Abstract
STUDY QUESTION
Can a generally applicable morphokinetic algorithm suitable for Day 3 transfers of time-lapse monitored embryos originating from different culture conditions and fertilization methods be developed for the purpose of supporting the embryologist's decision on which embryo to transfer back to the patient in assisted reproduction?
SUMMARY ANSWER
The algorithm presented here can be used independently of culture conditions and fertilization method and provides predictive power not surpassed by other published algorithms for ranking embryos according to their blastocyst formation potential.
WHAT IS KNOWN ALREADY
Generally applicable algorithms have so far been developed only for predicting blastocyst formation. A number of clinics have reported validated implantation prediction algorithms, which have been developed based on clinic-specific culture conditions and clinical environment. However, a generally applicable embryo evaluation algorithm based on actual implantation outcome has not yet been reported.
STUDY DESIGN, SIZE, DURATION
Retrospective evaluation of data extracted from a database of known implantation data (KID) originating from 3275 embryos transferred on Day 3 conducted in 24 clinics between 2009 and 2014. The data represented different culture conditions (reduced and ambient oxygen with various culture medium strategies) and fertilization methods (IVF, ICSI). The capability to predict blastocyst formation was evaluated on an independent set of morphokinetic data from 11 218 embryos which had been cultured to Day 5.
PARTICIPANTS/MATERIALS, SETTING, METHODS
The algorithm was developed by applying automated recursive partitioning to a large number of annotation types and derived equations, progressing to a five-fold cross-validation test of the complete data set and a validation test of different incubation conditions and fertilization methods. The results were expressed as receiver operating characteristics curves using the area under the curve (AUC) to establish the predictive strength of the algorithm.
MAIN RESULTS AND THE ROLE OF CHANCE
By applying the here developed algorithm (KIDScore), which was based on six annotations (the number of pronuclei equals 2 at the 1-cell stage, time from insemination to pronuclei fading at the 1-cell stage, time from insemination to the 2-cell stage, time from insemination to the 3-cell stage, time from insemination to the 5-cell stage and time from insemination to the 8-cell stage) and ranking the embryos in five groups, the implantation potential of the embryos was predicted with an AUC of 0.650. On Day 3 the KIDScore algorithm was capable of predicting blastocyst development with an AUC of 0.745 and blastocyst quality with an AUC of 0.679. In a comparison of blastocyst prediction including six other published algorithms and KIDScore, only KIDScore and one more algorithm surpassed an algorithm constructed on conventional Alpha/ESHRE consensus timings in terms of predictive power.
LIMITATIONS, REASONS FOR CAUTION
Some morphological assessments were not available and consequently three of the algorithms in the comparison were not used in full and may therefore have been put at a disadvantage. Algorithms based on implantation data from Day 3 embryo transfers require adjustments to be capable of predicting the implantation potential of Day 5 embryo transfers. The current study is restricted by its retrospective nature and absence of live birth information. Prospective Randomized Controlled Trials should be used in future studies to establish the value of time-lapse technology and morphokinetic evaluation.
WIDER IMPLICATIONS OF THE FINDINGS
Algorithms applicable to different culture conditions can be developed if based on large data sets of heterogeneous origin.
STUDY FUNDING/COMPETING INTEREST(S)
This study was funded by Vitrolife A/S, Denmark and Vitrolife AB, Sweden. B.M.P.’s company BMP Analytics is performing consultancy for Vitrolife A/S. M.B. is employed at Vitrolife A/S. M.M.’s company ilabcomm GmbH received honorarium for consultancy from Vitrolife AB. D.K.G. received research support from Vitrolife AB.
Keywords: time-lapse, algorithm, decision support tool, prediction model, morphokinetic, implantation, embryo selection, blastocyst
Introduction
As single embryo transfers (SETs) become the desired standard of care world-wide in clinical IVF, a major challenge in the field of embryology is to develop quantitative methods of identifying which embryo, within a cohort, possesses the highest probability of resulting in a healthy live birth. Since the beginning of human IVF, embryo selection for transfer has relied on the assessment of morphology (Edwards et al., 1984; Claman et al., 1987), and over the intervening three decades, elegant grading systems have been created to assist in the classification of embryos at successive stages of development (Scott and Smith 1998; Gardner and Schoolcraft, 1999; Tesarik and Greco, 1999; Van Royen et al., 1999; Gardner et al., 2000; Scott et al., 2000, 2007; Montag and van der Ven, 2001; De Neubourg et al., 2004; Ahlström et al., 2011; Diamond et al., 2012). Furthermore, the significance of assessing embryos at key discrete times, thereby including the variable of time and rates of development, has also been included in embryo scoring systems (Sakkas et al., 2001; Gardner and Sakkas, 2003; Salumets et al., 2003).
In 2011 an expert meeting from Alpha and ESHRE resulted in a consensus paper on morphological criteria for embryo assessment (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011). In the same year the first study was published which described a morphokinetic algorithm based on parameters, determined by time-lapse imaging and using implantation as endpoint (Meseguer et al., 2011). Time-lapse technology enables continuous monitoring of embryo development in vitro. In 1997 Payne and colleagues successfully imaged the events following ICSI and were able to monitor polar body extrusion and pronuclear formation in the human embryo in vitro for the first time (Payne et al., 1997). In clinical embryology routine time-lapse imaging systems have only been available since 2008 (Pribenszky et al., 2010). The use of time-lapse allows for the mapping of morphological changes or events with the exact time-point of occurrence (Ciray et al., 2014). This in turn has initiated the search for morphokinetic parameters that are characteristic of implantation, blastocyst formation and aneuploidy, and subsequently several reports have been published on this subject (e.g. Lemmen et al., 2008; Wong et al., 2010; Meseguer et al., 2011, 2012; Azzarello et al., 2012; Cruz et al., 2012; Dal Canto et al., 2012; Rubio et al., 2012, 2014; Chamayou et al., 2013; Conaghan et al., 2013; Freour et al., 2013; Campbell et al., 2013a, 2013b; Aguilar et al., 2014; Liu et al., 2014, 2016; VerMilyea et al. 2014; Yalçınkaya et al., 2014; Basile et al., 2015; Cetinkaya et al., 2015; Gardner et al., 2015; Milewski et al., 2015; Yang et al., 2015).
Analysis of implantation-related morphokinetic characteristics has facilitated the development of algorithms aimed at being clinically applicable in embryo evaluation for prediction of aneuploidy risk (Campbell et al., 2013a), blastocyst formation or implantation (e.g. Meseguer et al., 2011; Conaghan et al., 2013; Campbell et al., 2013b; VerMilyea et al. 2014; Basile et al., 2015; Milewski et al., 2015; Goodman et al., 2016; Liu et al., 2016; Motato et al., 2016). Prediction algorithms have so far been developed on the basis of relatively small data sets, with numbers between 292 and 432 embryos in prediction of blastocyst formation and 132–754 embryos in prediction of implantation. These are relatively low numbers, which do not favour the development of a generally applicable algorithm. Further, variations in clinic-specific characteristics may not be represented in the data (e.g. incubation conditions, fertilization method, media, etc.).
The majority of the published morphokinetic algorithms are based on clinic-/chain-specific data and only a few have been tested in prospective randomized trials (Rubio et al., 2014; Adamson et al., 2016). Some algorithms have been tested on independent data sets or in other clinical settings (Kirkegaard et al., 2014; VerMilyea et al., 2014; Yalçınkaya et al., 2014; Basile et al., 2015; Freour et al., 2015) with varying outcomes. While some clinics have implemented morphokinetic algorithms based on data from their own clinics or chain of clinics, these algorithms may not be generally applicable without modification (Best et al., 2013; Kirkegaard et al., 2014; Yalçınkaya et al., 2014; Freour et al, 2015). One aspect of this may be related to the documented effect of clinical and laboratory conditions on embryo morphokinetics (Wale and Gardner, 2010, 2016; Ciray et al., 2012; Dal Canto et al., 2012; Kirkegaard et al., 2012, 2013, 2014; Yalçınkaya et al. 2014). Hence, an algorithm developed from data which covers too narrow a range of conditions is at risk of only performing adequately on the data set it was developed from, or on data that originates from very similar clinical conditions. This has been documented for one generally applicable algorithm for blastocyst prediction, which failed when tested in a multi-clinical outcome study (Kirkegaard et al., 2014).
Morphokinetic time ranges representing optimum implantation may vary between clinics (Freour et al., 2015). Therefore an algorithm aiming at avoiding embryos of low implantation potential rather than selecting embryos of the highest potential is more likely to be generally applicable. In addition, such an algorithm reduces the risk of rejecting embryos which may still have potential for implantation. This further underlines that when creating an algorithm capable of embracing a broad range of clinical variation, it is important to have a sufficient amount of data that includes multiple clinics and different culture conditions.
