Abstract
The connectivity architecture of the human brain varies across individuals. Mapping functional anatomy at the individual level is challenging, but critical for basic neuroscience research and clinical intervention. Using resting-state functional connectivity, we parcellated functional systems in an “embedding space” based on functional characteristics common across the population, while simultaneously accounting for individual variability in the cortical distribution of functional units. The functional connectivity patterns observed in resting-state data were mapped in the embedding space and the maps were aligned across individuals. A clustering algorithm was performed on the aligned embedding maps and the resulting clusters were transformed back to the unique anatomical space of each individual. This novel approach identified functional systems that were reproducible within subjects, but were distributed across different anatomical locations in different subjects. Using this approach for intersubject alignment improved the predictability of individual differences in language laterality when compared with anatomical alignment alone. Our results further revealed that the strength of association between function and macroanatomy varied across the cortex, which was strong in unimodal sensorimotor networks, but weak in association networks.
Keywords: functional parcellation, individual differences, resting-state fMRI
Introduction
The connectivity architecture of the human brain exhibits an extraordinary level of complexity. Its development is shaped by genetic and environmental factors that are variable across individuals. Whereas some brain systems are relatively consistent across the population, many networks exhibit substantial interindividual differences (Hill et al. 2010; Zilles et al. 2013). Recent findings suggest that interindividual variability in functional connectivity is not uniformly distributed across the cortex: the association regions, including language, executive control, and attention networks, are likely more variable than the unimodal regions, such as the visual and sensorimotor cortices (Mueller et al. 2013). Crucially, individual differences in functional networks exist not only in the connectivity patterns among the nodes supporting those functions, but also in the morphological locations of these nodes (Rajkowska and Goldman-Rakic 1995) and their cytoarchitecture (Brett et al. 2002). Consequently, individual differences observed in functional connectivity measures are often a combination of both functional and anatomical variability (Brett et al. 2002).
Currently, fMRI studies often rely on intersubject normalization based on global brain morphology, brain areas derived from cytoarchitectural segmentation of a template (Fischl et al. 1999), or functional atlases derived from a population (Yeo et al. 2011; Baker et al. 2014). These approaches implicitly assume a static relation between anatomy and function, and they can confound intersubject variability in functional characteristics with variability in anatomy. The mixture of variability sources can thus reduce the specificity of functional connectivity markers related to cognitive capability (Seeley et al. 2007; van den Heuvel et al. 2009; Cole et al. 2012) or psychiatric diseases (Fox and Greicius 2010), because the targeted networks are often those that are most variable across individuals, both anatomically and functionally (Fox et al. 2012; Mueller et al. 2013). The same challenge is faced by genomewide association studies that aim to unveil the genetic underpinnings of specific brain functions (Meyer-Lindenberg et al. 2006; Potkin et al. 2009). A brain parcellation technique that can establish valid functional correspondences across individuals will facilitate the investigation of individual differences in fine-grained functional characteristics, especially those that are not tightly coupled to macroscopic brain structures. More broadly, mapping functional networks at the individual level will not only improve group-level analyses in basic and clinical neuroscience research, but will also lead to direct clinical applications, such as preoperative functional mapping and noninvasive brain stimulation.
Here, we demonstrate a novel approach to mapping functional networks in individual subjects. By representing function in a reference space that is decoupled from global morphology of the individual brain, we investigated the multivariate function across subjects. Correspondence across subjects was established by aligning these low-dimensional “embedding maps” of the whole-brain functional connectivity patterns derived from individual subjects. Brain parcellation was performed using these aligned embedding maps and the resulting networks were projected back to each subject's anatomical space (e.g., vertices on the cortical surface). Intrasubject reproducibility of the functional networks and intersubject variability in their anatomical distribution were evaluated.
Materials and Methods
Participants and Data Collection
Two separate resting-state fMRI datasets were used in the current study; both datasets have been previously reported (Mueller et al. 2013; Wang et al. 2013, 2014). The first dataset consisted of 23 healthy subjects (age 51.8 ± 6.99, 9 females) who were recruited as the healthy control cohort for a longitudinal stroke recovery study. Each subject underwent 5 resting-state fMRI scanning sessions within 6 months (7, 14, 30, 90, and 180 days from enrollment) and had at least 2 good resting-state runs (temporal signal to noise ration > 100) in each session (6 min and 12 s per run, mean = 2.02 runs). The second dataset consisted of 55 subjects who participated in a task-based fMRI semantic classification paradigm, as well as resting-state scans. The design of the semantic classification task is described in Wang et al. (2014). Each subject had one or two resting-state (eyes open) fMRI runs (6 min and 12 s per run, mean = 1.7 runs). All participants provided written, informed consent in accordance with guidelines established by the Institutional Review Boards of Harvard University, Partners Healthcare, or Xuanwu Hospital.
Both datasets were acquired on 3-Tesla TimTrio scanners (Siemens, Erlangen, Germany) using the 12-channel phased-array coils supplied by the vendor. Functional data were obtained using the same gradient echo-planar pulse sequence (TR, 3000 ms; TE, 30 ms; 3 mm isotropic voxels; transverse orientation; 47 slices that fully covered the cerebral cortex and the cerebellum). Structural images were acquired using a sagittal MP-RAGE 3D T1-weighted sequence.
fMRI and Structural MRI Data Processing
Resting-state fMRI data were preprocessed using previously described procedures (Van Dijk et al. 2010; Yeo et al. 2011). The processing included the following steps: 1) slice timing correction; 2) rigid body correction for head motion with the FSL package; 3) normalization for global mean signal intensity across runs; and 4) low-pass temporal filtering, head motion regression, whole-brain signal regression, and ventricular and white matter signal regression. Anatomical data were processed using the FreeSurfer version 4.5.0 software package (http://surfer.nmr.mgh.harvard.edu). Surface mesh representations of the cortex for each individual subject were reconstructed and registered to a common spherical coordinate system (Fischl et al. 1999). Anatomical and functional images were aligned using boundary-based registration (Greve and Fischl 2009). Resting-state BOLD fMRI data were then aligned to the common spherical coordinate system by sampling from the middle of the cortical ribbon in a single interpolation step (Yeo et al. 2011). To evaluate functional laterality, a symmetric surface template of the cerebral cortex was constructed (Greve et al. 2013). The symmetric surface template was downsampled to 2562 vertices in each hemisphere, with an average distance of 4.3 mm between any 2 neighboring vertices. fMRI data of each individual were then registered to this template and smoothed on the surface with a 6-mm full-width half-maximum (FWHM) kernel.
