Abstract
Resveratrol (RES) has been studied for its effects on the lifespan extension of Caenorhabditis elegans, but controversy still remains on its mechanism related with SIR-2. In this study, longevity assay was performed to confirm SIR-2-dependent lifespan extension of C. elgeans with RES and oxyresveratrol (OXY), an isomer of hydroxylated RES using loss-of-function mutants of C. elegans including sir-2.1 mutant. The results showed that OXY and RES significantly (P < 0.05) extended the lifespan of C. elegans compared with the control. OXY and RES also significantly (P < 0.05) increased the mRNA expression levels of sir-2.1 and aak-2 in a dose-dependent manner and increased the protein expression levels of SIR-2.1. OXY and RES treatment extended the lifespan in daf-16 loss-of-function mutants, which suggested that lifespan extension was not occurring via the activation of DAF-16. However, OXY and RES failed to extend the lifespan in loss-of-function mutants of sir-2.1 and aak-2. Therefore, OXY and RES extend the lifespan of C. elegans by overexpression of SIR-2.1, which is related to lifespan extension through calorie restriction and the AMP-activated protein kinase (AMPK) pathway, although this process is independent of the FOXO/DAF-16 pathway.
Keywords: Oxyresveratrol, SIR-2.1, lifespan, Caenorhabditis elegans, resveratrol
Introduction
In Caenorhabditis elegans, SIR-2.1 is a member of the Sir-2 family (NAD+-dependent protein deacetylases) and regulates nematode ageing by interacting with various transcription factors including DAF-16.1 SIR2 protein has been associated with lifespan regulation in C. elegans, yeast, Drosophila melanogaster and even mammals.2 In mammals, there are seven Sir2 orthologs (SIRT1−7), and they are involved in diverse metabolic processes including adipogenesis, myogenesis and apoptosis.1 In addition, mammalian SIRT has been shown to interact with FOXO3a to modulate the balance between cell survival and apoptosis, and the FOXO transcription factor is known as an important regulator of C. elegans ageing.3
Dietary restriction (DR) is the most influential environmental intervention to extend lifespan and health span in a wide range of species.4 There are several methods that restrict the diet, and a number of chemical compounds have been suggested to act as DR mimetics to extend lifespan.5 A well-known antioxidant, resveratrol (RES), has been proposed to act as one such mimetic, as demonstrated in yeast, worms, flies and other organisms.6 Although the mechanism remains unclear, the positive effects of DR on lifespan have been suggested.7 The energy-sensing AMP-activated protein kinase (AMPK)/aak-2 pathway upstream of FOXO/DAF-16 mediates lifespan extension induced by DR in worms.4 However, for the DR mimetic RES, AMPK without FOXO was shown to extend lifespan in worms.5
Oxidative stress is known to be involved in ageing, and antioxidants from natural sources have been shown to extend lifespans in mice,8 yeast and human fibroblast cells.9 Although RES has been studied for its effects on the lifespan extension of C. elegans, controversy still remains on its mechanism related with SIR-2.10,11 RES extends the lifespan of C. elegans via Sir-2.1-dependent manner.2 On the other hand, a slight increase in lifespan of C. elegans by RES is Sir-2.1-independent.10 Oxyresveratrol (OXY) is the aglycone of a hydroxystilbene compound, mulberroside A, and is found in the white mulberry (Morus alba), and is an isomer of hydroxylated RES. OXY inhibits the activity of dihydroxyphenylalanine (DOPA) oxidase, cyclooxygenase and rat liver mitochondrial ATPases; indeed, OXY showed higher antioxidative activity than RES.12 In this study, to clarify OXY- and RES-mediated lifespan extension of C. elegans, we investigated the lifespan extension effect of OXY and RES on C. elegans and elucidated the mechanism of OXY-dependent lifespan extension in C. elegans using several loss-of-function mutants of C. elegans including sir-2.1 mutant.
