Abstract
Complex I (NADH:ubiquinone oxidoreductase), one of the largest membrane-bound enzymes in the cell, powers ATP synthesis in mammalian mitochondria by using the reducing potential of NADH to drive protons across the inner membrane. Mammalian complex I1 contains 45 subunits, comprising 14 core subunits that house the catalytic machinery and are conserved from bacteria to humans, and a mammalian-specific cohort of 31 supernumerary subunits1,2. Knowledge about the structures and functions of the supernumerary subunits is fragmentary. Here, we describe a 4.2 Å resolution single-particle cryoEM structure of complex I from Bos taurus. We locate and model all 45 subunits to provide the entire structure of the mammalian complex. Furthermore, computational sorting of the particles identified different structural classes, related by subtle domain movements, which reveal conformationally-dynamic regions and match biochemical descriptions of the ‘active-to-deactive’ enzyme transition that occurs during hypoxia3,4. Thus, our structures provide a foundation for understanding complex I assembly5 and the effects of mutations that cause clinically-relevant complex I dysfunctions6, insights into the structural and functional roles of the supernumerary subunits, and new information on the mechanism and regulation of catalysis.
Starting with the structures determined for bacterial complex I7–9, the highly-conserved core subunits and their nine cofactors (a flavin mononucleotide and eight iron–sulphur (FeS) clusters) were identified previously in medium-resolution structures of the mammalian enzyme from B. taurus10 and the yeast enzyme from Yarrowia lipolytica11. The arrangement and structures of the supernumerary subunits (constituting half the mammalian complex) are far less well defined. The 5 Å resolution electron cryo-microscopy (cryoEM) structure of B. taurus complex I revealed the supernumerary ensemble wrapped around the core, and fourteen supernumerary subunits were assigned10. Subsequently, eight further assignments were proposed using the crystallographic structure of subcomplex Iβ (part of the membrane domain)12. Thus, nine subunits remain unlocated, and models for the supernumerary subunits are fragmentary. The complete structure of mammalian complex I is crucial for elucidating the roles of the supernumerary subunits in complex I function and dysfunction.
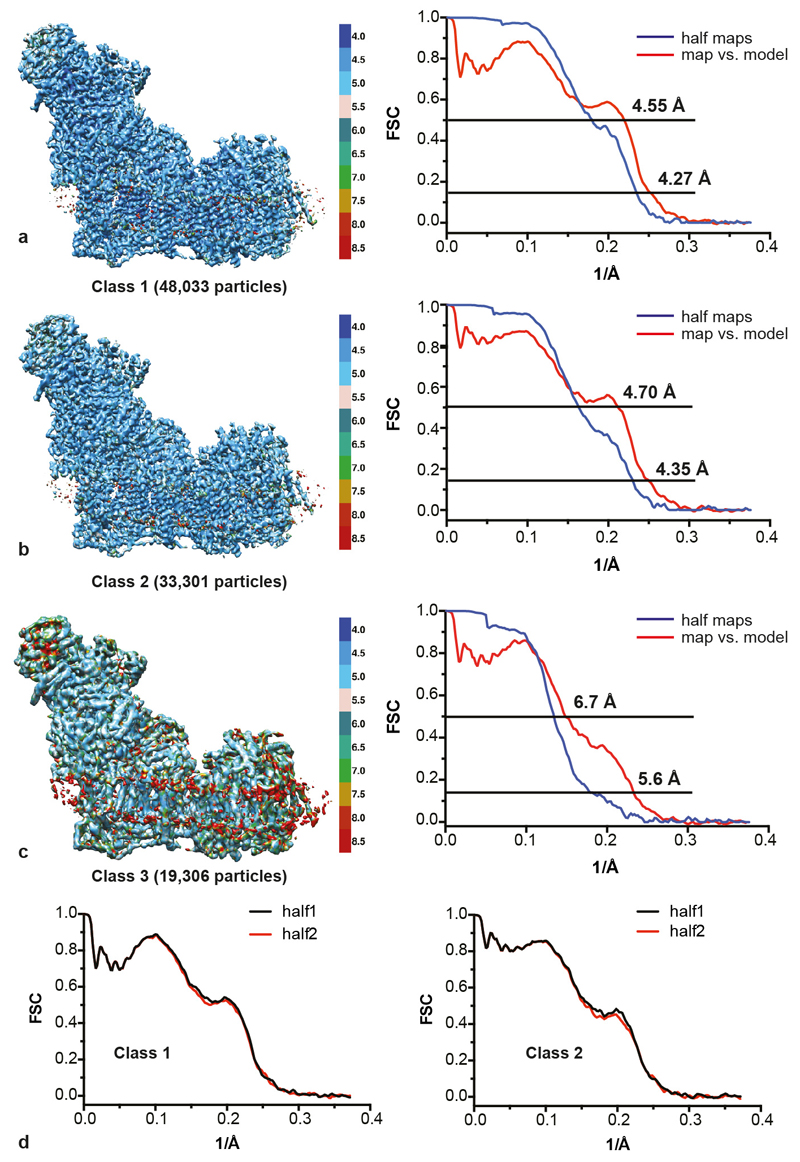
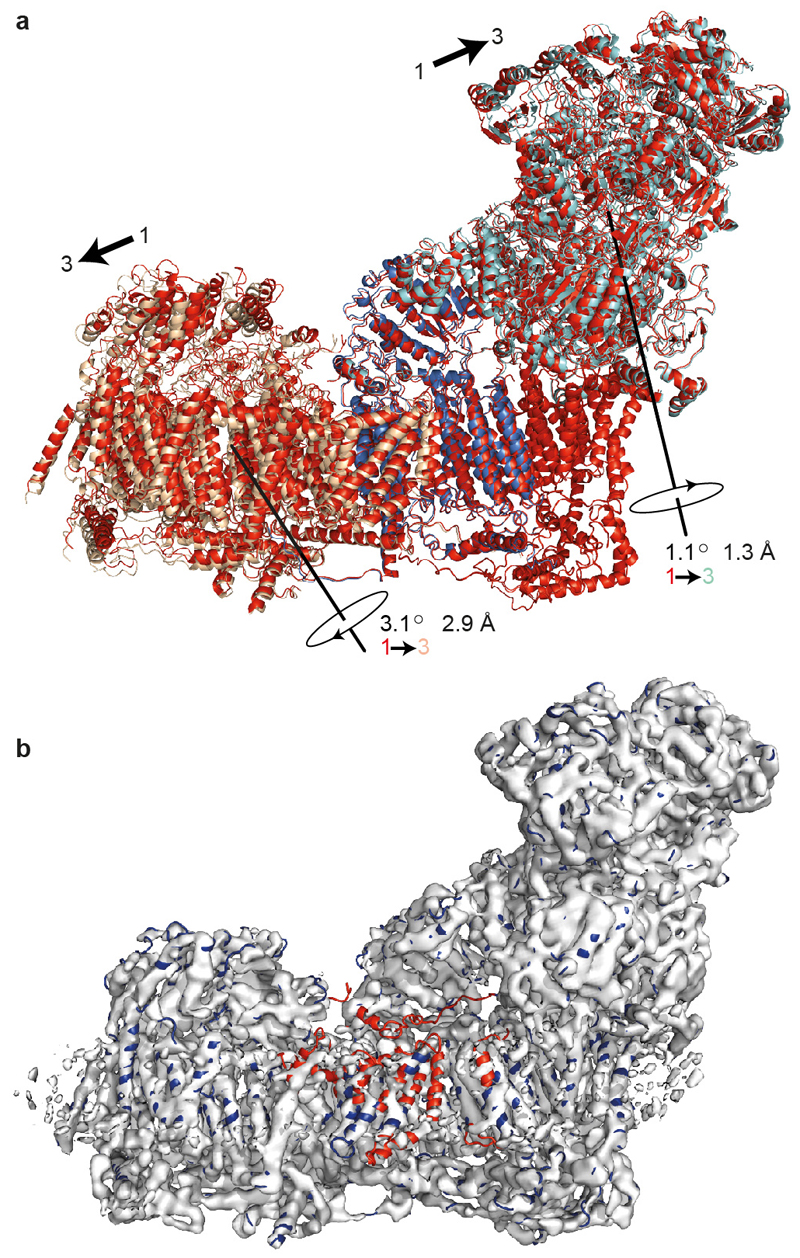
Here, we describe a cryoEM map for B. taurus complex I with 4.16 Å resolution overall (Figure 1A and ED Figure 1) that enabled us to model all its 45 subunits and 93% of its 8515 residues (ED Tables 1 and 2). Computational sorting of the particles revealed three major classes with overall resolutions 4.27 (class 1), 4.35 (class 2) and 5.60 Å (class 3) (ED Figure 2) for which the quality of the map in several regions was improved significantly (see below). ED Figures 3 and 4 present example densities and we use class 2 to describe the structure, unless stated otherwise. The different classes represent different states and their analysis provides new insights into the elusive mechanism of complex I catalysis.
