Abstract
Choroidal neovascularization (CNV) commonly occurs in age related macular degeneration and pathological myopia patients. In this study we conducted a case-control prospective study including 431 participants. The aim of this study was to determine the potential association between 10 single nucleotide polymorphisms (SNPs) located in 4 different genetic regions (CFI, COL8A1, LIPC, and APOE), and choroidal neovascularization in age-related macular degeneration and the development of choroidal neovascularization in highly myopic eyes of a Caucasian population. Univariate and multivariate logistic regression analysis adjusted for age, sex and hypertension was performed for each allele, genotype and haplotype frequency analysis. We found that in the univariate analysis that both single-nucleotide polymorphisms in COL8A1 gene (rs13095226 and rs669676) together with age, sex and hypertension were significantly associated with myopic CNV development in Spanish patients (p<0.05). After correcting for multiple testing none of the polymorphisms studied remained significantly associated with myopic CNV (p>0.05); however, analysis of the axial length between genotypes of rs13095226 revealed an important influence of COL8A1 in the development of CNV in high myopia. Furthermore we conducted a meta-analysis of COL8A1, CFI and LIPC genes SNPs (rs669676, rs10033900 and rs10468017) and found that only rs669676 of these SNPs were associated with high myopia neovascularization.
Introduction
Myopia is the most common ocular disorder worldwide [1] with an approximate prevalence rate of 27% in the United States and Western Europe [2], and has increased in the last 50–60 years [3,4]. The prevalence of myopia in Asian populations is even higher [1,2,5–9]. In some East and Southeast Asian countries, nearly 80% of the population has myopia, whereas 10–20% has high myopia (HM) [10–12].
HM is defined as having an as axial length ≥26.5 mm or ≤−6 diopters (D) of myopic refractive error [13,14]. Myopic axial length elongation can appear as retinal pathological changes and ocular comorbidities that may lead to blinding disorders, including chorioretinal atrophy and choroidal neovascularization (CNV), premature cataracts, retinal detachment, and glaucoma. HM is a common vision-threatening disease that affects 0.5–5.0% of the population worldwide [1,3,15]. It is now considered the fourth-most common cause of irreversible blindness in developed countries after age-related macular degeneration (AMD), glaucoma, and diabetic retinopathy [16], leading to a substantial impact on the public health economy. Therefore, it is crucial to elucidate the pathological mechanisms underlying HM to develop new therapies for this disabling pathology.
Epidemiological studies have shown that both genetic and environmental factors contribute to myopia development [17–23]. However, after years of intensive research, the precise mechanisms controlling ocular growth and development of refractive errors remain unclear. Genetic associations with HM have been investigated for several decades. Twin and family studies have shown that myopia, particularly HM, has a high heritability [24,25]. It is now generally accepted that there are major genetic contributions to HM, in contrast non-pathological myopia appears to be multifactorial, possibly involving a large number of genes with small individual effects, and major environmental factors. Several HM susceptibility genes have been identified in linkage and candidate gene studies, such as paired box gene 6 (PAX6) [26], collagen type II alpha 1 (COL2A1) [27], collagen type I alpha 1 (COL1A1) [28,29], myocilin (MYOC) [30,31], hepatocyte growth factor (HGF) [32], transforming growth factor beta 1 (TGFB1) [33,34], transforming growth-induced factor (TGIF) [35,36], and lumican (LUM) [37,38].
CNV beneath the fovea is a common complication of HM and AMD and is a common cause of visual impairment [39]. Myopic CNV (mCNV) typically occurs in the fourth and fifth decades of life; HM is the most common cause of CNV in patients younger than 50 years of age. Moreover, CNV can develop in highly myopic eyes in the elderly population [40]; however, most highly myopic eyes never develop mCNV. In the elderly population, the most frequent cause of CNV is AMD. Both AMD and HM are caused by interactions between genetic and environmental factors, and genetic factors are thought to strongly contribute to CNV development in AMD patients [41–44]. Our previous studies [45]; suggested that the development of AMD-CNV may have a common genetic origin with mCNV, but we did not find a significant association between two-single nucleotide polymorphisms (SNPs) strongly associated with AMD (rs1061170 [Y402H] in CFH and rs10490924 [A69S] in ARMS2) and myopic CNV in a Spanish population. These results were confirmed in Japanese population by Nakanishi et al [46], who found no association between mCNV development and the three most important SNPs associated with AMD development in the Japanese population (rs1061170 (Y402H), rs10490924 (A69S) and rs11200638 of HTRA1). More recently, Leveziel et al [47] conducted a case-control study analyzing 15 genes associated with AMD in a North American Caucasian population of European origin, and found that only one SNP located in the gene for the complement factor I (CFI) significantly associated with mCNV. Despite the similarities between wet AMD and mCNV in the growth of macular new microvasculature in the choroid layer, only this SNP involved in the pathogenesis of wet AMD was previously reported to be associated with mCNV development.
Because the prevalence of HM varies among the populations, there may also be differences between the genetic variants associated with CNV, and potential associations in other races and other SNPs recently found to be associated with AMD require further analysis. The purpose of this study was to determine the contribution of some of the most important CNV-AMD-associated SNPs in a Spanish population and several recently described AMD-associated SNPs located in 4 genetic regions (COL8A1, LIPC, CFI and APOE) to the development of mCNV in a Spanish population.
Materials and Methods
Study subjects
Two hundred fifty unrelated highly myopic Spanish Caucasian patients (147 HM patients with mCNV, 103 HM patients without mCNV (HMnoCNV) patients) and 181 controls were recruited from various centers of the Red Temática de Investigación Cooperativa (RETICS). The inclusion criteria for the HM group were spherical refractive error ≤ -6.00 diopters (D) or an axial length ≥ 26 mm, and age over 50 years at the time of inclusion in the study. HM patients were divided into two groups: one group with mCNV in at least 1 eye and the other group without mCNV, who did not present any chorioretinal degeneration and had a visual acuity over 20/32 in both eyes. The population based control group was composed of patients aged over 50 years with spherical refractive error > -6.00 D without any pathology in the retina who came to annual revision. The exclusion criteria were tractional maculopathy, and/or epiretinal membrane in the OCT or other diseases different from HM that may occur with CNV (e.g. wet AMD, diabetic retinopathy, pseudoxantoma elasticum, presumed ocular histoplasmosis syndrome, lacquer cracks due to trauma). Patients presenting macular drusen or patients with a family history of AMD, known genetic diseases associated with myopia, such as Stickler or Marfan syndrome, and any ocular surgery, were also excluded, with the exception of cataract surgery.
All procedures used in this study conformed to the guidelines of the Declaration of Helsinki. The Institutional Review Board and the Ethics Committee of Clínica Universidad de Navarra approved the protocols used in this study. All patients were fully informed of the purpose and procedures, and written consent was obtained from each patient. All cases underwent detailed ophthalmologic examination, including visual acuity assessment, dilated slit-lamp biomicroscopy, automatic objective refraction, measurement of the axial length by A-scan ultrasound (UD-6000, Tomey, Nagoya, Japan) or partial coherence interferometry (IOLMaster, Carl Zeiss Meditec, Jena, Germany), color fundus photography, optical coherence tomography, and fluorescein angiography. Controls underwent visual acuity assessment, mydriasis fundus examination, and measurement of refractive error and axial length.
