Abstract
Intraflagellar transport (IFT) is a motile process critical for building most cilia, including those of mammalian cells. Defects in IFT lead to short or missing cilia, and in animals can cause defects in development, for example in hedgehog mediated signaling, as well as disease symptoms such as polycystic kidney disease or retinal degeneration. Understanding how IFT works is thus a high priority in ciliary biology. Imaging of living cells has played a key role in understanding the mechanism of IFT, and this is particularly the case in mammalian cells where biochemical analysis of IFT is extremely difficult, due to the difficulty of isolating cilia away from the rest of the cell. Imaging IFT in living mammalian cells requires solution to several problems: constructing cell lines that express fluorescent protein tagged IFT proteins, obtaining cell populations with a high degree of ciliation, confocal or TIRF imaging with sufficient time resolution and signal to noise ratio to observe the majority of IFT particles as they travel back and forth inside the cilium, and analyzing the image data to extract quantitative measurements of IFT. We describe optimized solutions to each of these technical challenges. Using the approaches described here, mammalian cultured cells become powerful platforms for quantitative analysis of IFT dynamics.
Keywords: primary cilium, IFT, TIRF, live-imaging
Introduction
The assembly of cilia and flagella (i.e. ciliogenesis) is a molecular assembly process of tremendous complexity (reviewed in Ishikawa and Marshall, 2011; Avasthi and Marshall, 2012). Ciliogenesis in most cell types relies on Intraflagellar transport (IFT), a kinesin-based motile process that takes place within cilia and flagella (Rosenbaum and Witman, 2002; Scholey 2003; Cole 2003; Pedersen and Rosenbaum 2008; Qin 2012; Bhogaraju et al., 2013a). During IFT, a set of proteins called IFT proteins associate into a multi-protein complex called an IFT particle. These particles dock onto the basal body (Deane et al., 2001) and then enter the interior of the cilium where they self-assemble into linear arrays known as IFT trains (Pigino et al., 2009). These trains are moved processive from the base to the tip of the cilium driven by kinesin-2. At the tip, the trains re-arrange into smaller trains which are then pulled by cytoplasmic dynein back to the base. The movement of the IFT trains out to the tip is called “anterograde” transport, and their movement back to the base is called “retrograde” transport. IFT is thought to be necessary for building and maintaining cilia, and the IFT proteins are able to bind and transport cargo proteins (Qin et al., 2004; Hao et al., 2011; Bhogaraju et al., 2013b; Ishikawa et al., 2014). The IFT system shows a tremendous degree of dynamics, with train size, speed, and frequency all being variable in a dynamic fashion even within a single cilium (Engel et al., 2009; Ludington et al., 2013), and it is thought that the regulation of IFT dynamics may play an important role in regulating the length of cilia and flagella (Wemmer and Marshall, 2007).
Imaging has played a central role in the discover and understanding of IFT. Indeed, the initial discovery of IFT resulted from video enhanced DIC microscopy of Chlamydomonas flagella, in which it was seen that blob-like objects appeared to be moving back and forth from base to tip (Kozminski et al., 1993; Iomini et al., 2001). Following the biochemical identification of the IFT proteins (Cole 1998), live imaging was used to show that the same motile process occurred in C. elegans (Orozco et al., 1999) and in mammalian cells (Follitt et al., 2006). Imaging continues to be a central tool in studies of IFT, and with the growing interest in the biology of mammalian cilia, inspired by interest in ciliopathies, there is a pressing need for reliable methods to image and analyze IFT in living mammalian cells. Imaging IFT in mammalian culture cells requires that several technical hurdles must be overcome. First, it is necessary to obtain cells expressing fluorescent tagged IFT protein constructs. This step benefits from powerful expression construct systems for mammalian cells. Second, it is necessary to grow cells under conditions in which a large fraction of cells contain cilia. Degree of ciliation can be quite variable in cultured cells and care must be taken to ensure sufficient ciliated cells are available, in order to avoid spending most of ones time in a futile search for something to image. Third, it is necessary to image the living cells under conditions that allow the fast-moving IFT particles to be reliably detected with high enough signal to noise to allow interpretation of the images but without bleaching the signal over the timecourse of a typical experiment. Collection of live cell image data of IFT has benefitted greatly from recent advances in digital camera technology, particularly the widespread adoption of emCCD cameras. Finally, the image data needs to be analyzed to extract meaningful quantitative measures of IFT. Without such measurements, imaging is just an exercise in making pretty pictures. Quantitative measurements, in contrast, allow rigorous statistical analysis of differences between samples, for example to determine whether a particular gene knockdown experiment perturbs the system. We present a relatively simple analysis based on freely available kymograph software.