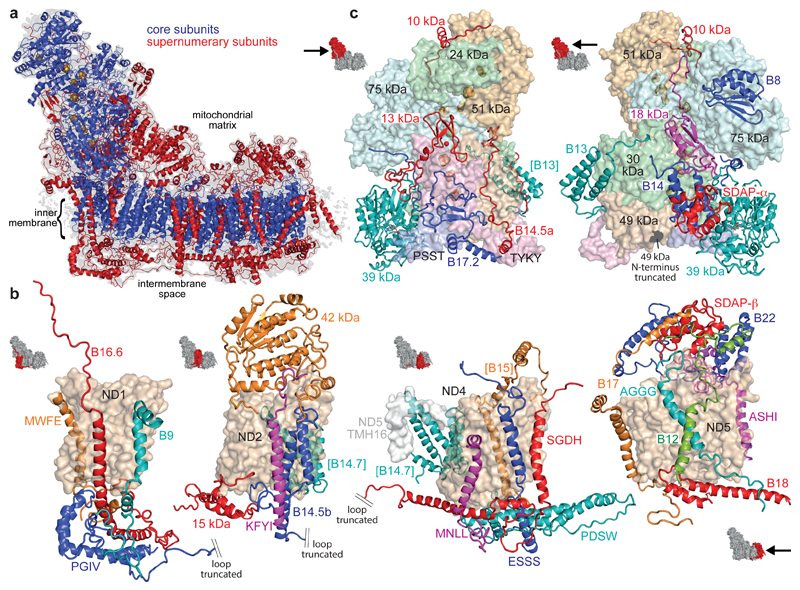
Figure 1. The supernumerary subunits of mammalian complex I.
a) Overview of the complex with the 14 core subunits in blue (FeS clusters in yellow/orange), the 31 supernumerary subunits in red, and the cryoEM density in grey. b-c) Arrangement and structures of the supernumerary subunits around the core membrane (b) and hydrophilic (c) subunits. The core subunits are in surface representation and the supernumerary subunits in cartoon; icons show viewpoints and locations in the complex. Subunits in brackets are behind the domain. The assignments of B12 and AGGG may be reversed. See ED Tables 1 and 2 for the subunit nomenclatures in other species.
Figure 1 presents the structures and locations of all 31 supernumerary subunits in mammalian complex I (see ED Table 3 for subunit-subunit interactions and additional details). The supernumerary subunits are central to the structure, stability and assembly of the complex, and some also have regulatory or independent metabolic roles.
The 18 supernumerary transmembrane helices (TMHs) (Figure 1b) establish a cage around the core membrane domain. Three TMH-containing subunits, B9 (NDUFA3), B16.6 (NDUFA13) and MWFE (NDUFA1), interact extensively with PGIV (NDUFA8) on the intermembrane space (IMS) face, enclosing core subunit ND1. Subunit B14.5b (NDUFC2), bound to ND2, contains two different-length TMHs and attaches KFYI (NDUFC1) to the complex. Three TMHs that interact with ND4 are assigned to MNLL (NDUFB1), ESSS (NDUFB11) and SGDH (NDUFB8). Four TMHs, assigned to B17 (NDUFB6), AGGG (NDUFB2), B12 (NDUFB3) and ASHI (NDUFB8), are bound to ND5. The TMHs of ASHI and B15 (NDUFB4, on the side of ND4) cross the ND5 transverse helix, and the four TMHs of B14.7 (NDUFA11) appear to support ND5-TMH16 in anchoring it against ND2. Four subunits confined to the IMS (PGIV, 15 kDa (NDUFS5), PDSW (NDUFB10) and B18 (NDUFB7)) form a helix latticework (together with SGDH and B16.6) on the IMS face (Figure 2a). PGIV, 15 kDa and B18 contain CHCH domains (pairs of helices linked by two disulphides)13 and are canonical substrates for the Mia40 oxidative-folding pathway14; PDSW probably contains two further disulphides. The disulphides form during complex I biogenesis and are probably important for enzyme stability. Thus, the supernumerary cage has evolved to become integral to the structure/stability of the membrane domain.
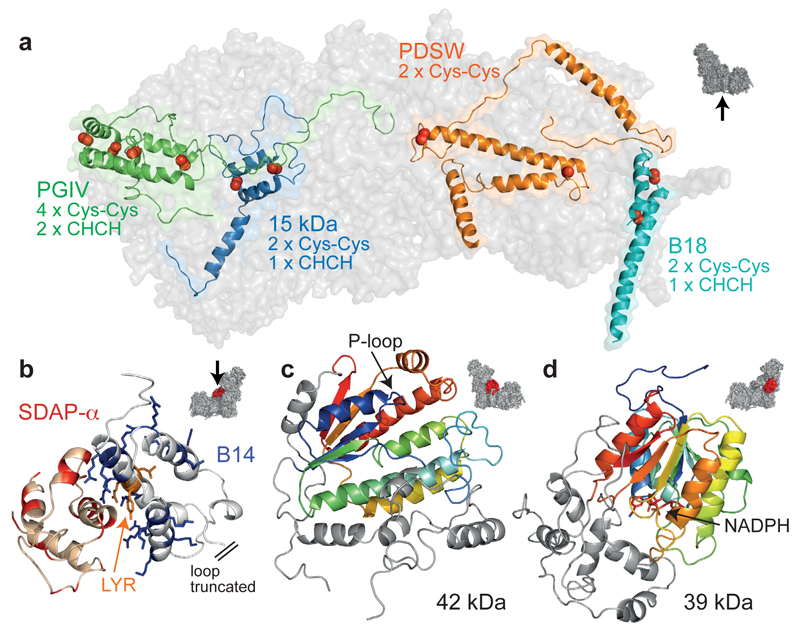
Figure 2. Details of some of the supernumerary subunits.
a) Subunits confined to the IMS face with disulphide bonds indicated by red spheres. b) Positively-charged residues in the LYR-protein B14 (blue) interact with negatively-charged residues in SDAP-α (red); B22 and SDAP-β exhibit the same structural motif. c) 42 kDa with the core nucleoside-kinase fold in rainbow; the extensions (grey) dock it to ND2. d) 39 kDa with the core dehydrogenase-reductase fold in rainbow and bound nucleotide; the C-terminal domain (grey) approaches the membrane interface. Icons indicate viewpoints and locations in the complex.
Subunits B14 (NDUFA6) and SDAP-α (NDUFAB1), and B22 (NDUFB9) and SDAP-β (NDUFAB1), constitute matching subdomains on the hydrophilic domain and matrix face of the membrane domain, respectively (Figure 1)10,12,15. SDAP-α/β are identical to the mitochondrial acyl-carrier protein (ACP) and exhibit densities consistent with the pantetheine-4’-phosphate that covalently attaches an acyl chain to Ser-4416,17 (ED Figure 4e). Their ACP ‘recognition’ helices interact with Arg/Lys-rich helices in the LYR proteins B14 and B22 (Figure 2b and ED Figure 4e) as canonical ACPs interact with the enzymes of fatty-acid biosynthesis18. The 42 kDa subunit (NDUFA10, Figure 2c) contains a central α/β nucleoside kinase fold with a parallel 5-strand β-sheet, plus three extensions that dock it to the matrix face of ND2. Although the active site is accessible and the key nucleoside kinase residues are present19 no activity has been reported. The 39 kDa subunit (NDUFA9, Figure 2d) is attached to core subunits PSST (NDUFS7) and 30 kDa (NDUFS3) in the hydrophilic arm. The N-terminal domain of 39 kDa comprises an α/β short-chain dehydrogenase/reductase fold20 containing an NAD(P)-binding Rossmann-fold with a parallel 7-strand β-sheet and density for a bound nucleotide, modelled as NADPH21 (ED Figure 4). The separate C-terminal domain interacts with the long matrix loop of ND3 (see below).
The final seven supernumerary subunits adorn the hydrophilic domain (Figure 1c). Thioredoxin-like B8 (NDUFA2) is attached to the 75 kDa subunit (NDUFS1), and the three-helix bundle of B13 (NDUFA5) to 30 kDa. The remaining five subunits are located at interfaces. The zinc-binding domain of 13 kDa (NDUFS6)22 and the 18 kDa subunit (NDUFS4) four-strand β-sheet and helix are located where the NADH dehydrogenase domain meets the rest of the complex. All five subunits (the other three are B14.5a (NDUFA7), B17.2 (NDUFA12) and 10 kDa (NDUFV3)) contain long loops running over the domain surface. A striking example is the extensive loop in B14.5a, which arches up along the TYKY-49 kDa (NDUFS8-NDUFS2) interface, across 49 kDa, along its interface with 75 kDa and onto 30 kDa. The role of the supernumerary subunits in stabilising interfaces in the hydrophilic domain contrasts sharply with their arrangement into a rigid cage to stabilise the membrane domain. In summary, our structure describes the locations and structures of all 31 supernumerary subunits in mammalian complex I.
The structures of the mammalian core subunits (Figure 3a) closely match those of the bacterial subunits7–9, and contain corresponding mechanistically-relevant features.
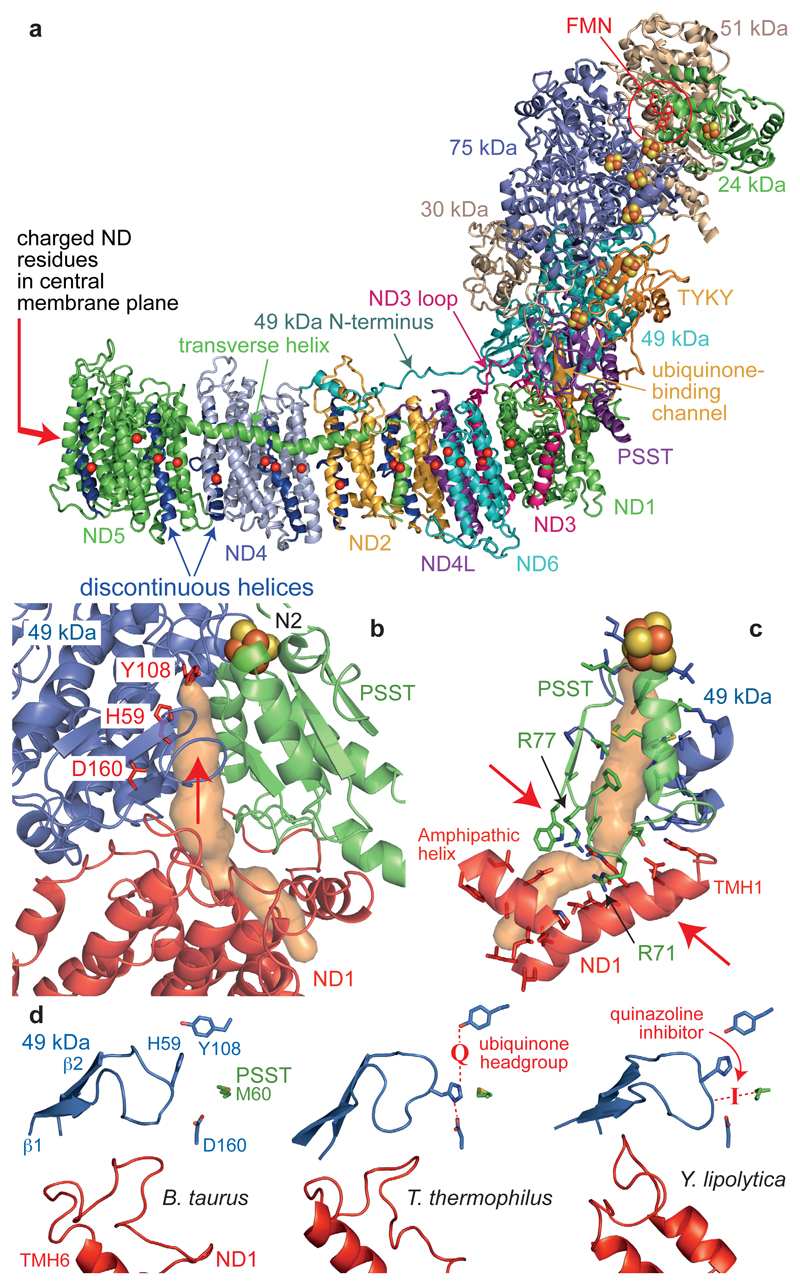
Figure 3. The core subunits and ubiquinone-binding site of mammalian complex I.