Materials and methods
Bacteria and nematodes culture
Escherichia coli OP50 was provided by the Caenorhabditis Genetics Centre, University of Minnesota (CGC), and used as a food source. E. coli OP50 was grown in Luria-Bertani (LB) broth (Difco, Detroit, MI, USA) at 37℃ for 18−24 h with shaking. Bacteria were harvested and adjusted to a final concentration of 0.1 mg (wet weight) per microliter in M9 buffer.13
Caenorhabditis elegans Bristol strain N2 (wild-type) and mutant strains were provided by the CGC. Bristol strain N2 was used for all measurements, and mutant strains were used only for the longevity assay. The mutants used for lifespan measurements included CF1038 daf-16 (mu86), RB754 aak-2 (ok524) and VC199 sir-2.1 (ok434). Nematodes were maintained and propagated at 25℃ according to standard techniques.14 The E. coli OP50 suspension was spread onto nematode growth medium (NGM) in 90-mm-diameter plates to feed the worms. Eggs were harvested from adult worms after they were exposed to a sodium hypochlorite-sodium hydroxide solution as previously described.15 The egg suspension in M9 buffer was incubated overnight at 25℃ to allow the eggs to hatch, and the suspension of L1 stage worms was centrifuged at 1200 × g for 2 min. After removing the supernatant, the remaining larvae were transferred onto fresh NGM plates seeded with E. coli OP50 and incubated at 25℃ for 2 day to synchronise pubescence. All experiments were conducted with 3-day-old young-adult (day 1 of adulthood) wild-type worms (except for mutant survival tests).
Longevity assay
For the longevity assay, OXY (Sigma–Aldrich, St. Louis, MO, USA) and RES (Sigma–Aldrich) were dissolved in dimethyl sulphoxide (DMSO) to final concentrations of 100, 500 and 1000 µM. OXY and RES did not show any toxicity to C. elegans within the concentration of 1000 µM (data not shown). DMSO alone was used as a control, and the solutions or control were added at a ratio of 3:1000 to NGM/FUdR during the preparation of the plates, which were supplemented with 5-fluoro-2′-deoxyuridine (FUdR, Sigma–Aldrich) (50 µM).10 Escherichia coli OP50 cells in M9 buffer (3 mg wet weight) were spread onto each NGM/FUdR plate (35 mm diameter) as the food source. The longevity assay was started at the L4 stage of N2 nematodes and mutants, at which time the worms were transferred to the seeded plates using a platinum wire. For each assay, approximately 30 worms were assayed on three plates (10 worms per plate) for each compound. The plates were incubated at 25℃, and live and dead worms were counted every 24 h. The longevity assay was carried out at least three times independently, and over 100 worms were used. The mean lifespan (MLS) was estimated using the following formula16
where j is the age (day), dj is the number of worms that died in the age interval (xj, xj+1) and N is the total number of worms. The standard error (SE) of the estimated MLS was calculated using the following equation
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Worms fed for 24 h with E. coli OP50 on NGM/FUdR plates including RES or OXY were collected in M9 buffer, and total RNA was isolated from whole worms as previously reported.17 Total RNA was converted to cDNA using the RevertAid First Strand cDNA Synthesis kit according to the manufacturer’s instructions (Thermo Scientific, Wilmington, DE, USA), followed by quantitative RT-PCR (qRT-PCR) using the SYBR Green (KAPA Biosystems, Wilmington, MA, USA) and StepOnePlus real time PCR systems (Applied Biosystems, Foster City, CA, USA). The primer sequences are shown in Table 1.5,18 Relative expression levels were calculated using the 2−ΔΔCT method.19 The control gene act-2 was used to normalise gene expression data. Three independent experiments were performed.
Table 1.
