labeled. The RNA complexes were resolved on a denaturing SDS-PAGE and transferred to a nitrocellulose membrane. RNA was extracted and reverse transcribed into cDNA libraries. The cDNA libraries were sequenced by MiSeq as 150bp single-end read or as HiSeq2500 RapidRun 50bp single-end read and mapped to the mm10 mouse genome. iCLIP data was generated from two biological replicates for C57BL/6 and one for DCKO thymocytes. C57BL/6 data was merged during iCOUNT analysis. Unless specified, only binding to 3’UTR was considered. The analysis was performed as described in (25). Highly significant ZFP36L1 binding sites were identified using the iCount pipeline and a FDR was assigned to each crosslink site as previously described (32). We only considered ZFP36L1 crosslink sites with a FDR < 0.05 in our analysis. iCount was also used to determine the kmer content of the cross link sites.
RCLIP primers for C57BL/6 sample 1, 2 and DCKO:
RCLIP A: 5’-phosphate-NNNNGTTAGATCGGAAGAGCGTCGTGGATCCTGAACCGCTC-3′
RCLIP B: 5’-phosphate-NNNNGCCAGATCGGAAGAGCGTCGTGGATCCTGAACCGCTC-3′
RCLIP C: 5’-phosphate-NNNNATCAGATCGGAAGAGCGTCGTGGATCCTGAACCGCTC-3′
Gene set enrichment analysis
Differentially expressed mouse transcripts identified using DESeq2 (padj<0.01) and >1 iCLIP hits in the 3’UTR (FDR<0.05), were analyzed for enriched GO biological processes using Toppfun (33). A false discovery rate cut off of 0.05 was applied. DNA damage checkpoint (GO:0000077) genes were obtained from the GO biological pathways database.
Palbociclip in vivo experiment
Palbociclib (Selleck Chemicals) was dissolved in 50mM sodium lactate (Sigma) and administered once to mice by oral gavage at a dose of 150mg/kg. Mice were culled and analyzed 1 day after palbociclib administration.
Statistics
Except for data generated by high throughput sequencing, where statistical tests are described elsewhere, all statistical analyses were made using prism software (graphpad). The test used and sample sizes are indicated in the figure legends.
GEO Series accession number
GSE79179 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79179)
Results
Thymocytes deficient in Zfp36l1 and Zfp36l2 are committed to the T cell lineage but fail β-selection
To investigate how the loss of Zfp36l1 and Zfp36l2 leads to a bypass of β-selection we first performed detailed flow cytometry analysis of thymocytes from Zfp36l1fl/fl; Zfp36l2fl/fl; CD2Cre double conditional knockout (DCKO) and Cre-negative Zfp36l1fl/fl; Zfp36l2fl/fl (control) mice. CD44 expression within the CD4- and CD8-double negative (DN) population of the DCKO mice was increased (Figure 1A) as well as surface expression of Sca1 (Figure 1B and Figure S1A), a marker of the more immature developmental stages of thymocytes. Innate lymphocyte (ILC2s) are characterized by expression of Sca1, CD44 and CD25 (34–36) and have been shown to originate from hematopoietic stem cells in culture (37). We therefore investigated the possibility that ILC2s are aberrantly populating the thymus of DCKO mice. Staining for ST2positive, Sca1hghi ILC2s showed that their percentage and number was significantly increased in the thymus of DCKO mice compared to control mice (Figure S1B, C). However, the absolute number of these cells was less than 10,000. An overlay of the ILC2 population with DN subsets based on CD44 and CD25 surface expression showed that ILC2s did not contribute to the aberrant CD44high cells seen in DCKO mice, as they were CD44high and CD25intermediate (Figure S1D, E). We next determined whether these cells were committed to the T cell lineage. Commitment is marked by low levels of Kit and the expression of intracellular CD3ε (icCD3ε (38)). Within DCKO mice, the abnormal CD44high population expresses icCD3ε (Figure 1C) and therefore represents DN3-like committed thymocytes. They exhibit higher expression of icCD3ε than control DN3-4 cells (Figure S1F), but fail to express icTCRβ. During development, DCKO thymocytes are not efficiently selected for icTCRβ expression until they become mature T cells (Figure 1D) (24). As a consequence the number of DP cells is decreased in the DCKO (Figure 1E and Figure S3F). The percentage of DN1, DN2 or DN3 populations is not changed in the DCKO compared to control, however the percentage of DN4 cells is reduced (Figure 1F). Since total thymocyte numbers are reduced, the numbers of DN3 and DN4 cells are decreased in the DCKO thymus (Figure 1G, H). This data demonstrates that DCKO thymocytes are committed to the T cell lineage, but development is retarded from the DN3 stage onwards. To assess the contribution of the previously published Zfp36l1/l2 mRNA target Notch1 (24) to the phenotype, we assessed differentiation of control and DCKO bone marrow (BM)-derived hematopoietic stem cells (HSCs) in a co-culture system either with Notch ligand (OP9-DL4) or without (OP9) (27). DCKO HSCs differentiated faster into DP cells on OP9-DL4 feeder cells compared to control HSCs and were also able to differentiate into DP cells in the absence of Notch ligand (Figure 1I). We therefore employed high-throughput approaches to identify additional pathways affected by loss of Zfp36l1/l2.
iCLIP and RNAseq identify molecular pathways in thymocytes regulated by Zfp36l1 and Zfp36l2
Since Zfp36l1 and Zfp36l2 promote RNA decay, biologically relevant targets of the RBPs were expected to be more abundant in DCKO cells. We therefore performed RNAseq to measure mRNA abundance. As pre- and post-selected DCKO DN3 cells cannot be subdivided on the basis of CD98 or icTCRβ (Figure 1D and S1G), we compared DCKO DN3 cells with sorted DN3a and DN3b cells from control animals (Figure S1H). Cluster analysis showed that the DN3 cells from DCKO mice (DCKO-DN3) were more closely related to wild-type DN3b (Ctrl-DN3b) than to DN3a cells (Ctrl-DN3a), despite their lack of icTCRβ (Figure 2A). 3,044 genes in the DCKO were differentially expressed compared to control DN3a cells and 519 genes compared to control DN3b cells (Figure 2B, Table S1 tab1 and tab2). To study in more detail how similar DCKO DN3 cells are to control cells, the DN3a and DN3b signature genes (genes differentially regulated between wild-type DN3a and DN3b cells, Figure 2B, Table S1 tab3) were plotted against the genes that were differentially regulated in the DCKO DN3 cells compared to either the control DN3a (Figure 2C, Table S1 tab4) or DN3b cells (Figure 2D, Table S1 tab5). DCKO DN3 cells showed strong positive correlation with the signature of DN3b when compared to DN3a cells (R=0.8) and none when compared to DN3b cells (R=-0.094). Strikingly, many DN3b genes missing in the DCKO were Trbv genes (approx. 30%), consistent with absence of icTCRβ protein expression. Notably, we found that the Trbv genes were expressed at considerably lower levels compared to control DN3a cells. These findings suggest that DCKO cells have progressed in development without the formation of the pre-TCR.
Figure 2. Zfp36l1/l2 control mRNA targets which are involved in the cell cycle.
(A) Cluster analysis of RNAseq samples showing the distance between sorted DN3a (Ctrl-DN3a) or DN3b (Ctrl-DN3b) cells from control mice (Zfp36l1/l2fl/fl) and DN3 cells from Zfp36l1/l2fl/fl; CD2Cre (DCKO) mice (DCKO-DN3) (for gating strategy see Figure S1H).
(B) MA plots of differentially expressed genes in RNAseq of DCKO DN3 vs Ctrl-DN3a (top) or DN3b (middle), and Ctrl-DN3b vs Ctrl-DN3b (bottom) cells.
(C-D) Log2Fold Change values of DCKO samples compared to the DN3a (C) or DN3b (D) control samples are plotted against the comparison of control DN3b and DN3a samples. Blue line indicates correlation of the comparisons.
(E) Density enrichment of iCLIP hits, normalized to gene feature length. Y axis represents percentage of enrichment.