a) The core subunits with the FMN, FeS clusters, conserved charged residues (Cα) in the membrane (overlaid for clarity), and proposed ubiquinone-binding channel (orange). b) The ubiquinone-binding channel. 49 kDa-Tyr108 and 49 kDa-His599 form hydrogen bonds to the bound ubiquinone at the top of the cleft (indicated by an arrow) between 49 kDa and PSST. The channel entrance is between three helices in ND1. c) Structural elements forming the channel and bottleneck (between the arrows). PSST-Arg77 is hydroxylated23. d) Conformations of the 49 kDa (β1-β2) and ND1 (TMH5-6) loops observed in different species. In T. thermophilus9 the ubiquinone headgroup binds between Tyr108 and His59 and His59 hydrogen bonds to 49 kDa-Asp160. In Y. lipolytica11 a quinazoline inhibitor is bound between PSST-Met60 and the tip of the 49 kDa loop.
NADH is oxidized by a flavin mononucleotide in the 51 kDa subunit (NDUFV1) (ED Figure 3), then electrons transfer along a chain of FeS clusters to the terminal cluster (N2) and to ubiquinone-10. In mammalian complex I an unusual di-methylated arginine (49 kDa-Arg85)23 close to N2 probably contributes to its relatively high potential24. In the hydrophilic domain, the large domain of mammalian 75 kDa differs from that of T. thermophilus7 because the fourth sub-domain contains just two short helices separated by a ~30 residue loop. The core subunit N- and C-terminal extensions also vary between species; in B. taurus the 30 kDa C-terminus (which loops into a cleft between PSST, 39 kDa and B14) and the TYKY N-terminus (which wraps around the hydrophilic/hydrophobic domain interface) recapitulate the stabilising role of the supernumerary subunits. Notably, our cryoEM maps reveal the 49 kDa N-terminus forms a long loop on the surface of the membrane domain (Figure 3a). The extended conformation explains its susceptibility to proteases25 but it is unlikely to be central to the mechanism because it is not conserved in T. thermophilus, and in Escherichia coli is fused to the 30 kDa C-terminus. The long matrix loop in ND3, which lies across the front of the hydrophilic domain and is central to the active/deactive transition in mammalian complex I3, is also resolved (Figure 3a).
Four proton-transfer routes (in ND2, ND4, ND58, and ND1+ND4L+ND69) have been proposed for the four protons that complex I is generally considered to translocate for each NADH oxidised. ND2, ND4 and ND5 each contain two TMHs interrupted by loops in the central membrane plane (TMHs 4 and 9 in ND2, and 7 and 12 in ND4 and ND5, Figure 3a and ED Figure 3). The chain of conserved Asp, Glu, Lys and His residues that runs along the middle of the membrane domain is now well-defined in the mammalian enzyme (ND1-Glu143, ND3-Asp66-Glu68; ND4L-Glu34-Glu70; ND2-Glu34-Lys105-Lys135-Lys263; ND4-Glu123-Lys206-Lys237-Glu378; ND5-Glu145-Lys223-His248-Lys392, Figure 3a). Distortions of the helical structure are observed in ND6-TMH3, ND2-TMH5, ND4-TMH8 and ND5-TMH8 (ED Figure 3). They resemble the π–bulge in bacteriorhodopsin but do not satisfy its technical definition26, perhaps due to the intermediate resolution of the maps. The distortions are centred on Gly pairs in ND6 (62-3) and ND4 (239-40), on a Ser pair in ND5 (249-50), and on Trp167 (flanked by two Gly pairs) in ND2. Notably, ND6-TMH3 is more distorted in the mammalian structure than in T. thermophilus (which contains only one Gly)9, such that ND6-Phe67 is displaced around the helical axis.
Ubiquinone-10 binds with its redox-active headgroup close to cluster N2, at the top of a cleft between 49 kDa and PSST7,27 (Figure 3b), and T. thermophilus complex I co-crystallized with decylubiquinone showed it forms hydrogen bonds with 49 kDa-His59 and 49kDa-Tyr1089. Here, the Tyr108 and His59 side-chains are poorly resolved, and the conformation of the β1-β2 His59-containing loop is different to in T. thermophilus (Figure 3d). Thus, it appears the binding site is flexible, allowing it to organise around substrates and inhibitors, neither of which are present here. The putative ubiquinone-access channel, identified first in T. thermophilus9, connects the cleft to an entrance in ND1 (between TMH1, an amphipathic helix, and TMH6) and can also be detected here (minimum diameter 2.9 Å). Alternative entrances, between ND1 TMHs 1 and 7, and 5 and 6, are also evident but narrower (minimum diameters 1.9–2.2 Å). However, the planar ubiquinone ring is ~6 Å across so all the channels in the static structure would have to open to allow it to enter: a structure containing ubiquinone-10 (or a long-chain analogue) is required to confirm its access pathway.
In the mammalian complex, further consideration of the most plausible (widest) channel for ubiquinone reveals a ‘bottleneck’ at the base of the cleft (Figure 3c). Ubiquinone-10 is highly hydrophobic, so most of the channel-lining residues are uncharged and hydrophobic. In contrast, the bottleneck is formed by charged and polar residues including Glu24 and Arg25 (ND1-TMH1), Arg274 (ND1-TMH7), and Arg71 and hydroxy-Arg7723 (PSST α2-β2 loop). Nearby, the ND1 TMH5-6 loop, with many acidic residues, contributes more significantly to the channels in T. thermophilus and Y. lipolytica9,11. This cluster of charged residues suggests the presence of water molecules and appears incompatible with an ubiquinone-10 binding channel. However, the PSST loop was modelled incompletely in T. thermophilus and Y. lipolytica, and the ND1 loop is poorly resolved here, indicating their flexibility. It is possible that conformational changes at the bottleneck, linked to ubiquinone binding/dissociation, contribute to coupling the redox reaction to proton translocation.
When the particles comprising the whole data set were subjected to 3D classification three major, slightly-different classes emerged. Class 3, the smallest, lowest resolution class, is closer to class 1 than class 2 and characterized by movement of the ND4-ND5 subdomain (relative to class 1, ED Figure 5 and ED Table 4) and disorder in the ND5 transverse helix and its anchor (ND5-TMH16). Similar disorder was observed in subcomplex Iβ12, which comprises the ND4-ND5 subdomain. Thus, we attribute class 3 to molecules in the first stages of dissociation and do not discuss it further.
Classes 1 and 2 are related (Figure 4a) by opposing rotations of the hydrophilic domain and a large section of the membrane domain, relative to the ND1 subdomain (ED Table 4). In class 1, 42 kDa has moved towards B14+SDAPα (Figure 4b), and 39 kDa has moved relative to ND1. Notably, the long ND3 TMH1-2 loop is partially disordered in class 1 (Figure 4d). This loop is symptomatic of decreased order in class 1 at the hydrophilic-membrane domain junction: the ND1 TMH5-6 loop, the 49 kDa β1-β2 loop containing His59, and parts of the 39 kDa C-terminal domain are also disordered (Figure 4c). In addition, the distortion in ND6-TMH3 is less pronounced in class 1 than class 2.