Here we report a morphokinetic algorithm developed on a data set of 3275 embryos transferred on Day 3. For all embryos, the development up to Day 3 was documented by time-lapse and for every embryo the fate after transfer (implanted versus non-implanted) was known, leading to the so-called known implantation data (KID). The resulting algorithm, KIDScore D3 Basic hereafter called KIDScore, was based on six annotations, consisting of one morphological and five morphokinetic events, and the output is a ranking of relative implantation potential from a score of 1 to 5. One of the aims with KIDScore was to achieve a deselection algorithm, so that the higher score groups contain a large proportion of the embryos, subsequently leaving the final choices for transfer or freeze to the embryologist. The algorithm was validated on an independent data set of embryos cultured to Day 5 to test its capability to predict blastocyst formation. It was also compared with other published morphokinetic algorithms.
Materials and Methods
Data used for developing the KIDScore algorithm
Data were collected from 24 clinics between 2009 and 2014 which agreed to share data for time periods from one to several years. No specific methods were applied in the selection of the clinics. No demographic patient data were available except for patient age and type of treatment (IVF or ICSI). Data were subjected to the following criteria and filtering: Patients aged 40+ were excluded and only Day 3 transfers were included, and a total of 3275 embryos with known outcome after transfer remained (see Table I for details).
Table I.
Overview of data sets: Day 3 transfer data set (24 clinics) and the Day 5 incubation data set (31 clinics), used for development of KIDScore and blastocyst predictions, respectively, according to fertilization (Fert.) method and oxygen level during incubation.
| Fert. method/oxygen | Day 3 transfers | Day 5 incubation | ||
|---|---|---|---|---|
| Embryos (n) | Treatments (n) | Embryos (n) | Treatments (n) | |
| ICSI/reduced | 1042 | 682 | 9294 | 1977 |
| IVF/reduced | 478 | 314 | 915 | 322 |
| ICSI/ambient | 1364 | 791 | 800 | 163 |
| IVF/ambient | 280 | 191 | 53 | 19 |
| Unknown | 111 | 71 | 156 | 36 |
| Total | 3275 | 2049 | 11218 | 2517 |
Data used for developing the algorithm originated from embryos with known implantation after uterine transfer. As the number of gestational sacs was the most widely used outcome in this data set, this was chosen as the endpoint. In cases where this information was missing it was substituted by the number of foetal heart beats. If two embryos were transferred, only cases where either both or none of the embryos implanted were used, according to the KID definition. Treatments with more than two embryo transfers were not included. The references for the recorded timings are the time of insemination defined by adding sperm to oocytes (IVF) or by injecting sperm into the oocyte (ICSI).
Methods for evaluating predictive power
Receiver operating characteristics (ROCs) curves were calculated and the area under the curve (AUC) was used as determination of the predictive value (e.g. Fawcett, 2006). AUC is a commonly used quantifier of the overall predictive capability of an algorithm. Algorithms with zero predictive capability will have AUC values of 0.5 on average and algorithms with perfect prediction will have AUC values of 1. In this study, AUC values were used as the primary quantification method for predictive capability. A relative predictive value (RPV), given by the linear rescaling of AUC values to a Gini Index, calculated as 2AUC-1 (Hand and Till, 2001), was used as a relative measure of the capability of the algorithms to predict blastocyst formation and blastocyst quality. This measure provides a more comprehensible scale, where 0 signifies no predictive ability and 1 signifies perfect prediction. The predictive power of the respective algorithms was tested against a common reference using the method for estimation of the AUC for clustered data proposed by Obuchowski (1997). P-values were adjusted using Holm–Bonferroni correction (Holm, 1979). In order to counter false claims of significant differences, a conservative significance level of P < 0.001 was chosen.
Methods used for developing the KIDScore algorithm
As an initial criterion, the presence of two pronuclei in the embryo was a prerequisite for inclusion in the analysis. In agreement with common embryological practice (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011) this criterion was applied prior to developing the algorithm.
After testing a range of algorithm types, a decision tree approach was selected. This type of algorithm was derived by automatically splitting the entire population of embryo timings into smaller subgroups. This procedure is known as recursive partitioning where all splits are dichotomous and each subgroup may in turn be split. The ‘rpart’ software package (Therneau et al., 2015) was used in R (R Core Team, 2015). The specific settings applied in the rpart procedure ensured that only the largest subgroup would be further split in order to create a deselection rather than a selection algorithm. Each of the subgroups that were not further split represents a population of embryos which were given a score.
Further, we aimed at developing an algorithm based on variables which can be consistently and objectively annotated, thereby creating an algorithm which is easy to implement as a decision support tool in clinical settings. A large number of annotation types and derived equations were tested in order to have a biologically meaningful algorithm with good numerical performance. The first condition is partly subjective.
Five-fold cross-validation
A five-fold cross-validation approach was applied to evaluate the robustness of the parameters. The Day 3 transfer data were randomly divided into five groups and five algorithm calibration–validation runs were performed. For each run, we performed a split procedure in a fixed order that followed the structure of the obtained classification tree in order to generate a calibration of the splitting value for each variable in 4/5 of the data. The remaining 1/5 of the data which was not part of the calibration was then used for validation. The calibrated splitting values and the AUCs for calibrations and validations were recorded.
Algorithm robustness with respect to incubation environment and fertilization method
The Day 3 transfer data included a four level grouping comprising the combinations of the two variables insemination (IVF or ICSI) and oxygen level (reduced (~5%) or ambient (~20%)), which have been shown to impact morphokinetics (Kirkegaard et al., 2013; Bodri et al., 2015). The robustness in relation to these different factors was tested as follows: similar to five-fold validation, all of the data except for those in a given environmental group were used to optimize the splitting values according to the structure of the obtained classification tree. Data from the excluded environmental group were then used for validation and the splitting values and AUC were recorded.
Data used for predicting blastocyst formation and quality
The KIDScore algorithm was tested on morphokinetic data from a separate group of 11 218 embryos cultured to Day 5, using AUC as the principal measure of predictive capability of blastocyst formation and quality. Data were collected from 31 clinics between 2010 and 2014 and were not overlapping with any of the data used in the development of the algorithms investigated in this study, see Table I for details. As for the data used for developing the KIDScore algorithm, the clinics agreed to share data for time periods from one to several years and no specific methods were applied in the selection of these clinics. Only data from embryos exhibiting at least one cell division were included.
There was overlap at the clinic level which provided data from Day 3 cycles used for developing the KIDScore algorithm and from Day 5 cycles for predicting blastocyst development, but independence at the patient and treatment levels. Here, blastocyst formation was defined as whether or not an embryo had reached the stage of a fully developed blastocyst at 120 h post insemination. A subset of the embryos (n = 4136) was classified at 114–120 h post insemination using Gardner's scoring criteria (Gardner and Schoolcraft, 1999). For simplification, three groups were created for inclusion in the analysis: top quality (TQ), good quality (GQ) and poor quality (PQ) blastocysts. The TQ group was comprised of 3AA, 4AA and 5AA blastocysts, and the GQ group was comprised of 3/4/5BB, AB or BA. Blastocysts with scorings below those of the GQ group were classified as PQ. The above three groups are in accordance with Cetinkaya et al. (2015). The blastocyst quality was further categorized in two groups; TQ and GQ blastocysts versus PQ blastocysts.
Algorithms used for comparing capability to predict blastocyst formation and quality
A number of published algorithms were included in the analysis of blastocyst formation and quality for evaluation and comparison purposes. Three of the evaluated algorithms originally included morphology assessments. In order to make this a pure morphokinetic algorithm comparison, we have excluded morphological assessments from the models in question, hereby only evaluating the morphokinetic component. Adding or removing morphology based assessments to the algorithms is hereby implicitly assumed to give each algorithm comparable increase or reduction in predictive capability. However, the comparison based on this assumption can be biased if the morphological components omitted not are statistically independent of the morphokinetic components included.
The algorithm described in Meseguer et al. (2011) (referred to as ‘Meseguer’), and the algorithm described in Basile et al. (2015) (referred to as ‘Basile’) are both based on symmetrical decision trees with eight morphokinetic scoring levels, from A+ as the highest to E as the lowest. This implies that the morphological group F (‘clearly not viable’) cannot be explored here as this seems to be a partially subjective criterion. The E group could only partially be obtained here, using the division from 1 to 3 cells (for Meseguer this timing was not specified but referred to as ‘abrupt division’ interpreted in this study as progression from 1 to 3 cells within 1 h, and for Basile direct cleavage from 1 to 3 cells within 5 h according to Basile et al., 2015), while uneven blastomere size at the 2-cell stage and multinucleation at the 4-cell stage were not available in the current study.
The algorithms in Conaghan et al. (2013) and VerMilyea et al. (2014) are based on two early time intervals comprising the 4-cell stage. The algorithm described in Conaghan et al. (2013) (referred to as ‘Eeva I’) has two possible scores. Building upon the same two intervals, VerMilyea et al. (2014) described an algorithm with three scores (referred to as ‘Eeva II’).
The algorithm described in Milewski et al. (2015) (referred to as ‘Milewski’) has a score derived by multiplication of factors that differ depending on whether morphokinetic effects occur inside or outside given intervals. The calculated score is here shown with the quartile grouping from the original paper.