In this study, we have compared different parcellation methods. In all comparisons, fMRI data were processed in the same way, i.e. the data were downsampled to the same number of vertices on the same brain surface generated using FreeSurfer. The fMRI signal on each vertex was thus identical for different parcellation methods.
Spectral Embedding of Global Intrinsic Connectivity Patterns
For each fMRI session, we constructed a correlation matrix using the fMRI signal time courses extracted from the 5124 cortical vertices in 2 hemispheres, where each entry in the matrix represented the correlation coefficient value between 2 vertices. Correlation values smaller than a certain threshold were set to zero. We then performed diffusion map embedding (Coifman et al. 2005) of the cortical vertices, by defining non-negative symmetric weights, , among pairs of vertices, where <i,j> are the entries in the correlation matrix, and defines the weight-decay (Langs et al. 2014). Diffusion map embedding treats the resulting graph as the basis of the diffusion process, and projects the vertices to an embedding space. The probability of transition between nodes in a given time defines the Euclidean distance between the corresponding points in the embedding space (Coifman and Lafon 2006). The diffusion time parameter of the embedding steers the granularity of the representation. Parameters for the embedding process, including correlation threshold, dimensionality, and diffusion time, were selected based on intrasubject test–retest reliability and sensitivity to individual differences of the resulting parcellation maps (details described in a later section).
Each point in the embedding map represented a cortical vertex. The distribution of points in the embedding map characterized the global functional connectivity pattern, with cliques of vertices that exhibit coherent activity forming clusters of points, and regions exhibiting different signals being mapped to positions that are far apart in the embedding space. Although neighboring vertices on the cortical surface tend to have a coherent fMRI signal and may be positioned closely in the embedding space, spatial relation in the embedding space is otherwise decoupled from global anatomical characteristics and is determined solely by functional relations among vertices. The key property of the embedding space is that it represents the pairwise functional connectivity relationships among cortical points in a global map, allowing for the quantification of relationships among groups of points. We used this property to cluster the points in the embedding space. Clustering the points in the embedding space corresponds to grouping vertices on the cortical surface that exhibit a coherent fMRI signal. The one-to-one correspondence between the data points in the embedding space and the surface vertices allows direct transformation of the clustering results back to each subject's anatomical space. For each fMRI session, we obtained an embedding map that formed the basis for intersubject functional alignment.
Functional Alignment across Subjects
We performed intersubject alignment in the embedding space where the functional connectivity patterns of each subject had been mapped.
The embedding maps containing the embedded vertices of both hemispheres were treated as a representation of the whole cortex and were aligned across all subjects using the following procedure. Embedding points were paired across subjects if they represented the same vertex on the cortical surface. Embedding maps of each subject were then aligned orthonormally to a reference subject by minimizing the sum of squared distances between paired embedding points that corresponded to the same surface vertex. The alignment procedure allowed for rotation, translation, permutation of dimensions, change of axis, and change of axis signs, i.e., the resulting transformation was an isometry that left distances within each individual map unchanged (Langs et al. 2014). This alignment resulted in a joint map, with data points corresponding to all surface vertices acquired in all scan sessions. It represented the shared connectivity architecture observed in the entire population. Each point in the joint map corresponded to one vertex in one subject.
Parcellation in the Joint Embedding Space
To identify functional networks, we performed a single clustering in the joint embedding map. We fitted a Gaussian mixture model to the distribution. Each Gaussian had a diagonal covariance matrix with independent diagonal entries. After the clustering, each point in the embedding map was assigned the cluster label corresponding to the Gaussian component with the highest a posteriori probability at this position. To obtain the network parcellation in each individual, the cluster labels were projected back to the individual subject's corresponding surface vertex. To reduce noise, we removed clusters with fewer than 40 vertices (∼0.78% of the cortex) from the result. To enable the comparison with parcellations established in the literature (Yeo et al. 2011), we evaluated 2 Gaussian mixture models that resulted in 7 and 17 clusters. Because a single clustering was performed on embedding points derived from all subjects, this population-based analysis is less sensitive to noise that can significantly affect individual-level analysis. Importantly, individual subject information is well preserved in the data because the procedure does not require averaging across subjects. The parcellation results in the embedding space can be directly transformed back to the anatomical space to inform the unique functional architecture of each individual.
Evaluating Intrasubject and Intersubject Variability
To facilitate research on individual differences, a mapping technology must achieve high intrasubject test–retest reliability and high sensitivity to intersubject variability simultaneously. Here, we defined a metric, termed variability signal-to-noise ratio (vSNR), to quantify the potential usefulness of a functional mapping technology in individual differences research. In the context of individual differences research, the signal of interest is intersubject variability of functional measures. Intersubject variability of parcellation maps reflects not only true individual differences in functional organization, but also variability caused by technical artifacts and dynamic brain states. Variability due to artifacts and dynamic brain states can be approximated by intrasubject variation. Assuming variability due to different sources is additive, then
Therefore,
Intersubject variability and intrasubject variability of the brain parcellation maps were evaluated based on the first dataset. For each subject and each fMRI session, the parcellation algorithm generated a network label for each cortical vertex.
To obtain intrasubject variability (i.e. the inverse of reliability), we first applied a binary matching between any 2 scans from the 5 scans (i.e. possible combinations for each subject). The resulting 10 binary matching maps were then averaged within the subject. Finally, the variability maps were averaged across 23 subjects.
To obtain intersubject variability, we randomly selected 5 fMRI sessions, each from a different subject. Variability was quantified similarly at each vertex by counting the fraction of cluster labels that were not identical among all possible comparisons ( possible combinations). This permutation was repeated 23 times to match the number of subjects, and the resulting intersubject variability maps were then averaged across the 23 permutations.