Genotyping
We examined the available literature regarding genetic associations in AMD and the SNPs showing the most significant associations were selected. Genomic DNA was extracted from oral swabs using QIAcube (Qiagen, Hilden, Germany) and processed in the Laboratory of Experimental Ophthalmology at the University of Navarra (Spain). A set of 10 SNPs in 4 previously identified AMD-associated genes (shown in S1 Table) were genotyped in the control, HMnoCNV and mCNV cohorts using genomic DNA by allelic discrimination with validated assays (TaqMan; Applied Biosystems, Foster City, CA, USA; S1 Table) and real-time PCR (PE7300; Applied Biosystems), according to the manufacturer's instructions. None of the SNPs were in linkage disequilibrium except the SNPs in CFI, which have been classified as risk or protective haplotypes [48,49]; therefore the haplotypes of this gene were analyzed.
Literature search strategy for meta-analysis
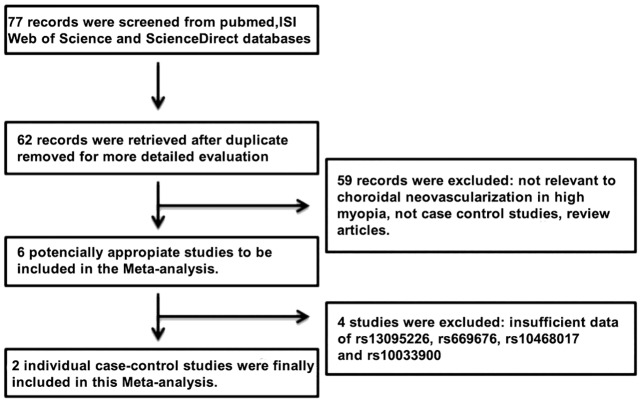
For the comparative study between our results and previous studies related with mCNV, we conducted a meta-analysis of principal SNPs analyzed in this study. Potential articles were identified by a systematic search of the ISI Web of Science, PubMed, and ScienceDirect databases through June 09, 2016 using combinations of the following search terms: "collagen type VIII alpha 1" OR " COL8A1", "polymorphism" OR "SNP" AND "CNV High myopia" without language or publication date restrictions. All relevant publications and their reference were manually screened to identify eligible studies (Fig 1). Selection criteria papers identified during the literature search had to meet the following inclusion criteria for inclusion in our study: 1) case-control or cohort design studies of humans; 2) evaluation of the association of COL8A1, CFI and LIPC genes polymorphisms (rs13095226, rs669676, rs10033900, and rs10468017) with neovascularization high myopia; 3) sufficient published data available for our team to estimate the odds ratios (ORs) of different genotype frequencies; and 4) published original full-text literature. Studies meeting the following criteria were excluded from the analysis: 1) insufficient reported data; 2) abstracts, review papers, and case-only studies; 3) duplication of previously published literature. Three investigators (Velazquez, Fernandez-Robredo, and Recalde) independently extracted the data. Discrepancies between different investigators were resolved by 2 other investigators (Garcia and Hernandez) to reach a consensus. The collected data included: name of the first author, publication date, geographical location, numbers of cases and controls, ethnicity, genotyping methods and matching variables (Table 1).
Fig 1. Flow chart for the selection of studies according to the criteria of this Meta-analysis.
Table 1. Main characteristics of eligible studies included in the Meta-analysis.
| First Author | Year | mCNVn | Controlsn | Region | Ethnicity | Genotyping Method | Matching |
|---|---|---|---|---|---|---|---|
| Leveziel et al. [47] | 2012 | 71 | 196 | France/USA | Caucasian | ABI | Sex and Ethnicity |
| Miyake et al.[50] | 2013 | 478 | 557 | Japan | Asian | ABI | Age, sex, and axial length |
| Velazquez Villoria et al. | 2016 | 147 | 103 | Spain | Caucasian | TaqMan | Age,sex and ethnicity |
Statistical analyses
General characteristics (age, gender, refractive error, tobacco, HT and hypercholesterolemia) of patients with HM (mCNV and HMnoCNV) and controls were tested using Student´s t-test for the continuous variables age and refractive error, or chi-square test for the categorical variables sex, HTA, HC, and tobacco history. The frequencies of alleles, genotypes, and haplotypes were calculated in all the groups and were compared using a chi-square test and Fisher’s exact test and corresponding ORs were calculated. All SNPs analyzed in this study were in Hardy-Weinberg equilibrium (S1 Table).
Univariable logistic regression adjusted for all covariates was used to estimate the ORs and 95% confidence intervals (95% CI) using SNPStats software (Sole X et al., 2006). Analyses were performed for each genetic variant independent of other variants using codominant, dominant, recessive, and/or overdominant genetic models in base Akaike information, which chooses the inheritance model that best fits the data. We also used a multivariate logistic regression model to take into account all SNPs studied and environmental variables. Correction for multiple testing was performed using the Bonferroni method. For SNPs showing a significant association (rs13095226 and rs669676), to verify the influence of genotype on ophthalmological variables (refractive power and axial length), independent regression analysis was performed. Statistical analysis was conducted with SPSS 20.1 Software (SPSS Inc., Chicago, IL, USA). For all statistical tests, corrected p values of < 0.05 (two-tailed) were considered statistically significant. Separate meta-analyses were conducted on each of the 3 polymorphisms (rs669676, rs10033900 and rs10468017) determined from the articles during the systematic review. We used Z test to judge the significance of the pooled ORs, and statistical significance was considered when p < 0.05. Heterogeneity among included studies was evaluated by the 2-based test and the 2 index; < 0.10 or 2 > 50% were considered statistically significant. The random effects model was used to estimate pooled ORs when obvious heterogeneity was present; otherwise the fixed-effects model was used. Significant publication bias was considered when p < 0.05. All statistical analyses were performed using STATA 12.0 software (StataCorp LP, College Station, TX, USA).
Results
The demographic characteristics of the study population are shown in Table 2. The mCNV group showed no significant differences in age, gender, refractive error, axial length, tobacco history, and hypertension with respect to the HMnoCNV group. Compared to the control group, the mCNV group showed significant differences in gender (p < 0.05), very significant differences in hypertension (p < 0.01), and highly significant differences in age, axial length, and refractive power (p < 0.001).
Table 2. Characteristics of the study population.
| High myopia | P-value 1 | control group | P-value 2 | ||
|---|---|---|---|---|---|
| mCNV | HMnoCNV | No CNVNo HM-AMD | |||
| Number of patients | 147 | 103 | 181 | ||
| Age (Mean ± SD) | 60.68 (±14.07) | 58.58 (±13.85) | ns | 73.39 (±5.31) | *** |
| Female Gender % | 67.35% | 69.90% | ns | 53.59% | * |
| Axial Lenght (mm ± SD) | 30.74 (±2.60) | 29.81 (±2.61) | ns | 23.46 (±0.49) | *** |
| Refractive power (Diopters ± SD) | -13.30D (±4.25D) | -12.84 (±4.21D) | ns | -1.77 (±0.93D) | *** |
| Tobacco smokers | 31.29% | 27.22% | ns | 23.76% | ns |
| Hypertension % | 29.25% | 23.33% | ns | 46.96% | ** |
CNV, Choroidal Neovascularization; NA, not available; SD, Standard deviation.
1) P-value comparing HM-CNV+ with HM-CNV-
2) P-value comparing HM-CNV+ with Population based control group.