(F-G) Log2 fold change of RNAseq mRNA expression from DCKO cells compared to control DN3a (F) or DN3b (G) and split into mRNA bound or not bound based on iCLIP results.
(H) Venn diagram depicting the number of differentially regulated pathways in gene set enrichment analyses of genes bound in the iCLIP and differentially regulated (log2 fold change) in RNAseq of DCKO cells compared to control DN3a or DN3b cells.
RNAseq data is from 4 biological replicates each and iCLIP data from 2 biological replicates.
To identify those RNAs directly bound by Zfp36l1 and Zfp36l2 we applied high-resolution iCLIP to mouse thymocytes (25). To this end, we used an antibody (BRF1/2), which recognizes mouse Zfp36l1 strongly and Zfp36l2 weakly on Western blots, but is selective for mouse Zfp36l1 when used for immunoprecipitation (Figure S2A). To further validate antibody specificity, we compared immunoprecipitations from C57BL/6 and DCKO thymocytes after UV crosslinking. After immunoprecipitation and partial RNA digestion by RNase I, the samples were labelled with 32P-γ-ATP. As expected, the C57BL/6 samples showed a greater signal on the autoradiograph than the DCKO (Figure S2B). Although there was little material in the DCKO sample, a DCKO library was prepared and sequenced for quality control purposes. BRF1/2 immunoprecipitations from C57BL/6 thymus showed that crosslinks are present to a high degree in introns and intergenic regions. This is consistent between data from two individual and merged biological replicates (Figure S2C). After normalizing to segment lengths of each genomic feature, density enrichment was greatest for binding to 3’untranslated regions (3’UTRs) as well as non-coding RNAs (ncRNAs) (Figure 2E and S2D). To verify specific binding, we sought to identify the presence of ARE binding motifs in 3’UTRs and ncRNAs. Bound sequences from the two C57BL/6 replicates were plotted against each other and showed that AREs were highly enriched above the mean in bound 3’UTRs but not in bound ncRNA (Figure S2E, F). Moreover, this enrichment of ARE motifs was only present in gene features from C57BL/6, and not in DCKO samples (Figure S2G), indicating the specificity of the interaction between Zfp36l1 and AREs within 3’UTRs.
Combining iCLIP and RNAseq datasets is a powerful approach to identify both the direct targets of the RBPs and the effects on the transcriptome caused by loss of Zfp36l1 and Zfp36l2. We therefore compared RNAseq data from DCKO DN3 to either DN3a or DN3b control samples, divided on whether the RNA was bound or unbound in the iCLIP (Figure 2F, G). There was a positive correlation between mRNA with increased expression in DCKO DN3 cells and bound iCLIP-identified targets. Thus in the absence of Zfp36l1 and Zfp36l2 direct targets show increased expression of their RNA.
Gene set enrichment analysis (GSEA) of transcripts bound by Zfp36l1 in the iCLIP and differentially up- or down-regulated compared to DN3a or DN3b cells identified 364 pathways that were changed in the DCKO DN3 cells compared to control DN3a, and 26 pathways that were changed compared to DN3b (Figure 2H). 20 pathways are distinct from either comparison and include the cell cycle and RNA metabolic processes (Table S2). DCKO DN3 differ from control DN3b cells in VDJ recombination, dephosphorylation and T cell activation pathways (Table S2). Together the data show that DCKO DN3 cell phenocopy wild-type DN3b cells but differ in expression of genes that are part of the cell cycle and VDJ recombination pathways.
Increased cell cycling in early DN thymocyte subsets from DCKO mice
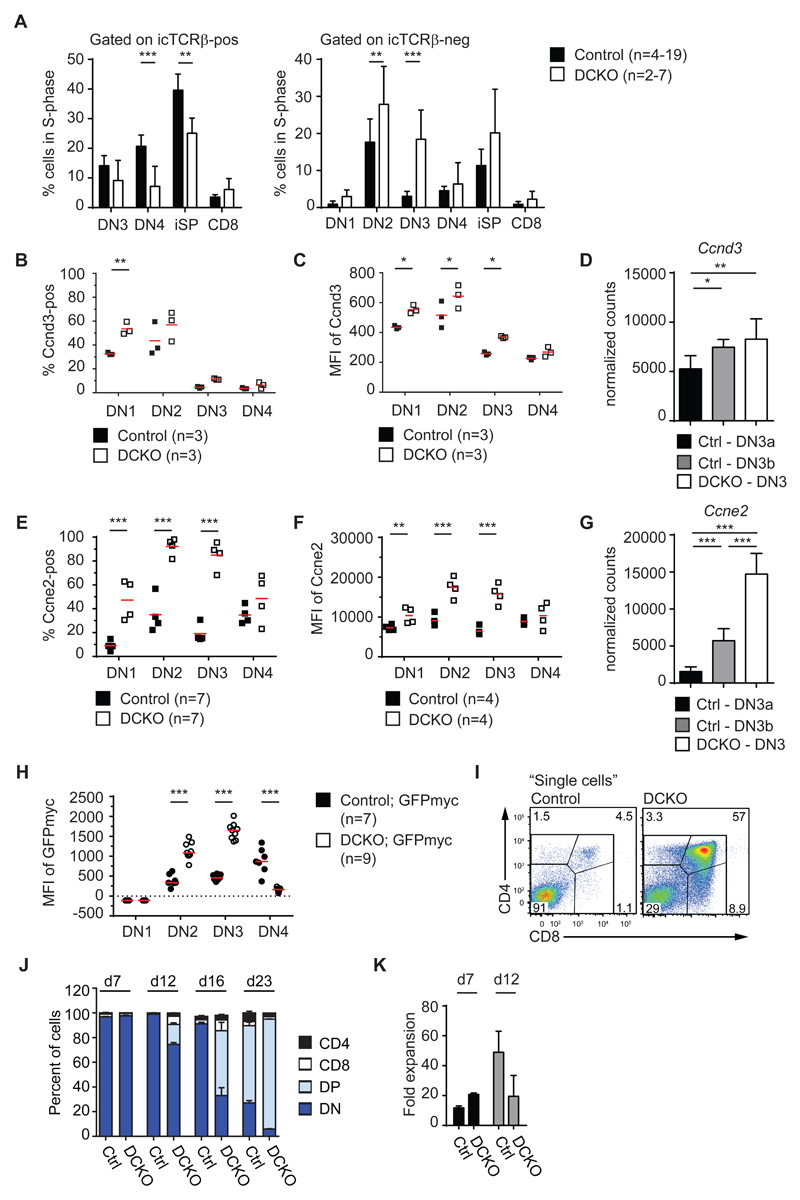
The strong enrichment for cell cycle mRNA targets in the GSEA of the DCKO DN3 transcriptome could reflect a direct role of Zfp36l1 and ZFP36l2 in cell cycle control. Therefore, we assessed the cycling properties of DN thymocytes by measuring 2-Bromodeoxyuridine (BrdU) incorporation into the DNA of dividing cells 2.5 hours after administration. DCKO DN4 and immature CD8 single positive thymocytes (iSP) that are icTCRβpositive showed a decrease in the percentage of cells in S-phase (Figure 3A, left). By contrast, icTCRβnegative DN2 and DN3 cells from DCKO mice show a significantly increased percentage of cells in S-phase (Figure 3A, right). Strikingly, Zfp36l1 and Zfp36l2 deficient DN3 cells exhibit a more than five-fold increase in cycling cells over control DN3 thymocytes, which undergo cell cycle arrest to enable V-DJ recombination (2). Flow cytometric analysis showed the proportion of DN1, DN2 and DN3 cells from DCKO thymi positive for the cell cycle regulators Cyclin D3 (Ccnd3) and Cyclin E2 (Ccne2) were elevated. Moreover, these cells contained more Ccnd3 and Ccne2 mRNA and protein (Figure 3B-G). Representative histograms are shown in Figure S2H, I. Myc, another key component promoting cell cycle progression, is also increased at the protein level in DN2 and DN3 cells as assessed by a GFPmyc knockin allele (27) crossed to the DCKO (Figure 3H).
