Abstract

Whole brain radiotherapy (WBRT) produces unwanted sequelae, albeit via unknown mechanisms. A deacetylase expressed in the central nervous system, Sirtuin 2 (SIRT2), has been linked to neurodegeneration. Therefore, we sought to challenge the notion that a single disease pathway is responsible for radiation-induced brain injury in Sirt2 wild-type (WT) and knockout (KO) mice at the proteomic level. We utilized isobaric tag for relative and absolute quantitation to analyze brain homogenates from Sirt2 WT and KO mice with and without WBRT. Selected proteins were independently verified, followed by ingenuity pathway analysis. Canonical pathways for Huntington’s, Parkinson’s, and Alzheimer’s were acutely affected by radiation within 72 h of treatment. Although loss of Sirt2 preferentially affected both Huntington’s and Parkinson’s pathways, WBRT most significantly affected Huntington’s-related proteins in the absence of Sirt2. Identical protein expression patterns were identified in Mog following WBRT in both Sirt2 WT and KO mice, revealing a proteomic radiation signature; however, long-term radiation effects were found to be associated with altered levels of a small number of key neurodegeneration-related proteins, identified as Mapt, Mog, Snap25, and Dnm1. Together, these data demonstrate the principle that the presence of Sirt2 can have significant effects on the brain proteome and its response to ionizing radiation.
Keywords: SIRT2, brain, radiation, neurotoxicity, iTRAQ, neurodegeneration
1. Introduction
Radiation therapy (RT) is one of the most accepted and widely used treatments for primary and metastatic intracranial tumors. Recent technical advances in RT and multimodality approaches to treatment have increased the life span of many patients; however, a major concern remains as to radiation’s effect on long-term neurotoxicity and cognitive impairment. Approximately 100 000 primary and metastatic brain tumor patients per year in the U.S. survive long enough (>6 months) to experience radiation-induced brain injury.1−4 Acute and subacute damage that produces symptoms such as nausea, headache, and vomiting following WBRT appears to be reversible, but in some patients a delayed irreversible neurotoxicity manifests as cognitive impairment and memory deficits.1,5,6 The exact nature and cause of the subsequent biochemical alterations in the central nervous system (CNS) microenvironment responsible for the irreversible neurotoxicity are currently unknown. Some reports suggest that a few crucial factors play significant roles in radiation-induced neurotoxicity, such as damage to vasculature, loss of oligodendrocyte type 2 astrocyte progenitor cells, as well as mature oligodendrocytes, which manifest in abnormal myelin production, loss of neuronal stem cells, and altered expression of several cytokines.7−11 A prevailing hypothesis is that the pathogenesis of long-term radiation-induced neurodegeneration is similar to the pathogenesis of Alzheimer’s; however, clinical attempts to target this end point have been met with limited success.12,13 Thus, it is necessary to better define the molecular events following RT so that better and targeted approaches could be exploited in this regard.
Sirtuin family members (SIRT1–7) are highly conserved, structurally and functionally related proteins, falling into two types that have either NAD+-dependent enzymatic activity (SIRT1, SIRT2, SIRT3, SIRT5, and SIRT7) for deacetylation of proteins or ADP ribosyl transferase activity (SIRT4 and SIRT6).14 Their functions in metabolic processes and chromatin remodeling have been conserved throughout evolution.15 Among seven known family members, SIRT1 localizes to the nucleus and cytoplasm, SIRT2 primarily to cytoplasm, SIRT 3, 4, and 5 mainly inside mitochondria, and SIRT 6 and 7 inside the nucleus. Sirtuins have been implicated in several processes like aging, cell cycle regulation, cancer, metabolism, DNA repair, and neurobiology.16−18 Some important studies also show that SIRT1 and SIRT2 play crucial roles in neurodegeneration. While SIRT1 appears to have limited expression localized primarily to cortex, hippocampus, cerebellum, and hypothalamus in the mouse brain,19,20 SIRT2 is abundantly expressed in the CNS, particularly within oligodendrocytes, hippocampal neurons, and olfactory neurons.21 Whereas there are reports suggesting overexpression or activation of SIRT1 is protective against Alzheimer’s, Parkinson’s, and Huntington’s by targeting several key molecules such as Rarβ, Tau (Mapt), Pgc1α, Lxr, NFκB, HSF1, and TORC1,22−28 preliminary and very limited studies in murine and in vitro models have demonstrated that inhibition of SIRT2 reduces the aggregation of α-synuclein via modulation of tubulin activity resulting in the suppression of Parkinson’s mediated toxicity.29,30 Inhibition of SIRT2 has also been shown to mitigate Huntington’s toxicity by reducing cholesterol levels via transcriptional regulation of SREBP2.31,32 While the exact role of SIRT2 in Huntington’s related toxicity is a matter of some controversy,33 SIRT2 does appear to accumulate in aging rodent brain.34
WBRT in some patients produces a clinical pattern of neurodegeneration that is reminiscent of certain aspects of Parkinson’s, Huntington’s, and Alzheimer’s phenotypes and is dependent on the volume of brain treated as well as the total dose and fractionation schedule received.1,7,35,36 By exploiting a global proteomic analysis of whole brain samples from Sirt2 WT and KO mice, followed by targeted validation of proteins whose levels were observed to change, we demonstrate that long-term radiation toxicity in the brain is a complex process involving multiple neurodegenerative pathways. In doing so, we shed light on novel molecular and cellular details following whole brain radiation treatment and considered whether SIRT2 is an important mediator of radiation-induced neurotoxicity.
2. Materials and Methods
2.1. Sirt2 WT and KO Mice and Cell Culture
Mouse embryonic fibroblasts (MEFs) were derived from Sirt2 knockout (KO) mice generated by genomic deletion in a C57BL/6 background. These mice were a kind gift of David Lombard and Fred Alt of Children’s Hospital, Boston, Massachusetts. All animal care and experiments were conducted in accordance with guidelines set by, and with approval of, the Animal Care and Use Committee at the National Cancer Institute. Cells were harvested after trypsin digestion of E12.5–13.5 embryos collected from sacrifice of timed pregnant females. Cells were cultured in Dulbecco’s modified essential medium supplemented with 1% penicillin (100 units/mL)–streptomycin (100 μg/mL), 15% FBS, and 1% nonessential amino acid (Invitrogen) in a humidified 37 °C incubator with 6% O2. ATP levels were assayed in MEFs using CellTiter-Glo 2.0 (Promega) as per the manufacturer’s instructions. Unless otherwise indicated, analyses were performed in triplicate.
2.2. Rotarod
Mice were placed individually on an accelerating rotating cylinder (Rotarod, Boston Scientific) to evaluate coordination. This was performed once a day at the same daily time until scheduled euthanasia. The diameter of the cylinder was 3 cm, covered with scored plastic. Mice were confined to a 6.5 cm long section of the cylinder by Plexiglas dividers. Four mice were placed on the cylinder at once. The rotation rate of the cylinder was increased from 4 to 40 rpm over a 6 min period. The latency time (in seconds) of each mouse to fall off the rotating cylinder onto soft bedding was recorded. Mice were trained from days 1–4 and tested on days 5–17, with all measurements recorded.
2.3. Irradiation
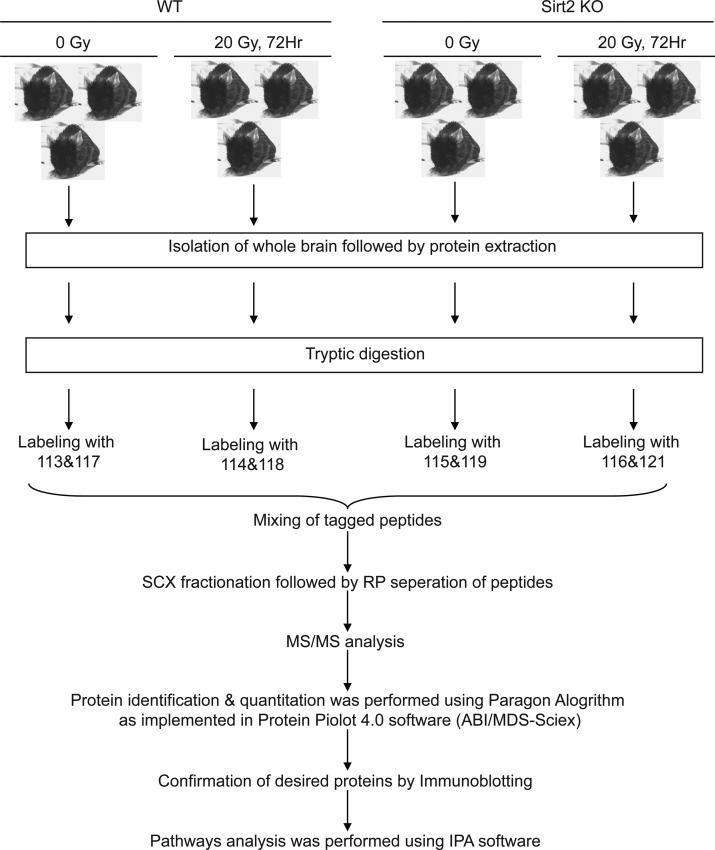
For in vitro experiments, MEFs were irradiated as monolayers at room temperature in an X-Rad 320 biological irradiator (Precision X-ray) operated at 320 kV and 12.5 mA with 2 mm Al filtration to achieve a dose rate of 2.1 Gy/min. After exposure, cells were returned to 37 °C. For in vivo irradiation, 6 month old female Sirt2 WT and KO mice were anesthetized using ketamine (90 mg/kg)/xylazine (10 mg/kg) intraperitoneally for immobilization and placed in well-ventilated Plexiglas jigs designed with an aperture to allow exposure for whole brain radiation, and to provide shielding for the mouse along the torso and critical normal structures of the head. Irradiation was conducted with a Pentak X-irradiator (Inspection Systems) at 300 kV, 10 mA at a dose rate of 253 cGy/min. WBRT was administered as 20 Gy in a single fraction to 3 Sirt2 WT mice and 3 Sirt2 KO mice. This dose-fractionation schedule was chosen to mimic previous work demonstrating maximum inflammatory changes,37,38 induction of gene expression profiles by microarray,39 and decreased oligodendrocyte density.40 An additional three mice from each group were sham-irradiated as controls. Triplicate sets of mice were treated in this fashion, with one set sacrificed at 72 h after WBRT for iTRAQ analysis, another set sacrificed at 72 h to evaluate acute radiation changes as a biological replicate for immunoblot validation, and a third set was sacrificed at one year following WBRT to evaluate long-term changes as an additional biological replicate set for immunoblot validation.