Selecting Optimal Parameters for Embedding
To investigate the influence of embedding parameters on the performance of our parcellation algorithm, we parcellated brain networks using the first dataset by varying the correlation threshold, the dimensionality of the embedding space, and the diffusion time. The parameters were evaluated based on vSNR of the resulting parcellation maps. Intersubject variability and intrasubject variability of the parcellation maps were quantified for different combinations of parameters (see Supplementary Fig. 1). The results indicated that the correlation threshold had the most significant impact on vSNR. Based on this experiment, we chose a diffusion time of t = 0.5, a dimension of d = 30, and a correlation threshold of c = 0.1 for the subsequent analyses.
Comparison with Subject-Level Clustering Using Traditional Approaches
An alternative method to obtaining subject-specific functional parcellation (FP) is to perform individual-level clustering in the anatomical space using traditional approaches. For comparison purposes, we applied the k-means clustering based on the cortico-cortical connectivity profiles (Yeo et al. 2011) of each subject. To allow comparison between subjects, this approach requires matching the results across subjects by maximizing the overlap of the corresponding clusters. We used the Hungarian algorithm (Kuhn 1955) to obtain an optimal matching of clusters across all sessions and subjects. The Hungarian algorithm for maximum-weight bi-partite matching, finds correspondences between 2 sets of points. Using this method to find corresponding parcellation networks between 2 subjects ensures a globally optimal matching with regard to Dice overlap measure. Intersubject variability and intrasubject variability were then calculated using the same approach described above.
Correlation Between Language Laterality and Connectivity Laterality
For the second group of subjects who underwent both a language-task fMRI scan and a resting-state fMRI scan, we first performed a network parcellation based on the resting-state data in the embedding space. For each vertex, we calculated the degree of within-hemisphere connectivity (Wang et al. 2014) by counting the number of vertices that were strongly correlated with the seed vertex (r > 0.25). A laterality index was computed for each network by contrasting the within-hemisphere connectivity degree summed across the vertices in its left and right hemisphere portions. To obtain the task-based language laterality index, task fMRI data were analyzed using the general linear model in each participant's native fMRI space (Wang et al. 2013). The language laterality index was calculated for each individual subject based on the asymmetric activations in the 2 hemispheres, using an approach previously described (Liu et al. 2009). For each of the 17 (or 7) brain networks, the Pearson correlation coefficient between the task-based language laterality index and the connectivity-based laterality index was computed. For comparison, we also computed a laterality index for each of the 17 brain networks derived from 1000 subjects whose data were aligned in anatomical space (Yeo et al. 2011). The laterality index of a network was computed using the same approach as described above.
Visualization
For the purpose of visualization, maps were displayed on the left and right inflated PALS cortical surfaces using the Caret software (Van Essen 2005).
Results
Brain Parcellation Based on Functional Alignment in the Embedding Space
Functional networks were identified based on a joint analysis of the resting-state fMRI data of 23 subjects who were scanned 5 times during a period of 6 months. The intrinsic functional connectivity of the cerebral cortex was transformed into a low-dimensional embedding space and then aligned across individuals (see Supplementary Materials and Methods). A single clustering was performed in this transformed space (Fig. 1, right column) to identify the common networks within the entire study population. The spatial distributions of the networks in each individual brain were then obtained by projecting network markers from the embedding space back to the subject-specific anatomical space. This resulted in a FP for each individual and each fMRI acquisition, enabling the quantification of intrasubject reproducibility and intersubject variability.
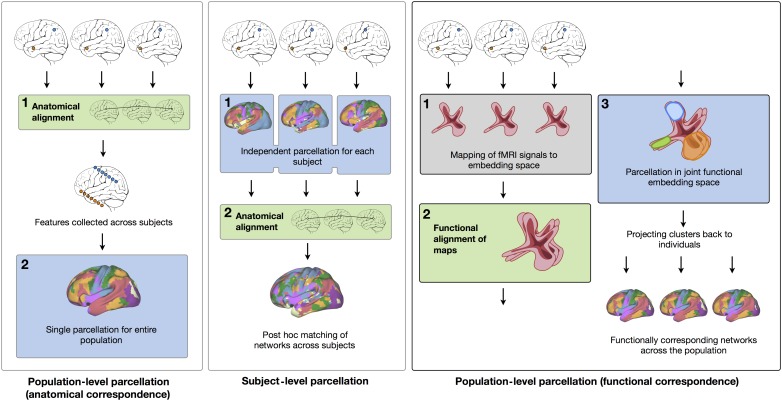
Figure 1.
Brain parcellation based on functional alignment in the embedding space. Parcellation can be performed at the population level when assuming anatomical consistency (left panel). The results reflect the general organization principle of brain networks, but lack subject-specific details. Subject-level parcellation in anatomical space is more sensitive to noise and lacks consistency across subjects (middle panel). Projecting individual subject data into a low-dimensional embedding space, and aligning the maps in this functional space enables population-level clustering and can reflect the interindividual variability of the spatial distribution of networks (right panel).
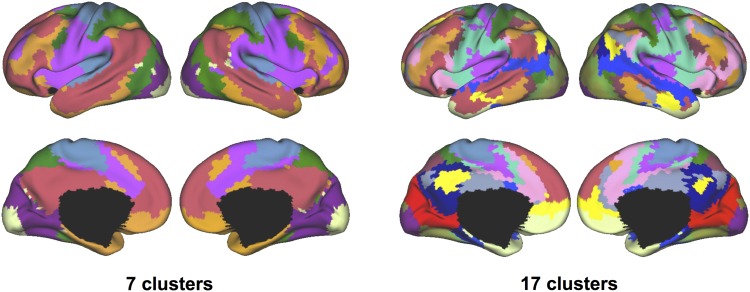
The cerebral cortex was parcellated into 7 and 17 networks in 2 different experiments (Fig. 2). Since the network distributions could vary across individuals, in order to compare with the previous parcellation results based on the anatomical space alignment of a large subject cohort (Yeo et al. 2011), a consensus map was generated in the anatomical space using majority voting of cluster labels across all individuals in this study (Fig. 2). At the group level, the consensus parcellation resembled the previous reports and identified the default, frontoparietal, dorsal attention, ventral attention, sensorimotor, and visual (including central and peripheral visual systems) networks. The consensus map (Fig. 2) and the group-level maps derived after anatomical alignment (Yeo et al. 2011) (Fig. 1, left column) both illustrate the general organizational patterns of functional systems in the population.