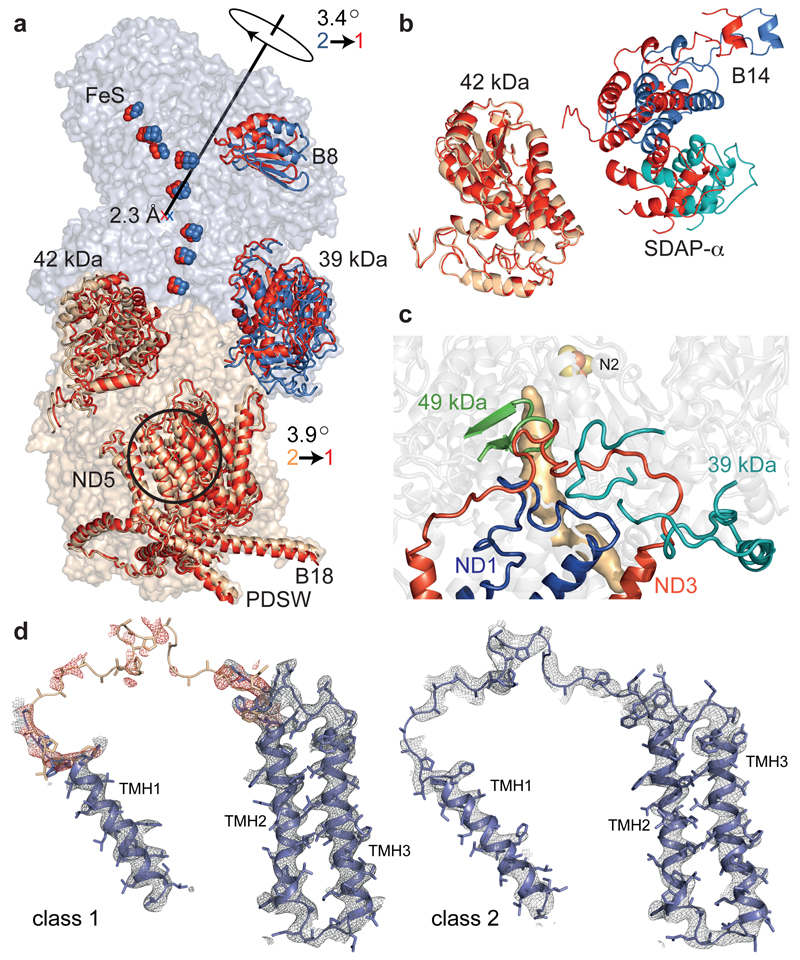
Figure 4. Relationships between classes 1 and 2.
a) Class 1 (red) and 2 (blue/wheat) were superimposed using ND1+ND3 and viewed along the axis of rotation for ND4+ND5. See ED Table 4 for details of the transformations. b) Change of ~10 Å in the relative positions of B14+SDAPα (hydrophilic domain) and 42 kDa (membrane domain); class 1 in red, viewed from the matrix. c) Loops (ND1 TMH5-6, ND3 TMH1-2, 49 kDa β1-β2 and in 39 kDa) in class 2 that are disordered in class 1, with the ubiquinone-binding site; adjacent TMHs/strands are shown for clarity. The site cannot be detected in class 1 as it appears open. d) Densities for the loop connecting ND3 TMH1-2. For class 1, the loop from the class 2 model (white) was used to identify the density in red.
Mammalian complex I exists in different states, according to its catalytic status. In the absence of substrates to sustain turnover (such as during hypoxia) it converts spontaneously to the ‘deactive’ state4, a profound resting state that requires slow, reactivating turnovers to regain ‘active’ status. The deactive state is characterized by the ability of cysteine-modifying reagents (such as N-ethylmaleimide) to derivatise Cys39 in the ND3 TMH1-2 loop3. Approximately half the preparation discussed here is susceptible to modification by N-ethylmaleimide. In class 2, the side-chain of ND3-Cys39 is inaccessible to modifying reagents, suggesting class 2 represents an active state. In contrast, the disordered loop in class 1 (Figure 4d) is probably mobile and accessible, suggesting class 1 represents a deactive state. Increased disorder in the 39 kDa C-terminal domain, and its altered position relative to ND1, support this assignment because both subunits are more exposed to lysine-modifying reagents in the deactive state28. However, the structures of biochemically-defined samples are required to confirm the assignments.
Different conformations of the ND1 TMH5-6 loop and the 49 kDa β1-β2 loop in Y. lipolytica (relative to in T. thermophilus) were proposed previously as characteristic of the deactive state11, but they vary between class 2 and T. thermophilus also (Figure 3d), and are disordered in class 1. Notably, Y. lipolytica complex I was co-crystallised with a quinazoline inhibitor (Figure 3d), and cross-linking studies have shown quinazolines interact with sections of 49 kDa and ND1 that contain the β1-β2 and TMH5-6 loops25. We propose quinazoline binding orders these loops, and the quinazoline-binding site overlaps (but does not superimpose) the ubiquinone-binding site. Our interpretation supports biochemical proposals for non-identical but overlapping sites for the myriad inhibitors of ubiquinone reduction29, but does not support an alternative, occluded ubiquinone-binding site in the deactive complex11.
The two states of mammalian complex I described support the idea that dynamic, flexible regions at the hydrophilic/membrane domain interface are important for coupling ubiquinone reduction to proton translocation. The class1-disordered loops in ND1, ND3 and 49 kDa all contribute to the ubiquinone-binding site (Figure 4c). Therefore, we attribute lack of catalytic activity in the deactive state to reversible disruption of this site, which can be recovered when the ubiquinone-binding site in the NADH-reduced enzyme reforms around its substrate. During catalysis the ND3 loop, which originates in the membrane and interacts extensively with the hydrophilic domain, may restrict conformational changes at the interface. Changes in the 49 kDa loop conformation may trigger proton translocation: molecular simulations were used to outline a mechanism in which the ubiquinol dianion deprotonates Tyr108 and His59, breaking a His59-Asp160 hydrogen bond and displacing Asp160 towards the membrane30. In ND1, TMHs 2 to 6 replicate the antiporter-like half-channel motif of ND2, ND4 and ND59. TMH5 resembles a discontinuous TMH, but with its matrix half-TMH unstructured and continuous with the TMH5-6 loop at the base of the ubiquinone-binding cleft (Figure 3d) that, like the α2-β2 loop in PSST (Figure 3c), may change conformation upon ubiquinone binding. Furthermore, this loop carries many conserved acidic residues that may collect protons for ND1-Glu1439. In turn, Glu143 is connected, by ND3-Asp66 and the dynamic distortion in ND6-TMH3, to the chain of charged residues along the membrane domain (Figure 3a). Thus, a cascade of events, originating from the ubiquinone-binding cleft, may couple ubiquinone reduction/protonation to proton translocation. Although all such mechanisms for complex I are currently hypothetical, cryoEM now provides a powerful tool to either study individual trapped conformations or separate mixed states computationally in order to determine how conformational changes are initiated, co-ordinated and propagated.
Methods
Protein preparation
Complex I was purified from B. taurus heart mitochondrial membranes by solubilisation and anion exchange chromatography in n-dodecyl-β-D-maltoside (DDM), and size-exclusion chromatography in 7-cyclohexyl-1-heptyl-β-D-maltoside (Cymal 7), as described previously10,31.
CryoEM specimen preparation, imaging and image processing
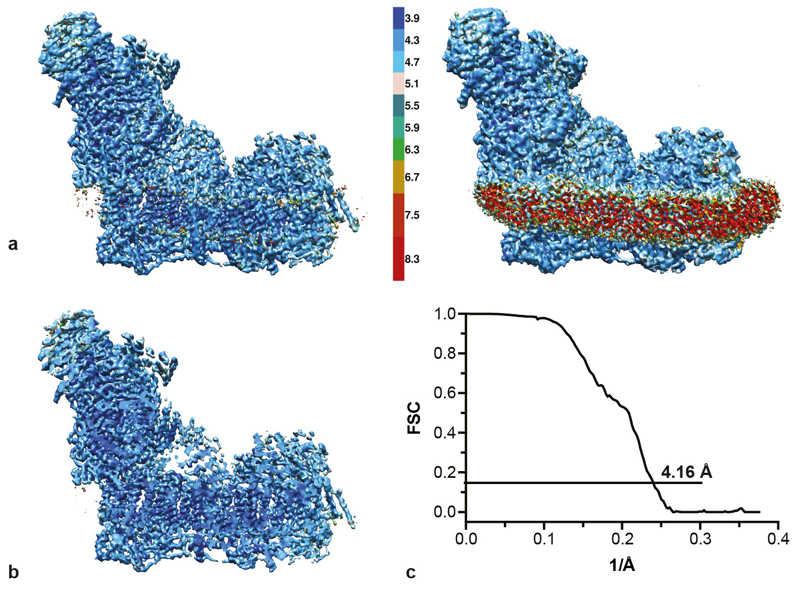
CryoEM grids were prepared as described previously10. Images were recorded using a 300 keV FEI Titan Krios electron microscope with EPU software, with the specimen temperature at -186 °C. A Falcon II CMOS (complementary metal oxide semiconductor) direct electron detector was used for imaging at 105,263x magnification (nominally 59,000x), corresponding to a sampling density of 1.33 Å pixel-1, at 2.4-4.2 μm under-focus in 0.3 μm steps, with the autofocus routine performed every 8 μm to give a range of defocus. Each image was exposed for 2 s (total dose ~35 e- Å-2) and an in-house protocol was used to capture 34 movie frames. The frames were aligned using Unblur (without dose filtering)32 and the CTF was determined with CTFFIND433. A total of 139,456 particles were picked manually and extracted using a box of 360 pixels. Further processing was performed with RELION34. Following an initial 2D classification to discard ‘bad’ particles, 115,966 particles were used for refinement. The 5 Å resolution map described previously10 was low-pass filtered to 60 Å and used as the reference map. The whole data set was subjected to the auto-refine routine in RELION, followed by modelling of the beam-induced movement (using a running average of 7 frames) and B-factor weighting. All the resolutions described here are defined at FSC = 0.143 following application of a shape mask, phase randomisation to check for effects of the mask, and correction for the modulation transfer function of the detector. The resolution of the data set containing all the particles after B-factor weighting and refinement was 4.16 Å, with an estimated orientation accuracy of 0.93 degrees.
The B-factor weighted particles were subjected to 3D classification into eight classes using an angular sampling of 0.9° for 25 iterations, with the resolution limited to 8 Å. Three major classes were identified, containing 48,033 (class 1), 33,301 (class 2) and 19,306 (class 3) particles. Each class was refined individually, providing maps with overall resolution of 4.27 Å for class 1, 4.35 Å for class 2 and 5.6 Å for class 3. The maps were sharpened with B-factors of -114 for class 1, -110 for class 2 and -125 for class 3. Local analysis of the resolution was performed using ResMap35 (ED Figures 1 and 2). Note that the map used as the reference for refinement is a class 1 map, which we described previously as the major class10, and that the number of particles required to achieve the reported resolutions indicates the need for future improvements in both the biochemical homogeneity and specimen preparation of the samples.