Oligonucleotide primers used in this study
| Target (accession #) | Primer | Sequence | Product size (bp) |
|---|---|---|---|
| act-2 (NM_073417) | Forward | 5′-CCCACTCAATCCAAAGGCTA-3′ | 168 |
| Reverse | 5′-GGGACTGTGTGGGTAACACC-3′ | ||
| daf-16 (AF032112) | Forward | 5′-TCCTCATTCACTCCCGATTC-3′ | 175 |
| Reverse | 5′-CCGGTGTATTCATGAACGTG-3′ | ||
| aak-2 (NM_001029697) | Forward | 5′-TGCTTCACCATATGCTCTGC-3′ | 219 |
| Reverse | 5′-GTGGATCATCTCCCAGCAAT-3′ | ||
| sir 2.1 (NM_001268556) | Forward | 5′-TGGCTGACGATTCGATGGAT-3′ | 179 |
| Reverse | 5′-ATGAGCAGAAATCGCGACAC-3′ |
Primers against age-related gene were designed using NCBI and Primer 3 software.
Western blot analysis
Protein was prepared from synchronous nematodes raised for 24 h on E. coli OP50-seeded plates that included OXY or RES. Worms were collected in dH2O and washed with dH2O several times and protein was extracted using an ultrasonic processor (Cole-Parmer, Vernon Hills, IL, USA). Equal amounts (30 µg) of protein from each sample were loaded into lanes, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) (9.4%) was performed. Then, proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bed-ford, MA, USA) with a Trans-Blot semi-dry transfer cell (Bio-Rad, Hercules, CA, USA). The membrane was blocked using 5% non-fat milk (Blotting-Grade Blocker, Bio-Rad) in phosphate buffered saline (PBS)-Tween 20 (PBS-T) for 1 h at room temperature and subsequently incubated with either anti-SIR-2.1 rabbit polyclonal antibody (Thermo Scientific, PA1-16933, diluted 1:1500 in PBS-T) or anti-GAPDH mouse monoclonal antibody (Thermo Scientific, MA5-15738, diluted 1:5000 in PBS-T) for 1 h. Bound antibodies were detected with peroxidase-conjugated secondary antibodies. Goat anti-rabbit IgG (H + L) horseradish peroxidase-conjugated antibody (Thermo Scientific, 31460, diluted 1:10000) and goat anti-mouse IgG (H + L) horseradish peroxidase-conjugated antibody (Thermo Scientific, 31430, diluted 1:200,000) were used as secondary antibodies. The protein bands were visualised using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific). Blot images were obtained and protein expression was determined with the imaging system FluorChem E (ProteinSimple, Santa Clara, CA, USA). More than three independent worm cultures were used.