Figure 2.
Functional alignment in the embedding space captured the general organization of functional systems in the human brain. The cerebral cortex was parcellated into 7 networks and 17 networks in 2 experiments. Population-level functional atlases were derived from the brain networks of all individual subjects through a majority voting approach. Each network was represented by a different color.
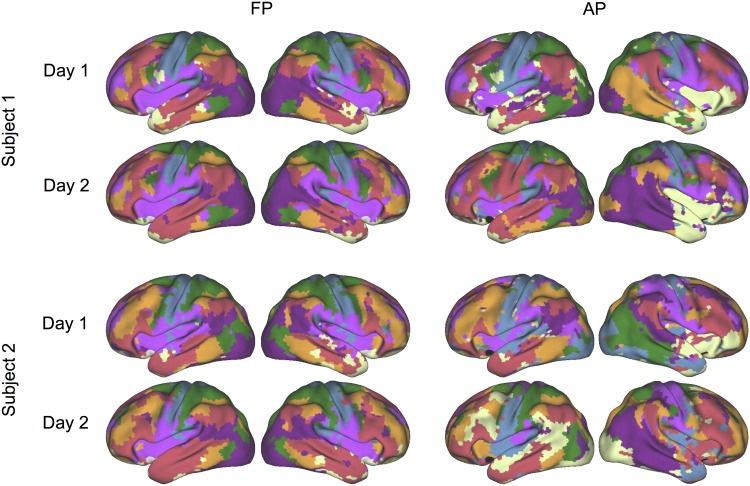
In individual subjects, networks derived from FP in the embedding space exhibited high reproducibility in repeated sessions (Fig. 3, left). Critically, the maps also demonstrated substantial intersubject variability in spatial distribution, suggesting that the same brain function could involve different brain structures in different subjects (see Supplementary Fig. 2 for the results from all 23 subjects). In contrast, individual-level parcellation in the anatomical space (AP) based on traditional k-means clustering (analogous to the population clustering in (Yeo et al. 2011)) suffered from overall weaker stability and required a post hoc matching of identified clusters across subjects, which was quite challenging (Fig. 3, right, also see Supplementary Fig. 2).
Figure 3.
The functional parcellation (FP) in the embedding space captured differences across subjects and achieved high reproducibility within subjects. The maps show the test–retest reliability of parcellation results in 2 subjects (left 2 columns). As a comparison, results of subject-level parcellation in anatomical space (AP) are also shown (right 2 columns). Parcellation results based on functional space alignment are consistent within the same subject, but vary between 2 different subjects. See also Supplementary Figure 2 for the results from all 23 subjects.
Parcellation in the Embedding Functional Space Captures Interindividual Variability
The performance of brain parcellation methods can be evaluated based on their capability to capture the differences across subjects, while, at the same time, reliably recovering the connectivity architecture of each subject in repeated measurements. Unless an imaging measure is highly reproducible within subjects, the variance observed across individuals cannot be fully attributed to individual differences. Here, we quantified parcellation reliability based on 5 fMRI sessions for each individual subject and evaluated the variability across 23 subjects. The overall performance was assessed using vSNR, which reflected the sensitivity to individual differences, while intrasubject variability was controlled (see Supplementary Materials and Methods).
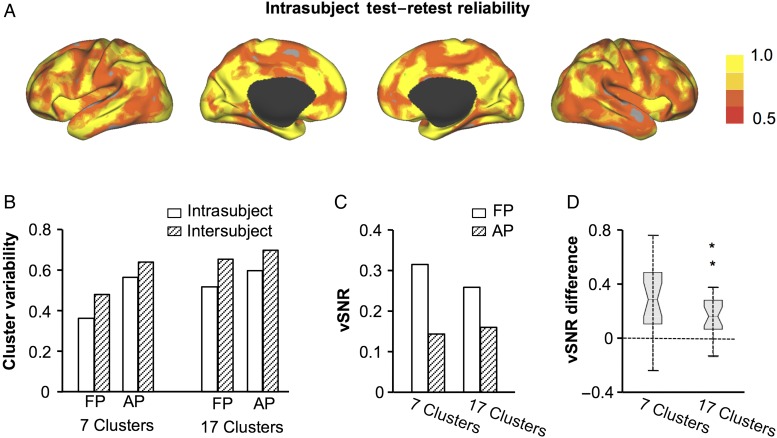
The performance of FP was compared with that of AP. Using the 5 scans of each subject, intrasubject test–retest reliability was evaluated at each vertex (see Fig. 4A for the reliability of the seven-clusters solution). FP yielded a mean reliability of 0.64 for the seven-clusters solution, whereas AP had a reliability of 0.45 (Fig. 4B). Intersubject variability was also evaluated for both methods. Compared with FP, AP showed both higher inter- and intrasubject variability, suggesting that the results of AP were inherently unstable (Fig. 4B).
Figure 4.
Performance of 2 different brain parcellation methods. Intrasubject test–retest reliability of the seven-network functional parcellation in the embedding space was calculated at each vertex, based on the 5 scans of each subject. Reliability maps were then averaged across the 23 subjects (A). Intersubject variability was also computed at each vertex. Intersubject variability and intrasubject variability values were then averaged across all vertices on the brain surface (B). The overall performance of the 2 different parcellation methods was evaluated based on vSNR, which was derived from the intersubject variability and intrasubject variability. FP resulted in a higher vSNR than AP (C). To test whether vSNR was significantly higher for FP than AP, intrasubject variability was computed for each of the 23 subjects. vSNR was then evaluated for each subject. Pairwise comparison of this subject-level vSNR indicated a significantly higher vSNR for FP than AP, in both the 7-clusters and 17-clusters solutions (both P < 0.01). The increases in vSNR for FP compared with AP are illustrated by the boxplots (D).