Significance:
ns p > 0.05;
* p < 0.05;
** p < 0.01;
***p < 0.001.
Tables 3 and 4 show the allele and genotype frequencies, ORs, p-values, and corrected p-values for the candidate SNPs, associations with the mCNV vs HMnoCNV group and vs control population. Univariate logistic regression analyses results were adjusted for age, gender, and HT. The SNPs rs13095226 and rs669676 in COL8A1 showed a significant association with mCNV. In SNP rs13095226, the C allele showed a significant difference between mCNV patients and the HMnoCNV group [p = 0.034, OR = 2.0 (1.1–3.9)] but was no longer significant after Bonferroni correction was performed (p = 0.30). Genotype analysis showed also significant results in rs13095226 in the mCNV vs HMnoCNV groups in the recessive model [p = 0.0097, OR NA (0.0-NA)], but were no longer significant after multiple testing correction (p = 0.08). Furthermore, when mCNV patients were compared with the population-based control group (Table 4), both SNPs (rs13095226 and rs669676) were significantly associated with high myopia, but no longer significant when multiple testing corrections were performed. For the SNP rs669676, the recessive model of genotype analysis, the minor allele homozygotes (AA versus GG-GA), showed significant differences in the mCNV group vs controls without the HM group [p = 0.023, OR = 2.48 (1.12–5.49); however, Bonferroni correction revealed no association (p corrected = 0.20). Similarly, for rs13095226, allele C was related to a significant risk of mCNV vs. in controls [p uncorrected = 0.016; OR = 2.2 (1.2–4.2); p corrected = 0.14]. Genotype analysis also showed significant results for the TT-CT genotypes versus CC, [p uncorrected = 0.0096 and OR = NA (0.0–NA) p corrected = 0.08]. The remaining SNPs evaluated did not show a significant association with mCNV after adjusting for confounding factors.
Table 3. Comparison of the genotype frequencies between mCNV and HMnoCNV group.
| SNPs | Gene | Chr | Genotype | HM mCNV Genotype freq (%) | MAF | HMnoCNV Genotype freq (%) | MAF | Allele *Uncorrected P-value | Allele OR (95% CI) | Allele **Corrected P-value | Genetic model Co/Do/Re/Ov | Gen *Uncorrected P-value | Genotype OR (95% CI) | Gen **Corrected P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11728699 [48] | CFI | Chr4 | GG/GT/TT | 24.1/48.1/27.8 | 0.48 | 32.6/44.2/23.3 | 0.55 | 0.20 | 0.8 (0.5–1.1) | - | Do | 0.13 | 1.66 (0.87–3.18) | |
| rs13117504 [48] | CFI | Chr4 | CC/CG/GG | 28.8/44.6/26.6 | 0.49 | 21.1/48.9/30.0 | 0.54 | 0.25 | 0.8 (0.5–1.1) | - | Re | 0.13 | 1.66 (0.85–3.23) | |
| rs10033900 [48] | CFI | Chr4 | CC/CT/TT | 35.2/46.0/18.7 | 0.42 | 41.1/45.6/13.3 | 0.36 | 0.24 | 1.3 (0.7–2.2) | - | Re | 0.26 | 1.57 (0.71–3.48) | |
| rs11726949 [48] | CFI | Chr4 | CC/CT/TT | 90.7/8.5/0.8 | 0.05 | 86.5/12.4/1.1 | 0.07 | 0.41 | 0.7 (0.3–2.2) | - | Do | 0.47 | 0.72 (0.30–1.76) | |
| rs6854876 [48] | CFI | Chr4 | GG/CG/CC | 29.3/47.4/23.3 | 0.47 | 23.5/44.7/31.8 | 0.54 | 0.17 | 0.7 (0.5–1.3) | - | Re | 0.12 | 0.60 (0.31–1.15) | |
| rs7439493 [48] | CFI | Chr4 | GG/GA/AA | 28.6/45.1/26.3 | 0.49 | 22.4/45.9/31.7 | 0.55 | 0.24 | 0.8 (0.5–1.1) | - | Re | 0.19 | 1.56 (0.80–3.05) | |
| rs13095226 [47,51] | COL8A1 | Chr3 | TT/CT/CC | 77.8/17.0/5.2 | 0.14 | 85.6/14.4/0.0 | 0.07 | 0.034 | 2.0 (1.1–3.9) | 0.30 | Re | 0.0097 | NA (0.00-NA) | 0.08 |
| rs669676[47,51] | COL8A1 | Chr3 | GG/GA/AA | 35.5/40.3/24.2 | 0.44 | 30.4/54.4/14.2 | 0.41 | 0.69 | 1.1 (0.7–1.9) | - | Ov | 0.15 | 0.66 (0.37–1.17) | |
| rs10468017 [52] | LIPC | Chr15 | CC/CT/TT | 46.6/42.1/11.3 | 0.32 | 49.4/40.2/10.3 | 0.30 | 0.75 | 1.1 (0.6–2.0) | - | Co | 1 | 1.01 (0.37–2.73) | |
| rs769455*** [53] | APOE | Chr19 | CC | 100.0/0.0/0.0 | - | 100.0/0.0/0.0 | - | - | NA | - | - | - | - |
Chr: Chromosome; Genotype freq: Genotype frequency; MAF: minor allele frequency, OR: Odds ratio; CI: Confidence interval; Co: Codominat; Do: Dominant; Re: Recessive; Ov: Overdominant;
*Uncorrected P-value: value from logistic regression model; P value significance < 0.05;
**P-value corrected for multiple testing using Bonferroni method.
***rs769455 was excluded from the analysis because it was monomorphic SNP.
Table 4. Comparison of the genotype frequencies between mCNV and population based control group.
| SNPs | HM mCNV Genotype freq (%) | MAF | control group Genotype freq (%) | MAF | Allele *Uncorrected P-value | Allele OR (95% CI) | Allele **Corrected P-value | Genetic model Co/Do/Re/Ov | Gen *Uncorrected P-value | Gen OR (95% CI) | Gen **Corrected P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11728699 | 24.1/48.1/27.8 | 0.48 | 24.7/51.7/23.6 | 0.50 | 0.78 | 0.9 (0.6–1.3) | Re | 0.41 | 1.34 (0.66–2.71) | - | |
| rs13117504 | 28.8/44.6/26.6 | 0.49 | 22.7/58.0/19.3 | 0.48 | 0.53 | 1.0 (0.6–1.8) | Re | 0.072 | 1.92 (0.94–3.91) | - | |
| rs10033900 | 35.2/46.0/18.7 | 0.42 | 30.2/52.1/17.6 | 0.43 | 0.65 | 0.9 (0.5–1.6) | Ov | 0.25 | 0.70 (0.38–1.29) | - | |
| rs11726949 | 90.7/8.5/0.8 | 0.05 | 88.2/11.8/0.0 | 0.06 | 0.70 | 0.8 (0.4–2.0) | Do | 0.12 | 0.43 (0.14–1.26) | - | |
| rs6854876 | 29.3/47.4/23.3 | 0.47 | 24.1/50.9/25.0 | 0.50 | 0.47 | 0.9 (0.6–1.3) | Co | 0.79 | 1.32 (0.56–3.11) | - | |
| rs7439493 | 28.6/45.1/26.3 | 0.49 | 24.1/57.8/18.1 | 0.47 | 0.37 | 1.1 (0.7–1.6) | Co | 0.13 | 2.06 (0.84–5.03) | - | |
| rs13095226 | 77.8/17.0/5.2 | 0.14 | 86.7/13.3/0.0 | 0.07 | 0.016 | 2.2 (1.2–4.2) | 0.14 | Re | 0.0096 | NA (0.00-NA) | 0.08 |
| rs669676 | 35.5/40.3/24.2 | 0.44 | 34.9/48.6/16.5 | 0.41 | 0.25 | 1.1 (0.8–1.7) | Re | 0.023 | 2.48 (1.12–5.43) | 0.20 | |
| rs10468017 | 46.6/42.1/11.3 | 0.32 | 50.6/33.1/16.3 | 0.33 | 0.93 | 0.9 (0.6–1.4) | Ov | 0.063 | 1.15 (0.65–2.05) | - | |
| rs769455*** | 100/0.0/0.0 | - | 100/0.0/0.0 | - | SD | - | - | Co | 0.68 | 1.45 (0.61–3.44) | - |
Genotype freq: Genotype frequency; MAF: minor allele frequency, OR: Odds ratio; CI: Confidence interval; Co: Codominat; Do: Dominant; Re: Recessive; Ov: Overdominant;
*Uncorrected P-value: value from logistic regression model; P value significance < 0.05;