Statistical analysis
The nematode survival rate was calculated with the Kaplan–Meier method, and the significance of survival differences among the whole sample and between groups were tested using the log-rank test.13 In other experiments, the mean values of the control (DMSO) and chemical-treated (RES and OXY) groups were determined using Student’s t-tests. A P-value of 0.05 or less was considered statistically significant, and error bars depict the standard deviation.
Results and discussion
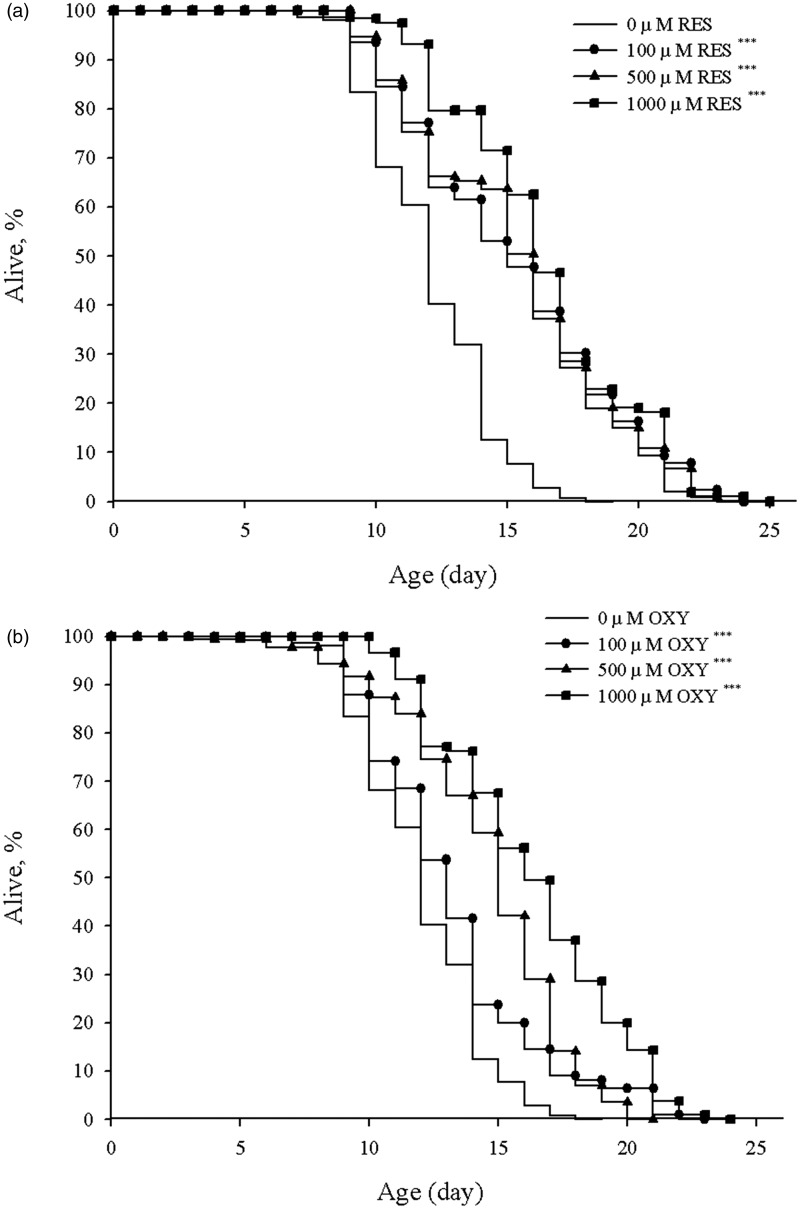
Caenorhabditis elegans (N2) longevity assays were performed on NGM/FUdR plates containing OXY or RES at the designated concentrations. OXY and RES significantly (P < 0.05) increased the MLS of worms in a dose-dependent manner (Figure 1, Table 2). The MLSs of worms fed E. coli OP50 on NGM plates containing 100, 500 and 1000 µM of RES were 16.5 ± 0.35 (22.2%), 16.7 ± 0.35 (23.7%) and 17.6 ± 0.25 (30.4%), respectively, which were greater than those of control worms fed E. coli OP50 on NGM/FUdR plates without RES or OXY. The MLSs of worms fed E. coli OP50 on NGM plates containing 100, 500 and 1000 µM of OXY were 14.5 ± 0.32 (7.4%), 15.9 ± 0.31 (17.8%) and 17.7 ± 0.28 (31.1%), respectively, which were also greater than those of the control group. The MLSs in the 100 and 500 µM RES-treated groups were higher than those in the 100 and 500 µM OXY-treated groups, although the MLSs were similar in the 1000 µM OXY- and RES-treated worms.
Figure 1.
Effect of RES or OXY on Caenorhabditis elegans (N2) lifespan. After 3 days of feeding with Escherichia coli OP50 on NGM plates, young adult worms were transferred to NGM/FUdR plates seeded with E. coli OP50 and containing RES (a) or OXY (b). The lifespans of nematodes fed on NGM/FUdR plates containing RES or OXY were significantly (P < 0.05) extended compared to those fed on NGM/FUdR plates containing DMSO only
Table 2.