The vSNR of both methods was then compared (Fig. 4C). For the seven-clusters solution, FP yielded a vSNR of 0.32, whereas AP had a vSNR of 0.15. For the 17-clusters solution, FP had a vSNR of 0.26 and AP had a vSNR of 0.16. To statistically test whether vSNR was significantly higher for FP than AP, intrasubject variability was computed for each of the 23 subjects. vSNR was then evaluated at the individual subject level using intrasubject variability and the intersubject variability derived from the 23 subjects. Pairwise comparison of this subject-level vSNR indicated a significantly higher vSNR for FP than AP, in both the 7-clusters and 17-clusters solutions (both P < 0.01, Fig. 4D). These results suggest that FP can better capture individual differences in network distribution when variability due to noise is controlled.
Parcellation in the Embedding Space Reflects Individual Differences in Functional Activity
To illustrate that parcellation in the embedding space can be translated into improved measurements of individual functional differences, we studied language lateralization in a group of subjects. The language network is known as one of the most variable networks in the human brain. While the left hemisphere is dominant for language processing in most individuals, atypical language lateralization is estimated to occur in 4%–6% of healthy right-handed individuals, with a rare few showing complete reversal of brain asymmetry (Rasmussen and Milner 1977). Here, we investigated whether aligning subjects in the embedding space and identifying the networks can help to capture the variability of language lateralization across individuals. Fifty-five subjects performed a semantic decision task in the scanner and the language laterality of each individual was estimated based on the asymmetric activation evoked by the task (Wang et al. 2014). Resting-state fMRI data collected from the same subjects were used for the network parcellation. The degree of within-hemisphere functional connectivity (Wang et al. 2014) was computed for each vertex. The asymmetry of connectivity degree was then computed for each brain network.
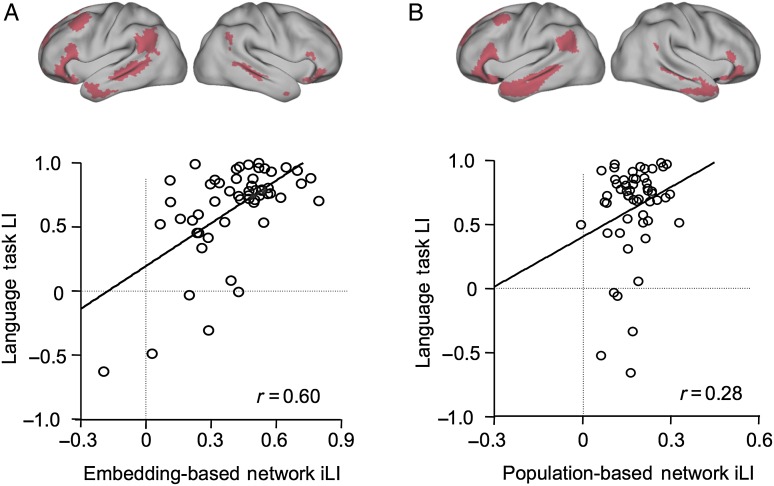
Applying the FP in the embedding space, we identified a strongly left-lateralized network whose connectivity laterality was significantly correlated with task-based language laterality (r = 0.60, P < 0.001). This network was localized to the inferior frontal gyrus, the inferior parietal lobule, and the superior/middle temporal gyrus in most individuals (see Fig. 5A for this network in the group consensus map), which overlapped with the traditional language regions (Binder et al. 1997).
Figure 5.
Parcellation based on functional alignment in the embedding space reflected individual differences in functional activity. The degree of within-hemisphere connectivity (Wang et al. 2014) was computed at each vertex and the laterality of the connectivity degree was then evaluated for each network. The network with the strongest leftward asymmetry was identified, which involved the inferior frontal gyrus and the superior temporal gyrus. The consensus map of this network is shown in Figure 5A. Note that this network is highly variable across different subjects. The laterality of connectivity degree in this network was significantly correlated with the language laterality index calculated from the task data (r = 0.60, P < 0.001, Fig. 5A). The laterality of the degree of connectivity was also computed for 17 networks in the population-level atlas derived from 1000 subjects (Fig. 5B). The most left-lateralized network was also identified. However, the laterality of the degree of connectivity in this network showed only a weak correlation with the language laterality index calculated from the task data (r = 0.28, P < 0.05, Fig. 5B).
As a comparison, connectivity laterality was also estimated in the group-level brain parcellations, derived from 1000 healthy subjects, that were aligned in the anatomical space (Yeo et al. 2011). The network that best predicted the task-based language lateralization fell within the similar frontal, parietal, and temporal regions (Fig. 5B), but showed a weaker correlation with the task-based language laterality index (r = 0.28, P < 0.05). These results suggest that decoupling functional characteristics from spatial variability enables detection of network boundaries that can better reflect individual differences in brain function, as manifested in the greater predictability of language lateralization during tasks. In contrast, a static parcellation that assumes anatomical correspondence of functional networks in multiple individuals is less predictive of individual differences in functional laterality.
The Spatial Distribution of Association Networks Is Highly Variable
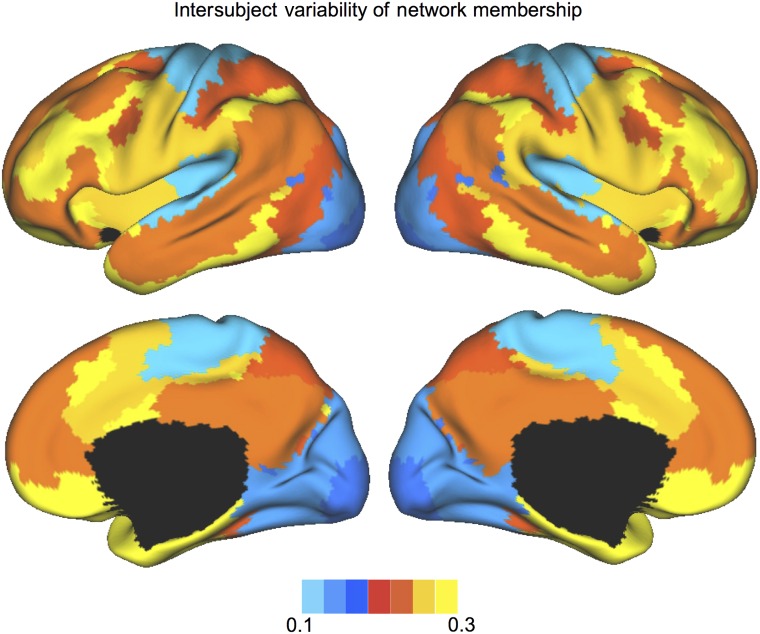
Intersubject variability of the network distribution was assessed after projecting the FPs from the embedding space back to each subject's native space. The disagreements in network membership across individual subjects were computed for each vertex and then averaged within each network (based on the consensus map shown in Fig. 2). The association areas, including the frontal, parietal, and temporal regions, exhibited particularly high intersubject variability in network membership (Fig. 6). The uni-modal areas, including the motor, sensory, and visual cortices, showed minimal spatial distribution variability. These results are reminiscent of the intersubject variability observed in the correlation profiles of individual surface vertices (Mueller et al. 2013), which, however, are a mixture of variability in macroanatomy and variability in functional coupling. The stronger variability in network membership in the association areas suggests a weaker link between macroanatomical structure and functional role, i.e., macroscopic structures in a given location can be assigned to a particular functional network in one subject, but to a different network in another subject. These results suggest that group averaging based on macroanatomical alignment could be most confounded when studying the higher-order association functions.