1. Preparation of cells for IFT imaging
When IFT was first discovered, it was imaged in Chlamydomonas flagella using differential interference contrast (DIC) microscopy. Generation of contrast in DIC is accomplished by sending two parallel beams of light through the sample with different polarization and with a slight offset, and then recombining them to detect differences in optical path length between the two offset beams. Because of its mode of contrast generation, DIC microscopy is highly sensitive to any variation in refractive index within a sample, even outside of the imaging plane, since such variations lead to wavefront distortion. DIC works well with Chlamydomonas flagella because these flagella stick to the glass coverslip and extend well beyond the cell body, so that they can be imaged by DIC without any cellular structures overlying the flagellum itself. This would not work for mammalian cells, which are shorter than the cell radius so that any field of view that contains the cilium also contains a portion of the cell body. The mammalian cell body is full of refractile organelles that will scatter light and introduce wavefront distortions into the DIC light path. It is thus generally not possible to see cilia by DIC on mammalian cultured cells, let alone resolve tiny IFT particles in motion. A second disadvantage of DIC microscopy is that while it is able to show the position of objects, it does not provide information about the size of small objects and cannot be used to quantify the size of IFT trains. For these reasons, it is necessary to image IFT in mammalian cilia using fluorescence microscopy, in which one or more IFT proteins are tagged with a fluorescent protein like GFP, allowing them to be detected and tracked over a cell background that is hopefully much less intense. Thus the first important challenge is to identify cell lines that express specific fluorescent markers of IFT. Unless sufficient attention is paid to this first step, subsequent imaging attempts will not be successful.
To observe IFT movement in mammalian primary cilia efficiently, many cells must have cilia with high-expression of fluorescent-tagged IFT protein. Although strictly speaking IFT could be visualized even in a sparsely ciliated cell population, the time required to search for cells that have cilia can be substantial. In contrast, when the majority of cells in a population are ciliated, it becomes much easier to find cells for imaging. For mammalian cells, the key to obtaining a large fraction of ciliated cells is to control the cell cycle state. Formation of cilia is tightly coupled with the cell cycle and occurs in interphase during the cell cycle (Plotnikova, Pugacheva, & Golemis, 2009; Wheatley, 1971). Especially in quiescent G0 phase, most cells can form primary cilia. In order to introduce cells into G0 phase, cells are cultured until confluence so that cell proliferation should be suppressed by contact inhibition. Moreover, it is known that serum starvation induce ciliogenesis (Tucker, Pardee, & Fujiwara, 1979). These culture conditions increase the frequency of cilia.
For optimal imaging of IFT, it is important that many cells stably and highly express an IFT protein fused with a fluorescent tag. Therefore, we use enhanced green fluorescent protein (EGFP), which is relatively high-intensity and photostable among fluorescent proteins, for a fluorescent tag of IFT. An EGFP-tagged IFT protein (IFTGFP) is stably expressed by the CAG promoter, a high-expression promoter (Niwa, Yamamura, & Miyazaki, 1991). The stable cell lines were selected by expression level of IFT-GFP.
We have observed IFT behavior in several cell lines including mIMCD-3, NIH3T3, hTERT-RPE1 and MDCK cells. Here, we describe a method for mIMCD-3 cells, but our method can be broadly applicable to ciliated mammalian cell lines.
1.1 Materials
mIMCD-3 cells (ATCC: CRL-2123) that are stably expressing EGFP-tagged IFT protein (IFT-GFP)
Dulbecco's modified Eagle's high-glucose 50%/F-12 50% mix, medium (DMEM/F-12; UCSF Cell Culture Facility)
Heat inactivated fetal bovine serum (FBS; UCSF CCF)
DMEM/F-12, no phenol red (Gibco)
0.25% Trypsin with EDTA in Saline A (trypsin/EDTA ; UCSF CCF)
6.5-mm Transwell with 0.4 μm pore polyester membrane insert, TC-treated (Corning)
35-mm glass bottom dish, thickness: No. 1.5 (P35G-1.5-14-C, MatTek)
Sterile humid chamber (we use 150-mm TC-treated culture dish; Corning)
1.2 Procedure
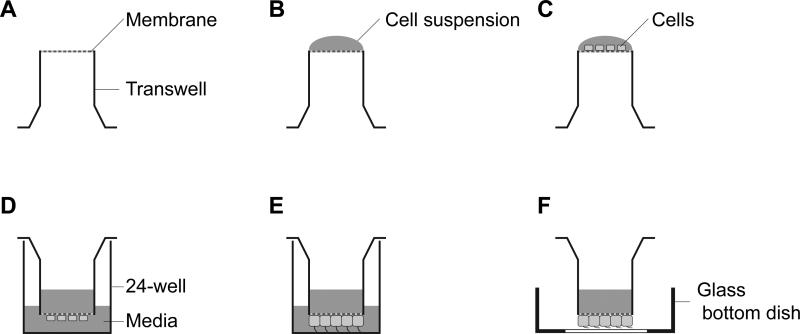
A transwell cup is put upside down in a humid sterile chamber (Figure 1A).