The algorithm described in Liu et al. (2016) (referred to as ‘Liu’) has a score derived by a decision tree with five strictly and one partially morphokinetic scoring levels, from A+ as the highest to E as the lowest, excluding the morphological group F as the required data were not available in this study. The E score contains direct cleavage, which is implemented as stated in Liu et al. (2016), but also reverse cleavage and intercellular contact points at the end of the 4-cell stage, and data for the latter two variables were not available in this study.
As elaborated above the Meseguer, Basile and Liu algorithms were not implemented fully as described in the original studies.
The algorithm used for common reference, referred to as ‘Alpha/ESHRE’, was derived by creating an additive score using the Day 3 transfer timings from the Istanbul consensus workshop (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011). Initially all embryos are given a score of 1. Fulfilling the criteria of the assessments referred to as early cleavage check (exactly two blastomeres at 26 h post ICSI and 28 h post IVF), Day 2 embryo assessment (exactly four blastomeres at 44 h post insemination) and Day 3 embryo assessment (minimum developmental stage: eight blastomeres 68 h post insemination), each, respectively, adds 1 to the initial score. These criteria resulted in a 4-score algorithm.
Results
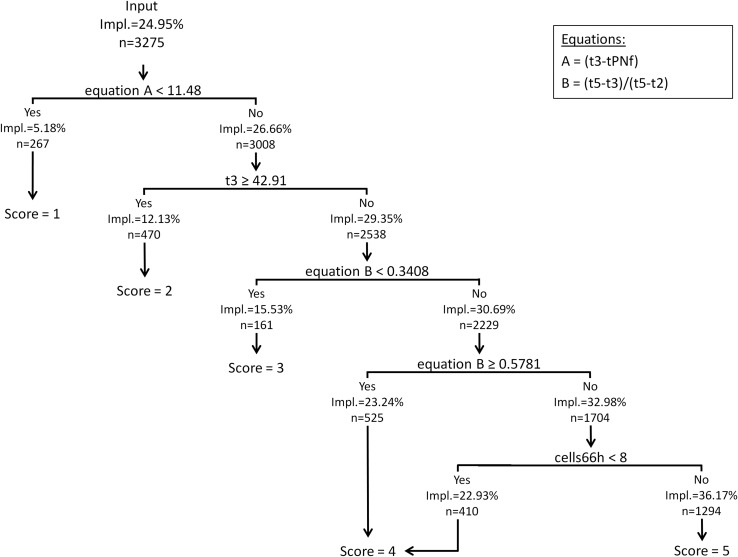
The annotations used in the final KIDScore algorithm are 2PN (the number of pronuclei equals 2 at the 1-cell stage), tPNf (time from insemination to pronuclei fading at the 1-cell stage), t2 (time from insemination to the 2-cell stage), t3 (time from insemination to the 3-cell stage), t5 (time from insemination to the 5-cell stage) and t8 (time from insemination to the 8-cell stage). Only t3 is used directly. The utilization of t8 was derived from the criteria for number of cells at 66 h (cells66h). Therefore, the criterion of cells66h < 8 can be substituted with t8 > 66 h, which can be used in implementations of the algorithm.
The overall implantation rate of all 3275 embryos utilized for development of the algorithm was 24.95%. Using recursive partitioning we obtained a decision tree algorithm which ranked embryos according to their implantation potential between 5.18% (Score 1) and 36.17% (Score 5) as shown in Fig. 1. In the first split, embryos which had a very fast development into the 3-cell stage were deselected using the equation A = t3−tPNf (Score 1). In the second split, embryos that were very slow to develop were deselected by measuring t3 (Score 2). In the third and fourth splits, embryos with irregular division were deselected, measured by equation B = (t5−t3)/(t5−t2). Equation B was applied twice with a low and a high threshold (Scores 3 and 4, respectively). The last split deselected embryos which had developed beyond t5 but not reached the 8-cell stage at 66 h after insemination (Gardner and Sakkas, 2003). In KIDScore, this is an additional way for an embryo to obtain the Score 4. All embryos which were not deselected in any of the above splits were given the Score 5. Sub-optimum groups of embryos were identified in chronological order of development, their implantation potential increasing in each step. The corresponding scores signified a seven-fold difference in implantation potential between Score 1 and Score 5.
Figure 1.
Classification tree diagram illustrating the KIDScore algorithm, which was based on morphokinetic information from embryo culture up until Day 3. The Scores 1–5 are indicated for each tree split with the corresponding number of embryos and implantation rate (Impl.). tPNf, time (h) from insemination to pronuclei fading at the 1-cell stage. t2, t3, t5, time (h) from insemination to the 2, 3 or 5-cell stage, respectively. cells66h, number of cells 66 h after insemination.
The robustness of the algorithm parameters was then tested by applying it to different randomly generated subsets of the complete data set by conducting a five-fold validation test (Table II) as well as by testing the robustness in relation to incubation conditions and fertilization method (Table III). When 1/5 of the data set was removed during the five-fold validation process, the splitting values (parameters) of the variables used for KIDScore did not display any substantial differences. The implantation rate varied from 22.0% to 27.6% between these subsets. Overall, the splitting values were comparable although a small variation was evident in the split using equation A and the low split of equation B (Table II). By excluding the incubation and fertilization groupings one at a time, very similar values were obtained for all splits comprised by the algorithm (Table III). The splitting values were only mildly impacted by the incubation and fertilization groupings where the low split of the equation B parameter (Table III) displayed the largest variation relative to its value when using the complete calibration data set. The AUC values did not differ substantially between calibration and validation data, with the exception of the subset validation where oxygen level and fertilization method were unknown. Here the AUC value was quite high compared to the calibration, presumably due to uncertainty caused by the relatively small size of this particular subset (n = 111). The implantation rate varied from 21.6% to 28.4% between these subsets. Additionally the predictive performances obtained for SET and double embryo transfer (DET) were equivalent, with AUC = 0.642 for SET and AUC = 0.658 for DET. Combined, the five-fold validation test and the evaluation of the culture and fertilization conditions indicate that the KIDScore algorithm captures a structure which is robust across data that spans multiple conditions.
Table II.
Five-fold validation.
| Data subset used for validation | Calibration subset (n) | Validation subset (n) | Splitting values | AUC calibration | AUC validation | ||||
|---|---|---|---|---|---|---|---|---|---|
| Equation A | t3 (h) | Equation B Low | Equation B High | cells66h | |||||
| All data | 3275 | 11.481 | 42.905 | 0.341 | 0.578 | 7.5 | 0.653 | ||
| 1 | 2620 | 655 | 11.481 | 42.885 | 0.441 | 0.577 | 7.5 | 0.655 | 0.648 |
| 2 | 2620 | 655 | 11.330 | 43.085 | 0.359 | 0.578 | 7.5 | 0.653 | 0.645 |
| 3 | 2620 | 655 | 11.481 | 42.905 | 0.341 | 0.578 | 7.5 | 0.656 | 0.640 |
| 4 | 2620 | 655 | 9.511 | 42.910 | 0.341 | 0.602 | 7.5 | 0.654 | 0.646 |
| 5 | 2620 | 655 | 11.999 | 42.905 | 0.441 | 0.578 | 7.5 | 0.656 | 0.642 |
| Mean | 11.16 | 42.94 | 0.38 | 0.58 | 7.50 | ||||
| SD | 0.96 | 0.08 | 0.05 | 0.01 | 0.00 | ||||
The stability of the algorithm structure was verified by performing five calibration procedures on the same variables and in the same order as the final output obtained by the rpart routine. Each split was first calibrated by excluding 1/5 of the data at a time (calibration subset). The calibrated parameters obtained from the 4/5 of the data which was not excluded were used to generate scores for both the calibration subset and the remaining 1/5 of the data (validation subset). The AUC values are shown both for the scored calibration subset (AUC calibration) and the validation subset (AUC validation). Equation A, (t3−tPNf); Equation B, (t5−t3)/(t5−t2); cells66h, number of cells at 66 h. AUC, area under the curve.
Table III.