Figure 3. The cell cycle is increased in Zfp36l1/l2fl/fl; CD2Cre (DCKO) mice.
(A) Percentage of BrdU+ cells in S-phase (gated as in Figure 1D with DN1 = Lin-, CD44+, CD25-) and split into icTCRβ+ and icTCRβ- cells. Data is depicted as mean + SD and collated from 4-19 control (Zfp36l1/2fl/fl) and 2-7 DCKO mice.
(B) Percentage of Cyclin D3 (Ccnd3)-positive DN1-4 cells from control and DCKO mice.
(C) Median fluorescence intensity (MFI) of Ccnd3 as depicted in (B).
(D) Normalized counts of Ccnd3 mRNA in control DN3a (Ctrl-DN3a) or DN3b (Ctrl-DN3b) and DN3 cells from DCKO mice (DCKO-DN3). RNAseq data was obtained from sorted thymocytes as described in Figure S1H. Data depicted as mean + SD of 4 biological replicates with statistical significances calculated by differential expression analysis (*p < 0.05, **p < 0.01).
(E) Percentage of Cyclin E2 (Ccne2)-positive DN1-4 cells from control and DCKO mice.
(F) MFI of Ccne2 as depicted in (E).
(G) Normalized counts of Ccne2 mRNA as described in (D), (***p < 0.001).
(H) MFI of GFPmyc in DN subsets from control; GFPmyc and DCKO; GFPmyc mice which were crossed with GFPmyc reporter mice. Data are from 2 pooled experiments.
(I) Representative FACS plots showing CD4/CD8 expression of bone-marrow derived hematopoietic stem cells (BM-HSCs) from control and DCKO mice after 16 days of co-culture on OP9-DL4 cells.
(J) Percentage of BM-HSCs from control (Ctrl) and DCKO mice expressing CD4, CD8 or CD4/CD8 (DP) markers or no CD4/CD8 (DN); after 7, 12, 16 and 23 days of co-culture on OP9-DL4 cells.
(K) Fold expansion of bone-marrow derived hematopoietic stem cells (BM-HSCs) from control and DCKO mice after 7 and 12 days of co-culture on OP9-DL4 cells in the presence of 1ng/ml IL-7.
Data is representative for 2 (B, C, E, K), 3 (F) or 4 (I, J) independent experiments with 6 (B, E), 7 (C), 11 (F) and 10-11 (I, J) mice per genotype. Horizontal lines indicate mean and statistical significances were calculated by 2way ANOVA with Holm-Sidak's multiple comparisons test (*p < 0.05, **p < 0.01, ***p < 0.001).
Consistent with the DN3a to DN3b transition being linked to mitogenesis, control thymocytes show increased expression of both Ccnd3 and Ccne2 mRNA at the DN3b stage when compared to DN3a (Figure 3D, G). However, DN3 cells from DCKO mice show an even greater elevation of Ccne2 mRNA (Figure 3G). Both Ccnd3 and Ccne2 mRNA were bound by Zfp36l1 in the iCLIP indicating that they are direct targets in thymocytes (Figure S2J, K). Binding of Zfp36l1/l2 to an ARE in Cyclin E2 mRNA was further validated by luciferase assays (23). Myc mRNA was not differentially expressed in DCKO thymocytes (Table S1), but was bound in the iCLIP, which suggests that Myc may be controlled by Zfp36l1/l2 as well as the cell cycle. To corroborate these results, we investigated if the increased cell cycle properties of DCKO cells facilitate T cell differentiation. As sorted DN3 cells from DCKO mice rapidly died in culture (data not shown), we seeded BM-HSCs from control and DCKO mice onto OP9-DL4 cells (28). DCKO HSCs differentiated into DPs more rapidly than control cells (Figure 3I, J). DCKO cells consistently expanded more until the first cell culture passage (day 7), but expanded similarly or less than control cells with following passages (day 12-onwards; Figure 3K). This is consistent with the hypothesis that cell division is linked to differentiation in wild-type cells (39).
The cell cycle is inhibited in GFPZFP36L1 transgenic mice
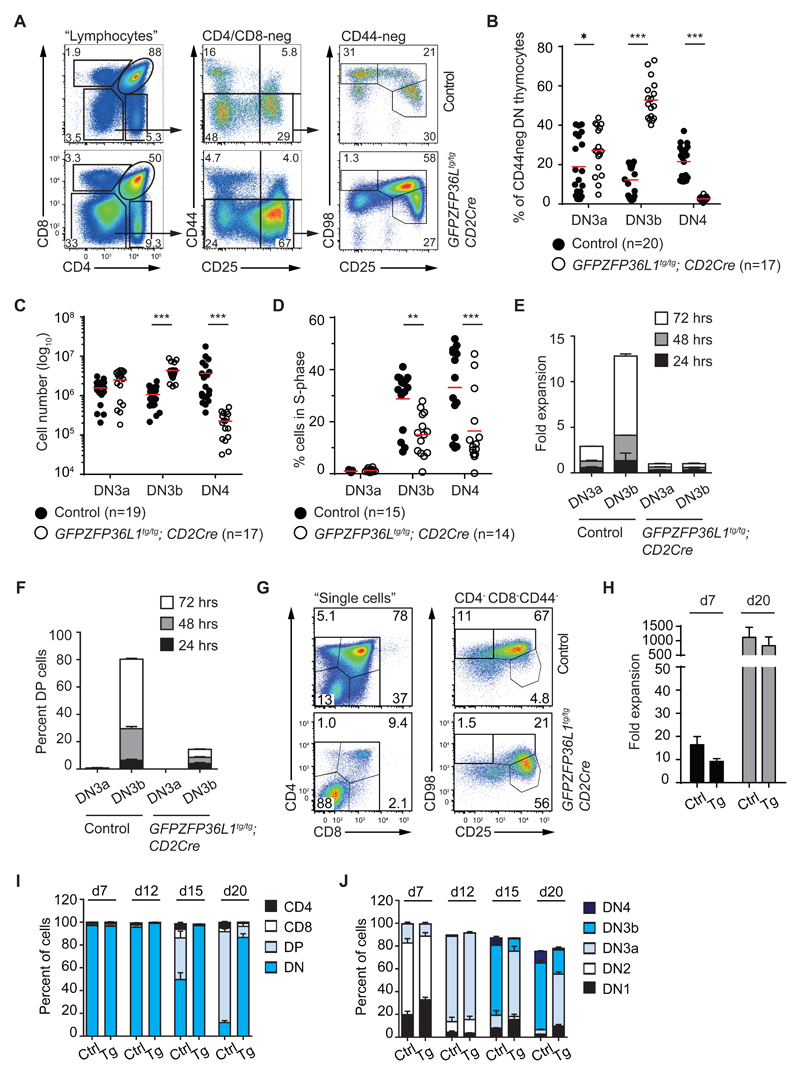
To examine if overexpression of ZFP36L1 can oppose the DCKO phenotype, we bred mice in which the endogenous Rosa26 locus had been engineered to express a cDNA encoding a GFPZFP36L1 fusion protein in a Cre-dependent manner with CD2Cre mice. The expression of the fusion protein and endogenous Zfp36l1 was then assessed by Western blotting (Figure S3A). GFPZFP36L1 was of the predicted molecular size and mice homozygous for the modified Rosa26 locus had higher fusion protein abundance than did heterozygotes. Endogenous Zfp36l1 protein appeared to be expressed less in transgenic animals than in controls, which we interpret to be due to suppression of the endogenous Zfp36l1 transcript by the fusion protein through binding to its ARE (40). Flow cytometry for GFPZFP36L1 indicated that this protein was expressed in each of the DN populations, but greatest expression was in DN3 thymocytes (Figure S3B, C). To establish whether the transgene was functional in DN thymocytes we next measured the surface expression of Notch-1 on DN thymocytes from GFPZFP36L1tg/tg; CD2Cre mice as Notch1 mRNA was previously found to be targeted by Zfp36l1 and Zfp36l2 (24). Reduced surface Notch-1 expression was found on DN2, DN3 and DN4 transgene-expressing thymocytes (Figure S3D), suggesting the GFPZFP36L1 fusion protein was functional. Furthermore, introduction of the GFPZFP36L1 transgene into the DCKO mice substantially restored the cellularity of the DP compartment (Figure S3E, F). Most notably, the percentage of icTCRβ expressing cells in the DN3 and DN4 subset was restored to wild-type levels in GFPZFP36L1/DCKO mice, showing that the GFPZFP36L1 fusion protein can functionally complement loss of endogenous Zfp36l1 (Figure S3G).