Model building and analysis
Model building was performed using Coot36. The first model was built using the map from the complete dataset, with cross-referencing to the maps from classes 1 and 2, using the 5 Å model for B. taurus complex I described previously (4UQ8.pdb10) as the initial template. This unrefined polyalanine model contains models for the fourteen core subunits that are structural homologues of the subunits of the bacterial enzyme9, partial models for fourteen assigned supernumerary subunits, and a further 21 polypeptide chains from unassigned supernumerary structures. Assignments to some of these chains were subsequently proposed for a further eight supernumerary subunits using the 6.8 Å X-ray crystallographic structure of subcomplex Iβ from B. taurus complex I12. The new maps show clear connectivity within the density features, allowing many of the previously traced chains to be extended significantly, and some of them to be joined together. Furthermore, the helical pitches of most of the TMHs and of many of the helices in the globular subunits are now clear, and the β-strands are well separated. These significant improvements in the density, together with information from secondary structure analyses and homologous structures, allowed improved and more complete models to be built for all 45 subunits (see below). Note that the former subunit MLRQ (NDUFA4) is no longer considered a subunit of complex I37 and that there are two copies of subunit SDAP10. The only substantial unmodelled protein densities are underneath the tip of the membrane domain and are accounted for by the termini of two supernumerary subunits (B18 and ASHI).
In well-resolved regions of the map, protruding densities of the side chains of the bulky aromatic residues Phe, Tyr and Trp, along with some side chain densities from Arg and His, are clearly visible (ED Figures 3 and 4). For those subunits which had already been assigned these side chain features were used as landmarks for assigning the sequences. Side chains were added in well-resolved regions, but omitted when their density features are unclear. The assignments of four subunits that were previously assigned in pairs (B9 and MWFE10, and PDSW and B1812) were also confirmed. For three subunits (B8, SDAP-α and SDAP-β) models of the human homologues are available in the PDB (1S3A.pdb38 and 2DNW.pdb) and were used to assign the residues. For highly-conserved regions of the 51 kDa, 24 kDa (NDUFV2) and 75 kDa subunits, residue assignments were supported by the structure of complex I from T. thermophilus9. In some less well-resolved regions of the map it is not possible to assign the sequence confidently using the current data. In these regions the polyalanine model has been retained (residue names UNK), but the residues for each subunit have been numbered as accurately as possible, to provide a guide to the location of individual residues. The exceptions are subunits B12, 10 kDa and part of B14.5a, for which residue numbers cannot be confidently proposed. In summary, the models described are mixed models in which the residues in some subunits are fully assigned, some partly assigned, and others not assigned at all (ED Tables 1 and 2). Next, the hitherto-unknown subunits were assigned. The patterns of the bulky residues observed in the TMH-containing regions of unknown densities were compared with the amino acid sequences of candidate subunits. By combining this information with information from secondary structure analyses, supported with biochemical knowledge, the seven hitherto-unassigned TMHs were assigned and the subunits modelled as described above. Subunits B12 and AGGG were assigned to the two TMHs on the tip of the membrane domain, but lack of clear features in the densities means that our specific assignment of B12 to chain k and AGGG to chain j is less confident, so we cannot exclude the possibility that they have been reversed. The two additional TMH-like densities observed in the structure of subcomplex Iβ12 are clearly absent from the cryoEM maps, so they are attributed to an artefact produced by crystal contacts with either dissociated subunits or the reorganised transverse helix. Remaining polypeptide chains, located on the outside of the hydrophilic domain and the IMS face of the membrane domain, were assigned by combining secondary structure analyses with biochemical knowledge and by using the densities from bulky residues, and the residues and side chains assigned and modelled where possible. Maps with different B-factor sharpening were used to help with chain tracing and assignment of residues, and the model was checked for consistency with the individual maps from the three different classes. The geometry of the model was improved by cycles of manual adjustment in Coot36, real-space refinement by Phenix39 and refinement by REFMAC40, with secondary structure restraints.
Separate models were subsequently created for classes 1 and 2 by rigid-body fitting of each subunit, manual adjustment to account for significant local differences identified in the density maps, and cycles of adjustment and refinement as described above. To provide some reassurance of the refinement, the coordinates for classes 1 and 2 were shifted by 0.1 Å and the B-factors re-set to 75, then the coordinates were refined against one of the half maps. The resulting models were then used to calculate Fourier-shell correlation (FSC) curves for both half maps; as shown in ED Figure 2 the curves display very little evidence of overfitting. The class 3 model was created from the class 1 model by rigid-body fitting. It was created purely for comparison with classes 1 and 2 so the individual subunit models were retained unchanged from class 1, the model was not deleted in the regions of poor density reported in ED Figure 5, and no refinement was performed. The refinement statistics for classes 1 and 2 are summarized in ED Table 5.
The sequences of all the subunits are numbered starting from residue 1 of the mature proteins2. The naming of the chains has been retained as much as possible from our previous model (4UQ8.pdb). The names are unchanged for A to Z and a to n, except that the previous chains d and e have been combined to form new chain d (B14.5b) and chain e has been reallocated (15 kDa). Previous chains o to w all represented sections of subunits that have now been combined; new chains n to s have been reallocated.
After modelling the protein, we observed two additional short, elongated densities located at interfaces between core membrane-domain subunits, which may represent phospholipid molecules. They are between ND2-TMH10 (residues 291-295), ND4-TMH5 (residues 144-147) and the ND5 transverse helix (residues 564-567), and between ND4-TMH11 (residues 356-360) and ND5-TMH4 (residues 116-123). These densities have not been modelled, because similar densities are observed elsewhere within the detergent/phospholipid belt but at lower contour levels, so we cannot exclude the possibility that they are due to noise in the density map at the current resolution.
Bioinformatics
Secondary structure analyses were carried out using PSIPRED41 and raptorX42. The identification of TMHs in the sequences and the structures of homologous proteins were described previously10. Cavities and channels in the structures were investigated using CAVER43. Figures were created using the PyMOL Molecular Graphics System. The subunit interactions in ED Table 3 were calculated with NCONT in CCP444, and defined as a centre-to-centre distance of less than 5 Å between any two atoms. For some subunits, such as B8 and SDAP-α, the interactions are limited to one subunit, while other subunits with long loops and extended structures, such as SGDH, form multiple interactions. Some of the residues in our current model lack side chains so the number of interactions detected may increase in future models.
Extended Data
ED Figure 1. Resolution estimation and ResMap analysis of the density map for complex I before classification.
a) The map, shown at two different contour levels, is coloured according to the local resolution, as determined by ResMap35. At the higher contour level (left), the majority of the protein is resolved to 3.9–4.7 Å; only the very peripheral regions (parts of the 51 kDa, 24 kDa and 75 kDa subunits in the matrix arm, and the distal end of the membrane arm) are at lower resolutions of 5–6 Å. At the lower contour level (right), the detergent/lipid belt dominates the 7–9 Å resolution range. b) A slice through the map shows that large portions of the central, core regions are resolved to 4 Å or better. c) The FSC curve defines an estimated overall resolution of 4.16 Å at FSC = 0.143.
ED Figure 2. Resolution estimation and ResMap analysis of the classes of complex I.
Panels a, b and c show data on classes 1, 2 and 3, respectively. Classes 1 and 2 display similar distributions in local resolution, with the majority of the protein in the range 4–5 Å. In class 3 the majority of protein displays a resolution of 4.5–5 Å. In all three cases the refined models agree very well with the maps as shown by comparison of the FSC curves (red) and the FSC curves from the half-maps (blue), and the similarity of the resolution values at FSC 0.143 and 0.5. The estimated resolutions, defined where the line at FSC = 0.143 crosses the blue curve, are 4.27 Å for class 1, 4.35 Å for class 2 and 5.6 Å for class 3. d). Cross-validation of the refinement parameters, confirming lack of over-fitting. For classes 1 and 2, one of the half maps was used for refinement then the FSC curves were calculated for each of the two half maps using the same model.
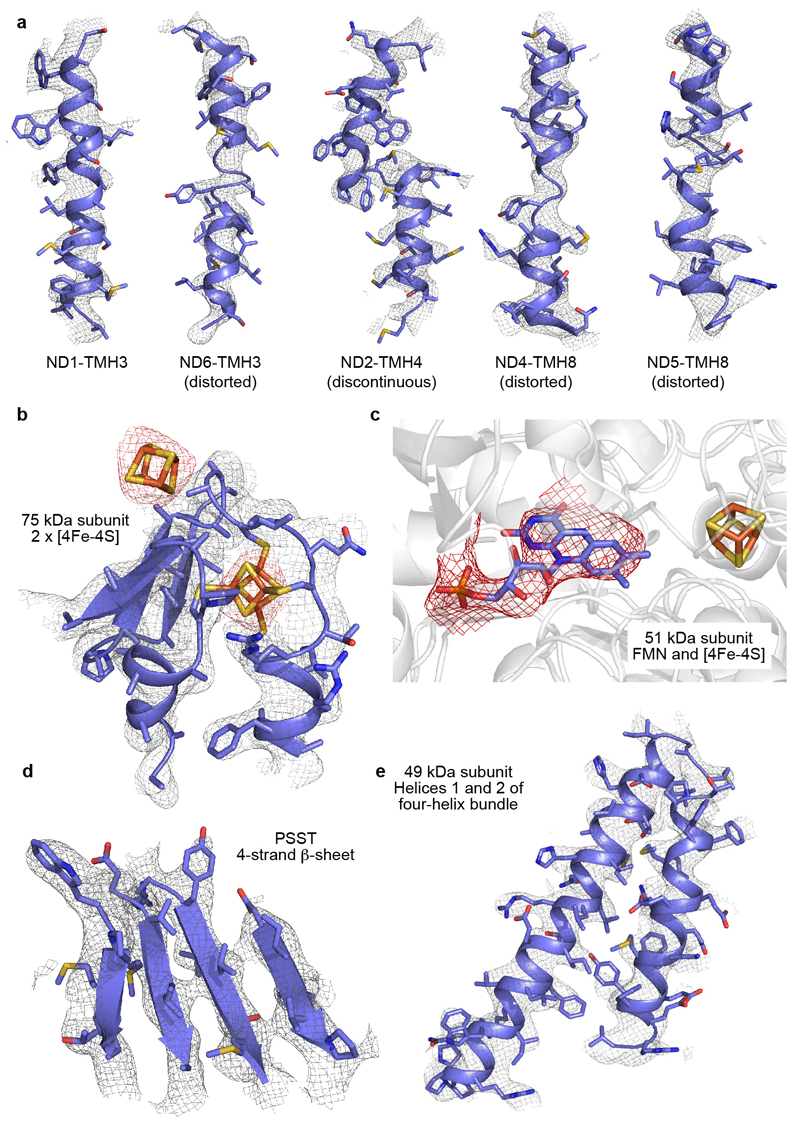
ED Figure 3. Example regions of the cryoEM density map for the core subunits, and the model fitted to the map.