2.4. Whole Brain Isolation and Protein Extraction
Whole brains from each group were minced in RIPA buffer (Thermo Scientific) and homogenized with FastPrep Lysing matrix bead D (MP Biomedicals) for 20 s, followed by incubation on ice for 5 min. After gentle shaking for 1 h at 4 °C, samples were centrifuged at 13 000 rpm in an Eppendorf 5417R microcentrifuge for 20 min at 4 °C. Clarified supernatants were collected, and pooled specimens from each group were used as samples for iTRAQ labeling. Protein concentrations of each sample were measured using DC Protein Assay Kit per manufacturer’s instructions (Bio Rad). One hundred micrograms of protein from each set was selected for isobaric tag labeling.
2.5. iTRAQ Labeling
iTRAQ 8plex kit was obtained from Applied Biosystems (MDS Sciex), and labeling was done according to protocol prescribed by Pennsylvania State University’s College of Medicine Proteomics and Mass Spectrometry Core Research Facility (http://www.pennstatehershey.org/web/core/proteinsmassspectometry/protocols/itraq). In brief, protein from each treatment group was precipitated with TCA, and 100 μg of dried protein was dissolved in 20 μL of dissolution buffer (0.5 M triethylammonium bicarbonate) to give a final concentration of 5 μg/μL. One microliter of 2% SDS was added to each sample and reduced with 1 μL of 110 mM TCEP (tris(2-carboxyethyl) phosphine) at 60 °C for 1 h. Samples were then alkylated with 1 μL of 84 mM iodoacetamide, and reactions were left for completion in the dark at room temperature for 30 min. Ten micrograms of Promega Sequencing grade trypsin was added to each sample, and reactions were left overnight at 48 °C. Eight plex reagents were reconstituted with 50 μL of 100% isopropanol (Sigma-Aldrich). Trypsinized samples were added individually to the desired isobaric tag and kept for 2 h at room temperature to allow interaction between reactive tags and peptide N-termini and free amino groups of lysine side chains. Tagging was performed as follows: tags 113 and 117 for WT sham-irradiated; tags 114 and 118 for WT 20 Gy-irradiated; tags 115 and 119 for Sirt2 KO sham-irradiated; and tags 116 and 121 for Sirt2 KO 20 Gy-irradiated. Reactions were stopped by the addition of 100 μL of Milli-Q water. All samples were combined and vacuum-dried, followed by resuspension in 100 μL of Milli-Q water. This process was repeated a total of three times. Dried combined samples were then subjected to 2D-LC separation and MS analysis at the Proteomics and Mass Spectrometry Core Research Facility of the College of Medicine, Pennsylvania State University.
2.6. 2D-LC Separations
Dried sample was resuspended in 500 μL of cation exchange buffer (12 mM ammonium formate in 25% acetonitrile at pH 2.5 to 3.0) and subjected to SCX separation. SCX separations were performed on a passivated Waters 600E HPLC system, using a 4.6 × 250 mm PolySULFOETHYL aspartamide column (PolyLC) at a flow rate of 1 mL/min. Buffer A contained 10 mM ammonium formate, pH 2.7, in 20% acetonitrile/80% water. Buffer B contained 666 mM ammonium formate, pH 2.7, in 20% acetonitrile/80% water.
The gradient was Buffer A at 100% (0–22 min following sample injection), 0% → 40% Buffer B (16–48 min), 40% → 100% Buffer B (48–49 min), then isocratic 100% Buffer B (49–56 min), then at 56 min switched back to 100% A to re-equilibrate for the next injection. The first 26 mL of eluant (containing all flow-through fractions) was combined into one fraction; then, 14 additional 2 mL fractions were collected. All 15 of these SCX fractions were dried down completely to reduce volume and to remove the volatile ammonium formate salts then resuspended in 9 μL of 2% (v/v) acetonitrile, 0.1% (v/v) TFA and filtered prior to reverse-phase C18 nanoflow-LC separation.
For second-dimension separation by reverse-phase nanoflow LC, each SCX fraction was autoinjected onto a Chromolith CapRod column (150 × 0.1 mm, Merck) using a 5 μL injector loop on a Tempo LC MALDI Spotting system (ABI-MDS/Sciex). Buffer C was 2% acetonitrile, 0.1% TFA, and Buffer D was 98% acetonitrile, 0.1% TFA.
The elution gradient was 95% C/5% D (2 μL per minute flow rate from 0 to 3 min, then 2.5 μL per minute from 3 to 8.1 min), 5% D → 38% D (8.1–40 min), 38% D → 80% D (41–44 min), 80% D → 5% D (44–49 min) (initial conditions). Flow rate was 2.5 μL/min during the gradient, and an equal flow of MALDI matrix solution was added postcolumn (7 mg/mL recrystallized CHCA (a-cyano-hydroxycinnamic acid), 2 mg/mL ammonium phosphate, 0.1% trifluoroacetic acid, 80% acetonitrile). The combined eluant was automatically spotted onto a stainless-steel MALDI target plate every 6 s (0.6 μL per spot) for a total of 370 spots per original SCX fraction.
2.7. Mass Spectrometry Analysis
After the sample spots were dried, 13 calibration spots (ABI 4700 Mix) were added to each plate manually. MALDI target plates (15 per experiment) were analyzed in a data-dependent manner on an ABI 5800 or 800 MALDI TOF-TOFs. As each plate was entered into the instrument, a plate calibration/MS default calibration update was performed; then, the MS/MS default calibration was updated. MS spectra were then acquired from each sample spot using newly updated default calibrations, with 500 laser shots per spot, at a laser intensity of 3200. A plate-wide interpretation was then automatically performed, choosing the highest peak of each observed m/z value for subsequent MS/MS analysis. Up to 2500 laser shots at laser power 4200 were accumulated for each MS/MS spectrum.
2.8. Identification and Quantification of Proteins
Protein identification and relative quantification for the iTRAQ experiments were performed with the Paragon algorithm implemented in the ProteinPilot 4.0 software program (AB Sciex) and the Paragon (Applied Biosystems) protein database search algorithm using a species-specific search from the National Center for Bioinformatics (NCBI) database. The concatenated target-decoy database-search strategy was used to check the false-positive rate.41 The Paragon algorithm42,43 in ProteinPilot v4.0 software was used as the default search program with digestion enzyme set as trypsin and methylmethanethiosulfonate as cysteine modification. Data were normalized for possible loading errors by bias correction calculated with ProGroup algorithm and identified proteins with at least 95% confidence with an overall labeling efficiency of >95%. A summary of the MS/MS analysis has been mentioned in Figure S1A. A total of 14 601 MS/MS spectra were produced and 74.5% of those spectra (10 854) were identified at 95% confidence or higher including redundant identifications. After removing redundant IDs, there were 7502 distinct peptides identified at 95% confidence or higher. 1792 proteins were identified before grouping using the ProteinPilot Unused Score >1.3 (95% confidence) as a cutoff threshold. This number of identified proteins was further reduced to 815 proteins by the combination of the ProGroup protein grouping algorithm and accepting as identified only proteins with a local FDR estimation of 5% or lower based on the rate of “identifications” in the decoy database at lower and lower unused scores. Quality of MS/MS data were also checked for distributions of the acquired signals (discussed in the Results section). The results obtained from ProteinPilot v4.0 software were exported to Microsoft Excel and to R for further analysis.