**P-value corrected for multiple testing using Bonferroni method.
***rs769455 was excluded from the analysis because it was monomorphic SNP.
rs13095226 was not included in the multivariate analysis because none of the HMnoCNV participants showed the minor allele homozygote genotype (CC). Thus, only age and hypertension showed a significant association in the multivariable analysis between mCNV and HMnoCNV [p = 0.027 OR = 0.45 (0.22–0.91), p = 0.011 OR = 0.43 (0.22–0.82) respectively] (Table 5).
Table 5. Multivariable logistic regression analysis between mCNV and HMnoCNV.
| B | Sig. | OR | 95% CI | ||
|---|---|---|---|---|---|
| Lower | Upper | ||||
| mCNV vs HM No CNV | |||||
| Tobacco (1) | 0.848 | 0.006 | 2.33 | 1.27 | 4.28 |
| Age | -0.558 | 0.096 | .57 | .29 | 1.10 |
Reference: Good response.
B: multivariate logistic regression value, Sig: P-value significance (< 0.05), OR: Odds Ratio, CI 95%: 95% confidence interval.
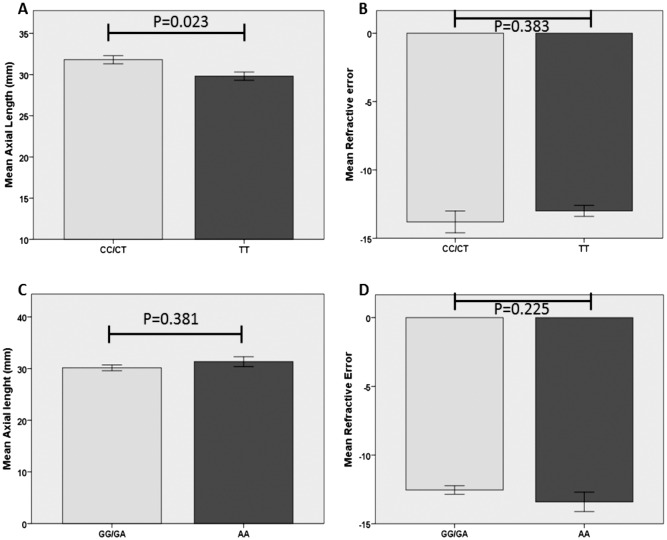
Based on the significant association of rs13095226 and rs669676 before applying Bonferroni correction, we independently analyzed the ophthalmological parameters refractive power and axial length to determine the influence of the genotype on the distribution of these variables (Fig 2). Our results showed a significant association (p = 0.023) between the CT/CC genotype of rs13095226 and a longer axial length (Fig 2A), but this association was not found with the refractive power (Fig 2B). rs669676 showed no significant association either with axial length or number of diopters (Fig 2C and 2D).
Fig 2. Axial length and refractive error of Col8A1 gene SNPs (rs13095226 and rs669676) genotypes.
(A-B) Axial length showed statistically significant differences between CC/CT vs TT genotypes of rs13095226, whereas no differences were found for Refractive error. (C-D) Axial length and Refractive error (respectively) did not show differences between GG/GA vs AA genotypes of rs669676.
Furthermore, we evaluated the inferred haplotype frequencies in the CFI gene showing a strong association with AMD in previous studies between the mCNV and HMnoCNV groups, which are shown in Table 6. None of the haplotype frequencies were significantly different after multiple testing corrections. Haplotype analyses between the mCNV group and the population-based control group for the CFI gene showed no significant associations (data not shown).
Table 6. Haplotype analyses of LD blocks in the CFI gene.
| SNPs | Haplotype | Freq mCNV | Freq HMnoCNV | P-Value | OR |
|---|---|---|---|---|---|
| rs11728699/rs6854876/rs7439493/rs13117504 | GCAG | 0.444 | 0.507 | 1.00 | |
| TGGC | 0.484 | 0.424 | 0.17 | 1.32 (0.89–1.97) | |
| GCGC | 0.045 | 0.029 | 0.46 | 1.55 (0.48–5.01) | |
| TGAG | 0.027 | 0.029 | 0.77 | 0.70 (0.05–9.05) | |
| rs10033900/rs11726949 | CC | 0.568 | 0.614 | 1.00 | |
| TC | 0.356 | 0.382 | 0.35 | 1.22 (0.81–1.85) | |
| TT | 0.045 | 0.035 | 0.8 | 0.86 (0.27–2.72) | |
| CT | 0.015 | 0.024 | 0.78 | 0.79 (0.16–4.02) | |
| rs13117504/rs10033900 | GC | 0.4412 | 0.501 | 1 | |
| CT | 0.370 | 0.331 | 0.32 | 1.25 (0.81–1.92) | |
| CC | 0.139 | 0.137 | 0.49 | 1.23 (0.68–2.22) | |
| GT | 0.049 | 0.031 | 0.72 | 1.23 (0.39–3.90) |
Freq mCNV: Frequency of Myopic Choroidal Neovascularization, Freq HMnoCNV: Frequency of High Myopic eyes without Choroidal Neovascularization, P-value significance (<0.05), OR: Odds Ratio
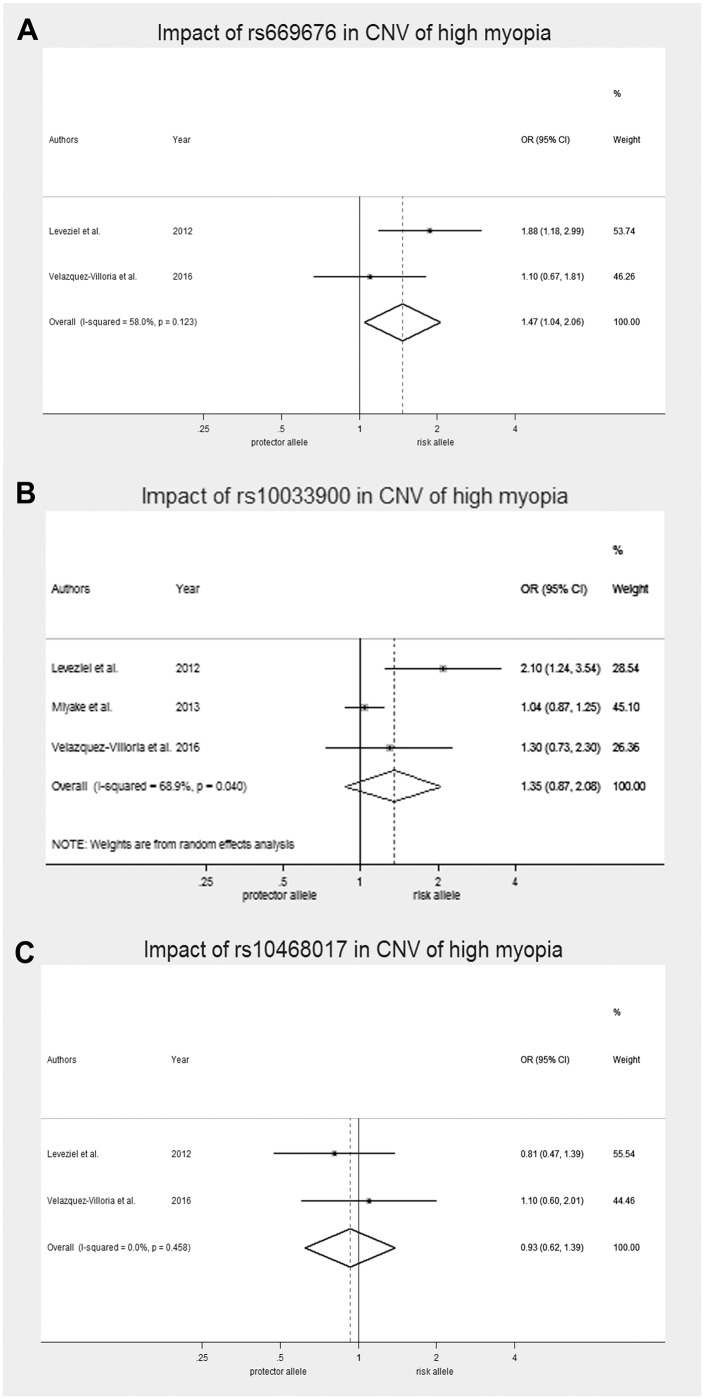
We also conducted a meta-analysis of the 3 principal SNPs identified in previous studies (rs669676, rs10033900, and rs10468017). After systematic review, only two studies (Leveziel et al. [47] and Miyake et al. [50]) met the review requirements. A separate meta-analysis of these 3 SNPs revealed that only the rs669676 SNP presented significant association with mCNV (OR 1.47; IC95% 1.0–2.1) (Fig 3 and S2 Table). The rs10033900 showed high heterogeneity with I-squared values of 68.9% and we used the random-effect model to evaluate this SNP, while the fixed effect model was used for rs10468017 and rs669676.
Fig 3. Forest plots of the association of polymorphisms and susceptibility to mCNV.
A) Impact of rs669676 in mCNV. B) Impact of rs10033900. C) Impact of rs10468017.
Discussion
CNV is a major cause of visual impairment in highly myopic patients and is the second leading cause of neovascular maculopathy after AMD. CNV is a common process in both diseases [39] and it can be difficult or impossible to distinguish between CNV resulting from excessive axial elongation and CNV attributable to the etiology of wet-type AMD in highly myopic eyes [39,54–56]. Thus, we hypothesized that SNPs associated with neovascularization in wet AMD may have be related to mCNV.
Our previous study [45] of CFH and ARMS2 was conducted in a cohort > 30 years of age, with subjects showing no significant differences. Recent studies have suggested the importance of other genetic regions in the development of wet AMD. Thus, we evaluated 10 new AMD-associated SNPs located in 4 different genes (COL8A1, LIPC, CFI and APOE) in an older population.