a) A selection of TMHs from the membrane domain: TMH3 from ND1, the distorted TMHs in ND6, ND4 and ND5, and a discontinuous TMH from ND2. The series of TMHs from left to right illustrates the decrease in resolution along the domain. b) The two [4FeS4] clusters in the 75 kDa subunit (density in red, at higher contour level) with the protein ligating one of them. c) The FMN cofactor in the 51 kDa subunit. d) The β-sheet in subunit PSST, showing clear separation between the strands. e) Two helices from the 49 kDa subunit.
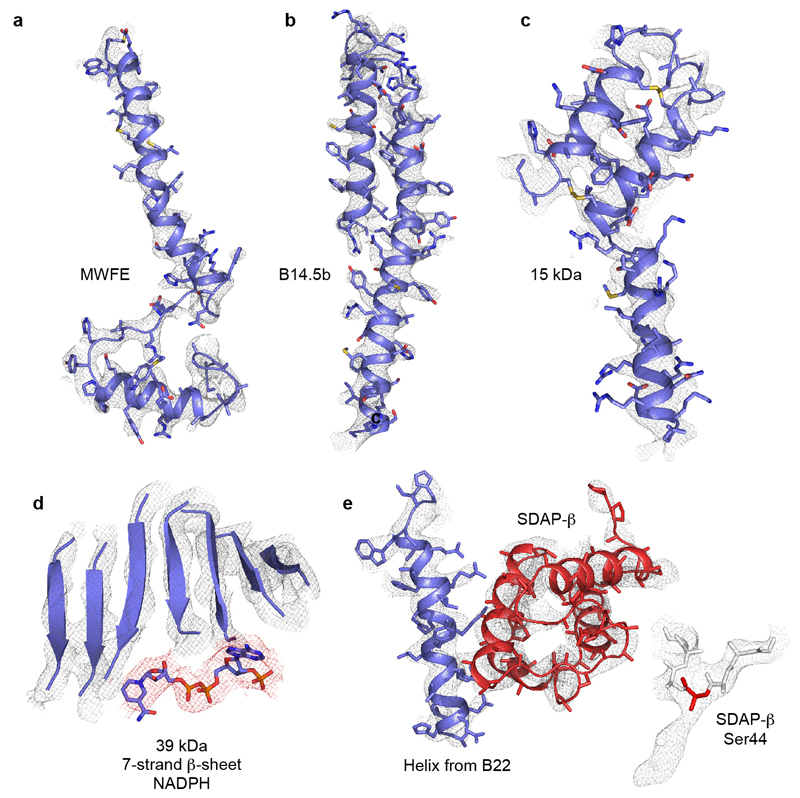
ED Figure 4. Example regions of the cryoEM density map for the supernumerary subunits, and the model fitted to the map.
a) Subunit MWFE, containing one TMH. b) Subunit B14.5, containing two TMHs. The N- and C-terminal loops are not shown. c) 15 kDa subunit on the IMS face, containing a CHCH domain with two disulphide bonds. The N- and C-terminal loops are not shown. d) The seven-strand β-sheet in the 39 kDa subunit, showing the separation of the strands, and the bound nucleotide (red density) modelled as NADPH. e) Helix 1, one of the arginine-rich helices, in B22, and SDAP-β, on the matrix side of the tip of the membrane domain. Inset: the weak density attached to Ser44 in SDAP-β attributed to the attached pantetheine-4'-phosphate group (side chain of Ser44 not shown).
ED Figure 5. Relationships between classes 1 and 3.
a) The structures for classes 1 and 3 have been superimposed using ND1 and ND3. In class 3, relative to class 1, the hydrophilic and distal membrane domains are both rotated and shifted, but the change in the membrane domain dominates. Although the changes appear to make the angle of the L-shaped molecule increase they do not originate from a hinge-like motion at the interface of the hydrophilic and membrane domains. Class 1 is in red, class 3 is in red (ND1 domain), wheat, blue and cyan. Details of the composition and movement of the domains are given in ED Table 4. b) The density for class 3 (white) is presented with the model for well-resolved regions of class 3 in blue (the model is enclosed in the density) and the model for poorly resolved regions in red (the model appears outside the density). The poorly resolved regions include the N-terminus of 49 kDa and the transverse helix in ND5, as well as elements of ND4, ND6, B14.7, ESSS and B15.
ED Table 1. Summary of the models for the core subunits of B. taurus complex I.
| Subunit | Other names* | Chain | Total residues | Modelled residues | Assigned residues | Unknown residues | % residues modelled | % residues assigned | % with sidechains | % unknown residues | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ND1 | Nqo8 | H | 318 | 3-200 | 3-200 | - | 93.1 | 93.1 | 89.0 | 0 | |
| class 1 | NuoH | 218-315 | 218-315 | ||||||||
| ND1 | Nqo8 | H | 318 | 3-315 | 3-315 | - | 98.4 | 98.4 | 89.0 | 0 | |
| class 2 | NuoH | ||||||||||
| ND2 | Nqo14 | N | 347 | 2-345 | 2-345 | - | 99.1 | 99.1 | 87.6 | 0 | |
| NuoN | |||||||||||
| ND3 | Nqo7 | A | 115 | 2-27 | 2-27 | - | 76.5 | 76.5 | 72.2 | 0 | |
| class 1 | NuoA | 51-112 | 51-112 | ||||||||
| ND3 | Nqo7 | A | 115 | 2-112 | 2-112 | - | 96.5 | 96.5 | 72.2 | 0 | |
| class 2 | NuoA | ||||||||||
| ND4 | Nqo13 | M | 459 | 3-459 | 3-459 | - | 99.6 | 99.6 | 96.3 | 0 | |
| NuoM | |||||||||||
| ND4L | Nqo11 | K | 98 | 2-96 | 2-96 | - | 96.9 | 96.9 | 96.9 | 0 | |
| NuoK | |||||||||||
| ND5 | Nqo12 | L | 606 | 2-605 | 2-605 | - | 99.7 | 99.7 | 88.1 | 0 | |
| NuoL | |||||||||||
| ND6 | Nqo10 | J | 175 | 2-172 | 2-172 | - | 97.7 | 97.7 | 79.4 | 0 | |
| NuoJ | |||||||||||
| 75 kDa | NDUFS1 | G | 704 | 8-692 | 8-209 | 210-692 | 97.3 | 28.7 | 8.9 | 68.6 | |
| Nqo3 | |||||||||||
| NuoG | |||||||||||
| 51 kDa | NDUFV1 | F | 444 | 14-438 | 14-438 | - | 95.7 | 95.7 | 18.2 | 0 | |
| Nqo1 | |||||||||||
| NuoF | |||||||||||
| 49 kDa | NDUFS2 | D | 430 | 5-50 | 5-50 | - | 96.7 | 96.7 | 87.0 | 0 | |
| class 1 | Nqo4 | 61-430 | 61-430 | ||||||||
| NuoCD | |||||||||||
| 49 kDa | NDUFS2 | D | 430 | 5-430 | 5-430 | - | 99.1 | 99.1 | 89.3 | 0 | |
| class 2 | Nqo4 | ||||||||||
| NuoCD | |||||||||||
| 30 kDa | NDUFS3 | C | 228 | 8-213 | 8-213 | - | 90.4 | 90.4 | 85.5 | 0 | |
| Nqo5 | |||||||||||
| NuoCD | |||||||||||
| 24 kDa | NDUFV2 | E | 217 | 8-193 | 8-193 | - | 85.7 | 85.7 | 1.8 | 0 | |
| Nqo2 | |||||||||||
| NuoE | |||||||||||
| PSST | NDUFS7 | B | 179 | 27-173 | 27-173 | - | 82.1 | 82.1 | 82.1 | 0 | |
| Nqo6 | |||||||||||
| NuoB | |||||||||||
| TYKY | NDUFS8 | I | 176 | 1-176 | 1-176 | - | 100 | 100 | 92.0 | 0 | |
| Nqo9 | |||||||||||
| Nuol | |||||||||||
The names of the human, T. thermophilus and E. coli subunits (if different to the names in B. taurus).