2.9. Technical Replicate Analysis and Cutoff Estimation
All statistical analysis was performed using R packages (http://www.bioconductor.org). To generate the quantitative proteome using iTRAQ labeling, we first determined the labeling efficiency, which exceeded 95% (99.4% of the possible lysine labeling sites were labeled with iTRAQ tags and 96.7% of the possible amino-terminus labeling sites were labeled with iTRAQ tags). Estimation of the quantitative false discovery rate was also controlled by using a target-decoy approach for multiple comparisons (Figure S1D). To identify a minimum p-value cutoff at which to accept a protein level as “changed”, we calculated individual p values for the change of level (ratio) of each individual protein between the samples whose ratio should not change (the weighted average and error of the log of all individual ratios for each of the peptides connected to a particular protein). The p values for the protein quantity changes from the experiment are then put in rank order, and a Global FDR is calculated by determining how many quantity changes from the two samples that should be the same would be included at different p-value cutoffs. Using a p-value cutoff of 0.05 produced a Global FDR estimate of 39%. (This is for the quantitative aspects, that is, this is independent of the FDR estimates for the identifications.) In the four comparison sets, the number of proteins was chosen based on the following criteria: Each protein identification had to be based on at least two unique peptides (>95%) and have calculated value <0.05 for the reporter ions (Figure S2A). The 815 proteins were used to monitor technical variations and confirm the threshold for meaningful differences.
Next, the cutoff for significant fold change was determined based on the four comparison sets WTCT (tags 113, 117) and WTRT (tags 114,118) of iTRAQ experiments. The technical variations for the 114/113 and 118/117 reporter ions were estimated by Pearson correlation coefficients and were found to be r = 0.86 and 0.95, respectively. Accordingly, >90% of the proteins observed in the technical replicates fell within 20% of the respective experimental variation (Figure S2B). Comparison of other technical replicates yielded similar results (data not shown). Given that the technical variations were very minimal between various comparisons, we then used very stringent criteria for the selection of the ratios for further analysis. For any comparison, criteria for acceptance as a true protein level change included the four ratio parameters, as depicted in Figure S2A: (1) one or more p values <0.05 and corresponding tags in same directions; (2) two p values should not be contradicted by each other with respect to expression patterns; (3) average only for selected tags if one or more p values are significant; and (4) for increased expression the ratio cut-off value was set at 1.10, and for lowered expression the ratio cut off was 0.9. The selected proteins that met the stringent significant threshold cutoffs, as previously described, were then subjected to clustering analysis using correlation distance metrics and unsupervised hierarchical method.
2.10. Ingenuity Pathway Analysis
The differentially expressed proteome was evaluated using Ingenuity Pathway Analysis (Ingenuity Systems; www.ingenuity.com). Proteins from the data set that were associated with a canonical pathway in the Ingenuity Pathways Knowledge Base were considered for the analysis. The significance of the association between the data set and the canonical pathway was measured in two ways: (1) A ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway is displayed. (2) Fischer’s exact test was used to calculate a p value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone. Network analysis was performed using the String 9.1 program.
2.11. Immunoblot Analysis
Tissues and MEFs were harvested and lysed in RIPA buffer (Pierce) containing complete mini EDTA-free protease inhibitor cocktail (Roche) plus phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich). Total lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes using the iBlot apparatus (Invitrogen). Immunoblot analysis was performed per standard procedures using commercially available antibodies (Atp5a, Hspa8, Pacsin1, Uchl1, Mog, ubiquitin (Santa Cruz Biotechnologies); Mapt, Ppia, Sirt2 (Sigma); Sod1 (Millipore); Dnm1 (Cell Applications); and Snap25, Stmn1, Parp (Cell Signaling). All of the primary antibodies were used at dilutions of 1:1000. Horseradish-peroxidase-conjugated secondary antibodies were used at dilutions of 1:2000 (Invitrogen). Proteins were visualized using ECL (Cell Signaling Technology) and autoradiography. Densitometry of immunoreactive bands was performed using ImageJ software. p values between two comparisons were calculated using a two-tailed t test from the average raw values of pixel densities of bands in triplicate where indicated. Pearson correlation coefficients were calculated from the average of the raw values of pixel densities of bands in triplicate using the ratio of various comparisons for the biological and technical replicates, where indicated.
3. Results
3.1. SIRT2 Is Correlated to Improved Cognitive Ability in Mice
To determine if SIRT2 is important for cognitive functioning, we evaluated Rotarod performance in Sirt2 WT and KO mice bred in a C57BL6 background over a consecutive 17 day testing period. The Rotarod, validated to correlate to function of dopaminergic pathways in rodents, was chosen to evaluate cognitive function.44 Mice in groups (as mentioned in Figure 1, legend) were subjected to daily Rotarod testing, and the amount of time each mouse remained on the rod was recorded. The compiled data are represented as the average time the mice in each group remained on the rod ± SD versus the day tested (Figure 1). The data demonstrate significant differences in the curves between WT and KO mice after Day 4 on every day the mice were evaluated until day 17 (p < 0.05). Consistently, WT mice remained on the rod longer than their Sirt2 KO counterparts, suggesting that the presence of SIRT2 correlates to improved learning curve performance, coordination, and cognitive ability.
Figure 1.
Neurocognitive evaluation of Sirt2 WT and KO mice. Age-matched, 6 month old female mice from each group (n = 6/group) were subjected to daily Rotarod testing for 17 consecutive days. The rotation rate of the cylinder was increased from 4 to 40 rpm over a 6 min period. The latency time (in seconds) of each mouse to fall off the rotating cylinder was recorded. Mice were trained from day 1–4, and tested on day 5–17, with all measurements recorded and plotted as time remained on the rod versus day tested.
3.2. MS/MS Analysis Reveals High Number of True Positive Proteins Identified
The initial data linking SIRT2 to neurocognitive ability prompted us to explore the possibility that SIRT2 could be important in mediating radiation-induced neurotoxicity and impaired cognition. Using Sirt2 WT and KO mice as a model system, we designed an experiment to evaluate the proteomic changes in brain following whole brain radiotherapy (WBRT). Figure 2 depicts the experimental setup for our study. Six month old female WT and KO mice (n = 6/group) were randomly selected and divided into two subgroups (composed of three mice in each subgroup). One subgroup served as a sham-irradiated (untreated) control set, and the other subgroup was subjected to whole brain irradiation delivered as 20 Gy in a single fraction. After 72 h, the mice were sacrificed by decapitation, and whole brains were isolated. Because iTRAQ methodology allows for quantitative analysis to changes in the proteome of heterogeneous tissues, brain proteins were extracted, digested with trypsin, pooled by condition, and iTRAQ-labeled as described in the Materials and Methods. Figure S1A shows the details of the statistics regarding protein identification by LC–MS/MS. As mentioned in Materials and Methods, MS/MS analysis revealed 815 independent proteins in the data set meeting both criteria of 95% confidence (unused peptide score of 1.3) and the local FDR estimation of <5%. The quality of the proteomic data was also tested by analyzing distributions of signals of all eight tags and protein ratios. Both parameters were found to be distributed normally (Figure S1B,C). Figure S1D represents the Volcano plot that demonstrates the significant changes of the proteomic ratios of replicate data. This was done using a target decoy approach. On the basis of analysis of significant p values calculated by ProteinPilot for 116/121 ratios (two samples that should not show differences), all ratios having a p value threshold below 0.001 have a 0% estimated probability of being false-positive changes, and all ratio p values below 0.01 have at worst a 10% estimated probability of being false-positive changes; that is, the 5% FDR threshold p value is somewhere between p < 0.01 and 0.001. Together, these data suggest that the MS/MS analysis yielded highly significant data for changes in protein levels.
Figure 2.
Schematic diagram depicting the experimental setup for iTRAQ labeling and analysis.
3.3. Acute Global Alterations at the Protein Level Show a Unique and Distinct Pattern in Irradiated Brain
The proteomic data were divided into four groups (Figure S2A) as follows: WTRT:WTCT (where average values of wild-type (WT) mice treated with radiation were divided by WT untreated controls); KOCT:WTCT (where average values of Sirt2 KO untreated controls were divided by WT untreated controls); and KORT:KOCT (where average values of Sirt2 KO mice treated with radiation were divided by Sirt2 KO untreated controls). To normalize samples to the effect of SIRT2 on irradiated brains, we generated the fourth comparison between KORT and WTRT groups as follows
A heatmap (Figure S2C) using an unsupervised hierarchical clustering and correlation distance metric, to compare altered protein levels among the four groups after filtering and processing of the iTRAQ data as depicted in Figure S2A, was generated (total numbers in all four groups: 233). The order of proteins from top to bottom in the heatmap is depicted in Supplementary Table 1, with red in the heatmap indicating increased expression and blue indicating decreased expression of proteins. Each group has a unique and distinct pattern, but interestingly, the clustering dendrogram displays the highest similarity between WTRT:WTCT and KOCT:WTCT (r = 0.99). This suggests that genomic deletion of Sirt2 alters the brain proteome in similar manner to radiation treatment of WT mice.