Our results showed that only the COL8A1 gene was significantly associated with mCNV in a Caucasian Spanish population before multiple analysis correction. In the SNP rs13095226, the C allele showed OR = 2.0 for developing CNV in Spanish high myopic patients. For SNP rs669676, the recessive model of genotype analysis, GG-GA versus AA showed similar risk ORs vs the control group (OR = 2.4). These results indicate that this gene is important in the development of myopic CNV. We verified the influence of this SNPs using quantitative ophthalmological variables. Our results showed that heterozygotes and minor allele homozygotes of rs13095226 (dominant model, CT/CC) had significantly higher axial length than patients presenting major allele homozygosis (TT). Furthermore our results showed that the rs13095226 genotype is associated with axial length (Fig 2A), but not with refractive power (Fig 2B). This may be because refractive power is influenced not only by the axial length of the eye, but also by the corneal keratometry and the lens. These results confirm the influence of this protein in axial growth and thus CNV development resulting from excessive axial elongation.
COL8A1 encodes one of the two alpha chains of collagen type VIII, a major component of ocular basement membranes such as Bruch's membrane and choroidal stroma [57]. Collagen VIII is produced by muscle cells and macrophages in the vascular wall. This protein regulates matrix metalloproteinases activity and endothelial cell migration mediated by vascular endothelial growth factor during angiogenesis. Col8A1 protein may lead to direct or indirect structural alterations in the Bruch’s membrane, which is a risk factor for mCNV. [58] Numerous studies have investigated the association between alterations in genes encoding the different collagen types and the development of HM, finding discrepant results between different populations[59–64]. Furthermore, many of the syndromes caused by alterations in collagen are associated with the development of HM resulting from excessive axial elongation [65–69]. Taken together, our results and those obtained in other studies [44,47,51,70,71] indicate the importance of COL8A1 in retinal angiogenesis, both in pathological myopia and AMD. This may be related to alterations in COL8A1 which can alter its expression to favor angiogenesis in these patients. This may be because proteins expressed by this gene stabilize the phenotype of endothelial cells [72]. During angiogenesis, where the differentiated phenotype is lost, the control and regulation of type VIII collagen may play a key role in processes in which new blood vessel formation is required and when excessive blood vessel formation is a pathological event (neovascularization).
Leveziel et al [47] previously studied the SNP rs669676 and only found a significant association with mCNV in multivariable analysis, which was no longer significant after correction for multiple testing. In our study, the recessive model of genotype analysis of the SNP rs669676 showed significant differences in mCNV vs controls without the HM group. Validating the results obtained by Leveziel et al, a comparison between highly myopic cohorts with or without CNV in rs669676 frequencies showed no statistically significant differences, but meta-analysis results of our study together with the Leveziel et al, showed a significant association of rs669676 with with mCNV. So, we confirmed a statistically significant association of the rs669676 validating the importance of the COL8A1 gene in the development of mCNV.
One of the most important systems related to the development of wet AMD is the complement system. In this study, we analyzed complement factor I (CFI), which showed no association both individually and as CFI SNPs haplotypes. This may be because the complement system is more closely associated with the etiology of AMD and less with that of HM. In AMD, waste substances excessively accumulate between the layers of the retina, leading to chronic activation of the complement system [73]. However, HM is caused by excessive axial eye growth and does not appear to be related to chronic activation of the complement system as in AMD.
Moreover, the results of the studies that analyzing the association between CFI and mCNV have not been replicated. Leveziel et al reported an increased risk of mCNV associated with the T allele of rs10033900, which is located 2781 base pairs upstream of the 3´-untranslated region of the CFI gene, in a Caucasian US population with European ancestry [47]. However, Miyake et al found no association between this SNP and mCNV in a Japanese population [50]. Because the results of the previous studies were not consistent, we evaluated all haplotypes in the CFI gene previously found to be associated with CNV in AMD [48,74]. We found no association between this gene and mCNV.