The mean lifespan of Caenorhabditis elegans (N2) and C. elegans loss-of-function mutants when fed Escheichia coli OP50 on NGM plates containing RES or OXY
| Nematode type | Chemical in NGM | No. of total worms (dead/censored) | MLS ± SE (days) |
|---|---|---|---|
| N2 | DMSO (0.3%, v/v) | 156 (144/12) | 13.5 ± 0.21 |
| Resveratrol (100 µM) | 146 (129/17) | 16.5 ± 0.35*** | |
| Resveratrol (500 µM) | 147 (121/26) | 16.7 ± 0.35*** | |
| Resveratrol (1000 µM) | 144 (105/39) | 17.6 ± 0.23*** | |
| Oxyresveratrol (100 µM) | 142 (110/32) | 14.5 ± 0.32*** | |
| Oxyresveratrol (500 µM) | 144 (114/30) | 15.9 ± 0.31*** | |
| Oxyresveratrol (1000 µM) | 149 (105/44) | 17.7 ± 0.28*** | |
| CF1038 daf-16 (mu86) | DMSO (0.3%, v/v) | 165 (155/10) | 10.8 ± 0.45 |
| Resveratrol (100 µM) | 168 (123/45) | 11.4 ± 0.53*** | |
| Resveratrol (500 µM) | 150 (105/45) | 12.1 ± 0.57*** | |
| Resveratrol (1000 µM) | 150 (105/45) | 12.1 ± 0.61*** | |
| Oxyresveratrol (100 µM) | 130 (119/11) | 12.2 ± 0.58*** | |
| Oxyresveratrol (500 µM) | 166 (110/56) | 12.1 ± 0.60*** | |
| Oxyresveratrol (1000 µM) | 172 (113/59) | 12.3 ± 0.60*** | |
| RB754 aak-2 (ok524) | DMSO (0.3%, v/v) | 201 (176/25) | 11.1 ± 0.45 |
| Resveratrol (100 µM) | 199 (160/39) | 10.8 ± 0.43** | |
| Resveratrol (500 µM) | 201 (149/52) | 10.8 ± 0.47 | |
| Resveratrol (1000 µM) | 200 (142/58) | 10.8 ± 0.47 | |
| Oxyresveratrol (100 µM) | 201 (164/37) | 10.8 ± 0.44 | |
| Oxyresveratrol (500 µM) | 203 (150/53) | 10.5 ± 0.44*** | |
| Oxyresveratrol (1000 µM) | 197 (133/64) | 10.3 ± 0.46*** | |
| VC199 sir-2.1 (ok434) | DMSO (0.3%, v/v) | 171 (134/37) | 11.6 ± 0.55 |
| Resveratrol (100 µM) | 166 (118/48) | 11.4 ± 0.59 | |
| Resveratrol (500 µM) | 163 (113/50) | 11.2 ± 0.60 | |
| Resveratrol (1000 µM) | 183 (136/47) | 11.4 ± 0.54 | |
| Oxyresveratrol (100 µM) | 173 (135/38) | 11.4 ± 0.54 | |
| Oxyresveratrol (500 µM) | 160 (126/34) | 11.5 ± 0.57 | |
| Oxyresveratrol (1000 µM) | 170 (140/30) | 11.1 ± 0.68 |
P value versus control (DMSO); *P < 0.05, **P < 0.01, ***P < 0.001.
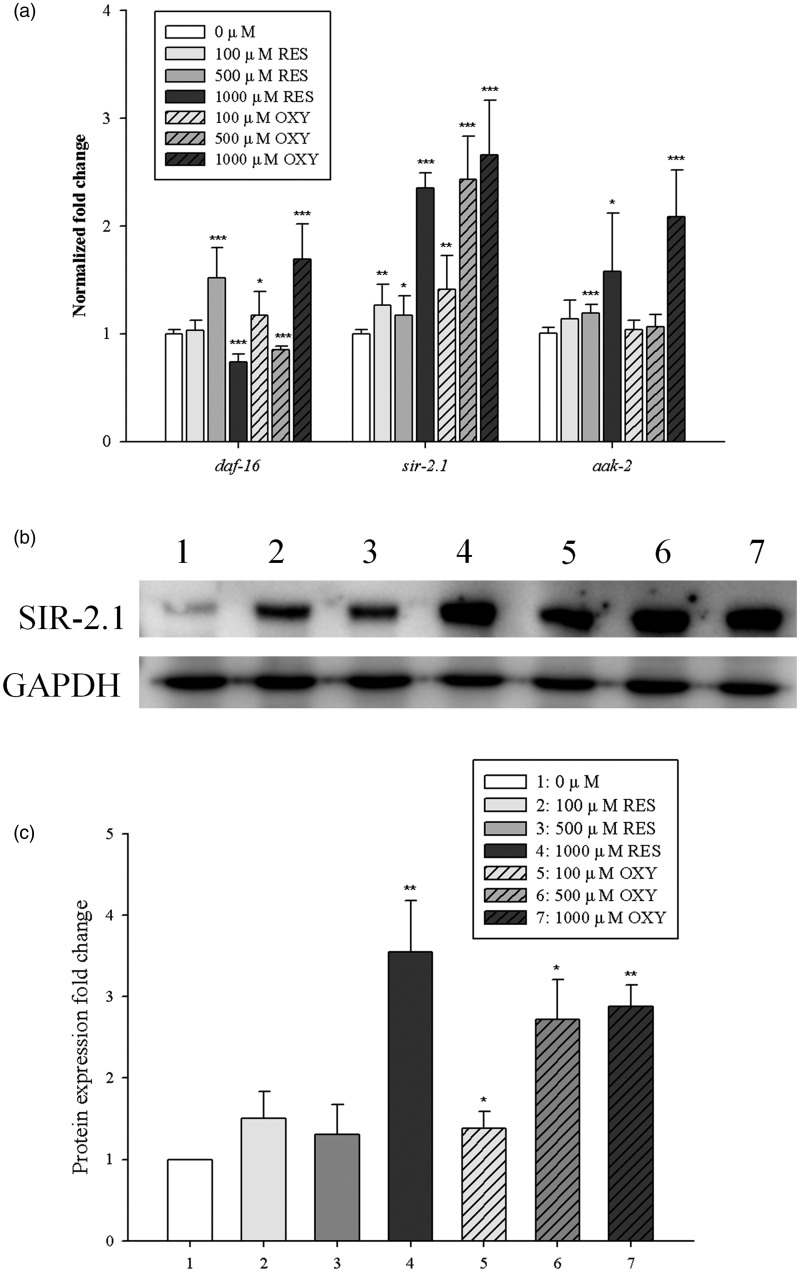