3.4. Immunoblots Independently Confirm the iTRAQ Data and Analysis
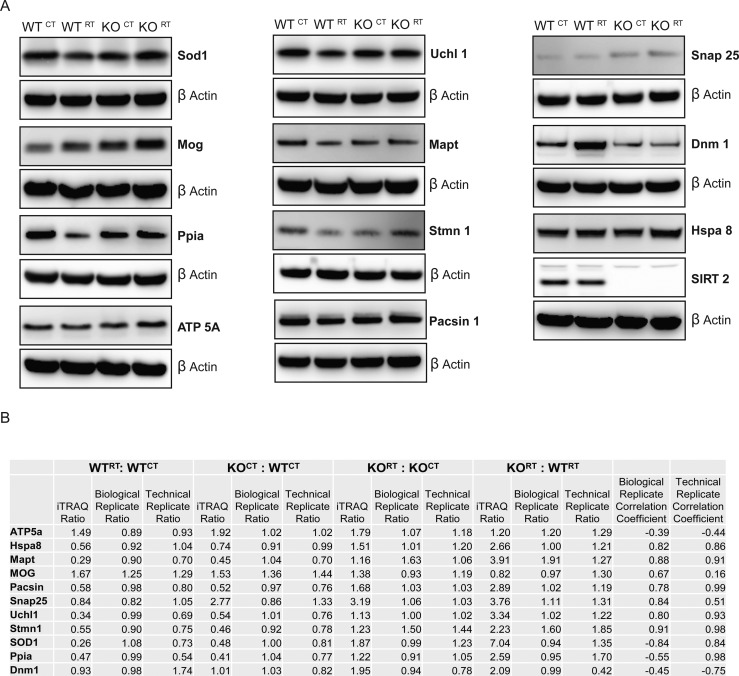
After filtering the iTRAQ data as described in the Materials and Methods section, we specifically selected key proteins involved in several neurological diseases and disorders and independently verified the iTRAQ results, derived from pooled technical replicates, by immunoblot. For equal loading control, we initially probed with an anti-β actin antibody; however, to rule out any discrepancy in our densitometric analysis due to any possible change in β actin level following radiation treatment, we also tested nitrocellulose membranes for equal proteins in each lane by staining with Pierce reversible protein stain followed by densitometric analysis. Analysis showed that levels of β actin remained unchanged among all four samples (Figure S3).45,46 With few exceptions, the majority of the selected proteins, Hspa 8, Stmn 1, Uchl 1, Mapt, Mog, Snap 25, Pacsin 1, and SIRT2, exhibited the same expression pattern by immunoblot as obtained via iTRAQ analysis (Figure 3A), with Sirt2 knock out samples serving as a negative control. To independently validate the results of the technical replicates, we performed a biological replicate experiment and analyzed for the same selected proteins. Densitometry of the immunoblots from the pooled technical replicates submitted for iTRAQ analysis and the biological replicates was performed using ImageJ. Pearson correlation coefficients were calculated and compared with the original iTRAQ ratios (Supplementary Table 2 and Figure 3B). There were 9 of 11 proteins that demonstrated identical correlation to the iTRAQ data, yielding 81.8% agreement in correlation between the technical and biological replicates. Two proteins demonstrated disconcordant results between the technical and biological replicates, Sod1 and Ppia. Pearson correlation analysis of protein profiles with agreement between immunoblot and iTRAQ data ranged from 0.16 to 0.98. As expected, iTRAQ revealed a significant multifold decreased expression level of SIRT2, confirmed by immunoblot of protein samples from KOCT: WTCT (Figure 3A). With those two exceptions, immunoblots of technical and biological replicates independently confirmed that the iTRAQ analysis was a highly valid data set that reliably demonstrated the direction and magnitude of protein level changes that were induced by the various conditions.
Figure 3.
Confirmation of iTRAQ data by immunoblot analysis. (A) Confirmation of candidate proteins obtained through MS/MS analysis and after filtration of data (as described in Materials and Methods). (B) Correlation between iTRAQ ratios, technical replicate validation densitometry, and biological replicate densitometry presented as ratios of the immunoblot band intensities. Pearson correlation coefficients were calculated for comparison of biological replicate versus iTRAQ and technical replicate versus iTRAQ, respectively.
3.5. Significantly Altered Protein Levels Contribute to Several Key Biologic Functions
After a stringent selection of the iTRAQ data, we identified 233 proteins with significantly (p < 0.05) altered expression levels in the different groups (Figure S2A). Figure 4A displays the number of proteins (upregulated and downregulated) in each group. Using Venn diagram analysis as depicted in Figure 4B (proteins are highlighted in Supplementary Table 1), 7 unique proteins were identified with altered expression only between the WTRT and WTCT groups, 5 unique proteins with altered expression only between the KOCT and WTCT groups, and 12 unique proteins with altered expression only between the KORT and KOCT groups. The molecular, cellular, and disease-related functional roles of the 233 significant proteins were further explored by IPA software. Figure 4C illustrates the percent contribution of different proteins within the biologic function of each group. Proteins with altered expression levels exhibit several biological and cellular functional classes within each group. There were 20, 24, 25, and 61 proteins, respectively, identified to each treatment comparison that were found to be associated with neurological disease and remained a focus of our interest for further analysis (Figure 4C and Supplementary Tables 3–6).
Figure 4.
Analysis of significantly altered proteins based on functional groups and roles in disease states. (A) Graph showing numbers of proteins with altered expression among different comparison groups. (B) Venn diagram showing unique proteins with altered expression levels and common proteins in all four groups. (C) Graph depicting top biological functions (including molecular, cellular, and disease-related) and their percent contribution to each group. Digits mentioned on the side of each bar represent the number of proteins involved in each molecular and cellular function.
3.6. IPA Analysis Exhibits Several Disease-Associated Canonical Signaling Pathways
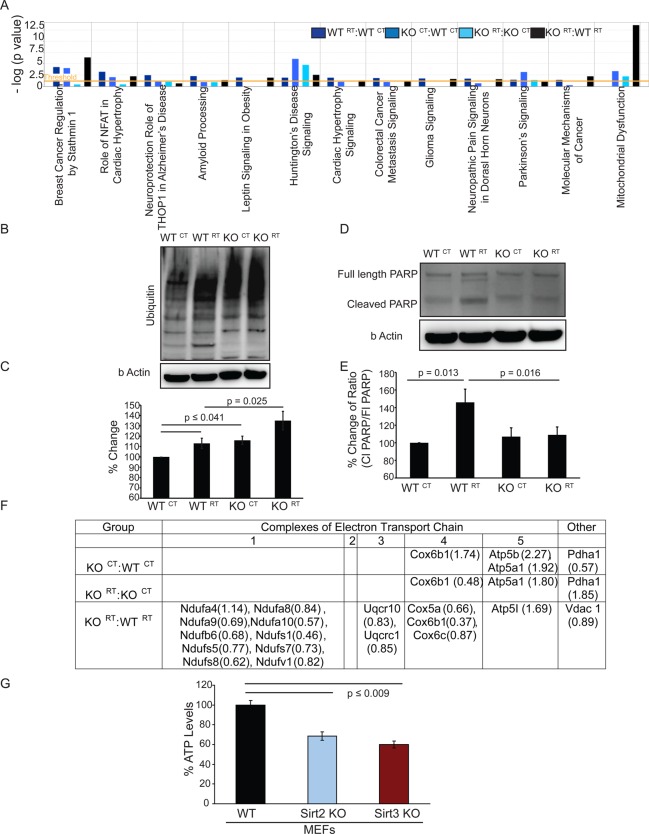
IPA analysis exploring the disease-oriented canonical signaling pathways resulted in multiple pathways after comparing the significant protein level changes observed between the four experimental groups (Figure 5A); however, our focus remained to analyze pathways associated with neurodegenerative disorders like Parkinson’s, Huntington’s, Alzheimer’s, and mitochondrial dysfunction. Radiation treatment of WT mice induced canonical signaling pathways such as the neuroprotective role of THOP1 (thimet oligopeptidase 1) and amyloid processing in Alzheimer’s disease more significantly compared with other groups, indicating a major effect of radiation toward Alzheimer’s disease-related signaling pathways in the acute phase following radiation exposure (Figure 5A). Also, in the WT RT:WT CT comparison, Mapt (microtubule-associated protein tau) was downregulated.
Figure 5.