Previous studies have not shown consistent results. We performed a meta-analysis based on previously published data, but found no significant associations. The large heterogeneity (I-squared 68.9%) observed in the meta-analysis can be explained by the different ethnic origins of the populations studied, which may have led to different responses of the SNPs. Additional studies are needed to elucidate the importance of these genes in the development of myopic CNV, particularly those focused on rs13095226 of COL8A1. This is the first study examining this SNP in mCNV; a previous study evaluated wet AMD and found no significant association in Chinese patients [75]. There are at least two main strengths to this study. First, all participants were from a similar ethnic background, reducing the heterogeneity in different populations. Second, the mCNV group was compared not only with HM without CNV patients, but also with a population-based control group. Both HM cohorts had a high myopic genetic profile; therefore, we compared these cohorts with a control group of very advanced age to rule out the genetic predisposition for developing CNV (both mCNV and AMD-CNV). However, there were some limitations to this study. First, the sample size was low, and no cases had the CC genotype for rs13095226 in the population-based control group or HM without CNV. Thus, multivariate logistic regression analysis of this genetic region could not be conducted. However, allele analysis showed significant differences in the C allele of SNP rs13095226 in mCNV patients vs the HMnoCNV group, and further studies with a larger sample size are needed to replicate these genes and other candidate loci for this important vision-threatening complication of HM. The second limitation is that the groups showed demographic differences particularly in terms of age, but these subjects were included to obtain a population-representative control group with lower expectations of having CNV. Furthermore, to minimize the possible influence of these differences, we adjusted for all confounding factors using logistic regression analysis and found no new significant associations.
In summary, the results of this study showed that age and HT were significantly associated with CNV development in highly myopic Caucasian patients and that COL8A1 plays an important role in axial elongation of the eye and possibly in the CNV. The function of COL8A1 in remodeling of the extracellular matrix of the sclera may involve genetic pathways in the development of CNV in highly myopic eyes in elderly Caucasian populations. Furthermore, additional studies are needed to determine whether any of the SNPs analyzed are associated with the size of the CNV.
Supporting Information
SNP: Single nucleotide polymorphism (dbSNP1D); MAF: Minor allele frequency, HWE: exact test for Hardy-Weinberg equilibrium. *Excluded from analysis because all patients showed CC genotype.
(DOCX)
OR; odds Ratio, D+L pooled OR; random effect model OR, I-V pooled ES; fixed effect model OR, d.f.; degree of freedom.
(DOCX)
Acknowledgments
We greatly thank all the patients who participated in this study. This work has been developed by members of the Spanish Vitreoretinal society (SERV), the RETICs: RD07-0062: “Age Related Ocular Diseases, Quality of Life, and Vision”, and RETICS OFTARED (RD12/0034) ‘‘Prevention, Early Detection, and Treatment of the Prevalent Degenerative and Chronic Ocular Pathology” from the Instituto de Salud Carlos III from the Ministerio de Economía y Competitividad, Spain. This work has been partly funded by the PN I + D + I 2008–2011; the ISCIII-Subdireccion General de Redes y Centros de Investigacion Cooperativa; and the European Program FEDER.
All of the authors declare no conflict of interest.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work has been developed by members of the Spanish Vitreoretinal society (SERV), the RETICs: RD07-0062: “Age Related Ocular Diseases, Quality of Life, and Vision’’, and RETICS OFTARED (RD12/0034) ‘‘Prevention, Early Detection, and Treatment of the Prevalent Degenerative and Chronic Ocular Pathology’’ from the Instituto de Salud Carlos III from the Ministerio de Economia y Competitividad, Spain. This work has been partly funded by the PN I + D + I 2008–2011; the ISCIII-Subdireccion General de Redes y Centros de Investigacion Cooperativa; and the European Program FEDER.
References
- 1.Sawada A, Tomidokoro A, Araie M, Iwase A, Yamamoto T, Tajimi Study Group. Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology. 2008;115: 363–370. e3 10.1016/j.ophtha.2007.03.075 [DOI] [PubMed] [Google Scholar]
- 2.He M, Zheng Y, Xiang F. Prevalence of myopia in urban and rural children in mainland China. Optom Vis Sci. 2009;86: 40–44. 10.1097/OPX.0b013e3181940719 [DOI] [PubMed] [Google Scholar]
- 3.Vitale S, Sperduto RD, Ferris FL 3rd. Increased prevalence of myopia in the United States between 1971–1972 and 1999–2004. Arch Ophthalmol. 2009;127: 1632–1639. 10.1001/archophthalmol.2009.303 [DOI] [PubMed] [Google Scholar]
- 4.Wolfram C, Hohn R, Kottler U, Wild P, Blettner M, Buhren J, et al. Prevalence of refractive errors in the European adult population: the Gutenberg Health Study (GHS). Br J Ophthalmol. 2014;98: 857–861. [DOI] [PubMed] [Google Scholar]
- 5.Wong TY, Ferreira A, Hughes R, Carter G, Mitchell P. Epidemiology and disease burden of pathologic myopia and myopic choroidal neovascularization: an evidence-based systematic review. Am J Ophthalmol. 2014;157: 9–25. e12 10.1016/j.ajo.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 6.Hrynchak PK, Mittelstaedt A, Machan CM, Bunn C, Irving EL. Increase in myopia prevalence in clinic-based populations across a century. Optom Vis Sci. 2013;90: 1331–1341. 10.1097/OPX.0000000000000069 [DOI] [PubMed] [Google Scholar]
- 7.Morgan I, Rose K. How genetic is school myopia? Prog Retin Eye Res. 2005;24: 1–38. [DOI] [PubMed] [Google Scholar]
- 8.Pan CW, Ramamurthy D, Saw SM. Worldwide prevalence and risk factors for myopia. Ophthalmic Physiol Opt. 2012;32: 3–16. 10.1111/j.1475-1313.2011.00884.x [DOI] [PubMed] [Google Scholar]
- 9.Lin LL, Shih YF, Hsiao CK, Chen CJ. Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singapore. 2004;33: 27–33. [PubMed] [Google Scholar]
- 10.Saw SM. A synopsis of the prevalence rates and environmental risk factors for myopia. Clin Exp Optom. 2003;86: 289–294. [DOI] [PubMed] [Google Scholar]
- 11.Woo WW, Lim KA, Yang H, Lim XY, Liew F, Lee YS, et al. Refractive errors in medical students in Singapore. Singapore Med J. 2004;45: 470–474. [PubMed] [Google Scholar]
- 12.Chan NS, Teo K, Cheung CM. Epidemiology and Diagnosis of Myopic Choroidal Neovascularization in Asia. Eye Contact Lens. 2015. [DOI] [PubMed] [Google Scholar]
- 13.Percival SP. Redefinition of high myopia: the relationship of axial length measurement to myopic pathology and its relevance to cataract surgery. Dev Ophthalmol. 1987;14: 42–46. [DOI] [PubMed] [Google Scholar]