We next investigated whether expression of the daf-16, sir-2.1 and aak-2 genes was related to lifespan extension in C. elegans using qRT-PCR in worms fed E. coli OP50 on plates containing OXY or RES. OXY significantly (P < 0.05) increased the sir-2.1 gene expression level in a dose-dependent manner (Figure 2(a)). The expression level of the sir-2.1 gene was up-regulated more than two-fold by the 500 and 1000 µM OXY treatments, while only the 1000 µM RES treatment up-regulated sir-2.1 more than two-fold compared with the control. Similarly, the aak-2 gene was up-regulated by the 500 and 1000 µM RES treatment and by the 1000 µM OXY treatment. Although 500 µM of RES and 1000 µM of OXY increased the daf-16 expression levels, a dose-dependent increase in expression levels was not observed in worms fed E. coli OP50 on plates containing OXY or RES. To confirm the increase in SIR-2.1 protein after exposure to RES and OXY, western blot analysis was performed. SIR-2.1 protein expression was significantly (P < 0.05) increased only in the 1000 µM RES treatment group compared with the control (DMSO), whereas OXY treatment significantly (P < 0.05) increased the expression level of SIR-2.1 protein in a dose-dependent manner (Figure 2(b) and (c)).
Figure 2.
Changes in gene expression and SIR-2.1 protein expression of Caenorhabditis elegans (N2) fed on NGM/FUdR plates containing RES or OXY. The expression level of the genes was used to evaluate the effect of RES and OXY on the lifespan of C. elegans (a). Western blot was conducted to investigate the effect of OXY on SIR-2.1 protein expression (b, c). The expression level of SIR-2.1 protein significantly increased in the 1000 µM RES and 100, 500, and 1000 µM OXY groups, as compared with the control, in a dose-dependent manner. Asterisks denote significant differences compared with worms fed on NGM/FUdR plates containing DMSO only. *P < 0.05; **P < 0.01; ***P < 0.001
To confirm the role of the aforementioned genes in lifespan extension by RES or OXY, C. elegans loss-of-function mutants were used to measure longevity. OXY and RES failed to extend lifespan in C. elegans aak-2 or sir-2.1 mutants, which suggests that these genes are essential to lifespan extension in C. elegans fed E. coli OP50 on NGM/FUdR plates containing RES or OXY (Table 2). Interestingly, the lifespan of aak-2 mutants was significantly (P < 0.05) decreased by treatment with 500 and 1000 µM of OXY compared with the control. Unexpectedly, the lifespan of daf-16 mutants was significantly (P < 0.05) increased by treatment with RES and OXY compared with the control. Taken together with the qRT-PCR results, these findings show that SIR-2.1 may play a key role in enhancing the longevity of C. elegans mediated by RES and OXY.