(A) Ingenuity Pathway Analysis of canonical pathway signaling related to disease and disorder in all four groups. (B) Representative immunoblot of ubiquitylation present in whole brain lysates used for iTRAQ analysis. (C) Relative expression of total ubiquitin levels by densitometric analysis of three different blots. (D) Representative immunoblot of uncleaved and cleaved Parp in whole brain lysates used for iTRAQ analysis. (E) Relative ratios of cleaved PARP/full length PARP by densitometric analysis of three different blots. (F) Table showing constituent proteins of the electron transport chain with altered expression. Numbers in parentheses denote ratios of altered expression. (G) ATP levels were assayed in Sirt2 WT and KO MEFs using CellTiter-Glo-2.0. Analysis was done in triplicate, with error bars denoting ± SD of the mean.
Canonical pathway analysis shows that radiation treatment also acutely affects proteins related to movement disorders such as Huntington’s and Parkinson’s diseases, shown when WTRT:WTCT samples are compared (Figure 5A). Expression levels of Pacsin 1 (protein kinase C and casein kinase substrate in neurons 1), part of the Huntington’s canonical pathway, were altered. Uchl1 (ubiquitin carboxyl–terminal esterase L1), a key protein in the Parkinson’s canonical signaling pathway, was found to be present at lower levels in whole brain tissue after radiation treatment (Supplementary Table 3 and Figure 3). As depicted in Figure 5B, ubiquitylation increases acutely following 20 Gy WBRT in wild-type brain samples and is moderately higher in Sirt2 knock out tissues. Densitometric analysis (Figure 5C) reveals a 10–15% increase in ubiquitylation in WTRT and KOCT samples when compared with WTCT, and the ubiquitylation level in KORT brain samples was ∼10% higher than WTRT.
Huntington’s and Parkinson’s canonical signaling pathways are significantly affected upon loss of Sirt2 (Figure 5A). In the Huntington’s canonical pathway, Pacsin1 and Snap 25 were found to be significantly altered. Uchl 1, which has been discussed previously (Figure 5B), was found to be decreased in Sirt2 KO brain samples, and decreased Uchl 1 hydrolase activity could potentially contribute to the observed increase in ubiquitylation.
The Huntington’s canonical pathway was most significantly affected following WBRT of Sirt2 KO mice. Unique proteins in this group with altered expression levels include Hspa 8 (heat shock 70 kDa protein 8). PARP, a maker of apoptosis, is downstream of Septin 5. Therefore, increased Septin 5 levels would be hypothesized to be neurotoxic and would theoretically be linked to increased apoptosis;47 however, in the absence of Sirt2, cleaved PARP is not increased in response to radiation, suggesting that radiation-induced apoptosis is blunted (Figure 5D,E).
Growing evidence has revealed that mitochondrial dysfunction is a key factor in the progression of many neurological disorders.48,49 Although SIRT2 is not a mitochondrial-associated Sirtuin isoform, our results demonstrate that loss of Sirt2 affects proteins in pathways related to mitochondrial dysfunction (Figure 5A). Pyruvate dehydrogenase, which is decreased in Sirt2 KO tissue samples (Figure 5F), is present inside the mitochondrial matrix and catalyzes oxidative decarboxylation of pyruvate to form acetyl-CoA, a key component for the initiation of Krebs cycle. Loss of Sirt2 also results in altered expression levels in components of the electron transport chain (ETC) (Figure 5F). Specifically, we observed increased levels of cytochrome c oxidase (Cox 6b) a factor of complex 4 of the ETC. Interestingly, our data shows decreased ATP levels (almost 50%, p ≤ 0.009) in Sirt2 KO cells (Figure 5G). This confirms that the absence of Sirt2 has a functional effect on the ETC. Likewise, when the Sirt2 KO brain is exposed to radiation, proteins associated with mitochondrial dysfunction were very significantly affected in comparison with radiation-exposed control animals (KORT:WTRT comparison, Figure 5A, black bar). Therefore, it appears that SIRT2 targets complexes of the ETC and makes cells susceptible toward radiation-induced mitochondrial dysfunction.
3.7. Common Protein Profile Changes Exist Following Whole Brain Radiation and Loss of Sirt2
The heat map analysis in Figure S2C illustrates similar proteins whose levels are changing comparing KO versus control (wild-type) animals (KOCT:WTCT) and by comparing radiation treated versus untreated control (wild-type) animals (WTRT:WTCT). Therefore, we examined whether proteins previously associated with neurologic disorders were also similarly changing between the two comparison groups. A Venn diagram (Figure S4A) illustrates 8 proteins whose levels were altered by WBRT in the Sirt2 wild-type animals (and not altered in the Sirt2 KO animals without WBRT) and 12 proteins whose levels were altered in the Sirt2 knock-out animals compared with wild-type animals (and not altered by WBRT in the wild-type animals). There were additional proteins that were similarly altered either by WBRT in wild-type animals or in Sirt2 KO animals without WBRT (Mog, Stmn 1, Uchl 1, Mapt, and Pacsin 1) (Figure S4B). The changes in the levels of these five latter proteins thus reflect a protein signature in the brain that is common between Sirt2 KO and irradiated WT animals.
3.8. Whole Brain Radiation Induces a Specific and Unique Protein Signature
To compare unique proteins that are up- or downregulated by radiation in either Sirt2 WT or KO phenotypes, we examined proteins associated with neurological disorders by comparing WTRT:WTCT and KORT:KOCT. There were 11 unique proteins whose levels were changed only with radiation exposure in wild-type brains and 16 unique proteins whose levels were changed only with radiation exposure in Sirt2 knock out brains (Figure S4C). However, nine proteins showed level changes following WBRT in either WT or Sirt2 KO mice. One of these nine common proteins, Mog, had identical expression patterns, suggesting a specific change in expression of these proteins acutely after radiation treatment and suggesting a specific protein signature exists following radiation to the brain, irrespective of the presence or absence of Sirt2. In contrast, other proteins identified as changing level following WBRT (Alb, Tpi1, Mapt, Sptan1, Pacsin1 and Sod1) do not follow the same expression patterns (Figure S4D).
3.9. Long-Term Radiation Effects in the Brain Are Associated with Altered Levels of a Small Number of Key Proteins
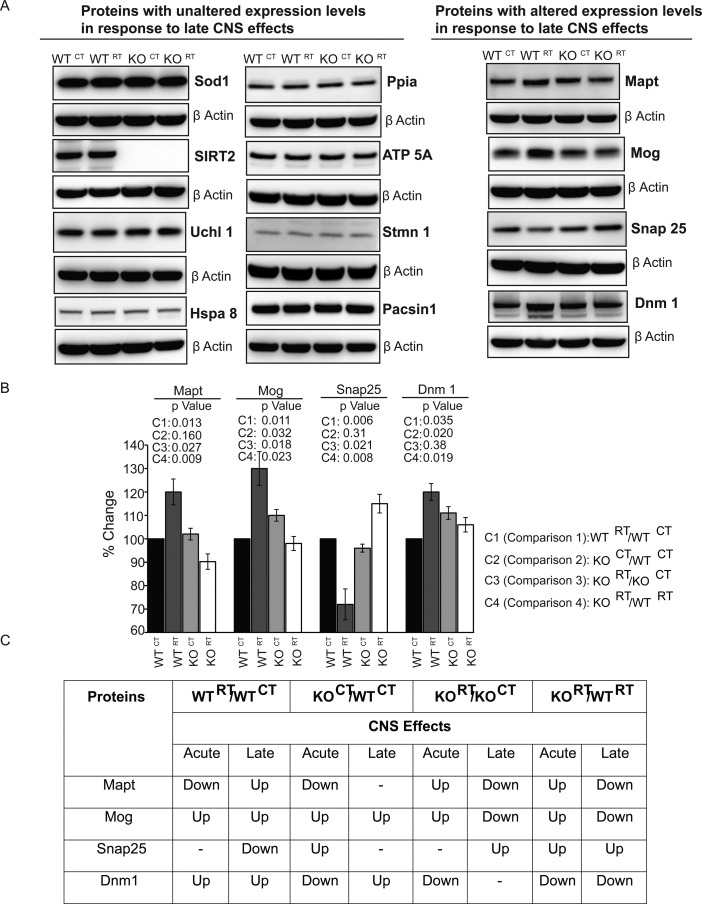
A duplicate set of WT and Sirt2 KO mice was treated identically to the mice in the acute radiation effects study above, but brain lysates were extracted from these animals 1 year after receiving a single dose of 20 Gy to the entire brain rather than after 72 h. These lysates were analyzed by immunoblot to identify which proteins from the protein signature observed in the acute phase following whole brain radiation remained up or down-regulated permanently (Figure S4D). While most proteins reverted back to their preradiation baseline over time, a small subset of proteins demonstrated expression levels that did not revert back to baseline at 1 year following whole brain radiation (Figure 6A–C). These proteins were identified as Mapt, Mog, Snap25, and Dnm1. These results suggest that these few key proteins may participate in driving long-term radiation toxicity in the brain.
Figure 6.