The GFPZFP36L1 fusion protein affected thymic development as GFPZFP36L1tg/tg; CD2Cre mice had reduced numbers of total thymocytes (Figure S3H), but a significantly increased percentage and number of DN3b cells (Figure 4A-C, Figure S3I). However, the proportion of these DN3b and of DN4 cells in S-phase was significantly less in GFPZFP36L1tg/tg; CD2Cre mice than in Cre-negative controls (Figure 4D) indicating the fusion protein inhibits the proliferation of developing thymocytes. This was consistent with slower expansion (Figure 4E) and differentiation (Figure 4F) of sorted DN3a and DN3b cells from GFPZFP36L1tg/tg; CD2Cre mice in vitro. Expansion was not significantly changed, but the appearance of DPs was delayed following co-culture of transgenic BM-derived HSCs on OP9-DL4 cells (Figure 4G-I). In these cultures GFPZFP36L1 expressing cells appeared mostly at the DN3a stage (Figure 4G, J). Taken together, these data suggest that the GFPZFP36L1 limits proliferation and differentiation at the β-selection checkpoint.
Figure 4. The cell cycle is inhibited in GFPZFP36L1 transgenic mice.
(A) Representative FACS plots showing lymphocyte subsets defined by CD4/CD8 (left panel), CD4/CD8-neg DN cells defined by CD44/CD25 (middle panel), and CD44-neg DN3a (CD25high, CD98low), DN3b (CD25int, CD98high) and DN4 (CD25-, CD98high) cells from Cre-negative control and GFPZFP36L1tg/tg; CD2Cre (Tg) mice.
(B) Proportions of DN3a, DN3b and DN4 cells (gated as in (A)) from control and GFPZFP36L1tg/tg; CD2Cre mice.
(C) Absolute cell numbers of DN3a, DN3b and DN4 cells as described in (B).
(D) Proportion of cells incorporating BrdU in S-phase within DN3a, DN3b and DN4 populations, defined as in (A).
(E) Fold expansion of sorted DN3a or DN3b cells from control and GFPZFP36L1tg/tg; CD2Cre mice after 24, 48 or 72 hrs of OP9-DL1 co-culture.
(F) Percentage of sorted DN3a or DN3b cells from control and GFPZFP36L1tg/tg; CD2Cre mice differentiating into CD4/CD8-pos DP cells after 24, 48 or 72 hrs of OP9-DL1 co-culture.
(G) Fold expansion of bone-marrow derived hematopoietic stem cells (BM-HSCs) from control and GFPZFP36L1tg/tg; CD2Cre (Tg) mice after 7 and 20 days of co-culture on OP9-DL4 cells in the presence of 1ng/ml IL-7.
(H) Representative FACS plots showing CD4/CD8 (left panel) and CD98/CD25 (right panel) expression of BM-HSCs from control and GFPZFP36L1tg/tg; CD2Cre mice after 20 days of co-culture on OP9-DL4 cells.
(I) Percentage of BM-HSCs from control (Ctrl) and GFPZFP36L1tg/tg; CD2Cre (Tg) mice expressing CD4, CD8, CD4/CD8 (DP) or no CD4/CD8 (DN) markers after 7, 12, 15 and 20 days of co-culture on OP9-DL4 cells.
(J) Percentage of BM-HSCs as in (H), in the developmental stages DN1, DN2, DN3a, DN3b or DN4.
Data are collated from 4 (B, C), and 3 (D) independent experiments. Horizontal lines indicate mean with statistical significances calculated by 2way ANOVA with Holm-Sidak's multiple comparisons test (*p < 0.05, **p < 0.01, ***p < 0.001) and number of mice indicated in the figure. (E-I) Data is representative for 2 independent experiments with 4 (E, F) and 6 (I, J) biological replicates per genotype. (E, F) 2-3 technical replicates are shown per genotype.
Pharmacological inhibition of cell cycle progression partially rescues icTCRβ expression
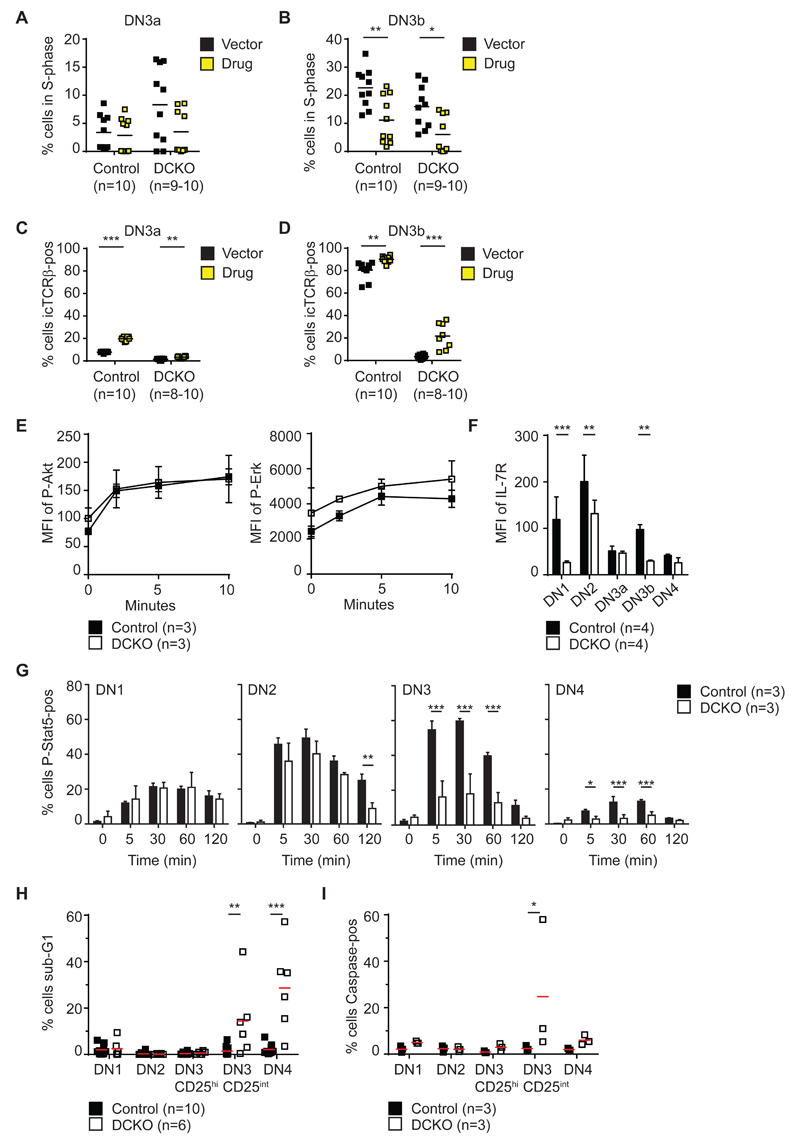
As cell cycle and VDJ recombination are linked in developing thymocytes (41) we tested whether icTCRβ expression could be increased in DCKO mice by pharmacological inhibition of the cell cycle in vivo. To this end, G1 to S progression was blocked with Palbociclib, a specific Cdk4/6 inhibitor by administering the drug to DCKO and control mice for 24 hours. Palbociclib treatment reduced the percentage of cells in S-phase in DCKO DN3a slightly (Figure 5A) but significantly diminished S-phase-positive DN3b cells from both DCKO and control animals (Figure 5B). Treatment also increased icTCRβ expression in DN3a cells and DN3b cells in both groups (Figure 5C, D). However, this increase was not to wild-type DN3b levels of icTCRβ, thus the rescue was incomplete. This suggests that inhibition of the cell cycle alone is insufficient to fully restore the icTCRβ positive DN3b population in DCKO mice.