In this study, no extension in lifespan was observed in sir-2.1 and aak-2 mutants, while the lifespan of daf-16 mutants was extended by OXY and RES. The Sir2 family and AMPK, but not FOXO, are necessary to extend lifespan in C. elegans through the DR mimetic RES,1,2,5,20 and the results of the present study are consistent with these results. Intriguingly, although the role of FOXO/DAF-16 remains unclear, the effect of OXY on lifespan extension in C. elegans was also dependent on SIR-2.1 and AMPK. The energy-sensing AMPK/aak-2 pathway and FOXO/DAF-16 are necessary for longevity induced by DR.5 In particular, AMPK can directly phosphorylate FOXO/DAF-16 to extend lifespan and aak-2 gene encodes AMPK.4 In worms treated with RES or OXY, aak-2 mutant decreased lifespan and daf-16 mutant extended lifespan. Although we do not have direct evidence, the results suggested that the lifespan extension of C. elegans via AAK-2 by RES and OXY might not be directly linked with DAF-16. Although the upstream activation of DAF-16 occurs via multiple factors, for instance, SIR-2.1, AAK-2 and JNK-1, nuclear translocation of DAF-16 triggers upregulation of several stress-response genes including longevity resulted in the lifespan extension of C. elegans. In worms, DR dose not increase the nuclear localization of DAF-16.4 Therefore, RES and OXY might not induce DAF-16 nuclear localization, which means that the lifespan extension of C. elegans by RES and OXY is DAF-16-independent. The results suggest that additional pathway(s) might exist downstream of Sir-2 and AMPK, distinct from FOXO/DAF-16.
Antioxidants extend the lifespan of C. elegans through a DAF-16/FoxO3a-dependent pathway.21 In contrast, C. elegans lifespan extension via the antioxidants OXY and RES was independent of the FOXO/DAF-16 pathway. These results suggest that stilbene antioxidants such as OXY and RES might extend the lifespan of C. elegans via Sir2 activation, which indicates that OXY and RES might induce caloric restriction. In addition, OXY and RES affect the lifespan extension of C. elegans via AMPK/aak-2 activation, which is involved in lifespan extension and resistance to oxidative stress.22 Furthermore, RES was shown to increase the lifespan of C. elegans in a SIR-2.1-dependent manner, specifically via SIR-2.1-dependent regulation of endoplasmic reticulum (ER) stress response genes in C. elegans.1 It was recently reported that overexpression of the sir2 gene by transgene insertion had no effect on lifespan extension in C. elegans and that sir2 did not mediate the effects of DR.18 Moreover, several reports have shown inconsistent results concerning the mechanism of lifespan extension by RES, including SIR-2.1-dependent2,3 and -independent,10 DAF-16-independent1,2 and AMPK/aak-2-dependent mechanisms.5 In this regard, RES might be involved in lifespan regulation via both negative and positive feedback mechanisms.5 Thus, further studies are needed to investigate additional factors and/or pathways contributing to the lifespan extension of C. elegans by OXY and RES.
In conclusion, OXY and RES extend the lifespan of C. elegans via a SIR-2.1 - and AMPK pathway-dependent manner but a FOXO/DAF-16-independent manner. DR is known to extend lifespan, and AMPK/aak-2 and Sir2 regulate caloric restriction; thus, OXY and RES may function as DR mimetics to extend the lifespan of C. elegans.