ED Table 2. Summary of the models of the supernumerary subunits of B. taurus complex I.
| Subunit | Human name | Chain | Total residues | Modelled residues | Assigned residues | Unknown residues | % residues modelled | % residues assigned | % with sidechains | % unknown residues |
|---|---|---|---|---|---|---|---|---|---|---|
| 42 kDa | NDUFA10 | O | 320 | 5-318 | 91-318 | 5-90 | 98.1 | 71.3 | 26.9 | 26.9 |
| 39 kDa | NDUFA9 | P | 345 | 2-186 | - | 2-186 | 82.0 | 0 | 0 | 82.0 |
| class 1 | 200-252 | 200-252 | ||||||||
| 280-324 | 280-324 | |||||||||
| 39 kDa | NDUFA9 | P | 345 | 2-336 | - | 2-336 | 97.1 | 0 | 0 | 97.1 |
| class 2 | ||||||||||
| 18 kDa | NDUFS4 | Q | 133 | 16-128 | - | 16-128 | 85.0 | 0 | 0 | 85.0 |
| 15 kDa | NDUFS5 | e | 105 | 6-94 | 6-94 | - | 84.8 | 84.8 | 41.9 | 0 |
| 13 kDa | NDUFS6 | R | 96 | 5-93 | 59-93 | 5-58 | 92.7 | 36.5 | 27. 1 | 56.3 |
| 10 kDa | NDUFV3 | s | 75 | 1-35† | - | 1-35† | 46.7 | 0 | 0 | 46.7 |
| AGGG | NDUFB2 | j* | 72 | 8-59 | - | 8-59 | 72.2 | 0 | 0 | 72.2 |
| ASHI | NDUFB8 | l | 158 | 5-122 | - | 5-122 | 74.7 | 0 | 0 | 74.7 |
| ESSS | NDUFB11 | g | 125 | 25-121 | 25-121 | - | 77.6 | 77.6 | 40.8 | 0 |
| KFYI | NDUFC1 | c | 49 | 1-46 | 1-46 | - | 93.9 | 93.9 | 55.1 | 0 |
| MNLL | NDUFB1 | f | 57 | 3-56 | 3-56 | - | 94.7 | 94.7 | 42. 1 | 0 |
| MWFE | NDUFA1 | a | 70 | 1-64 | 1-64 | 1-64 | 91.4 | 91.4 | 71.4 | 0 |
| PDSW | NDUFB10 | p | 175 | 4-172 | 76-142 | 4-75 | 96.6 | 38.3 | 30.9 | 58.3 |
| 143-172 | ||||||||||
| PGIV | NDUFA8 | X | 171 | 5-168 | 5-113 | 114-168 | 95.9 | 63.7 | 53.8 | 32.2 |
| SDAP-α | NDUFAB1 | T | 88 | 8-82 | 8-82 | - | 85.2 | 85.2 | 0 | 0 |
| SDAP-β | NDUFAB1 | U | 88 | 4-88 | 4-88 | - | 96.6 | 96.6 | 0 | 0 |
| SGDH | NDUFB5 | h | 143 | 7-140 | 7-45 | 46-140 | 93.7 | 27.3 | 21.0 | 66.4 |
| B22 | NDUFB9 | n | 178 | 10-175 | 10-136 | 137-175 | 93.3 | 71.3 | 35.4 | 21.9 |
| B18 | NDUFB7 | o | 136 | 57-114 | 57-114 | - | 42.6 | 42.6 | 2.9 | 0 |
| B17.2 | NDUFA12 | q | 145 | 2-139 | 2-139 | - | 95.2 | 95.2 | 0 | 0 |
| B17 | NDUFB6 | i | 127 | 6-32 | 6-32 | 40-118 | 83.5 | 21.3 | 14.2 | 62.2 |
| 40-118 | ||||||||||
| B16.6 | NDUFA13 | Z | 143 | 5-142 | 31-99 | 5-30 | 96.5 | 48.3 | 48.3 | 48.3 |
| 100-142 | ||||||||||
| B15 | NDUFB4 | m | 128 | 11-128 | 32-128 | 11-31 | 92.2 | 75.8 | 66.4 | 16.4 |
| B14.7 | NDUFA11 | Y | 140 | 1-138 | 1-138 | - | 98.6 | 98.6 | 98.6 | 0 |
| B14.5a | NDUFA7 | r | 112 | 4-70 | - | 4-70 | 77.7 | 0 | 0 | 77.7 |
| 100-119† | 100-119† | |||||||||
| B14.5b | NDUFC2 | d | 120 | 3-116 | 29-97 | 3-28 | 95.0 | 57.5 | 57.5 | 37.5 |
| 98-116 | ||||||||||
| B14 | NDUFA6 | W | 127 | 16-126 | 16-126 | - | 87.4 | 87.4 | 57.5 | 0 |
| B13 | NDUFA5 | V | 115 | 8-113 | 8-113 | - | 92.2 | 92.2 | 40.0 | 0 |
| B12 | NDUFB3 | k* | 97 | 1-74† | - | 1-74† | 76.3 | 0 | 0 | 76.3 |
| B9 | NDUFA3 | b | 83 | 1-80 | 1-45 | 46-80 | 96.4 | 54.2 | 54.2 | 42.2 |
| B8 | NDUFA2 | S | 98 | 16-95 | 16-95 | - | 81.6 | 81.6 | 0 | 0 |
The assignments of B12 and AGGG may be reversed
Arbitrary residue numbers
ED Table 3. Subunit-subunit interactions for the supernumerary subunits.
| Primary core subunit | Subunit interactions | Notes | |
|---|---|---|---|
| 42 kDa | ND2 | ND2, ND4, 49 kDa (N-terminus), ESSS, KFYI, B14.5b, B13 | Nucleoside kinase fold; does not contact hydrophilic core subunits or B14+SDAP-α |
| 39 kDa | PSST | ND1, ND3 (class 2 only), ND6, 75 kDa, 30 kDa, PSST, TYKY, 18 kDa, 13 kDa, B14 | Short-chain dehydrogenase/reductase fold20 with bound NAD(P)(H)21 |
| 18 kDa | 75 kDa | 75 kDa, 51 kDa, 49 kDa, 30 kDa, TYKY, 39 kDa, 10 kDa, B17.2, B14 | Four-strand β-sheet with helix located between 75 and 30 kDa |
| 15 kDa | ND2 | ND2, ND3, ND4L, ND6, PGIV, SGDH, B16.6, B14.5b | CHCH domain; IMS13 |
| 13 kDa | TYKY | 75 kDa, 49 kDa, TYKY, 39 kDa, B17.2 | Zinc-binding domain22 at the interface of TYKY, 75 and 49 kDa |
| 10 kDa | 24 kDa | 75 kDa, 51 kDa, 24 kDa, 18 kDa | Present in the three-subunit flavoprotein subcomplex with 51 and 24 kDa |
| AGGG | ND5 | ND5, B12 | 1 TMH; uncertain assignment of AGGG and B12 between chains j and k |
| ASHI | ND5 | ND4, ND5, B22, B15 | 1 TMH; large globular domain on the matrix face, TMH crosses transverse helix |
| ESSS | ND4 | ND4, ND5, 42 kDa, PDSW, SGDH, B22, B15, B14.5b | 1 TMH; poorly resolved N-terminus on matrix face; possible disulphide 112 to PDSW 154. |
| KFYI | ND2 | 42 kDa, B14.5b | 1 TMH; attached to complex by B14.5b |
| MNLL | ND4 | ND4, PDSW, SGDH | 1 TMH |
| MWFE | ND1 | ND1, ND6, TYKY, PGIV, B17.2, B16.6, B14.5a | 1 TMH; runs alongside ND1 TMH1 at proposed entrance to Q-binding site |
| PGIV | ND1 | ND1, ND2, ND4, 15 kDa, MWFE, SGDH, B16.6, B14.5b, B9 | Two CHCH domains; IMS13 |
| PDSW | ND4 | ND4, ND5, ESSS, MNLL, SGDH, B18, B17, B15, B14.5b | Extensive helix structure on IMS face; two likely internal disulphides (112-124, 76-83); possible disulphide 154 to ESSS-112 |
| SDAP-α | 30 kDa | B14 | ACP on the hydrophilic domain16 |
| SDAP-β | ND5 | ND5, ASHI, B22, B17, B12 | ACP on the membrane domain16 |
| SGDH | ND4 | ND2, ND4, ND5, ND6, 15 kDa, ESSS, MNLL, PDSW, PGIV, B22, B17, B16.6, B14.5b | 1 TMH; long helix running along the IMS face of the membrane domain |
| B22 | ND5 | ND4, ND5, ASHI, SDAP-β, SGDH, B17, B15, B12 | LYR protein that binds SDAP-β |
| B18 | ND5 | ND5, AGGG, PDSW, B17 | CHCH domain; IMS13 |
| B17.2 | TYKY | ND1, 75 kDa, PSST, TYKY, 18 kDa, 13 kDa, MWFE, B14.5a | Three-strand β-sheet and long loop running across hydrophilic domain |
| B17 | ND5 | ND5, PDSW, SDAP-β, SGDH, B22 | 1 TMH; helix on the matrix face, β-strand in IMS augments β-hairpin between ND5 TMHs |
| B16.6 | ND1 | ND1, ND3, ND6, 49 kDa, TYKY, 15 kDa, MWFE, PGIV, SGDH, B14.5a, B9 | 1 TMH; 6S-residue helix that crosses the membrane then turns along the IMS face |
| B15 | ND4 | ND4, ND5, ASHI, PDSW, B22, B14.7 | 1 TMH; long helix runs across the matrix face of the membrane domain |
| B14.7 | ND2 | ND2, ND4, ND5, SGDH, B15 | 4 TMHs; on the anchor of the transverse helix; likely disulphide 18-75 |
| B14.5a | 49 kDa | 75 kDa, 51 kDa, 49 kDa, 30 kDa, TYKY, MWFE, B17.2, B16.6, B13 | Long loop structure over hydrophilic domain |
| B14.5b | ND2 | ND2, ND4, 42 kDa, 15 kDa, ESSS, KFYI, PDSW, PGIV, SGDH, B15 | 2 TMHs; anchors KFYI to complex |
| B14 | 30 kDa | 75 kDa, 49 kDa, 30 kDa, PSST, 39 kDa, 18 kDa, SDAP-α, | LYR protein that binds SDAP-α15 |
| B13 | 30 kDa | 49 kDa, 30 kDa, 42 kDa, B14.5a | Three-helix bundle |
| B12 | ND5 | ND5, SDAP-β, AGGG, B22 | 1 TMH; uncertain assignment of AGGG and B12 between chains j and k |
| B9 | ND1 | ND1, ND3, ND6, TYKY, 15 kDa, PGIV, B16.6 | 1 TMH |
| B8 | 75 kDa | 75 kDa | Thioredoxin-like fold38. Possible disulphide 23-57 |
ED Table 4. Allocation of subunits to domains, and the relative movement of domains between classes 1, 2 and 3.