Figure 5. Lack of icTCRβ signaling correlates with increased apoptosis and is only partially rescued by pharmacological inhibition of cell cycle progression.
(A) Percentage of DN3a cells in S-phase (as gated on Dapi profile) from control (Zfp36l1/2fl/fl) and Zfp36l1/2fl/fl; CD2Cre (DCKO) mice, after 1-day of treatment with vector or palbociclib (drug).
(B) Percentage of DN3b cells in S-phase as described in (A).
(C) Percentage of DN3a cells positive for icTCRβ as described in (A).
(D) Percentage of DN3b cells positive for icTCRβ as described in (A).
(E) Median fluorescence intensity (MFI) of P-Akt and P-Erk in DN3 cells (CD44-, CD25+) from control (Zfp36l1/2fl/fl) and Zfp36l1/2fl/fl; CD2Cre (DCKO) mice after 0, 2, 5 and 10 min of PdBU stimulation. Depicted is the mean +/- SD.
(F) MFI of IL-7R on DN subsets from control and DCKO mice. Depicted is the mean + SD.
(G) Percentage P-Stat5-positive DN1-4 cells from control and DCKO mice after 0, 5, 30, 60 and 120 min of stimulation with IL-7. Depicted is the mean + SD.
(H) Percentage of cells in subG1 cell cycle phase of DN subsets from control and DCKO mice. Horizontal line represents the mean.
(I) Percentage of Caspase-pos cells in DN subsets as gated in (D) from control and DCKO mice. Horizontal line represents the mean.
Data are representative for 3 (E, P-Akt; G), 2 (E, P-Erk), 4 (F) and 1 (I) independent experiments with 6-9 (E), 12-14 (F), 9 (G) and 3 (I) biological replicates per genotype. (A-D, H) Data are collated from 2 independent experiments with 8-10 (A-D) and 6-10 (H) mice as indicated in the figure. The horizontal line represents the mean with statistical significances calculated by 2way ANOVA with Sidak's multiple comparisons test (A, B, C, D, F, G) or with Holm-Sidak’s multiple comparisons test (H, I) (*p < 0.05, **p < 0.01, ***p < 0.001).
Lack of pre-TCR signaling in thymocytes of DCKO mice correlates with increased apoptosis
Our observations are consistent with previously published work demonstrating a crucial but insufficient role of the cell cycle in β-selection; overexpression of several genes promoting cell cycle progression could not override β-selection in the absence of pre-TCR signaling (39). These were Ccnd3, Ccne1, Cdk2 and Cdk6, of which all but Ccne1 were detected as Zfp36l1 targets in our iCLIP. We therefore hypothesized that signaling events downstream of the pre-TCR could be activated by deregulation of direct Zfp36l1 or Zfp36l2 mRNA targets or through indirect feedback mechanisms. To assess pre-TCR activity in DN subsets, we measured levels of Akt and Erk phosphorylation by phospho-flow cytometry under basal conditions as well as after PdBU stimulation of thymocytes from control or DCKO mice to assess feedback mechanisms. There was no evidence of elevated Akt and Erk activation (Figure 5E).
IL-7 signaling is critical for thymocyte development. After transition from DN3a to DN3b, wild-type cells transiently up-regulate the IL-7 receptor (IL-7R) to receive survival signals (42). However, this up-regulation could not be detected in the DN3b-like population of DCKO mice (Figure 5F), likely reflecting the absence of the pre-TCR signal in the majority of these cells. In accordance with this finding, IL-7–dependent STAT-5 phosphorylation is diminished in DN3 and DN4 cells from DCKO mice (Figure 5G). Consistent with a lack of survival signal, we found increased apoptosis amongst late DN3 (CD25intermediate) and DN4 (CD25low) DCKO cells, as determined by the percentage of sub-G1 cells (Figure 5H). Increased total Caspase activity was also evident in late DN3 cells from DCKO animals (Figure 5I). We conclude that DCKO thymocytes die in part because of a failure to induce pre-TCR and IL-7R dependent survival signals.
Zfp36l1 and Zfp36l2 limit the DNA damage response in thymocytes
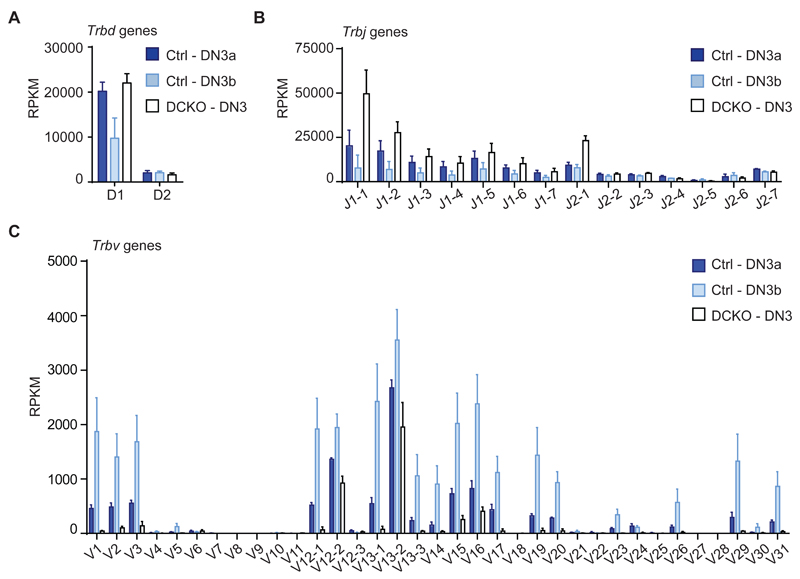
As described above, one of the most striking observations in the RNAseq data was the complete lack of Trbv transcripts in DCKO DN3 cells. We visualized D, J and V gene expression by plotting their RPKM values derived by RNAseq of control DN3a, DN3b and DCKO DN3 cells (Figure 6A-C). While Trbj gene transcripts were readily detectable in DCKO DN3 cells, there was hardly any detectable Trbv transcript expression (Figure 6B, C), suggesting a deficiency in V to DJ recombination.
Figure 6. Lack of Trbv gene signature in DCKO mice.
(A) RPKM values of Trbd genes in control (Zfp36l1/2fl/fl) DN3a (Ctrl-DN3a), DN3b (Ctrl-DN3b) and DN3 cells from Zfp36l1/2fl/fl; CD2Cre mice (DCKO-DN3). RNAseq data was obtained from sorted thymocytes as described in Figure 2A. Data is depicted as mean + SD of 4 biological replicates.
(B) RPKM values of Trbj genes as described in (A).
(C) RPKM values of Trbv genes as described in (A).