Incubation and fertilization method.
| Data subset used for validation | Calibration subset (n) | Validation subset (n) | Splitting values | AUC calibration | AUC validation | ||||
|---|---|---|---|---|---|---|---|---|---|
| Equation A | t3 (h) | Equation B Low | Equation B High | cells66h | |||||
| None | 3275 | 11.481 | 42.905 | 0.341 | 0.578 | 7.5 | 0.653 | ||
| red.ox_ICSI | 2233 | 1042 | 12.010 | 43.475 | 0.446 | 0.579 | 7.5 | 0.646 | 0.647 |
| red.ox_IVF | 2797 | 478 | 11.427 | 42.945 | 0.341 | 0.578 | 7.5 | 0.651 | 0.657 |
| amb.ox_ICSI | 1911 | 1364 | 9.517 | 42.175 | 0.341 | 0.578 | 7.5 | 0.664 | 0.621 |
| amb.ox_IVF | 2995 | 280 | 11.481 | 42.570 | 0.340 | 0.577 | 7.5 | 0.652 | 0.633 |
| Unknown | 3164 | 111 | 11.481 | 42.905 | 0.441 | 0.577 | 7.5 | 0.651 | 0.724 |
| Weighted mean | 10.82 | 42.76 | 0.38 | 0.58 | 7.50 | ||||
| SD | 1.12 | 0.56 | 0.05 | 0.00 | 0.00 | ||||
The stability of the algorithm structure is verified by performing a calibration on the same variables and in the same order as the final output obtained by the recursive partitioning routine. The splits were first calibrated by excluding one incubation type at a time (calibration subset). red.ox, reduced oxygen ~5% . amb.ox, ambient oxygen ~20%.The obtained calibrated parameters were used to generate scores for both the calibration subset and the excluded incubation type (Validation Subset). The AUC values are shown both for the scored calibration subset (AUC Calibration) and the validation subset (AUC Validation). Equation A, (t3−tPNf); Equation B, (t5−t3)/(t5−t2); cells66h, number of cells at 66 h after insemination.
Capability of algorithms to predict blastocyst formation and quality
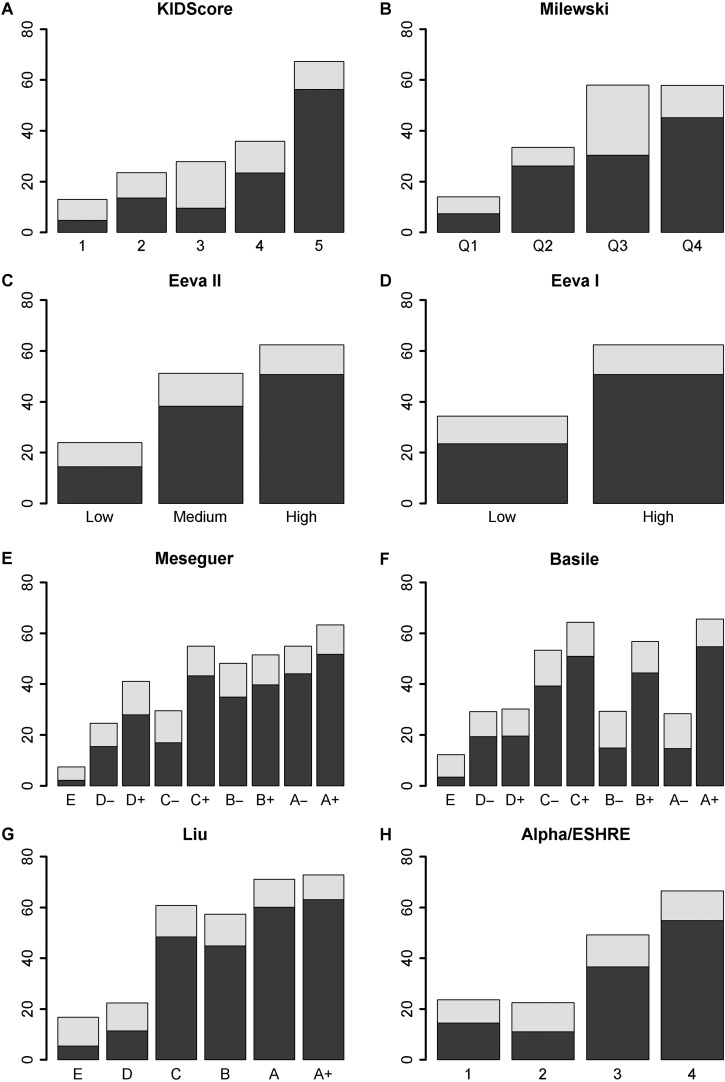
After developing KIDScore by using KID data from embryos transferred on Day 3, we wanted to test the capability of the algorithm to predict blastocyst formation and quality. When tested on an independent, multi-centric data set of 11 218 embryos cultured to Day 5, we found a proficient association in terms of AUC between the score derived by the KIDScore algorithm and the proportion of embryos that reached the blastocyst stage. Further, we found a proficient association between the algorithm score and the proportion of blastocysts assigned as TQ and GQ, using the Gardner score that is indicative of blastocyst quality on Day 5 (Table IV, Fig. 2A). The higher the score assigned by KIDScore, the more frequently the embryos reached the blastocyst stage on Day 5 and the better was the quality of the blastocysts. For each score group, the number of embryos and the proportion of these which reached the blastocyst stage within 120 h are listed in Table V.
Table IV.
The predictive capability of the algorithms was illustrated in terms of blastocyst formation and blastocyst quality in a multi-centric data set which was independent of the data used for developing the algorithms.
| Algorithm | Evaluation endpoint | AUC | 95% C.I. | RPV (Gini index) | P (adj) | Development endpoint | Information period |
|---|---|---|---|---|---|---|---|
| KIDScore | Blastocyst formation | 0.745 | 0.734–0.756 | 0.490 | «0.0001 | Implantation | Day 3 |
| Milewski | Blastocyst formation | 0.688 | 0.677–0.700 | 0.377 | 0.1198 | Blastocyst | Day 3 |
| Eeva II | Blastocyst formation | 0.685 | 0.673–0.697 | 0.370 | 0.1028 | Blastocyst | Day 2 |
| Eeva I | Blastocyst formation | 0.620 | 0.61–0.631 | 0.241 | «0.0001 | Blastocyst | Day 2 |
| Meseguer | Blastocyst formation | 0.676 | 0.665–0.688 | 0.353 | 0.0045 | Implantation | Day 3 |
| Basile | Blastocyst formation | 0.700 | 0.688–0.712 | 0.399 | 0.9370 | Implantation | Day 3 |
| Liu | Blastocyst formation | 0.753 | 0.743–0.764 | 0.507 | «0.0001 | Implantation | Day 3 |
| Alpha/ESHRE | Blastocyst formation | 0.700 | 0.687–0.714 | 0.400 | Ref. | Day 3 | |
| KIDScore | Blastocyst quality | 0.679 | 0.659–0.700 | 0.359 | «0.0001 | Implantation | Day 3 |
| Milewski | Blastocyst quality | 0.601 | 0.576–0.626 | 0.201 | 0.0428 | Blastocyst | Day 3 |
| Eeva II | Blastocyst quality | 0.611 | 0.589–0.633 | 0.222 | 0.3743 | Blastocyst | Day 2 |
| Eeva I | Blastocyst quality | 0.582 | 0.563–0.601 | 0.164 | 0.0028 | Blastocyst | Day 2 |
| Meseguer | Blastocyst quality | 0.603 | 0.579–0.628 | 0.207 | 0.1991 | Implantation | Day 3 |
| Basile | Blastocyst quality | 0.630 | 0.605–0.654 | 0.259 | 0.9679 | Implantation | Day 3 |
| Liu | Blastocyst quality | 0.695 | 0.672–0.717 | 0.389 | «0.0001 | Implantation | Day 3 |
| Alpha/ESHRE | Blastocyst quality | 0.629 | 0.606–0.652 | 0.258 | Ref. | Day 3 |
The AUC was used to generate a RPVs expressed as Gini Index (2×AUC-1). The Alpha/ESHRE timings algorithm was used as a common reference (Ref.) to which the algorithms were compared using a test for clustered receiver-operator characteristics curve (ROC) analysis, P-values <0.001 were considered significant. The endpoints on which the respective algorithms were developed (development endpoint) and the general incubation period from which the algorithms considers time-lapse variables (information period) are listed. Meseguer (Meseguer et al., 2011); Basile (Basile et al., 2015); Eeva I, (Conaghan et al., 2013); Eeva II (VerMilyea et al., 2014); Milewski (Milewski et al., 2015); Liu (Liu et al., 2016); Alpha/ESHRE (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011). ROC, receiver operating characteristics ; RPV, relative predictive value.
Figure 2.
Percentage of embryos that developed into blastocysts (light grey) and top or GQ blastocysts (Gardner: 3AA, 4AA, 5AA, 3/4/5BB, AB or BA; dark grey) shown according to their assigned score groups for all evaluated algorithms. GQ, good quality.
Table V.