Candidate proteins significant for delayed radiation changes in the brain proteome. (A) Representative immunoblots of whole brain lysates from animals 1 year after WBRT (20 Gy x 1 fraction). (B) Densitometric analysis of Mapt, Mog, Snap25, and Dnm1. P values are listed for each densitometric analysis for each indicated protein and each indicated comparison. (C) Table listing comparative expression profiles between acute and delayed response for selected proteins. (up = upregulated; down = downregulated in the respective comparisons).
3.10. SIRT2 Influences Key Proteins in Neurodegenerative Diseases Mainly via Indirect Modes
This proteomic analysis identified several key proteins involved in various neurodegenerative processes that may be involved in radiation-induced neurotoxic effects. Considering the importance of SIRT2 in this regard, we then explored the possible relationship between SIRT2 and the altered proteome obtained in KOCT:WTCT (highlighted with * in Figure 7). Network analysis using the String 9.1 program shows that none of the proteins are directly associated with SIRT2. Interestingly, the majority of proteins are associated with ubiquitin C, which is a precursor of polyubiquitin. Ubiquitin is involved mainly in targeting proteins for degradation through polyubiquitylation as well as in intracellular protein trafficking and activation through monoubiquitylation and alternate branching polyubiquitylation. This analysis shows that SIRT2 communicates with the altered proteome via SIRT1, FOXO1, TP3, and tubulin. The possibility of regulation of deacetylase activity of SIRT1 and or expression by SIRT2 cannot be ruled out. Interestingly, both SIRT1 and SIRT2 seem to act in a coordinated way.
Figure 7.
Network analysis of SIRT2 with proteins associated in various neurodegenerative disorders obtained in whole brain tissue sample of (KOCT:WTCT) using the String 9.1 program.
4. Discussion
Improving the efficacy and safety of RT for treatment of primary and metastatic tumors is dependent on having a better understanding of the mechanisms that drive acute and transient versus long-term, irreversible changes in the brain following radiation exposure. Because of the largely unexplored nature of this problem and the fact that there is not a homogeneous in vitro model to address this problem, we sought to analyze the brain proteome using iTRAQ as a methodology to reveal changes that occur in heterogeneous living tissue in an attempt to more accurately analyze biologic changes from WBRT. Additionally, we were interested in examining the role of the SIRT2 deacetylase on the brain proteome and its effect on radiation-induced proteomic changes within the brain. It should be noted that changes affecting specific regions cannot be detected in whole brain and future studies are needed to localize critical compartments. As such, an ongoing project is to determine the proteomic changes specifically within the neocortex and hippocampus following WBRT.
Preliminarily, we established that SIRT2 is important for neurocognitive ability in adult, middle-aged mice. This is important because it more accurately represents the stage of development where most adult patients receive brain radiation treatment and because SIRT2, the predominant sirtuin isoform in the CNS, increases expression with age34 and mutations in SIRT2 may lead to neurodegenerative phenotypes in rodents.50−53 For the first time, we have examined the proteome during the acute response following whole brain radiation to have a global insight toward long-term neurotoxicity and cognitive dysfunction using a murine model system.
WBRT of wild-type mice did indeed induce canonical signaling pathways of Alzheimer’s disease, indicating a major effect of WBRT in the acute phase toward Alzheimer’s disease-related signaling pathways. The THOP1 pathway has been shown to have a very crucial role in the clearance of amyloid beta and colocalizes with Mapt in neurofibrillary tangles;54 however, canonical pathway analysis also demonstrated that whole brain radiation treatment acutely affects proteins related to other neurodegenerative disorders, such as Huntington’s and Parkinson’s diseases. Expression levels of Pacsin 1, which is part of the Huntington’s canonical pathway, was altered. Likewise, Uchl1, a key protein in Parkinson’s canonical signaling, was found to be decreased in brain tissue 72 h after radiation treatment (Supplementary Table 3 and Figure S2).
Huntington’s and Parkinson’s canonical signaling pathways are significantly affected upon loss of Sirt2. Pacsin1 and Snap 25 (synaptosomal-associated protein 25 kDa) were found to have altered expression levels. Likewise, Uchl 1, decreased in Sirt2 KO tissue samples, was associated with increased ubiquitylation, suggesting a protective effect for dopaminergic neurons.47 Although elucidating the mechanism of how Sirt2 affects regulation of downstream effectors of Parkinson’s signaling is beyond the scope of the current investigation, we hope future investigations will more fully define their relationship.
Not surprisingly given previous data, the Huntington’s canonical pathway was found to be most significantly affected by WBRT in Sirt2 KO mice. In addition to Pacsin 1 and Snap25, Hspa 8 (heat shock 70 kDa protein 8) was found to have altered protein levels. One explanation as to why Sirt2 loss affects the brain proteome may rest with our observation that protein complexes within the ETC are likely altered by the loss of Sirt2, thereby increasing susceptibility to radiation-induced mitochondrial dysfunction.
iTRAQ analysis has revealed the presence of several important protein “signatures”. The loss of Sirt2 appears to proteomically mimic the effect of WBRT on wild-type animals. Therefore, it stands to reason that pharmacologic induction of SIRT2 activity in WT animals might mitigate some, but not all, of the proteomic changes induced by WBRT and if successful could prove to be a platform for future studies in human patients. Proteins that are associated with neurologic disorders and follow the same expression pattern in both Sirt2 KO and radiation treatment groups were identified as Mog, Stmn 1, Uchl 1, Mapt, and Pacsin 1. This suggests a proteomic link between radiation-induced changes in the brain and SIRT2 levels. Additionally, a specific and unique protein signature was discovered following radiation to the brain, irrespective of the presence or absence of Sirt2. Mog displayed an identical protein expression pattern following WBRT in both Sirt2 wild-type and KO mice.
Lastly, we sought to identify which brain proteins with altered levels in the acute phase following WBRT still had altered levels many months after exposure and therefore might be participating in, or correlated with, long-term phenotypic changes in the brain following WBRT. At 1 year following WBRT, Mapt, Mog, Snap25, and Dnm1 were found to have expression levels that did not revert back to baseline over time, suggesting that a few key proteins may participate in driving late radiation effects in the brain. Mapt is believed to colocalize with THOP1 in neurofibrillary tangles.54 Mog (myelin oligodendrocyte glycoprotein) is associated with the structural integrity of myelin,55 and Snap25 controls exocytosis of synaptic vesicles.56 Dnm1 (dynamin 1) is a GTPase responsible for endocytosis inside the cell,57 and in Huntington’s disease it is localized inside perinuclear inclusions along with other molecules like Hsp 70, cathepsin D, synuclein, huntingtin, Hsp 40, and Atp synthase.58
While these protein signatures require confirmation in human brain samples, these data represent an important first step in understanding the mechanisms behind acute and long-term changes in the brain following therapeutic radiation exposure and suggest a potentially critical role of Sirt2 in this process. Our investigation has revealed that long-term sequelae of CNS-induced radiation changes are likely not the result of a single disease process, such as the canonical Alzheimer’s signaling pathway, but instead are the consequence of disruptions in multiple neurodegenerative pathways, suggesting novel avenues to explore toxicity mitigation.
Acknowledgments
We gratefully acknowledge Anne Stanley and other members of the Proteomics and Mass Spectrometry Core Research Facility of the College of Medicine, Pennsylvania State University, Hershey, PA for technical assistance. This work was supported by the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute, project number ZIA BC 011222: CNS sirtuins in radiation-induced vascular permeability and neurodegeneration.
Glossary
Abbreviations
- SIRT
sirtuin
- WT
wild-type
- KO
knockout
- CNS
central nervous system
- RIPA
radioimmunoprecipitation assay
- SCX
strong cation exchange chromatography
- RP
reverse phase
- IPA
Ingenuity Pathway Analysis
- FDR
false discovery rate
- iTRAQ
isobaric tag for relative and absolute quantitation
- SILAC
stable isotope labeling by amino acids in cell culture
- ETC
electron transport chain
- WBRT
whole brain radiation therapy
- Gy
gray
- MEF
mouse embryonic fibroblast
- NCBI
National Center for Bioinformatics
- THOP1
thimet oligopeptidase 1
- Mapt
microtubule associated protein tau
- Pacsin 1
protein kinase C and casein kinase substrate in neurons 1
- Uchl1
ubiquitin carboxyl–terminal esterase L1
- Mog
myelin oligodendrocyte glycoprotein
- Snap 25
synaptosomal-associated protein 25 kDa
- Stmn 1
stathmin 1
- Dnm1
dynamin 1
- Hspa 8
heat shock 70 kDa protein 8
- PARP
poly ADP ribose polymerase
- Cox 6b
cytochrome c oxidase
- Alb
albumin
- Sptan1
spectrin alpha nonerythrocytic-1
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b00083.