Figure 6.
Intersubject variability of network distribution was higher in the association networks than in the sensorimotor and visual networks. The disagreement of network membership was computed for each vertex and then averaged within each network. The spatial distribution of the frontal-parietal control network showed the highest intersubject variability.
Discussion
Variability in resting-state functional connectivity has been related to individual differences in human behavior and cognition (van den Heuvel et al. 2009; Cole et al. 2012), as well as to neurological and psychiatric disorders (Fox and Greicius 2010). The methodology proposed in this paper can dissociate the interindividual variability in functional coupling and the interindividual variability in the spatial distribution of functional networks. The method identifies functional networks shared by a population even if those networks are located at different anatomical sites in different subjects. This method enables independent analysis of functional and spatial characteristics at the individual level. In 23 subjects with 5 scans each, the parcellation in functional space provided both high intrasubject reproducibility and high sensitivity to intersubject variability. In comparison, performing subject-level parcellation using a traditional approach in anatomical space yielded poor reproducibility. The parcellation in functional space captures the intersubject variability in network organization, which can lead to more accurate estimates of functional connectivity in each individual, and is informative about the anatomical underpinnings of functions. As a proof-of-principle, we showed that the functional space parcellation predicted individual differences in language lateralization more accurately than the static parcellation in anatomical space derived from a large population. Parcellation in the embedding space also revealed that the spatial distribution of association functions was highly variable, while uni-modal sensorimotor functions showed less spatial variability.
Dissociating Multiple Sources of Variability in Resting-State fMRI Data
Intersubject variability observed in resting-state fMRI data is a composition of multiple factors. When a specific brain function is studied across subjects, there are differences not only in the coupling pattern among the units anchoring the function, but also in their morphological location, segmental structure, relative size, or cytoarchitecture (Brett et al. 2002). The diverse sources of variability require modeling methods that can separate these sources and reveal their specific contributions to cognition and behavior.
To separate the spatial variability of functional units from the actual functional processes being investigated, some fMRI studies have used a functional localizer approach (Fedorenko et al. 2010). While a functional localizer is powerful when a specific function is analyzed, it cannot easily obtain the information regarding the cortical arrangement of multiple systems and their functional relations.
Based on resting-state fMRI, our study provides the proof-of-concept that variability in macroanatomy and variability in functional connectivity can be dissociated to improve the specificity of functional measures. Rather than assuming a static function-anatomy association, we represented the neural architecture by groups of points in the embedding space (similar to voxels or vertices in the anatomical space) that are functionally coupled during a specific state (e.g., rest or a task). These points may serve as the “intrinsic functional localizer” for the future investigation of individual functional differences. A reference system based on functional connectivity structure rather than macroanatomy might be more appropriate for studies of higher-order association functions. It is known that association networks exhibit the most widespread spatial distribution (Sepulcre et al. 2010), the highest connectivity variability across individuals (Mueller et al. 2013), and markedly complicated network organization when compared with hierarchical circuits, such as those that dominate the sensori-motor system (Buckner and Krienen 2013). The present study demonstrated that higher functional specificity could be achieved when anatomical variability was properly estimated and controlled. When we used connectivity measures derived from resting-state data to predict language lateralization, the prediction accuracy significantly improved when connectivity lateralization was computed using each subject's own network profile rather than a population-based brain atlas.
Implications for Group Analysis in Neuroscience Research
To establish correspondence across subjects and study group effects, registering individual brains to a common template is necessary. Aligning subjects based on brain morphology has become the standard procedure in imaging software, such as FreeSurfer, FSL, or SPM (Friston et al. 1996; Fischl et al. 1999; Woolrich et al. 2009). The key assumption of these morphologic registration technologies is the fixed function-anatomy relation across all individuals. This assumption is problematic if significant individual differences exist in this relation, particularly in higher-order cognitive functions or in nonstandard subject cohorts, such as patients or infants. Some studies have used population-specific templates rather than fixed templates during groupwise registration and achieved better performance (Klein et al. 2009). This indicates that the subject-specific, function-anatomy relation cannot be simply ignored. By incorporating the intersubject signal correlations into a cortical registration algorithm, a recent study also demonstrated substantial improvement in bringing functionally similar regions into correspondence during a movie-watching task (Sabuncu et al. 2010).
Owing to the complex correspondence between morphology and function, efforts have been made to align subjects directly in the signal space. For example, multi-dimensional scaling was employed to retrieve a low-dimensional representation of positron emission tomography (PET) signals in a set of activated regions (Friston et al. 1996). Dual regression on group-level independent component analysis (ICA) results has been used to project components back to individuals (Zuo et al. 2010). More recently, diffusion map embeddings of fMRI signals were matched across subjects to find the correspondences in fMRI language task data, independent of spatial location (Langs et al. 2010), and this approach was extended to a group-level model for language task data (Langs et al. 2014). Task-based data were also represented in a high-dimensional space that enabled decoding across subjects by matching response patterns to individual stimulus categories (Haxby et al. 2011). Collectively, these studies emphasize the importance of moving beyond cross-subject alignment based on macroanatomy. The methodology proposed in the present study provides a novel approach to aligning subjects based on resting-state fMRI, and can greatly benefit the group analyses of functional connectivity data.