Score group summary for the evaluated algorithms: The distribution of the embryos that ended up in each score group is given as numbers (n) and percentages, respectively.
| Algorithm | Score | Embryos (n) | Embryos (%) | Blastocysts (%) | |||
|---|---|---|---|---|---|---|---|
| KIDScore | 1 | 1893 | 17.9 | 13.0 | |||
| 2 | 1403 | 13.3 | 23.5 | ||||
| 3 | 645 | 6.1 | 27.9 | ||||
| 4 | 2235 | 21.1 | 35.9 | ||||
| 5 | 4401 | 41.6 | 67.3 | ||||
| Milewski | Q1 | 2163 | 20.5 | 14.0 | |||
| Q2 | 2680 | 25.3 | 33.5 | ||||
| Q3 | 2237 | 21.1 | 58.0 | ||||
| Q4 | 3497 | 33.1 | 57.8 | ||||
| Eeva II | Low | 4592 | 43.4 | 24.0 | |||
| Medium | 2828 | 26.7 | 51.2 | ||||
| High | 3157 | 29.8 | 62.5 | ||||
| Eeva I | Low | 7420 | 70.2 | 34.3 | |||
| High | 3157 | 29.8 | 62.5 | ||||
| Meseguer | E | 1079 | 10.2 | 7.4 | |||
| D− | 1027 | 9.7 | 24.6 | ||||
| D+ | 1457 | 13.8 | 41.1 | ||||
| C- | 1249 | 11.8 | 29.5 | ||||
| C+ | 2106 | 19.9 | 55.0 | ||||
| B- | 565 | 5.3 | 48.1 | ||||
| B+ | 813 | 7.7 | 51.5 | ||||
| A− | 879 | 8.3 | 54.9 | ||||
| A+ | 1402 | 13.3 | 63.3 | ||||
| Basile | E | 1780 | 16.8 | 12.2 | |||
| D− | 1917 | 18.1 | 29.2 | ||||
| D+ | 427 | 4.0 | 30.2 | ||||
| C- | 1067 | 10.1 | 53.3 | ||||
| C+ | 647 | 6.1 | 64.3 | ||||
| B- | 437 | 4.1 | 29.3 | ||||
| B+ | 912 | 8.6 | 56.8 | ||||
| A− | 645 | 6.1 | 28.4 | ||||
| A+ | 2745 | 26.0 | 65.5 | ||||
| Liu | E | 2909 | 27.5 | 16.8 | |||
| D | 2186 | 20.7 | 22.4 | ||||
| C | 1709 | 16.2 | 60.7 | ||||
| B | 1360 | 12.9 | 57.3 | ||||
| A | 1797 | 17.0 | 71.0 | ||||
| A+ | 616 | 5.8 | 72.7 | ||||
| Alpha/ESHRE | 1 | 2460 | 23.3 | 23.7 | |||
| 2 | 2105 | 19.9 | 22.5 | ||||
| 3 | 3087 | 29.2 | 49.1 | ||||
| 4 | 2925 | 27.7 | 66.5 | ||||
For each score group, the percentage of embryos that had formed a blastocyst <120 h post insemination is shown. Meseguer (Meseguer et al., 2011); Basile (Basile et al., 2015); Eeva I, (Conaghan et al., 2013); Eeva II (VerMilyea et al., 2014); Milewski (Milewski et al., 2015); Liu (Liu et al., 2016); Alpha/ESHRE (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011).
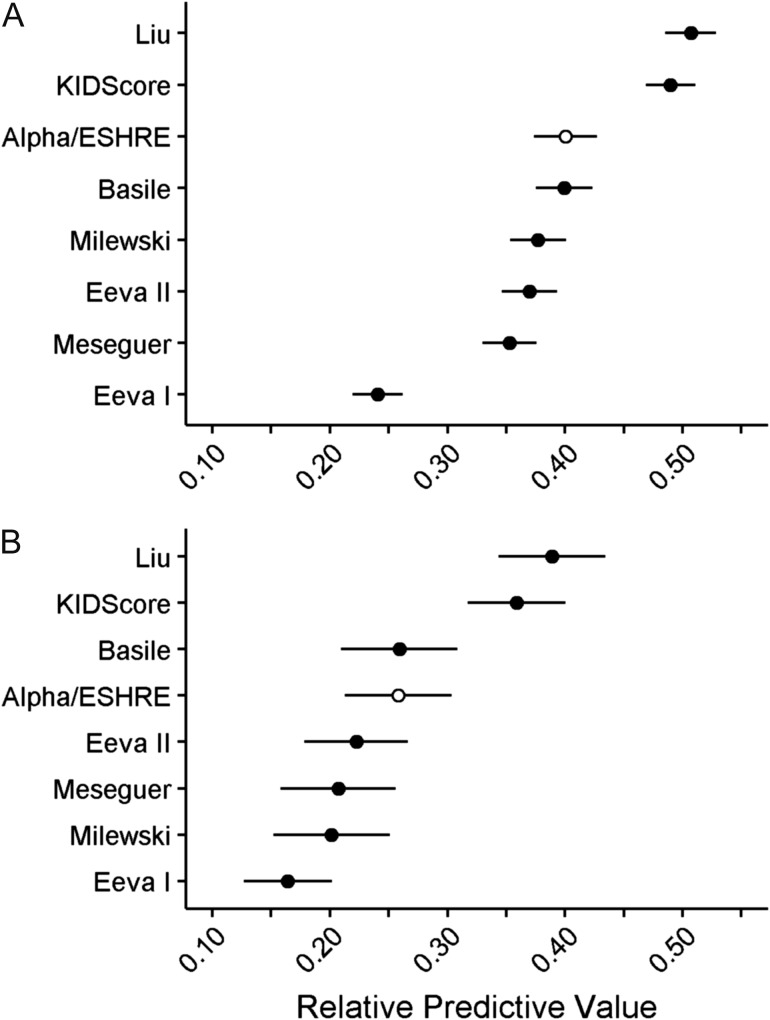
For the purpose of evaluating the relative capability to predict blastocyst formation and quality, we used the multi-centric data set cultured to Day 5 to compare the Alpha/ESHRE timings algorithm with three implantation based algorithms (KIDScore; Meseguer et al., 2011, Basile et al., 2015, Liu et al. 2016) and three blastocyst formation based algorithms (Conaghan et al., 2013; VerMilyea et al., 2014, Milewski et al., 2015). The blastocyst proportion and quality in the respective algorithm score groups are illustrated in Fig. 2A–G. The comparison based on the AUC of the obtained ROC curves revealed clear differences in the relative predictive capabilities of the various algorithms (Table IV, Fig. 3). Using the Alpha/ESHRE timings algorithm (Table IV, Fig. 2H) as a common reference to the other algorithms, KIDScore and Liu showed significantly higher predictive power regarding blastocyst formation. Milewski, Eeva II, Meseguer and Basile showed predictive power not significantly different from Alpha/ESHRE timings, whereas Eeva I was significantly below. For blastocyst quality KIDScore and Liu showed significantly higher predictive power, whereas Milewski, Eeva I, Eeva II, Meseguer and Basile here showed predictive power not significantly different from Alpha/ESHRE timings. The number of embryos in the score groups of the respective algorithms is given in Table V.
Figure 3.
The predictive capability of the evaluated algorithms is illustrated for blastocyst formation (A) and blastocyst quality (B). Predictive capabilities are shown as RPV (Gini Index; [2 × AUC]-1) with a theoretical range from 0 to 1, with 95% confidence intervals illustrated by error bars. The Alpha/ESHRE timings algorithm (open circle) was used as common reference. Meseguer (Meseguer et al., 2011); Basile (Basile et al., 2015); Eeva I, (Conaghan et al., 2013); Eeva II (VerMilyea et al., 2014); Milewski (Milewski et al., 2015); Liu (Liu et al., 2016); Alpha/ESHRE (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011). RPV, relative predictive value; AUC, area under the curve.
The inclusion of different variables in the various algorithms caused some embryos not to become a score (‘imputes’). Therefore the total number of embryos used in the comparison was 10 577 embryos which could obtain a score from all the algorithms included in the evaluation.
Discussion
Time-lapse imaging in human embryology has revealed developmental characteristics that are associated with lower implantation and pregnancy rates such as direct cleavage (Rubio et al., 2012) and reverse cleavage (Liu et al., 2014). Time-lapse has also revealed a higher incidence of multinucleation than has previously been reported through the use of static observations (Ergin et al., 2014; Aguilar et al., 2016). It has been documented in human IVF that the rate of embryo development, particularly that of the first cleavage division, is related to implantation potential (Sakkas et al., 2001; Salumets et al., 2003). Similarly, it has been shown through time-lapse that embryos with a specific pace of development have higher implantation rates than others (Dal Canto et al., 2012; Chamayou et al., 2013). Significantly, mapping the exact timing of developmental events and being able to access cell cycle length and cleavage patterns has resulted in the proposal of predictive algorithms (e.g. Meseguer et al., 2011; Conaghan et al., 2013; VerMilyea et al., 2014; Basile et al., 2015; Milewski et al., 2015; Liu et al., 2016) for different endpoints, including blastocyst formation or implantation. Despite implantation being a preferable endpoint over blastulation, implantation still remains subordinate to live birth, which obviously is the ultimate endpoint for analyses at present. Using implantation as endpoint for the model development is hence an inherent limitation of the present study. However, the development of a robust algorithm requires a large data set, and currently the largest amounts of data are available for blastocyst development, decreasing in amounts for embryo implantation and further decreasing for live born babies. In addition, the applicability of any algorithm relies on the data input and consequently using data from a single clinic may yield an algorithm that works in that clinic but cannot necessarily be transferred to a different clinic. One such example is the level of oxygen used during incubation, which has a substantial effect on embryo development (Wale and Gardner, 2010; Kirkegaard et al., 2013). An algorithm based on data from ambient oxygen conditions (Meseguer et al., 2011) failed when tested at reduced oxygen (Freour et al., 2015). This implies that considerations of aligned culture conditions should be done before the direct transfer of any algorithm, especially those based on single-centre data.