- 14.Shih YF, Ho TC, Hsiao CK, Lin LL. Visual outcomes for high myopic patients with or without myopic maculopathy: a 10 year follow up study. Br J Ophthalmol. 2006;90: 546–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noble KG, Carr RE. Pathologic myopia. Ophthalmology. 1982;89: 1099–1100. [DOI] [PubMed] [Google Scholar]
- 16.Bourne RR, Jonas JB, Flaxman SR, Keeffe J, Leasher J, Naidoo K, et al. Prevalence and causes of vision loss in high-income countries and in Eastern and Central Europe: 1990–2010. Br J Ophthalmol. 2014;98: 629–638. 10.1136/bjophthalmol-2013-304033 [DOI] [PubMed] [Google Scholar]
- 17.Pacella R, McLellan J, Grice K, Del Bono EA, Wiggs JL, Gwiazda JE. Role of genetic factors in the etiology of juvenile-onset myopia based on a longitudinal study of refractive error. Optom Vis Sci. 1999;76: 381–386. [DOI] [PubMed] [Google Scholar]
- 18.Feldkamper M, Schaeffel F. Interactions of genes and environment in myopia. Dev Ophthalmol. 2003;37: 34–49. [DOI] [PubMed] [Google Scholar]
- 19.Dirani M, Shekar SN, Baird PN. The role of educational attainment in refraction: the Genes in Myopia (GEM) twin study. Invest Ophthalmol Vis Sci. 2008;49: 534–538. 10.1167/iovs.07-1123 [DOI] [PubMed] [Google Scholar]
- 20.Lu B, Jiang D, Wang P, Gao Y, Sun W, Xiao X, et al. Replication study supports CTNND2 as a susceptibility gene for high myopia. Invest Ophthalmol Vis Sci. 2011;52: 8258–8261. 10.1167/iovs.11-7914 [DOI] [PubMed] [Google Scholar]
- 21.Kiefer AK, Tung JY, Do CB, Hinds DA, Mountain JL, Francke U, et al. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLOS Genet. 2013;9: e1003299 10.1371/journal.pgen.1003299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verhoeven VJ, Hysi PG, Wojciechowski R, Fan Q, Guggenheim JA, Hohn R, et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013;45: 314–318. 10.1038/ng.2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen SJ, Cheng CY, Li AF, Peng KL, Chou P, Chiou SH, et al. Prevalence and associated risk factors of myopic maculopathy in elderly Chinese: the Shihpai eye study. Invest Ophthalmol Vis Sci. 2012;53: 4868–4873. 10.1167/iovs.12-9919 [DOI] [PubMed] [Google Scholar]
- 24.Teikari JM, O'Donnell J, Kaprio J, Koskenvuo M. Impact of heredity in myopia. Hum Hered. 1991;41: 151–156. [DOI] [PubMed] [Google Scholar]
- 25.Lopes MC, Andrew T, Carbonaro F, Spector TD, Hammond CJ. Estimating heritability and shared environmental effects for refractive error in twin and family studies. Invest Ophthalmol Vis Sci. 2009;50: 126–131. 10.1167/iovs.08-2385 [DOI] [PubMed] [Google Scholar]
- 26.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75: 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mutti DO, Cooper ME, O'Brien S, Jones LA, Marazita ML, Murray JC, et al. Candidate gene and locus analysis of myopia. Mol Vis. 2007;13: 1012–1019. [PMC free article] [PubMed] [Google Scholar]
- 28.Inamori Y, Ota M, Inoko H, Okada E, Nishizaki R, Shiota T, et al. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007;122: 151–157. [DOI] [PubMed] [Google Scholar]
- 29.Metlapally R, Li YJ, Tran-Viet KN, Abbott D, Czaja GR, Malecaze F, et al. COL1A1 and COL2A1 genes and myopia susceptibility: evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009;50: 4080–4086. 10.1167/iovs.08-3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang WC, Yip SP, Lo KK, Ng PW, Choi PS, Lee SY, et al. Linkage and association of myocilin (MYOC) polymorphisms with high myopia in a Chinese population. Mol Vis. 2007;13: 534–544. [PMC free article] [PubMed] [Google Scholar]
- 31.Zayats T, Yanovitch T, Creer RC, McMahon G, Li YJ, Young TL, et al. Myocilin polymorphisms and high myopia in subjects of European origin. Mol Vis. 2009;15: 213–222. [PMC free article] [PubMed] [Google Scholar]
- 32.Han W, Yap MK, Wang J, Yip SP. Family-based association analysis of hepatocyte growth factor (HGF) gene polymorphisms in high myopia. Invest Ophthalmol Vis Sci. 2006;47: 2291–2299. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi T, Inoko H, Nishizaki R, Ohno S, Mizuki N. Exclusion of transforming growth factor-beta1 as a candidate gene for myopia in the Japanese. Jpn J Ophthalmol. 2007;51: 96–99. [DOI] [PubMed] [Google Scholar]
- 34.Zha Y, Leung KH, Lo KK, Fung WY, Ng PW, Shi MG, et al. TGFB1 as a susceptibility gene for high myopia: a replication study with new findings. Arch Ophthalmol. 2009;127: 541–548. 10.1001/archophthalmol.2008.623 [DOI] [PubMed] [Google Scholar]
- 35.Scavello GS, Paluru PC, Ganter WR, Young TL. Sequence variants in the transforming growth beta-induced factor (TGIF) gene are not associated with high myopia. Invest Ophthalmol Vis Sci. 2004;45: 2091–2097. [DOI] [PubMed] [Google Scholar]
- 36.Pertile KK, Schache M, Islam FM, Chen CY, Dirani M, Mitchell P, et al. Assessment of TGIF as a candidate gene for myopia. Invest Ophthalmol Vis Sci. 2008;49: 49–54. 10.1167/iovs.07-0896 [DOI] [PubMed] [Google Scholar]
- 37.Wang P, Li S, Xiao X, Jia X, Jiao X, Guo X, et al. High myopia is not associated with the SNPs in the TGIF, lumican, TGFB1, and HGF genes. Invest Ophthalmol Vis Sci. 2009;50: 1546–1551. 10.1167/iovs.08-2537 [DOI] [PubMed] [Google Scholar]
- 38.Lin HJ, Wan L, Tsai Y, Chen WC, Tsai SW, Tsai FJ. The association between lumican gene polymorphisms and high myopia. Eye (Lond). 2010;24: 1093–1101. [DOI] [PubMed] [Google Scholar]
- 39.Grossniklaus HE, Green WR. Choroidal neovascularization. Am J Ophthalmol. 2004;137: 496–503. [DOI] [PubMed] [Google Scholar]
- 40.Cohen SY, Laroche A, Leguen Y, Soubrane G, Coscas GJ. Etiology of choroidal neovascularization in young patients. Ophthalmology. 1996;103: 1241–1244. [DOI] [PubMed] [Google Scholar]
- 41.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51: 316–363. [DOI] [PubMed] [Google Scholar]
- 42.Gorin MB. A clinician's view of the molecular genetics of age-related maculopathy. Arch Ophthalmol. 2007;125: 21–29. [DOI] [PubMed] [Google Scholar]
- 43.Swaroop A, Branham KE, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits. Hum Mol Genet. 2007;16 Spec No. 2: R174–82. [DOI] [PubMed] [Google Scholar]
- 44.Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45: 433–9, 439e1–2. 10.1038/ng.2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Robredo P, Maestre SR, Zarranz-Ventura J, Mulero HH, Salinas-Alaman A, Garcia-Layana A. Myopic choroidal neovascularization genetics. 2008;115: 1632, 1632–e1.. [DOI] [PubMed] [Google Scholar]
- 46.Nakanishi H, Gotoh N, Yamada R, Yamashiro K, Otani A, Hayashi H, et al. ARMS2/HTRA1 and CFH polymorphisms are not associated with choroidal neovascularization in highly myopic eyes of the elderly Japanese population. Eye (Lond). 2010;24: 1078–1084. [DOI] [PubMed] [Google Scholar]