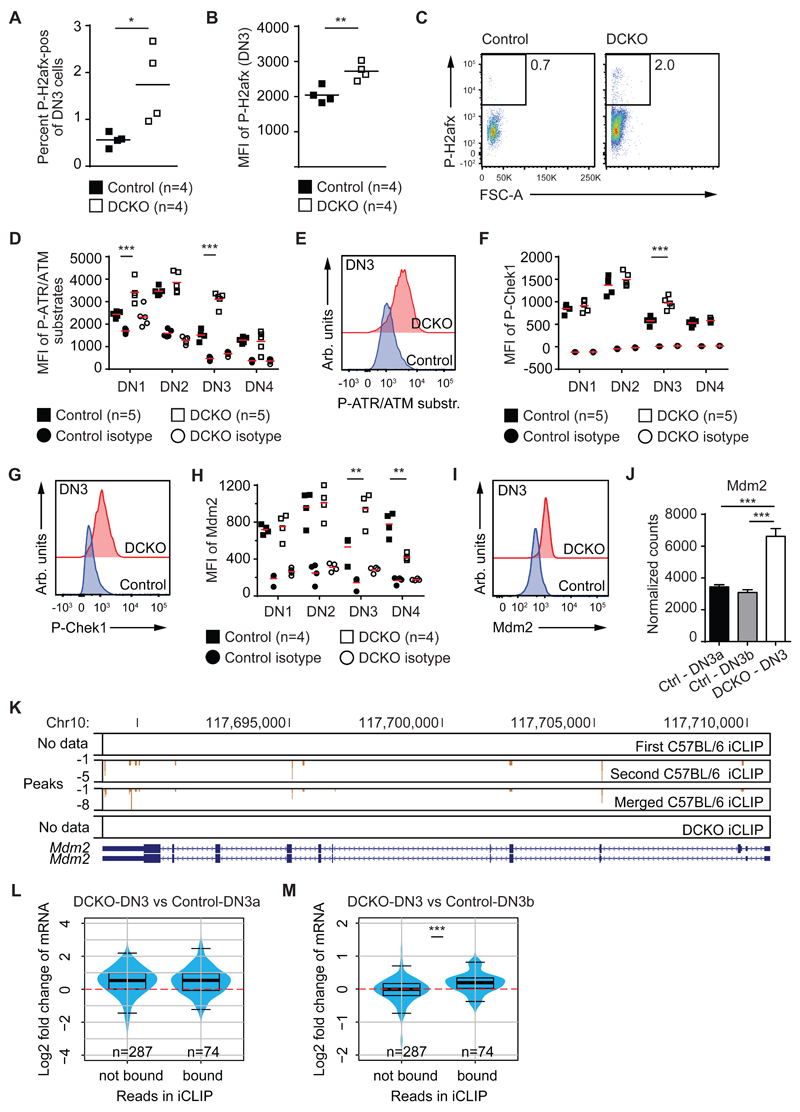
Several groups have demonstrated that the block in VDJ recombination seen in scid mice could be overcome by DNA damaging agents, which led to the development of thymic lymphomas (43–45). We therefore assessed the degree of DNA damage related signaling in DN cells from DCKO mice by staining for phosphorylated H2afx, a marker typically associated with DNA double-strand breaks (46). The percentage of P-H2afxpositive DN3 cells from DCKO mice was significantly increased compared to controls, as was the MFI of P-H2afx staining (Figure 7A-C). Furthermore, we found increased staining with an antibody detecting phosphorylated Atr/Atm substrates in DN1, and notably in DN3 cells from knockout mice (Figure 7D, E). The MFIs of P-Chek1 and Mdm2, which are Atr/Atm substrates, were also significantly increased in DCKO DN3 cells (Figure 7F-I). Furthermore, we found Mdm2 mRNA levels increased in DCKO DN3 cells (Figure 7J) and the 3’UTR of Mdm2 was bound by Zfp36l1 in the iCLIP (Figure 7K), suggesting Mdm2 is a potential direct target of Zfp36l1/l2.
Figure 7. Zfp36l1 and Zfp36l2 limit the DNA damage response in thymocytes.
(A) Percentage of P-H2afx-positive DN3 cells of control and DCKO mice. Horizontal line represents the mean.
(B) Median fluorescence intensity (MFI) of P-H2afx in DN3 cells as in (A).
(C) Representative FACS plots of P-H2afx expression in DN3 cells as in (A).
(D) MFI of P-ATR/ATM substrates in DN subsets from control and DCKO mice with respective isotype staining. Horizontal line represents the mean, significance calculated without including isotype staining.
(E) Representative histogram overlay of P-ATR/ATM substrates in DN3 cells from control and DCKO mice.
(F) MFI of P-Chk1 in DN subsets as in (D).
(G) Representative histogram overlay of P-Chek1 in DN3 cells from control and DCKO mice.
(H) MFI of Mdm2 in DN subsets as in (D).
(I) Representative histogram overlay of Mdm2 in DN3 cells from control and DCKO mice.
(J) Normalized counts of Mdm2 mRNA in control DN3a (Ctrl-DN3a) or DN3b (Ctrl-DN3b) and DN3 cells from DCKO mice (DCKO-DN3). RNAseq data was obtained from sorted thymocytes as described in Figure S1H. Data depicted as mean + SD of 4 biological replicates with statistical significances calculated by differential expression analysis (***p < 0.001).
(K) Genome tracks of Zfp36l1 binding sites in the Mdm2 gene with binding peaks of the individual and merged C57BL/6, and the DCKO iCLIP. Chromosomal location is indicated on the top and genome features of Mdm2 isoforms are shown on the bottom. The blue boxes depict exons connected by a line of arrows which are introns. Narrow boxes indicated 5’ or 3’ untranslated regions.
(L-M) Log2 fold change of mRNA expression of DNA damage genes from DCKO cells compared to control DN3a (L) or DN3b (M) and split into mRNA bound or not bound based on iCLIP data.
Data are representative of 5 (A-C), 1 (D-I) and 2 (L, M) independent experiments with 16 (A-C), 5 (D-G), 4 (H, I) and 1 (L, M) biological replicates per genotype. Statistical significances were calculated by unpaired T test (A, B), 2way ANOVA with Sidak's multiple comparisons test (D, F, H) or Wilcoxon test (L, M) (*p < 0.05, **p < 0.01, ***p < 0.001).
To confirm whether Zfp36l1 and Zfp36l2 may be directly regulating the DDR, we separated a list of DNA damage related genes on the basis of whether their transcripts were “bound” or “not bound” in the iCLIP and checked their expression in the RNAseq dataset. As expected, the expression of DNA damage related transcripts was greatest in DCKO DN3 when compared to control DN3a cells, irrespective of whether or not the transcript was bound in the iCLIP (Figure 7H). A comparison of DCKO DN3 cells with control DN3b cells revealed that the DNA damage related transcripts bound in the iCLIP were significantly up-regulated as opposed to unbound transcripts, which were unchanged (Figure 7I). Given the similarity of DCKO DN3 cells to control DN3b cells (Figure 2A), it is notable that only the DNA damage related transcripts bound in the iCLIP were increased in DCKO mice.
Taken together, these data demonstrate that Zfp36l1 and Zfp36l2 directly control cell cycle transcripts and play an important role in controlling DNA damage related transcripts connected to VDJ recombination to ensure the correct timing and progression of β-selection.
Discussion
In this study we demonstrated a novel post-transcriptional function of Zfp36l1 and Zfp36l2 in limiting the cell cycle and DNA damage response signaling in developing thymocytes at the β-selection checkpoint. We propose that these RBPs limit several signaling pathways, of which the cell cycle and DNA damage pathway are driving forces for ensuring the proliferation and differentiation associated with β-selection. The previously identified regulation of Notch1 mRNA by the RBPs is consistent with this model as Notch1 is an integral part of the mitogenic signal necessary for β-selection. Interestingly, overexpression of ICN1 can promote differentiation of Rag-deficient thymocytes (47), and activating mutations in ICN1 have been found in over 50% of human T-ALL (49). However, ICN1 overexpressing cells which were deficient in the proximal pre-TCR signaling component Lcp2 (Slp76), fail to develop into DPs. This suggests that ICN1 requires the pre-TCR signaling machinery (47). Likewise, it was shown that Notch3-driven T-ALL was dependent on a functional pre-TCR (48). In this respect it is possible that Notch signaling is not solely responsible for initiating the bypass of the β-selection checkpoint in DCKO mice, while it could still potentiate subsequent T-ALL development by a feed-forward-loop (50). This is consistent with our data showing differentiation of DCKO HSCs in the absence of Notch-ligand and the lack of mRNA up-regulation of Notch-target genes such as Myc, Hes1 and Dtx1 in our RNAseq dataset. Up-regulation of Notch1 (24) and Myc protein in the DCKO thymocytes may reflect an effect of the RBP on translation of these mRNAs, increased cell cycle feedback, lack of degradation by ubiquitinases such as Fbwx7 (51–53) or a combination thereof. The identification of translational targets of the RBP and their distinction form targets regulated principally by RNA decay will require the generation and integration of additional datasets such as ribosomal footprinting (54).