The KIDScore algorithm presented here is based on an extensive data set with implantation results of a large number of embryos derived from 24 clinics and the algorithm uses variables that are easy to annotate. Furthermore, the algorithm is biologically meaningful as it identifies embryos with a very fast development into the 3-cell stage (Split 1), embryos that are too slow to develop (Split 2), embryos that show an irregular cleavage pattern (Splits 3 and 4), and embryos that do not reach a desirable stage of development on Day 3 (Split 5). Significantly, the algorithm does not ‘reject’ any embryo for transfer, but rather ranks embryo potential compared with others in the same cohort and represents relative implantation potential based solely on morphokinetic behaviour. As the algorithm supports the decision on which embryo to transfer or freeze, it may be used in conjunction with morphology for making a final choice on Day 3 (Van Royen et al., 1999) according to morphological criteria applied by the individual clinics. The IVF clinics that provided the data have different culture conditions with regard to fertilization method, media, oxygen, carbon dioxide concentration, protein source, and also handling of oocytes prior to incubation and sperm preparation. The resulting general applicability of the KIDScore algorithm is reflected in the fact that the AUC of the algorithm did not change substantially when tested for different culture conditions such as reduced or ambient oxygen and when used with IVF or ICSI. Thus, and of importance, this algorithm can be applied under different conditions without the need to adapt the algorithm parameters to specific environmental conditions. This broad applicability is due to the utilization of a large, diverse data set for calibration; where possible using fractions instead of actual timings, compensating for differences relating to incubation conditions and fertilization method, robust parameterization methods using cross-validation, and the achievement of a deselection algorithm.
The study has focused on predictive ability, expressed in terms of AUC. In general, the most recent algorithms showed the highest predictive capabilities in terms of AUC, which may be owing to the overall enhanced knowledge and experience within the relatively new field of time-lapse embryology. But in order to provide a complete picture of how an algorithm performs, the actual relative numbers of embryos in each score group must also be considered, thereby not only taking into account the percentage of embryos within each group reaching the blastocyst stage. As an example, the Basile algorithm (Fig. 2F) displays a seemingly non-monotonous distribution of blastulation in its eight score groups. This algorithm nevertheless has a predictive capability at a similar level as the Alpha/ESHRE algorithm, mainly owing to the fact that the highest and lowest score groups (E, D− and A+) of the Basile algorithm comprise more than half of the embryos. Hence some of the seemingly irregular score groups contain quite few of the embryos.
Another important feature is the number of embryos in the highest group(s). Generally speaking, a low proportion of embryos in the highest score group(s) would designate a selection-oriented algorithm. Such an algorithm will have low capability to select embryos with a good to fair probability of becoming blastocysts as many embryos that could become blastocysts would be deselected. On the other hand, such an algorithm will be capable of deselecting embryos unlikely to develop into blastocysts. The Eeva I algorithm may serve as an example of this. With its two scores, the sensitivity of the Eeva I algorithm (i.e. its capability to identify embryos with a high probability of blastulating) can be calculated as 44%, which means that 56% of the blastulating embryos are in the low group. The capability to exclude non-blastulating embryos is designated by the specificity, which is 80%. Only 20% of non-blastulating embryos are thus placed in the high group. These values correspond to those reported in Conaghan et al. (2013) where a sensitivity of 38% and a specificity of 85% were found. Likewise, the Liu algorithm can largely be considered a selection-oriented model, with 5.8% of the embryos given the A+ score and 17.0% given the A score, showing that this algorithm is comparatively selection-orientated. The blastulation in those groups are 72.7% and 71.0%, respectively, which are the highest numbers recorded in the comparative study. While the Liu algorithm shares many characteristics with KIDScore, they are differently orientated with regards to selection/deselection. KIDScore utilizes a more conservative approach, retaining 62.7% of the embryos in the two highest score groups, and 41.6% of the embryos in the highest score group, which represents the highest proportion among the algorithms included in the comparison. This responds to creating a deselection algorithm rather than assigning the highest score to a small ‘top group’ of embryos. Even though the highest score is assigned to such a relatively large proportion, the percentage of blastulating embryos is 67.3%.
It has previously been reported that algorithms that are based solely on blastocyst development as endpoint do not correlate well with implantation (Kirkegaard et al., 2014). By comparing the algorithm presented here with other published algorithms, it is apparent that KIDScore, in comparison, has a satisfying explanatory power for blastocyst prediction, despite the algorithm being initially created for Day 3 implantation. This is plausibly due to the use of gestational sac and/or foetal heart beat as an endpoint, for which blastocyst formation is an inherent precondition. Consequently, this broadens the application of such an algorithm as it can be used in Day 3 as well as in Day 5 programs. In cases where it is difficult to determine on Day 3 whether culture can be continued to Day 5 and still have a transfer, the algorithm may be used to support such decisions. The suitability of the present KIDScore algorithm to support Day 5 decisions on which embryos to freeze or transfer is not investigated, though.
The predictive capability of KIDScore in terms of AUC for blastulation is higher than the predictive capability in terms of implantation. A number of reasons for this can be identified although not quantified. Firstly, the blastulation data set comprises all embryos which divided whereas the implantation data set only comprises embryos selected for transfer. Therefore, the implantation data set has been subjected to a stronger selection by embryologists than the data set comprising all embryos in the relevant treatments. Statistically, this could be categorized as a strong bias which reduces the variation in the data set. Secondly, the patient's uterus in a given treatment may not be receptive at the time of transfer, regardless of the characteristics of the transferred embryo(s). This may statistically be categorized as noise. Thirdly, the embryo characteristics resulting in implantation possibly differ slightly from the characteristics resulting in blastulation. This may statistically cause a difference of parameters or even a different structure for algorithms aimed at blastulation and implantation, respectively. Nevertheless, the present analysis demonstrates the suitability of an implantation algorithm to predict blastulation as well. Whether the reverse also holds true is not demonstrated by this analysis. A comparison of the algorithms using implantation data rather than the present Day 5 blastocyst data would be preferable. However, the KIDScore algorithm was developed on the basis of the available implantation data, and a comparison on the same implantation data would be biased, probably to the benefit of KIDScore.
The data set used in the evaluation has an overlap at the population level with the data that KIDScore was developed on, as some clinics have shared both 3-day incubation data used for calibration of the algorithm and 5-day incubation data used for validation of blastocyst prediction. Despite this, there is no treatment or patient overlap. The overlap at population level may introduce bias as the variation within those populations may be similar, which may put the KIDScore algorithm at an advantage regarding predictive capability. Many operators have been annotating the current morphokinetic parameters. As annotation by embryologists has a degree of judgement, the predictive power may be confounded by differences in annotation practice and experience. However, the early timings used in the current evaluations have low intra- and inter-operator variation (Sundvall et al., 2013), possibly minimizing this potentially confounding effect. The KIDScore algorithm was developed on KID data which presumably will affect the estimated implantation probabilities. Therefore, the estimated implantation probabilities (Fig. 1) should be interpreted with care.
The present study aimed at giving all investigated algorithms equal frames by not including conventional morphology assessments, even if specified in the respective composite algorithm. This cannot be fully achieved though, as the assessments omitted cannot be assumed statistically independent of those included. The algorithms by Meseguer, Basile, Liu and a complete Alpha/ESHRE consensus evaluation would all have behaved differently if there was access to the respective conventional morphology parameters required for each algorithm. Therefore, the present comparison presumably favours the algorithms that do not include conventional morphology parameters (KIDScore, Milewski, Eeva I and Eeva II). The present simple implementation of the timings part of Alpha/ESHRE consensus (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011) is one possible choice, but other algorithms might have been made on the same basis. An important future study could be an analysis of the interactions between morphokinetics and conventional morphology, thus facilitating inclusion of morphology in time-lapse algorithms.
As a quite notable outcome of this study the present attempt to mimic the pure timings part of conventional scoring (Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology, 2011), without the use of time-lapse, has higher or equal predictive power regarding blastulation than four of the six dedicated morphokinetic algorithms. Only Liu and KIDScore had significantly higher predictive power than the timing part of conventional embryo evaluation. As stated above, these two algorithms exhibit different orientations; KIDScore, embracing the majority of the embryos with the potential to form blastocysts, while Liu targets those with the highest chance of blastocyst formation.