Supplementary Figure S1: Statistics and quality of proteomic data obtained after mass spectrometry acquisition. Supplementary Figure S2: Filtering and processing methodology of iTRAQ data and Variability between replicates. Supplementary Figure S3: Total protein staining for equal loading control validation. Supplementary Figure S4: Comparison of proteins associated with neurological disorders. (PDF)
Supplementary Table 1: Order of proteins in heatmap (for Figure S2C). Supplementary Table 2: Immunoblot densitometry of biological replicate samples. Supplementary Table 3: Proteins associated with neurological disorders in WTRT:WTCT. Supplementary Table 4: Proteins associated with neurological disorders in KOCT:WTCT. Supplementary Table 5: Proteins associated with neurological disorders in KORT:KOCT. Supplementary Table 6: Proteins associated with neurological disorders in KORT:WTRT. (PDF)
Supplementary Table 7: Detailed IPA report of WTRT/WTCT. (XLS)
Supplementary Table 8: Detailed IPA report of WTRT/WTCT. (XLS)
Supplementary Table 9: Detailed IPA report of WTRT/WTCT. (XLS)
Supplementary Table 10: Detailed IPA report of WTRT/WTCT. (XLS)
Author Present Address
§ S.S.: Department of Biochemistry, Amity School of Applied Sciences, Amity University, Gurgaon-122413, India.
The authors declare no competing financial interest.
Supplementary Material
References
- Greene-Schloesser D.; Robbins M. E.; Peiffer A. M.; Shaw E. G.; Wheeler K. T.; Chan M. D. Radiation-induced brain injury: A review. Front. Oncol. 2012, 2, 73. 10.3389/fonc.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A. Z.; Choi M. Radiation-associated toxicities in the treatment of high-grade gliomas. Semin. Oncol. 2014, 41 (4), 532–40. 10.1053/j.seminoncol.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Begum N.; Wang B.; Mori M.; Vares G. Does ionizing radiation influence Alzheimer’s disease risk?. J. Radiat. Res. 2012, 53 (6), 815–22. 10.1093/jrr/rrs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant T. E.; Pollack I. F.; Loeffler J. S. Brain tumors across the age spectrum: biology, therapy, and late effects. Semin. Radiat. Oncol. 2010, 20 (1), 58–66. 10.1016/j.semradonc.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane N.; Quaghebeur G. CNS effects following the treatment of malignancy. Clin. Radiol. 2012, 67 (1), 61–8. 10.1016/j.crad.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Botturi M.; Fariselli L. Clinical results of unconventional fractionation radiotherapy in central nervous system tumors. Tumori 1998, 84 (2), 176–87. [DOI] [PubMed] [Google Scholar]
- Sano K.; Morii K.; Sato M.; Mori H.; Tanaka R. Radiation-induced diffuse brain injury in the neonatal rat model - Radiation-induced apoptosis of oligodendrocytes. Neurologia Medico-Chirurgica 2000, 40 (10), 495–500. 10.2176/nmc.40.495. [DOI] [PubMed] [Google Scholar]
- Li Y. Q.; Jay V.; Wong C. S. Oligodendrocytes in the adult rat spinal cord undergo radiation-induced apoptosis. Cancer Res. 1996, 56 (23), 5417–5422. [PubMed] [Google Scholar]
- Vandermaazen R. W. M.; Verhagen I.; Kleiboer B. J.; Vanderkogel A. J. Radiosensitivity of Glial Progenitor Cells of the Perinatal and Adult-Rat Optic-Nerve Studied by an Invitro Clonogenic-Assay. Radiother. Oncol. 1991, 20 (4), 258–264. 10.1016/0167-8140(91)90125-Z. [DOI] [PubMed] [Google Scholar]
- Yuan H.; Gaber M. W.; Boyd K.; Wilson C. M.; Kiani M. F.; Merchant T. E. Effects of fractionated radiation on the brain vasculature in a murine model: blood-brain barrier permeability, astrocyte proliferation, and ultrastructural changes. Int. J. Radiat. Oncol., Biol., Phys. 2006, 66 (3), 860–6. 10.1016/j.ijrobp.2006.06.043. [DOI] [PubMed] [Google Scholar]
- Cammer W. Effects of TNF alpha on immature and mature oligodendrocytes and their progenitors in vitro. Brain Res. 2000, 864 (2), 213–219. 10.1016/S0006-8993(00)02178-8. [DOI] [PubMed] [Google Scholar]
- Shaw E. G.; Rosdhal R.; D’Agostino R. B. Jr.; Lovato J.; Naughton M. J.; Robbins M. E.; Rapp S. R. Phase II study of donepezil in irradiated brain tumor patients: effect on cognitive function, mood, and quality of life. J. Clin. Oncol. 2006, 24 (9), 1415–20. 10.1200/JCO.2005.03.3001. [DOI] [PubMed] [Google Scholar]
- Rapp S. R.; Case L. D.; Peiffer A.; Naughton M. M.; Chan M. D.; Stieber V. W.; Moore D. F. Jr.; Falchuk S. C.; Piephoff J. V.; Edenfield W. J.; Giguere J. K.; Loghin M. E.; Shaw E. G. Donepezil for Irradiated Brain Tumor Survivors: A Phase III Randomized Placebo-Controlled Clinical Trial. J. Clin. Oncol. 2015, 33 (15), 1653–9. 10.1200/JCO.2014.58.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordo D. Structure and evolution of human sirtuins. Curr. Drug Targets 2013, 14 (6), 662–5. 10.2174/1389450111314060007. [DOI] [PubMed] [Google Scholar]
- Vassilopoulos A.; Fritz K. S.; Petersen D. R.; Gius D. The human sirtuin family: evolutionary divergences and functions. Hum. Genomics 2011, 5 (5), 485–96. 10.1186/1479-7364-5-5-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L.; Houtkooper R. H.; Auwerx J. NAD(+) metabolism: a therapeutic target for age-related metabolic disease. Crit. Rev. Biochem. Mol. Biol. 2013, 48 (4), 397–408. 10.3109/10409238.2013.789479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. Y.; Yuan H.; Su L. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. OncoTargets Ther. 2013, 6, 1399–1416. 10.2147/OTT.S37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G.; Outeiro T. F. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol. Med. 2013, 5 (3), 344–52. 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto J.; Miura T.; Shimamoto K.; Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004, 556 (1–3), 281–6. 10.1016/S0014-5793(03)01444-3. [DOI] [PubMed] [Google Scholar]
- Ramadori G.; Lee C. E.; Bookout A. L.; Lee S.; Williams K. W.; Anderson J.; Elmquist J. K.; Coppari R. Brain SIRT1: anatomical distribution and regulation by energy availability. J. Neurosci. 2008, 28 (40), 9989–96. 10.1523/JNEUROSCI.3257-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Zhang B.; Tang J.; Cao Q.; Wu Y.; Wu C.; Guo J.; Ling E. A.; Liang F. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J. Neurosci. 2007, 27 (10), 2606–16. 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Zhou Y. G.; Mueller-Steiner S.; Chen L. F.; Kwon H.; Yi S. L.; Mucke L.; Li G. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappa B signaling. J. Biol. Chem. 2005, 280 (48), 40364–40374. 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- Kim D.; Nguyen M. D.; Dobbin M. M.; Fischer A.; Sananbenesi F.; Rodgers J. T.; Delalle I.; Baur J. A.; Sui G.; Armour S. M.; Puigserver P.; Sinclair D. A.; Tsai L. H. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007, 26 (13), 3169–79. 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G.; Wang D.; Cohen D. E.; Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 2010, 142 (2), 320–32. 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li Y.; Xu W.; McBurney M. W.; Longo V. D. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008, 8 (1), 38–48. 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G.; Arun A.; Chung C. Y.; McLean P. J.; Lindquist S.; Guarente L. SIRT1 Protects against alpha-Synuclein Aggregation by Activating Molecular Chaperones. J. Neurosci. 2012, 32 (1), 124–132. 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jeong H.; Cohen D. E.; Cui L. B.; Supinski A.; Savas J. N.; Mazzulli J. R.; Yates J. R.; Bordone L.; Guarente L.; Krainc D. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat. Med. 2011, 18 (1), 159–165. 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M.; Wang J.; Fu J.; Du L.; Jeong H.; West T.; Xiang L.; Peng Q.; Hou Z.; Cai H.; Seredenina T.; Arbez N.; Zhu S.; Sommers K.; Qian J.; Zhang J.; Mori S.; Yang X. W.; Tamashiro K. L.; Aja S.; Moran T. H.; Luthi-Carter R.; Martin B.; Maudsley S.; Mattson M. P.; Cichewicz R. H.; Ross C. A.; Holtzman D. M.; Krainc D.; Duan W. Neuroprotective role of Sirt1 in mammalian models of Huntington’s disease through activation of multiple Sirt1 targets. Nat. Med. 2011, 18 (1), 153–8. 