Remarkably, even in the absence of productive VDJ rearrangements, activation of the DDR can promote the developmental progression of thymocytes. Irradiation induces differentiation and DP thymocyte development in both Rag-deficient and scid animals (43, 45, 55, 56). While sensing of DSBs and cell cycle arrest is important for enforcement of the β-selection checkpoint, proliferation is necessary but not sufficient for differentiation. Deletion of Ccnd3 or inhibition of Cdk4 and Cdk6 can block thymocyte development both in vitro and in vivo. However, forced cell-cycling cannot promote differentiation of Rag-deficient cells into DP thymocytes (39), indicating that the processes of proliferation and differentiation can be uncoupled.
Once the β-selection checkpoint is passed, Ccnd3 cooperates with the DDR machinery and inhibits VH transcription in order to ensure the allelic exclusion of Trb genes (41, 57). In B cells deficient for Zfp36l1/l2 (Zfp36l1fl/fl; Zfp36l2fl/fl; Mb1cre), loss of quiescence causes a failure to recombine and express Igµ (23). In vivo cell cycle inhibition in mutant B cells rescues Igµ VDJ recombination and the presence of pro- and pre-B cells containing excision circles. This is in contrast to our data showing only a partial rescue of icTCRβ expression in DCKO thymocytes upon cell cycle inhibition. We therefore propose that the feedback mechanisms between cell cycle and DDR are not functioning correctly in DCKO thymocytes. The gene list used to assess direct involvement of Zfp36l1/l2 targets in DDR, comprises genes of DNA repair mechanisms as well as DDR downstream signaling, which either recruits factors to damaged DNA or induces stress pathways. The nature and contribution of individual targets is not yet clear. We suggest Zfp36l1/l2 control transcripts involved in DDR resolution and signaling or the generation of double-strand breaks. Potential contributors could be Rag1 and Rag2, which were identified as iCLIP targets and could play a role in the generation of breaks at the DN3 stage. It was also shown that overexpression of the Zfp36l1/l2-target Mdm2 can inhibit DNA double strand break repair (58). While DNA repair could be delayed, we think it is likely to be functional, since mature T cells eventually express a TCR.
VDJ recombination is linked to apoptosis through the activation of p53 by DNA double strand breaks. While p53 and Atm are essential for the containment of double strand breaks in non-cycling cells (57), this process is finely balanced by a network of transcriptional and post-transcriptional mechanisms. In thymocytes undergoing VDJ recombination, the transcription factor Miz-1, for example, regulates the translation of p53 via the ribosomal protein Rpl22 which associates with p53 mRNA (59). We did not find a strong apoptosis gene signature in our RNAseq or iCLIP data. Apoptosis related genes such as Bbc3 and Bax were not increased in DCKO DN3 cells, however Mdm2 mRNA was bound in the iCLIP and elevated in DCKO DN3 cells. Consistent with this, Mdm2 was identified by iCLIP as a target of the highly related Zfp36 RBP in a mouse macrophage cell line (54). Mdm2 is a crucial inhibitor of p53 during lymphopoiesis (60), and p53 has been found down-regulated in thymic tumors of DCKO mice (Hodson, Ferrando and Turner unpublished). Moreover, p53 deficiency can promote the development of DP thymocytes when preTCR signaling is compromised (61). Taken together we suggest that increased Mdm2 could suppress p53-dependent cell death, arising in part from the lack of preTCR signaling, leading to DCKO cell survival in the presence of an increased DDR. Although we cannot fully rule out an involvement of Zfp36l1/l2 in apoptosis pathways, we think it is more likely that the increased apoptosis we have shown in late DN thymocytes is a consequence of the lack of pre-TCR and IL-7 dependent survival signals. This is supported by the fact that the majority of icTCRβnegative cells die at the late DN3/DN4 stage of development.
Our data suggests that Zfp36l1/l2 enforce the β-selection checkpoint and ensure its dependence on the pre-TCR signal. Firstly, the RBPs suppress cell cycle genes at the DN3a stage to allow time for productive VDJ recombination. In addition, they limit DDR gene expression to avoid bypass of the checkpoint due to the activation of this pathway. Upon the signaling of a productively rearranged pre-TCR, we speculate that Zfp36l1/l2 function is suppressed by an as yet unknown mechanism relieving inhibition of target transcripts that promote cell cycle progression and allowing differentiation into mature T cells. Part of this response may be related to cessation of IL-7R signal transduction which we showed to be dependent on Zfp36l1/l2. IL-7 signaling inhibits expression of key transcription factors required for progression to and survival of the DP stage of thymocyte development (62). These include TCF-1, LEF-1 and RORγt. Moreover, IL-7 coordinates proliferation and rearrangement of TCR genes, thus the phenotype may in part be influenced by altered IL-7 signaling (63). Exactly how the RBPs regulate IL-7 signaling is not known and could be the subject of future studies.
At the β-selection checkpoint the timing of proliferation and VDJ recombination is absolutely crucial to avoid sustained chromosomal damage. DN3b cells must not be allowed to proliferate until productive VDJ recombination and the cessation of recombinase and DDR activity. Since transcription is a relatively slow process, the regulation of mRNA stability by RBPs such as Zfp36l1/l2 can add dynamic flexibility to gene expression programs. Coupled with control of protein turnover this form of control allows gene expression to be regulated in a fast and effective way so that DN3b cells can burst into proliferation upon a positive pre-TCR signal.
Supplementary Material
Acknowledgements
We thank Vigo Heissmeyer, Manuel D. Díaz-Muñoz and Dan Hodson for advice and comments on the manuscript and James Di Santo for OP9 DL4 cells; Manuel D. Díaz-Muñoz, Alexander Saveliev, Ram Venigalla and Elisa Monzón-Casanova for technical advice; Sarah Bell, Kristina Tabbada, Arthur Davis and Kirsty Bates for expert technical assistance; Simon Andrews, Jernej Ule, Igor Ruiz de los Mozos and Tomaž Curk for help with iCLIP and Michaela Frye for providing GFPmyc mice.
Funding
This work was funded by the Biotechnology and Biological Sciences Research Council, an MRC centenary award and project grant from Bloodwise (A.G.) and the Federation of European Biochemical Societies (K.U.V.).