An algorithm developed in a given clinic cannot necessarily be transferred to another clinic (Best et al., 2013; Kirkegaard et al., 2014; Yalçınkaya et al., 2014; Freour et al, 2015). The strength and the general applicability of an algorithm is highly dependent on the included incubation conditions and the amount and quality of the data used as input for its development. Furthermore, the robustness of an algorithm depends on the variables that are included in it and the borders that are applied for selection or deselection. The availability of a robust algorithm, such as KIDScore, enables clinics which have not yet built a large data foundation to support their decision on which embryo(s) to transfer immediately upon implementing the technology, while accepting that the final decision is always with the embryologist. This is important, especially since embryologists who are only starting to use time-lapse technology will rely on traditional embryo scoring based on morphological characteristics that are not included as variables in the KIDScore algorithm, e.g. fragmentation, blastomere, size or multinucleation. Combining these traditional morphological assessments with morphokinetic information, only accessible through use of time-lapse, may allow for more well-informed decisions in embryo selection. The best combination of traditional morphological and morphokinetic information should be the subject of future studies, and even more important the gains in terms of increase in pregnancies from such a combination should be investigated in Randomized Controlled Trial studies.
Embryologists have preferred to transfer multiple embryos in order to obtain high pregnancy rates, which resulted in high rates of multiple births. The risk of a higher order pregnancy to both mother and foetus is well documented (Adashi et al., 2003). Therefore, there is a growing movement world-wide to achieve a single healthy pregnancy per IVF treatment by transferring only one embryo. Time-lapse imaging, in combination with decision support algorithms that are based on morphokinetic embryo assessment, may help to facilitate the implementation of an SET policy.
Keeping the above discussion and reservations in mind, the present study clearly indicates that there are substantial differences in the predictive power of the evaluated morphokinetic algorithms. It should still be emphasized that the ultimate verification of the advantage of time-lapse in terms of improved culture conditions and improved basis for selection can only be provided by a Randomized Controlled Trial (Kaser and Racowsky, 2014, Armstrong et al., 2015) of both sufficient size and suitable design, not by a retrospective study as the present.
KIDScore could be a suitable candidate for a generally applicable Day 3 algorithm which can be applied in different clinical settings. The predictive ability of the KIDScore algorithm primarily concerns implantation, but also relates to blastocyst development as well as blastocyst quality.
The current analyses suggest that conventional (non-time-lapse) timings evaluation has good predictive power for blastocyst formation. While many of the compared algorithms showed less or equal predictive power, the two most recent algorithms suggested a substantial benefit from using a time-lapse based algorithm, relative to the present algorithmic interpretation of a conventional timings evaluation. This we consider an important message from this study. While not serving as the ultimate answer on the question of the value of time-lapse, this study implies marked differences in predictive power of the evaluated algorithms.
Acknowledgements
The authors would like to acknowledge Reidun Kuhlmann, Betina Melgaard Rasmussen, Tine Qvistgaard Kajhøj and Francesca Anne Bahr for their invaluable practical support and suggestions to the manuscript. Further, we would like to acknowledge the reviewers for insightful input that has significantly improved this paper.
Authors’ roles
All authors took part in the analysis and interpretation of the results and have written substantial parts of the manuscript. B.M.P. was responsible for the algorithm development. M.B. was responsible for the statistical analyses.
Funding
This study was funded by Vitrolife A/S, Denmark and Vitrolife AB, Sweden.
Conflict of interest
B.M.P.’s company BMP Analytics is performing consultancy for Vitrolife A/S. M.B. is employed at Vitrolife A/S. M.M.’s company ilabcomm GmbH received honorarium for consultancy from Vitrolife AB. D.K.G. received research support from Vitrolife AB.
References
- Adamson GD, Abusief ME, Palao L, Witmer J, Palao LM, Gvakharia M. Improved implantation rates of day 3 embryo transfers with the use of an automated time-lapse-enabled test to aid in embryo selection. Fertil Steril 2016;105:369–375. [DOI] [PubMed] [Google Scholar]
- Adashi EY, Barri PN, Berkowitz R, Braude P, Bryan E, Carr J, Cohen J, Collins J, Devroey P, Frydman R et al. . Infertility therapy-associated multiple pregnancies (births): an ongoing epidemic. Reprod Biomed Online 2003;7:515–542. [DOI] [PubMed] [Google Scholar]
- Aguilar J, Motato Y, Escribá MJ, Ojeda M, Muñoz E, Meseguer M. The human first cell cycle: impact on implantation. Reprod Biomed Online 2014;28:475–484. [DOI] [PubMed] [Google Scholar]
- Aguilar J, Rubio I, Muñoz E, Pellicer A, Meseguer M. Study of nucleation status in the second cell cycle of human embryo and its impact on implantation rate. Fertil Steril 2016. doi:10.1016/j.fertnstert.2016.03.036. (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- Ahlström A, Westin C, Reismer E, Wikland M, Hardarson T. Trophectoderm morphology: an important parameter for predicting live birth after single blastocyst transfer. Hum Reprod 2011;26:3289–3296. [DOI] [PubMed] [Google Scholar]
- Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod 2011;26:1270–1283. [DOI] [PubMed] [Google Scholar]
- Armstrong S, Vail A, Mastenbroek S, Jordan V, Farquhar C. Time-lapse in the IVF-lab: how should we assess potential benefit?. Hum Reprod 2015;30:3–8. [DOI] [PubMed] [Google Scholar]
- Azzarello A, Hoest T, Mikkelsen AL. The impact of pronuclei morphology and dynamicity on live birth outcome after time-lapse culture. Hum Reprod 2012;27:2649–2457. [DOI] [PubMed] [Google Scholar]
- Basile N, Vime P, Florensa M, Aparicio Ruiz B, Garcia Velasco JA, Remohi J, Meseguer M. The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Hum Reprod 2015;30:276–283. [DOI] [PubMed] [Google Scholar]
- Best L, Campbell A, Duffy S, Montgomery S, Fishel S. Testing a published embryo selection algorithm on independent time-lapse data. Hum Reprod 2013;28:87–90.23081875 [Google Scholar]
- Bodri D, Sugimoto T, Serna JY, Kondo M, Kato R, Kawachiya S, Matsumoto T. Influence of different oocyte insemination techniques on early and late morphokinetic parameters: a retrospective analysis of 500 time-lapse monitored blastocysts. Fertil Steril 2015;104:1175–1181. [DOI] [PubMed] [Google Scholar]
- Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod BioMed Online 2013. b;27:140–146. [DOI] [PubMed] [Google Scholar]
- Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S, Hickman CFL. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod BioMed Online 2013. a;26:477–485. [DOI] [PubMed] [Google Scholar]
- Cetinkaya M, Pirkevi C, Yelke H, Colakoglu YK, Atayurt Z, Kahraman S. Relative kinetic expressions defining cleavage synchronicity are better predictors of blastocyst formation and quality than absolute time points. J Assist Reprod Genet 2015;32:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamayou S, Patrizio P, Storaci G, Tomaselli V, Alecci C, Ragolia C, Crescenzo C, Guglielmino A. The use of morphokinetic parameters to select all embryos with full capacity to implant. J Assist Reprod Genet 2013;30:703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciray HN, Aksoy T, Goktas C, Ozturk B, Bahceci M. Time-lapse evaluation of human embryo development in single versus sequential culture media-a sibling oocyte study. J Assist Reprod Genet 2012;29:891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciray HN, Campbell A, Agerholm IE, Aguilar J, Chamayou S, Esbert M, Sayed S. Proposed guidelines on the nomenclature and annotation of dynamic human embryo monitoring by a time-lapse user group. Hum Reprod 2014;29:2650–2660. [DOI] [PubMed] [Google Scholar]
- Claman P, Armant DR, Seibel MM, Wang TA, Oskowitz SP, Taymor ML. The impact of embryo quality and quantity on implantation and the establishment of viable pregnancies. J In Vitro Fert Embryo Transf 1987;4:218–222. [DOI] [PubMed] [Google Scholar]
- Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, Baker VL, Adamson GD, Abusief ME, Gvakharia M et al. . Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril 2013;100:412–419. [DOI] [PubMed] [Google Scholar]
- Cruz M, Garrido N, Herrero J, Perez-Cano I, Muñoz M, Meseguer M. Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod BioMed Online 2012;25:371–381. [DOI] [PubMed] [Google Scholar]
- Dal Canto M, Coticchio G, Renzini MM, De Ponti E, Novara PV, Brambillasca F, Comi R, Fadini R. Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod BioMed Online 2012;25:474–80. [DOI] [PubMed] [Google Scholar]
- De Neubourg D, Gerris J, Mangelschots K, Van Royen E, Vercruyssen M, Elseviers M. Single top quality embryo transfer as a model for prediction of early pregnancy outcome. Hum Reprod 2004;19:1476–1479. [DOI] [PubMed] [Google Scholar]