- 47.Leveziel N, Yu Y, Reynolds R, Tai A, Meng W, Caillaux V, et al. Genetic factors for choroidal neovascularization associated with high myopia. Invest Ophthalmol Vis Sci. 2012;53: 5004–5009. 10.1167/iovs.12-9538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17: 100–104. 10.1038/ejhg.2008.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lechanteur YT, van de Ven JP, Smailhodzic D, Boon CJ, Klevering BJ, Fauser S, et al. Genetic, behavioral, and sociodemographic risk factors for second eye progression in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2012;53: 5846–5852. 10.1167/iovs.11-7731 [DOI] [PubMed] [Google Scholar]
- 50.Miyake M, Yamashiro K, Nakanishi H, Nakata I, Akagi-Kurashige Y, Kumagai K, et al. Evaluation of pigment epithelium-derived factor and complement factor I polymorphisms as a cause of choroidal neovascularization in highly myopic eyes. Invest Ophthalmol Vis Sci. 2013;54: 4208–4212. 10.1167/iovs.13-12280 [DOI] [PubMed] [Google Scholar]
- 51.Seddon JM, Reynolds R, Yu Y, Rosner B. Three new genetic loci (R1210C in CFH, variants in COL8A1 and RAD51B) are independently related to progression to advanced macular degeneration. PLOS ONE. 2014;9: e87047 10.1371/journal.pone.0087047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y, Reynolds R, Fagerness J, Rosner B, Daly MJ, Seddon JM. Association of variants in the LIPC and ABCA1 genes with intermediate and large drusen and advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52: 4663–4670. 10.1167/iovs.10-7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Souied EH, Benlian P, Amouyel P, Feingold J, Lagarde JP, Munnich A, et al. The epsilon4 allele of the apolipoprotein E gene as a potential protective factor for exudative age-related macular degeneration. 1998;125: 353–9. [DOI] [PubMed] [Google Scholar]
- 54.Soubrane G. Choroidal neovascularization in pathologic myopia: recent developments in diagnosis and treatment. Surv Ophthalmol. 2008;53: 121–138. 10.1016/j.survophthal.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 55.Miller JW. Age-related macular degeneration revisited—piecing the puzzle: the LXIX Edward Jackson memorial lecture. Am J Ophthalmol. 2013;155: 1–35. e13 10.1016/j.ajo.2012.10.018 [DOI] [PubMed] [Google Scholar]
- 56.Augood CA, Vingerling JR, de Jong PT, Chakravarthy U, Seland J, Soubrane G, et al. Prevalence of age-related maculopathy in older Europeans: the European Eye Study (EUREYE). Arch Ophthalmol. 2006;124: 529–535. [DOI] [PubMed] [Google Scholar]
- 57.Tamura Y, Konomi H, Sawada H, Takashima S, Nakajima A. Tissue distribution of type VIII collagen in human adult and fetal eyes. Invest Ophthalmol Vis Sci. 1991;32: 2636–2644. [PubMed] [Google Scholar]
- 58.Ikuno Y, Sayanagi K, Soga K, Sawa M, Gomi F, Tsujikawa M, et al. Lacquer crack formation and choroidal neovascularization in pathologic myopia. Retina. 2008;28: 1124–1131. [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Zhou X, Qu X. Association between COL1A1 polymorphisms and high myopia: a meta-analysis. Int J Clin Exp Med. 2015;8: 5862–5868. [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D, Shi Y, Gong B, He F, Lu F, Lin H, et al. An association study of the COL1A1 gene and high myopia in a Han Chinese population. Mol Vis. 2011;17: 3379–3383. [PMC free article] [PubMed] [Google Scholar]
- 61.Metlapally R, Li YJ, Tran-Viet KN, Abbott D, Czaja GR, Malecaze F, et al. COL1A1 and COL2A1 genes and myopia susceptibility: evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009;50: 4080–4086. 10.1167/iovs.08-3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Otani A, Tsujikawa A, et al. Absence of association between COL1A1 polymorphisms and high myopia in the Japanese population. Invest Ophthalmol Vis Sci. 2009;50: 544–550. 10.1167/iovs.08-2425 [DOI] [PubMed] [Google Scholar]
- 63.Inamori Y, Ota M, Inoko H, Okada E, Nishizaki R, Shiota T, et al. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007;122: 151–157. [DOI] [PubMed] [Google Scholar]
- 64.Liang CL, Hung KS, Tsai YY, Chang W, Wang HS, Juo SH. Systematic assessment of the tagging polymorphisms of the COL1A1 gene for high myopia. J Hum Genet. 2007;52: 374–377. [DOI] [PubMed] [Google Scholar]
- 65.Snead MP, Yates JR. Clinical and Molecular genetics of Stickler syndrome. J Med Genet. 1999;36: 353–359. [PMC free article] [PubMed] [Google Scholar]
- 66.Haghighi A, Tiwari A, Piri N, Nurnberg G, Saleh-Gohari N, Haghighi A, et al. Homozygosity mapping and whole exome sequencing reveal a novel homozygous COL18A1 mutation causing Knobloch syndrome. PLOS ONE. 2014;9: e112747 10.1371/journal.pone.0112747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hufnagel SB, Weaver KN, Hufnagel RB, Bader PI, Schorry EK, Hopkin RJ. A novel dominant COL11A1 mutation resulting in a severe skeletal dysplasia. Am J Med Genet A. 2014;164A: 2607–2612. 10.1002/ajmg.a.36688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sergouniotis PI, Fincham GS, McNinch AM, Spickett C, Poulson AV, Richards AJ, et al. Ophthalmic and molecular genetic findings in Kniest dysplasia. Eye (Lond). 2015;29: 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terhal PA, Nievelstein RJ, Verver EJ, Topsakal V, van Dommelen P, Hoornaert K, et al. A study of the clinical and radiological features in a cohort of 93 patients with a COL2A1 mutation causing spondyloepiphyseal dysplasia congenita or a related phenotype. Am J Med Genet A. 2015;167A: 461–475. 10.1002/ajmg.a.36922 [DOI] [PubMed] [Google Scholar]
- 70.Seddon JM, Silver RE, Kwong M, Rosner B. Risk Prediction for Progression of Macular Degeneration: 10 Common and Rare Genetic Variants, Demographic, Environmental, and Macular Covariates. Invest Ophthalmol Vis Sci. 2015;56: 2192–2202. 10.1167/iovs.14-15841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y, Bhangale TR, Fagerness J, Ripke S, Thorleifsson G, Tan PL, et al. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20: 3699–3709. 10.1093/hmg/ddr270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shuttleworth CA. Type VIII collagen. Int J Biochem Cell Biol. 1997;29: 1145–1148. [DOI] [PubMed] [Google Scholar]
- 73.Pangburn MK. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology. 2000;49: 149–157. [DOI] [PubMed] [Google Scholar]
- 74.Ennis S, Gibson J, Cree AJ, Collins A, Lotery AJ. Support for the involvement of complement factor I in age-related macular degeneration. Eur J Hum Genet. 2010;18: 15–16. 10.1038/ejhg.2009.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu Y, Huang L, Wang B, Zhang C, Bai Y, Li X. COL8A1 rs13095226 polymorphism shows no association with neovascular age-related macular degeneration or polypoidal choroidal vasculopathy in Chinese subjects. Int J Clin Exp Pathol. 2015;8: 11635–11640. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNP: Single nucleotide polymorphism (dbSNP1D); MAF: Minor allele frequency, HWE: exact test for Hardy-Weinberg equilibrium. *Excluded from analysis because all patients showed CC genotype.
(DOCX)
OR; odds Ratio, D+L pooled OR; random effect model OR, I-V pooled ES; fixed effect model OR, d.f.; degree of freedom.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.