Clinical Relevance
Parcellating the individual brain into functional networks with high reliability has important clinical implications. A direct application for such techniques is preoperative functional mapping. To map the eloquent cortices, invasive cortical stimulation is often managed perioperatively, in the awake patient, or in the presurgical patient with subdural grids implanted (Penfield and Jasper 1954). More recently, fMRI has been suggested as a noninvasive alternative to map eloquent cortices (Desmond et al. 1995; Binder et al. 1997). However, obtaining robust maps in individual subjects with fMRI is still a daunting challenge. The recent advancement of functional connectivity fMRI techniques has made the goal of subject-level network parcellation much more attainable (Hacker et al. 2013; Wig et al. 2013). The parcellation methodology proposed in the present study could reliably identify the functional networks within the same subject and capture the difference between subjects. These characteristics are desirable in presurgical mapping. If validated using invasive measures, such as cortical stimulation, the individualized brain parcellation may serve as a fast and accurate technique for presurgical mapping, which could provide a functional layout across the entire cortex at once.
Decoupling functional characteristics from spatial variability also enables the removal of confounding factors in the study of clinical cohorts, such as schizophrenia patients, where disease affects both the morphology and function of the same structure (Breier et al. 1992). Our approach promises to improve the specificity of the functional markers of diseases, and enables quantification of the potential systematic differences in the network distribution between patients and controls. It will also enable the study of networks in individual subjects where atypical spatial configurations are present, including children, patients with lesions for whom the networks can be partially damaged or undergo substantial reorganization, or in patients with neurodegenerative diseases. In these cohorts, the comparison of functional network characteristics, independent of the spatial differences, is particularly difficult, but essential.
Coupling Between Anatomy and Function
The relationship between anatomy and function is complex, and not yet fully understood. The brain has long been viewed as a system of histologically differentiated segments (e.g., Brodmann areas) that fulfill specific functional roles. This anatomy-function model has led to the discovery of detailed maps of various functional systems, such as the visual processing pathways (Ishai et al. 1999), the sensorimotor processing units (Muellbacher et al. 2002), as well as some critical structures for higher-level functions, including memory (Young et al. 1997) and language (Price 2000). Overall, histological boundaries have strong functional implications and are an important subject of neuroscience investigations (Hinds et al. 2009). However, current neuroimaging techniques are limited in the ability to obtain microscopic histological information in vivo. As an initial step to probe the function-anatomy association, the present study explored the coupling between macroanatomical structure and functional organization. Our data demonstrated the potential advantage of decoupling functional organization from macroanatomy in the investigation of individual differences.
Nevertheless, these results should not be interpreted as an assertion of general dissociation between functional organization and brain anatomy. Interindividual differences in functional brain networks are likely to have a respective structural correlate at the microscopic level. It has been shown that interindividual variability exists in the locations of cytoarchitectonically defined brain areas (Van Essen et al. 2012). This variability in microanatomy can contribute to the variability observed in the functional data. To gain a comprehensive understanding of the function-anatomy relation will require a sophisticate resolution for both the functional and the structural measures. Future studies on the function-anatomy coupling at different levels of resolution will provide a broader view of this complex picture.
Supplementary Material
Supplementary Material can be found at http://www.cercor.oxfordjournals.org/online.
Funding
This work is supported by NIH grants K25NS069805 (H.L.), R01NS091604 (H.L.), and P50MH106435 (H.L.). NIH NICHD R01HD067312 (P.G., G.L.) and NIH NIBIB NAC P41EB015902 (P.G., G.L.), OeNB 14812 and 15929 (G.L.), and EU FP7 2012-PIEF-GA-33003 (G.L.). M.R.S. is supported by NIH NIBIB 1K25EB013649-01 and a BrightFocus Alzheimer's disease pilot research grant (AHAF A2012333). This research was partly supported by the Medical Imaging Cluster at Medical University of Vienna (G.L.).
Supplementary Material
Notes
Conflict of Interest: H.L. and D.W. are listed as inventors on submitted patents on mapping functional brain organization using fMRI. All other authors declare no conflict of interest.
References
- Baker JT, Holmes AJ, Masters GA, Yeo BT, Krienen F, Buckner RL, Öngür D. 2014. Disruption of cortical association networks in schizophrenia and psychotic bipolar disorder. JAMA Psychiatry. 71:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder JR, Frost JA, Hammeke TA, Cox RW, Rao SM, Prieto T. 1997. Human brain language areas identified by functional magnetic resonance imaging. J Neurosci. 17:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier A, Buchanan RW, Elkashef A, Munson RC, Kirkpatrick B, Gellad F. 1992. Brain morphology and schizophrenia. A magnetic resonance imaging study of limbic, prefrontal cortex, and caudate structures. Arch Gen Psychiatry. 49:921–926. [DOI] [PubMed] [Google Scholar]
- Brett M, Johnsrude IS, Owen AM. 2002. The problem of functional localization in the human brain. Nat Rev Neurosci. 3:243–249. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM. 2013. The evolution of distributed association networks in the human brain. Trends Cogn Sci. 17:648–665. [DOI] [PubMed] [Google Scholar]
- Coifman RR, Lafon SP. 2006. Diffusion maps. Appl Comput Harmon Anal. 21:5–30. [Google Scholar]
- Coifman RR, Lafon S, Lee AB, Maggioni M, Nadler B, Warner F, Zucker SW. 2005. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc Natl Acad Sci USA. 102:7426–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Yarkoni T, Repovs G, Anticevic A, Braver TS. 2012. Global connectivity of prefrontal cortex predicts cognitive control and intelligence. J Neurosci. 32:8988–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond JE, Sum JM, Wagner AD, Demb JB, Shear PK, Glover GH, Gabrieli JD, Morrell MJ. 1995. Functional MRI measurement of language lateralization in Wada-tested patients. Brain. 118 (Pt 6):1411–1419. [DOI] [PubMed] [Google Scholar]
- Fedorenko E, Hsieh PJ, Nieto-Castanon A, Whitfield-Gabrieli S, Kanwisher N. 2010. New method for fMRI investigations of language: defining ROIs functionally in individual subjects. J Neurophysiol. 104:1177–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. 1999. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 9:195–207. [DOI] [PubMed] [Google Scholar]