- Diamond MP, Willman S, Chenette P, Cedars MI. The clinical need for a method of identification of embryos destined to become a blastocyst in assisted reproductive technology cycles. J Assist Reprod Genet 2012;29:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RG, Fishel SB, Cohen J, Fehilly CB, Purdy JM, Slater JM, Steptoe PC, Webster JM. Factors influencing the success of in vitro fertilization for alleviating human infertility. J In Vitro Fert Embryo Transf 1984;1:3–23. [DOI] [PubMed] [Google Scholar]
- Ergin EG, Caliskan E, Yalcinkaya E, Oztel Z, Cokelez K, Ozay A, Ozornek HM. Frequency of embryo multinucleation detected by time-lapse system and its impact on pregnancy outcome. Fertil Steril 2014;102:1029–1033. [DOI] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett 2006;27:861–874. [Google Scholar]
- Freour T, Dessolle L, Lammers J, Lattes S, Barrière P. Comparison of embryo morphokinetics after in vitro fertilization-intracytoplasmic sperm injection in smoking and nonsmoking women. Fertil Steril 2013;99:1944–1950. [DOI] [PubMed] [Google Scholar]
- Freour T, Le Fleuter N, Lammers J, Splingart C, Reignier A, Barrière P. External validation of a time-lapse prediction model. Fertil Steril 2015;103:917–922. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril 2000;73:1155–1158. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Meseguer M, Rubio C, Treff N. Diagnosis of the human preimplantation embryo. Hum Reprod Update 2015;21:727–747. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Sakkas D. Assessment of embryo viability: the ability to select a single embryo for transfer. Placenta 2003;24:S5–S12. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Schoolcraft WB.. In vitro culture of human blastocyst In: Janson R, Mortimer D (eds). Towards Reproductive Certainty: Infertility and Genetics Beyond. Carnforth: Parthenon Press, 1999, 378–388. [Google Scholar]
- Goodman LR, Goldberg J, Falcone T, Austin C, Desai N. Does the addition of time-lapse morphokinetics in the selection of embryos for transfer improve pregnancy rates? A randomized controlled trial. Fertil Steril 2016;105:275–285. [DOI] [PubMed] [Google Scholar]
- Hand DJ, Till RJ. A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn 2001;45:171–186. [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand Stat Theory Appl 1979;6:65–70. [Google Scholar]
- Kaser DJ, Racowsky C. Clinical outcomes following selection of human preimplantation embryos with time-lapse monitoring: a critical review. Hum Reprod Update 2014;20:617–631. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K, Campbell A, Agerholm I, Bentin-Ley U, Gabrielsen A, Kirk J, Sayed S, Ingerslev HJ. Limitations of a time-lapse blastocyst prediction model: a large multicentre outcome analysis. Reprod BioMed Online 2014;29:156–158. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K, Hindkjaer JJ, Ingerslev HJ. Effect of oxygen concentration on human embryo development evaluated by time-lapse monitoring. Fertil Steril 2013;99:738–744. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K, Hindkjaer JJ, Ingerslev HJ. Human embryonic development after blastomere removal: a time-lapse analysis. Hum Reprod 2012;27:97–105. [DOI] [PubMed] [Google Scholar]
- Lemmen JG, Agerholm I, Ziebe S. Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod BioMed Online 2008;17:385–391. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chapple V, Feenan K, Roberts P, Matson P. Time-lapse deselection model for human day 3 in vitro fertilization embryos: the combination of qualitative and quantitative measures of embryo growth. Fertil Steril 2016;105:656–662. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chapple V, Roberts P, Matson P. The prevalence, consequence, and significance of reverse cleavage by human embryos viewed with the use of the EmbryoScope time-lapse video system. Fertil Steril 2014;102:1295–1302. [DOI] [PubMed] [Google Scholar]
- Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod 2011;26:2658–2671. [DOI] [PubMed] [Google Scholar]
- Meseguer M, Rubio I, Cruz M, Basile N, Marcos J, Requena A. Embryo incubation and selection in a time-lapse monitoring system improves pregnancy outcome compared with a standard incubator: a retrospective cohort study. Fertil Steril 2012;98:1481–1489. [DOI] [PubMed] [Google Scholar]
- Milewski R, Kuć P, Kuczyńska A, Stankiewicz B, Łukaszuk K, Kuczyński W. A predictive model for blastocyst formation based on morphokinetic parameters in time-lapse monitoring of embryo development. J Assist Reprod Genet 2015;32:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag M, van der Ven H. Evaluation of pronuclear morphology as the only selection criterion for further embryo culture and transfer: results of a prospective multicentre study. Hum Reprod 2001;16:2384–2389. [DOI] [PubMed] [Google Scholar]
- Motato Y, de Los Santos M, Escriba M, Ruiz BA, Remohí J, Meseguer M. Morphokinetic analysis and embryonic prediction for blastocyst formation through an integrated time-lapse system. Fertil Steril 2016;105:376–384. [DOI] [PubMed] [Google Scholar]
- Obuchowski NA. Nonparametric Analysis of Clustered ROC Curve Data. Biometrics 1997;53:567–578. [PubMed] [Google Scholar]
- Payne D, Flaherty SP, Barry MF, Matthews CD. Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum Reprod 1997;12:532–541. [DOI] [PubMed] [Google Scholar]
- Pribenszky C, Matyas S, Kovacs P, Losonczi E, Zadori J, Vatja G. Pregnancy achieved by transfer of a single blastocyst selected by time-lapse monitoring. Reprod Biomed Online 2010;21:533–536. [DOI] [PubMed] [Google Scholar]
- R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2015. URL https://www.R-project.org/. [Google Scholar]
- Rubio I, Galán A, Larreategui Z, Ayerdi F, Bellver J, Herrero J, Meseguer M. Clinical validation of embryo culture and selection by morphokinetic analysis: a randomized, controlled trial of the EmbryoScope. Fertil Steril 2014;102:1287–1294. [DOI] [PubMed] [Google Scholar]
- Rubio IR, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escriba MJ, Bellver J, Meseguer M. Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil Steril 2012;98:1458–1463. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Percival G, D'Arcy Y, Sharif K, Afnan M. Assessment of early cleaving in vitro fertilized human embryos at the 2-cell stage before transfer improves embryo selection. Fertil Steril 2001;76:1150–1156. [DOI] [PubMed] [Google Scholar]
- Salumets A, Hyden-Granskog C, Makinen S, Suikkari AM, Tiitinen A, Tuuri T. Early cleavage predicts the viability of human embryos in elective single embryo transfer procedures. Hum Reprod 2003;18:821–825. [DOI] [PubMed] [Google Scholar]
- Scott L, Alvero R, Leondires M, Miller B. The morphology of human pronuclear embryos is positively related to blastocyst development and implantation. Hum Reprod 2000;15:2394–2403. [DOI] [PubMed] [Google Scholar]
- Scott L, Finn A, O'Leary T, McLellan S, Hill J. Morphologic parameters of early cleavage-stage embryos that correlate with fetal development and delivery: prospective and applied data for increased pregnancy rates. Hum Reprod 2007;22:230–240. [DOI] [PubMed] [Google Scholar]
- Scott LA, Smith S. The successful use of pronuclear embryo transfers the day following oocyte retrieval. Hum Reprod 1998;13:1003–1013. [DOI] [PubMed] [Google Scholar]
- Sundvall L, Ingerslev HJ, Knudsen UB, Kirkegaard K. Inter- and intra-observer variability of time-lapse annotations. Hum Reprod 2013;28:3215–3221. [DOI] [PubMed] [Google Scholar]
- Tesarik J, Greco E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear stage morphology. Hum Reprod 1999;14:318–323. [DOI] [PubMed] [Google Scholar]
- Therneau T, Atkinson B, Ripley B. rpart: Recursive Partitioning and Regression Trees. 2015. R package version 4.1-10. http://CRAN.R-project.org/package=rpart.
- Van Royen E, Mangelschots K, De Neubourg D, Valkenburg M, Van de Meerssche M, Ryckaert G, Eestermans W, Gerris J. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod 1999;14:2345–2349. [DOI] [PubMed] [Google Scholar]
- VerMilyea MD, Tan L, Anthony JT, Conaghan J, Ivani K, Gvakharia M, Boostanfar R, Baker VL, Suraj V, Chen AA et al. . Computer-automated time-lapse analysis results correlate with embryo implantation and clinical pregnancy: a blinded, multicentre study. Reprod BioMed Online 2014;29:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wale P, Gardner DK. A comprehensive review of the adverse effects of chemical and physical factors on mammalian embryos and their importance for the practice of human assisted reproductive technology. Hum Reprod Update 2016;22:2–22. [DOI] [PubMed] [Google Scholar]
- Wale P, Gardner DK. Time-lapse analysis of mouse embryo development in oxygen gradients. Reprod BioMed Online 2010;21:402–410. [DOI] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, Reijo Pera RA. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol 2010;28:1115–1121. [DOI] [PubMed] [Google Scholar]
- Yalçınkaya E, Ergin EG, Çalışkan E, Öztel Z, Özay A, Özörnek H. Reproducibility of a time-lapse embryo selection model based on morphokinetic data in a sequential culture media setting. J Turk Ger Gynecol Assoc 2014;15:156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ST, Shi JX, Gong F, Zhang SP, Lu CF, Tan K, Leng LZ, Hao M, He H, Gu YF et al. . Cleavage pattern predicts developmental potential of day 3 human embryos produced by IVF. Reprod BioMed Online 2015;30:625–634. [DOI] [PubMed] [Google Scholar]