| Subunits | Transformation | |
|---|---|---|
| Class 1 vs. class 2 | ||
| Heel domain | ND1, ND3 | None (reference domain) |
| MWFE, B9, PGIV, B16.6 | ||
| Membrane domain | ND2, ND4L, ND6, ND4, ND5, N-terminus of 49 kDa subunit* | 3.9° rotation |
| 42 kDa, 15 kDa, KFYI, B14.5b, B14.7, MNLL, AGGG, B12, B15, SGDH, B17, B18, ASHI, B22+SDAP-β, PDSW, ESSS | 0.9 Å shift | |
| Hydrophilic domain | 75 kDa, 51 kDa, 24 kDa, 30 kDa, 49 kDa (except its N-terminus*), PSST, TYKY | 3.4° rotation |
| B8, B13, B14+SDAP-α, B14.5a, 39 kDa, B17.2, 18 kDa, 13 kDa, 10 kDa | 2.3 Å shift | |
| Class 1 vs. class 3 | ||
| Heel domain | ND1, ND3 | None (reference domain) |
| MWFE, B9, PGIV, B16.6 | ||
| Proximal membrane domain | ND2, ND4L, ND6 | 0.8° rotation |
| 42 kDa, 15 kDa, KFYI, B14.5b | 0.4 Å shift | |
| Distal membrane domain | ND4, ND5 | 3.1° rotation |
| B14.7, MNLL, AGGG, B12, B15, SGDH, B17, B18, ASHI, B22+SDAP-β, PDSW, ESSS | 2.9 Å shift | |
| Hydrophilic domain | 75 kDa, 51 kDa, 24 kDa, 30 kDa, 49 kDa (except its N-terminus), PSST, TYKY | 1.1° rotation |
| B8, B13, B14+SDAP-α, B14.5a, 39 kDa, B17.2, 18 kDa, 13 kDa, 10 kDa | 1.3 Å shift | |
The N-terminus of 49 kDa (residues 5-39) is displaced in class 1 relative to class 2 when considered from the core fold of the subunit because it lies on the surface of the membrane domain and moves with ND2.
ED Table 5. Data collection, refinement and model statistics for classes 1 and 2.
| Class 1 | Class 2 | |
|---|---|---|
| Data collection | ||
| Pixel size (Å) | 1.33 | 1.33 |
| Defocus range (µm) | 1.8 - 5.5 | 1.8 – 5.5 |
| Voltage (kV) | 300 | 300 |
| No. of particles | 48,033 | 33,301 |
| Orientation accuracy (°) | 0.92 | 0.95 |
| Model composition | ||
| Non-hydrogen atoms | 51, 117 | 51,652 |
| Protein residues | 7,789 | 7,891 |
| % of total | 91.5 | 92.7 |
| Core subunit residues | 4,294 | 4,344 |
| % of total | 95.5 | 96.6 |
| Supernumerary subunit residues | 3,495 | 3,547 |
| % of total | 87.0 | 88.3 |
| Refinement | ||
| Resolution (Å) | 4.27 | 4.35 |
| Average B-factor (Å2) | 93.4 | 110.4 |
| RMS deviations | ||
| Bonds (Å) | 0.008 | 0.008 |
| Angles (°) | 1.38 | 1.40 |
| Validation | ||
| Molprobity score | 2.11 | 2.50 |
| Clashscore, all atoms | 2.91 | 3.35 |
| Ramachandran plot | ||
| Favoured (%) | 86.66 | 86.92 |
| Outliers (%) | 3.57 | 3.72 |
Acknowledgements
We thank Richard Henderson and Garib Murshudov for helpful discussions, Jake Grimmett and Toby Darling for computational help, Shaoxia Chen and Christos Savva for electron microscopy help, and James Blaza for quantifying the ratio of deactive and active enzymes in our preparation. This work was supported by The Medical Research Council, grant numbers U105184322 (KRV, in Richard Henderson’s group) and U105663141 (JH).
Footnotes
Author contributions: JZ prepared protein; KRV carried out electron microscopy data collection and analysis with help from JZ; JZ built the initial model; JZ, KRV and JH worked together, led by JH, to model and analyse the data; JH designed the project; JH wrote the paper with help from JZ and KRV.
The authors declare no competing financial interests.
Map and model deposition: The EM maps and models for each class have been deposited with accession numbers EMD-4040, EMD-4032 and EMD-4041, and 5LDW.pdb, 5LC5.pdb and 5LDX.pdb, for classes 1, 2 and 3, respectively.
Author Information: Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 2.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 3.Galkin A, et al. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J Biol Chem. 2008;283:20907–20913. doi: 10.1074/jbc.M803190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galkin A, Abramov AY, Frakich N, Duchen MR, Moncada S. Lack of oxygen deactivates mitochondrial complex I: implications for ischemic injury? J Biol Chem. 2009;284:36055–36061. doi: 10.1074/jbc.M109.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Caballero L, Guerrero-Castillo S, Nijtmans J. Unraveling the complexity of mitochondrial complex I assembly: a dynamic process. Biochim Biophys Acta. 2016;1857:980–990. doi: 10.1016/j.bbabio.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 6.Fassone E, Rahman S. Complex I deficiency: clinical features, biochemistry and molecular genetics. J Med Genet. 2012;49:578–590. doi: 10.1136/jmedgenet-2012-101159. [DOI] [PubMed] [Google Scholar]
- 7.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 8.Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 9.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zickermann V, et al. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, et al. Structure of subcomplex Iβ of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc Natl Acad Sci USA. 2015;112:12087–12092. doi: 10.1073/pnas.1510577112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk R, et al. NDUFB7 and NDUFA8 are located at the intermembrane surface of complex I. FEBS Lett. 2011;585:737–743. doi: 10.1016/j.febslet.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 14.Banci L, et al. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- 15.Angerer H, et al. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc Natl Acad Sci USA. 2014;111:5207–5212. doi: 10.1073/pnas.1322438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runswick MJ, Fearnley IM, Skehel JM, Walker JE. Presence of an acyl carrier protein in NADH:ubiquinone oxidoreductase from bovine heart mitochondria. FEBS Lett. 1991;286:121–124. doi: 10.1016/0014-5793(91)80955-3. [DOI] [PubMed] [Google Scholar]
- 17.Feng D, Witkowski A, Smith S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J Biol Chem. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan DI, Vogel HJ. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J. 2010;430:1–19. doi: 10.1042/BJ20100462. [DOI] [PubMed] [Google Scholar]
- 19.Johansson K, et al. Structural basis for substrate specificities of cellular deoxyribonucleoside kinases. Nat Struct Biol. 2001;8:616–620. doi: 10.1038/89661. [DOI] [PubMed] [Google Scholar]
- 20.Fearnley IM, Walker JE. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim Biophys Acta. 1992;1140:105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- 21.Abdrakhmanova A, Zwicker K, Kerscher S, Zickermann V, Brandt U. Tight binding of NADPH to the 39-kDa subunit of complex I is not required for catalytic activity but stabilizes the multiprotein complex. Biochim Biophys Acta. 2006;1757:1676–1682. doi: 10.1016/j.bbabio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Kmita K, et al. The accessory NUMM (NDUFS6) subunit harbors a Zn-binding site and is essential for biogenesis of mitochondrial complex I. Proc Natl Acad Sci USA. 2015;112:5685–5690. doi: 10.1073/pnas.1424353112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll J, Ding S, Fearnley IM, Walker JE. Post-translational modifications near the quinone binding site of mammalian complex I. J Biol Chem. 2013;288:24799–24808. doi: 10.1074/jbc.M113.488106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirst J, Roessler MM. Energy conversion, redox catalysis and generation of reactive oxygen species by respiratory complex I. Biochim Biophys Acta. 2016;1857:872–883. doi: 10.1016/j.bbabio.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murai M, Mashimo Y, Hirst J, Miyoshi H. Exploring interactions between the 49 kDa and ND1 subunits in mitochondrial NADH-ubiquinone oxidoreductase (complex I) by photoaffinity labeling. Biochemistry. 2011;50:6901–6908. doi: 10.1021/bi200883c. [DOI] [PubMed] [Google Scholar]
- 26.Cooley RB, Arp DJ, Karplus PA. Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J Mol Biol. 2010;404:232–246. doi: 10.1016/j.jmb.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tocilescu MA, Fendel U, Zwicker K, Kerscher S, Brandt U. Exploring the ubiquinone binding cavity of respiratory complex I. J Biol Chem. 2007;282:29514–29520. doi: 10.1074/jbc.M704519200. [DOI] [PubMed] [Google Scholar]
- 28.Babot M, et al. ND3, ND1 and 39 kDa subunits are more exposed in the de-active form of bovine mitochondrial complex I. Biochim Biophys Acta. 2014;1837:929–939. doi: 10.1016/j.bbabio.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murai M, Miyoshi H. Current topics on inhibitors of respiratory complex I. Biochim Biophys Acta. 2016;1857:884–891. doi: 10.1016/j.bbabio.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Sharma V, et al. Redox-induced activation of the proton pump in the respiratory complex I. Proc Natl Acad Sci USA. 2015;112:11571–11576. doi: 10.1073/pnas.1503761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharpley MS, Shannon RJ, Draghi F, Hirst J. Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry. 2006;45:241–248. doi: 10.1021/bi051809x. [DOI] [PubMed] [Google Scholar]
- 32.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 34.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balsa E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Brockmann C, et al. The oxidized subunit B8 from human complex I adopts a thioredoxin fold. Structure. 2004;12:1645–1654. doi: 10.1016/j.str.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Cryst. 2015;D71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 42.Källberg M, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protocols. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chovancova E, et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collaborative computational project. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]