IFT-GFP expressing mIMCD-3 cells are dissociated with trypsin/EDTA, washed with the DMEM/F-12 medium with 10% FBS and resuspended with the medium at 2 × 105 cells/mL.
Seed 50 μL of the cell suspension (1 × 104 cells) on the top of the inverted transwell cup (Figure 1B).
Incubate the cup in a humid sterile chamber for 3 hours at 37°C in 5% CO2 so that the cells attach to the membrane of the transwell cup.
The transwell cup is placed in the 24-well plates and 100 μL and 800 μL of media are added into the transwell cup and the 24-well, respectively.
Incubate the 24-well plate with transwell cup for 2~3 days at 37°C in 5% CO2 until 90% confluency. Cell confluency in the transwell can be checked by an inverted light microscope, as is the case with normal culture plate.
Change culture media to serum-free DMEM/F-12 without phenol red and incubate around 24 hours for uniform cilia formation.
Take the transwell cup with IFT-GFP expressing mIMCD-3 cells from the 24-well plate by tweezers and place the transwell cup on the center of the 35-mm glass bottom dish (transwell imaging chamber; Figure 1F).
Figure 1. Sample preparation for imaging IFT in mammalian primary cilia.
(A) The transwell cup is placed upside down in a humid sterile chamber. (B) Put 50 μL of the cell suspension (1 × 104 cells) on the membrane of the inverted transwell cup. (C) Culture cells for 3 hours at 37°C in 5% CO2. (D) The transwell cup is transferred to 24-well plate and media add into the transwell cup and the 24-well. (E) Culture cells until 90% confluency. (F) Put the transwell cup on a 35-mm glass bottom dish for imaging. We call it transwell imaging chamber.
2. Live imaging of IFT
Once cell lines have been obtained in which IFT particles are fluorescent with stable expression and low background, and growth conditions established under which a large fraction of cells are ciliated, the next step is to determine appropriate imaging conditions. Because IFT moves at a speed of approximately 1 μm/s inside of cilia in both directions in mammalian cells (He et al., 2014; Ishikawa et al., 2014), IFT imaging needs to acquire images at a fast enough rate that the motion of the trains does not cause image blurring. Moreover, although the size of IFT trains varies depending on the length of cilia, quantitative measurements of IFT trains in Chlamydomonas flagella show that the number of copies of each IFT protein in a train is small, usually less than 10 (Engel, Ludington, & Marshall, 2009; Ludington, Wemmer, Lechtreck, Witman, & Marshall, 2013). Thus, the number of photons emitted by a train per unit time is small. Normally one would image dim samples using long exposures to increase signal to noise (since the scales as the square root of the average number of photons per pixel), but for IFT the exposure time is limited by the fact that imaging needs to be relatively fast. Therefore, it is necessary to use a fast and sensitive camera for imaging IFT. Recent semiconductor manufacturing technology has developed and brought a new generation of image sensors, such as charge-coupled device (CCD), electron multiplying CCD (EMCCD), complementary metal-oxide-semiconductor (CMOS), and scientific CMOS. Most of these are good enough for imaging IFT providing the sensitivity is high enough. We use an EMCCD camera (iXon3, Andor Technology), which is specifically designed to provide high sensitivity for low light imaging.
We have observed IFT behavior using a wide-field, spinning disk confocal and total internal reflection fluorescence (TIRF) microscopy. Compared to standard wide-field microscopy, Spinning disk confocal and TIRF microscopy are more suitable for imaging IFT because of less bleaching of fluorescent signal and better signal-to-noise by omitting out-of-focus light. In this section, we describe IFT imaging method in mammalian primary cilia using a TIRF microscope.
2.1 Microscope setup
Eclipse Ti Microscope with Laser TIRF system (Nikon)
Apo TIRF 100× magnification, 1.49 numerical aperture (NA) oil objective (Nikon)
iXon3 EMCCD camera (Andor Technology)
Top stage incubator (Oko lab)
2.2 IFT imaging
The TIRF microscope must be adequately set up according to the manufacture's instruction. To maximize the spatial resolution of the digital images by ensuring that the image data is sampled at a frequency equaling or exceeding the Nyquist rate, we usually choose extra 1.5× magnification by the intermediate magnification selector. Temperature of the top stage incubator is set at 37°C and 5% CO2 with humid air is supplied to the incubator.
Put a drop of immersion oil (Refractive index ND = 1.515) on the Apo TIRF 100×/1.49 oil objective.
Place the imaging chamber with cells (from Section 1) on the stage of the top stage incubator.
Adjust focus on primary cilia.