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro T. F.; Kontopoulos E.; Altmann S. M.; Kufareva I.; Strathearn K. E.; Amore A. M.; Volk C. B.; Maxwell M. M.; Rochet J. C.; McLean P. J.; Young A. B.; Abagyan R.; Feany M. B.; Hyman B. T.; Kazantsev A. G. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 2007, 317 (5837), 516–9. 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- Liu L.; Arun A.; Ellis L.; Peritore C.; Donmez G. Sirtuin 2 (SIRT2) enhances 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced nigrostriatal damage via deacetylating forkhead box O3a (Foxo3a) and activating Bim protein. J. Biol. Chem. 2012, 287 (39), 32307–11. 10.1074/jbc.C112.403048. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Luthi-Carter R.; Taylor D. M.; Pallos J.; Lambert E.; Amore A.; Parker A.; Moffitt H.; Smith D. L.; Runne H.; Gokce O.; Kuhn A.; Xiang Z. M.; Maxwell M. M.; Reeves S. A.; Bates G. P.; Neri C.; Thompson L. M.; Marsh J. L.; Kazantsev A. G. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (17), 7927–7932. 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. M.; Balabadra U.; Xiang Z. M.; Woodman B.; Meade S.; Amore A.; Maxwell M. M.; Reeves S.; Bates G. P.; Luthi-Carter R.; Lowden P. A. S.; Kazantsev A. G. A Brain-Permeable Small Molecule Reduces Neuronal Cholesterol by Inhibiting Activity of Sirtuin 2 Deacetylase. ACS Chem. Biol. 2011, 6 (6), 540–546. 10.1021/cb100376q. [DOI] [PubMed] [Google Scholar]
- Bobrowska A.; Donmez G.; Weiss A.; Guarente L.; Bates G. SIRT2 Ablation Has No Effect on Tubulin Acetylation in Brain, Cholesterol Biosynthesis or the Progression of Huntington’s Disease Phenotypes In Vivo. PLoS One 2012, 7 (4), e34805. 10.1371/journal.pone.0034805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell M. M.; Tomkinson E. M.; Nobles J.; Wizeman J. W.; Amore A. M.; Quinti L.; Chopra V.; Hersch S. M.; Kazantsev A. G. The Sirtuin 2 microtubule deacetylase is an abundant neuronal protein that accumulates in the aging CNS. Hum. Mol. Genet. 2011, 20 (20), 3986–3996. 10.1093/hmg/ddr326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis L. M.; Delattre J. Y.; Posner J. B. Radiation-induced dementia in patients cured of brain metastases. Neurology 1989, 39 (6), 789–96. 10.1212/WNL.39.6.789. [DOI] [PubMed] [Google Scholar]
- Wang X. F.; Zhang S.; Ye Y. H.; Chen Y. P.; Liu X. Y. Clinicopathologic features of delayed radiation-induced brain injury after radiotherapy for brain tumor. Zhonghua Bing Li Xue Za Zhi 2012, 41 (4), 224–8. 10.3760/cma.j.issn.0529-5807.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Gaber M. W.; Sabek O. M.; Fukatsu K.; Wilcox H. G.; Kiani M. F.; Merchant T. E. Differences in ICAM-1 and TNF-alpha expression between large single fraction and fractionated irradiation in mouse brain. Int. J. Radiat. Biol. 2003, 79 (5), 359–66. 10.1080/0955300031000114738. [DOI] [PubMed] [Google Scholar]
- Wilson C. M.; Gaber M. W.; Sabek O. M.; Zawaski J. A.; Merchant T. E. Radiation-induced astrogliosis and blood-brain barrier damage can be abrogated using anti-TNF treatment. Int. J. Radiat. Oncol., Biol., Phys. 2009, 74 (3), 934–41. 10.1016/j.ijrobp.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Mahmoud-Ahmed A. S.; Atkinson S.; Wong C. S. Early gene expression profile in mouse brain after exposure to ionizing radiation. Radiat. Res. 2006, 165 (2), 142–54. 10.1667/RR3485.1. [DOI] [PubMed] [Google Scholar]
- Chow B. M.; Li Y. Q.; Wong C. S. Radiation-induced apoptosis in the adult central nervous system is p53-dependent. Cell Death Differ. 2000, 7 (8), 712–20. 10.1038/sj.cdd.4400704. [DOI] [PubMed] [Google Scholar]
- Tang W. H.; Shilov I. V.; Seymour S. L. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 2008, 7 (9), 3661–7. 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- Mi H.; Guo N.; Kejariwal A.; Thomas P. D. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007, 35 (Database issue), D247–52. 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov I. V.; Seymour S. L.; Patel A. A.; Loboda A.; Tang W. H.; Keating S. P.; Hunter C. L.; Nuwaysir L. M.; Schaeffer D. A. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 2007, 6 (9), 1638–55. 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Frank-Cannon T. C.; Tran T.; Ruhn K. A.; Martinez T. N.; Hong J.; Marvin M.; Hartley M.; Trevino I.; O’Brien D. E.; Casey B.; Goldberg M. S.; Tansey M. G. Parkin deficiency increases vulnerability to inflammation-related nigral degeneration. J. Neurosci. 2008, 28 (43), 10825–34. 10.1523/JNEUROSCI.3001-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C. P.; Marz S. X.; Reiss R.; Schulenborg T.; Friauf E. Epicocconone staining: a powerful loading control for Western blots. Proteomics 2014, 14 (2–3), 162–8. 10.1002/pmic.201300089. [DOI] [PubMed] [Google Scholar]
- Gilda J. E.; Gomes A. V. Stain-Free total protein staining is a superior loading control to beta-actin for Western blots. Anal. Biochem. 2013, 440 (2), 186–8. 10.1016/j.ab.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J. H.; Kawamata H.; Yoo M. S.; Kim D. J.; Lee Y. K.; Kim S.; Dawson T. M.; Zhang H.; Sulzer D.; Yang L.; Beal M. F.; Degiorgio L. A.; Chun H. S.; Baker H.; Peng C. Neurotoxicity and behavioral deficits associated with Septin 5 accumulation in dopaminergic neurons. J. Neurochem. 2005, 94 (4), 1040–53. 10.1111/j.1471-4159.2005.03257.x. [DOI] [PubMed] [Google Scholar]
- Pathak D.; Berthet A.; Nakamura K. Energy Failure: Does It Contribute to Neurodegeneration?. Ann. Neurol. 2013, 74 (4), 506–516. 10.1002/ana.24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L.; Motherwell M. S. The impact of reactive oxygen species and genetic mitochondrial mutations in Parkinson’s disease. Gene 2013, 532 (1), 18–23. 10.1016/j.gene.2013.07.085. [DOI] [PubMed] [Google Scholar]
- Suzuki K.; Koike T. Mammalian Sir2-related protein (SIRT) 2-mediated modulation of resistance to axonal degeneration in slow Wallerian degeneration mice: a crucial role of tubulin deacetylation. Neuroscience 2007, 147 (3), 599–612. 10.1016/j.neuroscience.2007.04.059. [DOI] [PubMed] [Google Scholar]
- Green K. N.; Steffan J. S.; Martinez-Coria H.; Sun X.; Schreiber S. S.; Thompson L. M.; LaFerla F. M. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J. Neurosci. 2008, 28 (45), 11500–10. 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Z. Z.; Maiese K. Enhanced tolerance against early and late apoptotic oxidative stress in mammalian neurons through nicotinamidase and sirtuin mediated pathways. Curr. Neurovasc Res. 2008, 5 (3), 159–70. 10.2174/156720208785425666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L.; Mucke L. Paths of convergence: sirtuins in aging and neurodegeneration. Neuron 2008, 58 (1), 10–4. 10.1016/j.neuron.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollio G.; Hoozemans J. J. M.; Andersen C. A.; Roncarati R.; Rosi M. C.; van Haastert E. S.; Seredenina T.; Diamanti D.; Gotta S.; Fiorentini A.; Magnoni L.; Raggiaschi R.; Rozemuller A. J. M.; Casamenti F.; Caricasole A.; Terstappen G. C. Increased expression of the oligopeptidase THOP1 is a neuroprotective response to A beta toxicity. Neurobiol. Dis. 2008, 31 (1), 145–158. 10.1016/j.nbd.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Quarles R. H. Myelin sheaths: glycoproteins involved in their formation, maintenance and degeneration. Cell. Mol. Life Sci. 2002, 59 (11), 1851–1871. 10.1007/PL00012510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P. P.; Battenberg E.; Wilson M. C. SNAP-25 and synaptotagmin involvement in the final Ca(2+)-dependent triggering of neurotransmitter exocytosis. Proc. Natl. Acad. Sci. U. S. A. 1996, 93 (19), 10471–6. 10.1073/pnas.93.19.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A. E.; Kurihara L. J.; Vallee R. B.; Rose M. D. Dnm1, a Dynamin-Related Gene, Participates in Endosomal Trafficking in Yeast. J. Cell Biol. 1995, 130 (3), 553–566. 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waelter S.; Boeddrich A.; Lurz R.; Scherzinger E.; Lueder G.; Lehrach H.; Wanker E. E. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 2001, 12 (5), 1393–1407. 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.