- Fox MD, Greicius M. 2010. Clinical applications of resting state functional connectivity. Front Syst Neurosci. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Liu H, Pascual-Leone A. 2012. Identification of reproducible individualized targets for treatment of depression with TMS based on intrinsic connectivity. Neuroimage. 66C:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Frith CD, Fletcher P, Liddle PF, Frackowiak RS. 1996. Functional topography: multidimensional scaling and functional connectivity in the brain. Cereb Cortex. 6:156–164. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B. 2009. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 48:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Van der Haegen L, Cai Q, Stufflebeam S, Sabuncu MR, Fischl B, Brysbaert M. 2013. A surface-based analysis of language lateralization and cortical asymmetry. J Cogn Neurosci. 25:1477–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker CD, Laumann TO, Szrama NP, Baldassarre A, Snyder AZ, Leuthardt EC, Corbetta M. 2013. Resting state network estimation in individual subjects. Neuroimage. 82:616–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Guntupalli JS, Connolly AC, Halchenko YO, Conroy BR, Gobbini MI, Hanke M, Ramadge PJ. 2011. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron. 72:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Dierker D, Neil J, Inder T, Knutsen A, Harwell J, Coalson T, Van Essen D. 2010. A surface-based analysis of hemispheric asymmetries and folding of cerebral cortex in term-born human infants. J Neurosci. 30 (6):2268–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds O, Polimeni JR, Rajendran N, Balasubramanian M, Amunts K, Zilles K, Schwartz EL, Fischl B, Triantafyllou C. 2009. Locating the functional and anatomical boundaries of human primary visual cortex. Neuroimage. 46:915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishai A, Ungerleider LG, Martin A, Schouten JL, Haxby JV. 1999. Distributed representation of objects in the human ventral visual pathway. Proc Natl Acad Sci USA. 96:9379–9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P et al. 2009. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 46:786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HW. 1955. The Hungarian method for the assignment problem. Nav Res Log Q. 2:83–97. [Google Scholar]
- Langs G, Sweet A, Lashkari D, Tie Y, Rigolo L, Golby AJ, Golland P. 2014. Decoupling function and anatomy in atlases of functional connectivity patterns: Language mapping in tumor patients. NeuroImage. 103:462–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langs G, Tie Y, Rigolo L, Golby AJ, Golland P. 2010. Functional geometry alignment and localization of brain areas. In: Advances in neural information processing systems 23. Curran Associates, Inc p. 1225–1233. [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stufflebeam SM, Sepulcre J, Hedden T, Buckner RL. 2009. Evidence from intrinsic activity that asymmetry of the human brain is controlled by multiple factors. Proc Natl Acad Sci USA. 106:20499–20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Nichols T, Callicott JH, Ding J, Kolachana B, Buckholtz J, Mattay VS, Egan M, Weinberger DR. 2006. Impact of complex genetic variation in COMT on human brain function. Mol Psychiatry. 11:867–877, 797. [DOI] [PubMed] [Google Scholar]
- Muellbacher W, Ziemann U, Wissel J, Dang N, Kofler M, Facchini S, Boroojerdi B, Poewe W, Hallett M. 2002. Early consolidation in human primary motor cortex. Nature. 415:640–644. [DOI] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H. 2013. Individual variability in functional connectivity architecture of the human brain. Neuron. 77:586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield W, Jasper HH. 1954. Epilepsy and the functional anatomy of the human brain. Oxford, England: Little, Brown & Co. [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, Mathalon D, Ford J, Lauriello J, Macciardi F et al. 2009. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 35:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CJ. 2000. The anatomy of language: contributions from functional neuroimaging. J Anat. 197 Pt 3.:335–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowska G, Goldman-Rakic PS. 1995. Cytoarchitectonic definition of prefrontal areas in the normal human cortex: II. variability in locations of areas 9 and 46 and relationship to the Talairach Coordinate System. Cereb Cortex. 5:323–337. [DOI] [PubMed] [Google Scholar]
- Rasmussen T, Milner B. 1977. The role of early left-brain injury in determining lateralization of cerebral speech functions. Ann N Y Acad Sci. 299:355–369. [DOI] [PubMed] [Google Scholar]
- Sabuncu MR, Singer BD, Conroy B, Bryan RE, Ramadge PJ, Haxby JV. 2010. Function-based intersubject alignment of human cortical anatomy. Cereb Cortex. 20:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. 2007. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 27:2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Liu H, Talukdar T, Martincorena I, Yeo BT, Buckner RL. 2010. The organization of local and distant functional connectivity in the human brain. PLoS Comput Biol. 6:e1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE. 2009. Efficiency of functional brain networks and intellectual performance. J Neurosci. 29:7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. 2010. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 103:297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. 2005. A population-average, landmark- and surface-based PALS atlas of human cerebral cortex. Neuroimage. 28:635–662. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J, Coalson T. 2012. Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb Cortex. 22:2241–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Liu H. 2013. Cerebellar asymmetry and its relation to cerebral asymmetry estimated by intrinsic functional connectivity. J Neurophysiol. 109:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Liu H. 2014. Functional specialization in the human brain estimated by intrinsic hemispheric interaction. J Neurosci. 34 (37):12341–12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Cohen AL, Power JD, Nelson SM, Glasser MF, Miezin FM, Snyder AZ, Schlaggar BL, Petersen SE. 2013. Parcellating an individual subject's cortical and subcortical brain structures using snowball sampling of resting-state correlations. Cerebral Cortex, bht056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, Beckmann C, Jenkinson M, Smith SM. 2009. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 45:S173–S186. [DOI] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BJ, Otto T, Fox GD, Eichenbaum H. 1997. Memory representation within the parahippocampal region. J Neurosci. 17:5183–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Palomero-Gallagher N, Amunts K. 2013. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 36 (5):275–284. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP. 2010. Reliable intrinsic connectivity networks: test-retest evaluation using ICA and dual regression approach. Neuroimage. 49:2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.