Acknowledgements
This research was supported by the High Value-added Food Technology Development Program, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (313028-3 and 114033-03).
Author contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; YHL and JL designed the research and JL, GK, JP and JKK conducted the experiments. YHL and JL wrote the manuscript. All authors read and approved the final manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell 2005; 9: 605–15. [DOI] [PubMed] [Google Scholar]
- 2.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 2004; 430: 686–89. [DOI] [PubMed] [Google Scholar]
- 3.Tatar M. SIR2 calls upon the ER. Cell Metab 2005; 2: 281–82. [DOI] [PubMed] [Google Scholar]
- 4.Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 2007; 17: 1646–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 2009; 8: 113–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchal J, Pifferi F, Aujard F. Resveratrol in mammals: effects on aging biomarkers, age-related diseases, and life span. Ann NY Acad Sci 2013; 1290: 67–73. [DOI] [PubMed] [Google Scholar]
- 7.Ruetenik A, Barrientos A. Dietary restriction, mitochondrial function and aging: from yeast to humans. Biochim Biophys Acta 2015; 1847: 1434–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang SQ, Cai WJ, Huang JH, Wu B, Xia SJ, Chen XL, Zhang XM, Shen ZY. Icariin, a natural flavonol glycoside, extends healthspan in mice. Exp Gerontol 2015; 69: 226–35. [DOI] [PubMed] [Google Scholar]
- 9.Sung B, Chung JW, Bae HR, Choi JS, Kim CM, Kim ND. Extract exhibits antioxidative and anti-aging effects via modulation of the AMPK-SIRT1 pathway. Exp Ther Med 2015; 9: 1819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech Ageing Dev 2007; 128: 546–52. [DOI] [PubMed] [Google Scholar]
- 11.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell S, Napper A, Curtis R, Distefano PS, Fields S, Bedalov A, Kennedy BK. Substrate specific activation of sirtuins by resveratrol. J Biol Chem 2005; 280: 17038–45. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz P, Roychowdhury S, Engelmann M, Wolf G, Horn TF. Oxyresveratrol and resveratrol are potent antioxidants and free radical scavengers: effect on nitrosative and oxidative stress derived from microglial cells. Nitric Oxide 2003; 9: 64–76. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, Zhao L, Zheng X, Fu T, Guo H, Ren F. Lactobacillus salivarius strain FDB89 induced longevity in Caenorhabditis elegans by dietary restriction. J Microbiol 2013; 51: 183–88. [DOI] [PubMed] [Google Scholar]
- 14.Stiernagle T. Maintenance of C. elegans. C. elegans: a practical approach, New York: Oxford University Press, 1999. [Google Scholar]
- 15.Sulston J, Hodgkin J. Methods. The nematode Caenorhabditis elegans, New York: Cold Spring Harbor Laboratory Press, 1988. [Google Scholar]
- 16.Wu D, Rea SL, Yashin AI, Johnson TE. Visualizing hidden heterogeneity in isogenic populations of C. elegans. Exp Gerontol 2006; 41: 261–70. [DOI] [PubMed] [Google Scholar]
- 17.Burdine RD, Stern MJ. Easy RNA isolation from C. elegans: a TRIZOL based method. Worm Breed Gaz 1996; 14: 10–10. [Google Scholar]
- 18.Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 2011; 477: 482–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 5: 402–08. [DOI] [PubMed] [Google Scholar]
- 20.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci 2004; 101: 15998–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Ishihara N, Lee TR. A DAF-16/FoxO3a-dependent longevity signal is initiated by antioxidants. Biofactors 2014; 40: 247–57. [DOI] [PubMed] [Google Scholar]
- 22.Lee H, Cho JS, Lambacher N, Lee J, Lee SJ, Lee TH, Gartner A, Koo HS. The Caenorhabditis elegans AMP-activated protein kinase AAK-2 is phosphorylated by LKB1 and is required for resistance to oxidative stress and for normal motility and foraging behavior. J Biol Chem 2008; 283: 14988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]