Footnotes
Author Contributions
K.U.V. and L.S.B. did most of the experiments with A.G. contributing to specific experiments. H.A. did the bioinformatics analyses and M.T. supervised and provided guidance on the project. K.U.V., L.S.B. and M.T. wrote the manuscript.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 2.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janas ML, Turner M. Stromal cell-derived factor 1α and CXCR4: newly defined requirements for efficient thymic β-selection. Trends Immunol. 2010;31:370–376. doi: 10.1016/j.it.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and Molecular Characterization of Emerging β- and γδ-Selected Pre-T Cells in the Adult Mouse Thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiloh Y. ATM: ready, set, go. Cell Cycle. 2003;2:116–117. doi: 10.4161/cc.2.2.342. [DOI] [PubMed] [Google Scholar]
- 7.Rogakou EP, Pilch DR, Orr aH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 8.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 9.Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: The histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 11.Yin B, Savic V, Juntilla MM, Bredemeyer AL, Yang-Iott KS, Helmink Ba, Koretzky Ga, Sleckman BP, Bassing CH. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J Exp Med. 2009;206:2625–2639. doi: 10.1084/jem.20091320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dozier C, Bonyadi M, Baricault L, Tonasso L, Darbon J-M. Regulation of Chk2 phosphorylation by interaction with protein phosphatase 2A via its B’ regulatory subunit. Biol Cell. 2004;96:509–517. doi: 10.1016/j.biolcel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Helmink Ba, Sleckman BP. The Response to and Repair of RAG-Mediated DNA Double-Strand Breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin B, Lee B-S, Yang-Iott KS, Sleckman BP, Bassing CH. Redundant and nonredundant functions of ATM and H2AX in αβ T-lineage lymphocytes. J Immunol. 2012;189:1372–9. doi: 10.4049/jimmunol.1200829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Bogue MA, Nguyen AP, Roth DB. Irradiation-induced rescue of thymocyte differentiation and V(D)J recombination in mice lacking the catalytic subunit of DNA-dependent protein kinase. J Immunol. 1999;163:6065–71. [PubMed] [Google Scholar]
- 16.Brooks Sa, Blackshear PJ. Tristetraprolin (TTP): Interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 19.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–23. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell SE, Sanchez MJ, Spasic-Boskovic O, Santalucia T, Gambardella L, Burton GJ, Murphy JJ, Norton JD, Clark AR, Turner M. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev Dyn an Off Publ Am Assoc Anat. 2006;235:3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 21.Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, Shelley WC, Richfield EK, Ray MK, Yoder MC, Aplan PD, et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–2410. doi: 10.1182/blood-2009-04-214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Prak L, Rayon-Estrada V, Thiru P, Flygare J, Lim B, Lodish HF. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013;499:92–96. doi: 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galloway A, Saveliev A, ukasiak S, Hodson DJ, Bolland D, Balmanno K, Ahlfors H, Monzon-Casanova E, Mannurita SC, Bell LS, Andrews S, et al. RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science (80-. ) 2016;352:453–459. doi: 10.1126/science.aad5978. [DOI] [PubMed] [Google Scholar]
- 24.Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, Ferrando Aa, et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 27.Huang C-Y, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 28.Holmes R, Zúñiga-Pflücker JC. The OP9-DL1 System: Generation of T-Lymphocytes from Embryonic or Hematopoietic Stem Cells In Vitro. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5156. pdb.prot5156. [DOI] [PubMed] [Google Scholar]
- 29.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu638. btu638–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu X-D, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–7. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnenberg GF, Mjösberg J, Spits H, Artis D. SnapShot: Innate Lymphoid Cells. Immunity. 2013;39:622–622.e1. doi: 10.1016/j.immuni.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 35.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 36.Walker JA, Barlow JL, McKenzie ANJ. Innate lymphoid cells — how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 37.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, Radtke F, et al. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson A, Capone M, MacDonald HR. Unexpectedly late expression of intracellular CD3ϵ and TCR γδ proteins during adult thymus development. Int Immunol. 1999;11:1641–1650. doi: 10.1093/intimm/11.10.1641. [DOI] [PubMed] [Google Scholar]
- 39.Kreslavsky T, Gleimer M, Miyazaki M, Choi Y, Gagnon E, Murre C, Sicinski P, von Boehmer H. β-Selection-Induced Proliferation Is Required for αβ T Cell Differentiation. Immunity. 2012;37:840–853. doi: 10.1016/j.immuni.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide Analysis Identifies Interleukin-10 mRNA as Target of Tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinel NC, Fisher MR, Yang-Iott KS, Bassing CH. The Ataxia Telangiectasia Mutated and Cyclin D3 Proteins Cooperate To Help Enforce TCRβ and IgH Allelic Exclusion. J Immunol. 2014;193:2881–2890. doi: 10.4049/jimmunol.1302201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trigueros C, Hozumi K, Silva-Santos B, Bruno L, Hayday AC, Owen MJ, Pennington DJ. Pre-TCR signaling regulates IL-7 receptor α expression promoting thymocyte survival at the transition from the double-negative to double-positive stage. Eur J Immunol. 2003;33:1968–1977. doi: 10.1002/eji.200323831. [DOI] [PubMed] [Google Scholar]
- 43.Danska JS, Pflumio F, Williams CJ, Huner O, Dick JE, Guidos CJ. Rescue of T cell-specific V(D)J recombination in SCID mice by DNA-damaging agents. Science. 1994;266:450–455. doi: 10.1126/science.7524150. [DOI] [PubMed] [Google Scholar]
- 44.Guidos CJ, Williams CJ, Wu GE, Paige CJ, Danska JS. Development of CD4+ CD8+ thymocytes in RAG-deficient mice through a T cell receptor β chain-independent pathway. J Exp Med. 1995;181:1187–1195. doi: 10.1084/jem.181.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zúñiga-Pflücker JC, Jiang D, Schwartzberg PL, Lenardo MJ. Sublethal gamma-radiation induces differentiation of CD4-/CD8-into CD4+/CD8+ thymocytes without T cell receptor beta rearrangement in recombinase activation gene 2-/-mice. J Exp Med. 1994;180:1517–1521. doi: 10.1084/jem.180.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuo LJ, Yang L-X. γ-H2AX - A Novel Biomarker for DNA Double-strand Breaks. In Vivo (Brooklyn) 2008;22:305–309. [PubMed] [Google Scholar]
- 47.Michie AM, Chan AC, Ciofani M, Carleton M, Lefebvre JM, He Y, Allman DM, Wiest DL, Zúñiga-Pflücker JC, Izon DJ. Constitutive Notch signalling promotes CD4- CD8 - thymocyte differentiation in the absence of the pre-TCR complex, by mimicking pre-TCR signals. Int Immunol. 2007;19:1421–1430. doi: 10.1093/intimm/dxm113. [DOI] [PubMed] [Google Scholar]
- 48.Bellavia D, Campese AF, Checquolo S, Balestri A, Biondi A, Cazzaniga G, Lendahl U, Fehling HJ, Hayday AC, Frati L, von Boehmer H, et al. Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc Natl Acad Sci U S A. 2002;99:3788–93. doi: 10.1073/pnas.062050599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 50.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–35. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, Sandy P, et al. The Ubiquitin Ligase FBXW7 Modulates Leukemia-Initiating Cell Activity by Regulating MYC Stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiedje C, Diaz-Muñoz MD, Trulley P, Ahlfors H, Laaß K, Blackshear PJ, Turner M, Gaestel M. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bogue Ma, Zhu C, Aguilar-Cordova E, Donehower La, Roth DB. p53 is required for both radiation-induced differentiation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes Dev. 1996;10:553–565. doi: 10.1101/gad.10.5.553. [DOI] [PubMed] [Google Scholar]
- 56.Guidos CJ, Williams CJ, Wu GE, Paige CJ, Danska JS. Development of CD4+CD8+ thymocytes in RAG-deficient mice through a T cell receptor beta chain-independent pathway. J Exp Med. 1995;181:1187–1195. doi: 10.1084/jem.181.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dujka ME, Puebla-Osorio N, Tavana O, Sang M, Zhu C. ATM and p53 are essential in the cell-cycle containment of DNA breaks during V(D)J recombination in vivo. Oncogene. 2009;29:957–965. doi: 10.1038/onc.2009.394. [DOI] [PubMed] [Google Scholar]
- 58.Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 Binds to Nbs1 at Sites of DNA Damage and Regulates Double Strand Break Repair. J Biol Chem. 2005;280:18771–18781. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- 59.Rashkovan M, Vadnais C, Ross J, Gigoux M, Suh W-K, Gu W, Kosan C, Möröy T. Miz-1 regulates translation of Trp53 via ribosomal protein L22 in cells undergoing V(D)J recombination. Proc Natl Acad Sci U S A. 2014;111:E5411–9. doi: 10.1073/pnas.1412107111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23:462–72. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haks MC, Krimpenfort P, van den Brakel JH, Kruisbeek AM. Pre-TCR signaling and inactivation of p53 induces crucial cell survival pathways in pre-T cells. Immunity. 1999;11:91–101. doi: 10.1016/s1074-7613(00)80084-9. [DOI] [PubMed] [Google Scholar]
- 62.Yu Q, Erman B, Park J-H, Feigenbaum L, Singer A. IL-7 Receptor Signals Inhibit Expression of Transcription Factors TCF-1, LEF-1, and RORγt Impact on Thymocyte Development. J Exp Med. 2004;200:797–803. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boudil A, Matei IR, Shih H-Y, Bogdanoski G, Yuan JS, Chang SG, Montpellier B, Kowalski PE, Voisin V, Bashir S, Bader GD, et al. IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte β-selection. Nat Immunol. 2015 doi: 10.1038/ni.3122. advance on. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.