Decrease TIRF angle until the signal from cell body is lost (Figure 2).
Acquire images at 100 ms intervals for 1 min without binning.
Save the time-lapse image sequence as TIFF (Tagged Image File Format) file format.
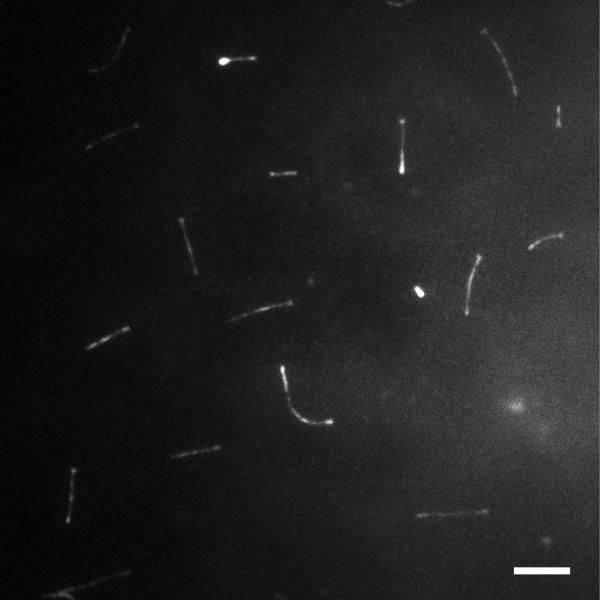
Figure 2. Image of cilia using transwell imaging chamber.
A representative image of mIMCD-3 cells expressing IFT88-EGFP on the transwell imaging chamber using a TIRF microscope. Most cilia are visible in a single plane. Bar: 5 μm.
Note: To reduce the effect of photo bleaching on IFT trains, we minimize the laser power for imaging. The cells can maintain a healthy state more than an hour in this imaging condition. We check the orientation of cilia every time from the nucleus position and cell shape by adjusting focus when we imaged them. Alternatively, the cilia orientation can be detected by expressing additional marker, such as a centrosomal protein fused with a red fluorescent protein. It is necessary to know the actual pixel size (μm/pixel) and image acquisition rate (frames per second; fps) for calculation of IFT velocity. We recommend that these image acquisition parameters be carefully recorded at every imaging session, especially when using microscopes in a shared access or core facility, where changes to the imaging setup can sometime occur.
3. Image processing and data analysis
Prior quantitative analysis of IFT in model systems such as Chlamydomonas has shown that even a simple characterization of IFT requires measurement of three key parameters: IFT speed, IFT frequency, and IFT train size (Engel et al., 2009). IFT frequency is defined as the frequency with which an IFT train passes a given point going in a given direction. Careful quantitative measurements show that IFT frequency and size are inversely correlated (Ludington et al., 2013). More detailed statistical analyses of IFT have been conducted which show that beyond simply measuring the overall frequency, it may in some cases be important to measure the distribution of time-intervals between successive IFT trains (Ludington et al., 2013). Without quantitative measurements of IFT, only crude comparisons are possible, so that only very extreme phenotypes such as complete loss of IFT may be detected. In contrast, quantitative analyses are able to detect more subtle effects such as the increase in total IFT entry into cilia when kinase pathways are activated (Ludington et al., 2013). For this reason it is important to have a way to conduct similar quantitative measurements on living mammalian cultured cells.
To measure the speed of IFT-GFP, we use kymographs. In a kymograph, a single line is imaged at sequential time points, and then these individual line images are stacked up to make a 2D image. Kymographs are a powerful and convenient tool for visualizing linear motion of particle-like objects, such as IFT trains, because a particle moving at s constant speed produces a straight line in a kymograph. For this reason, kymographs have long been used for visualizing intracellular motion. The advantage of a kymograph is two-fold. For one thing, time-lapse data is collapsed onto a single image, so that it is not necessary to view movies in order to visualize the motion. For another, the human eye is very good at picking out lines even in the presence of substantial noise, so that by making a kymograph, it can be much easier to see exactly how many moving particles are present in the cilium.
Kymographs can be generated from the time-lapse image sequence using image analysis software, such as MetaMorph (Molecular Devices), NIS-Elements (Nikon) and ImageJ (NIH). In this section, we explain how to make kymograph and measure IFT speed using free ImageJ software for two reasons. First, because it is a free software package, everyone can reproduce the method without paying additional cost. ImageJ can be downloaded from http://imagej.nih.gov/ij/. Second, because it is based on Java, ImageJ is platform-independent and so images can be analyzed in exactly the same way on any computer.
3.1 Make kymographs
To make kymograph in ImageJ, download the ImageJ plugin (KymoResliceWide) from the website (http://imagej.net/KymoResliceWide) and copy it into the “plugins” folder in the “ImageJ” folder in your computer.
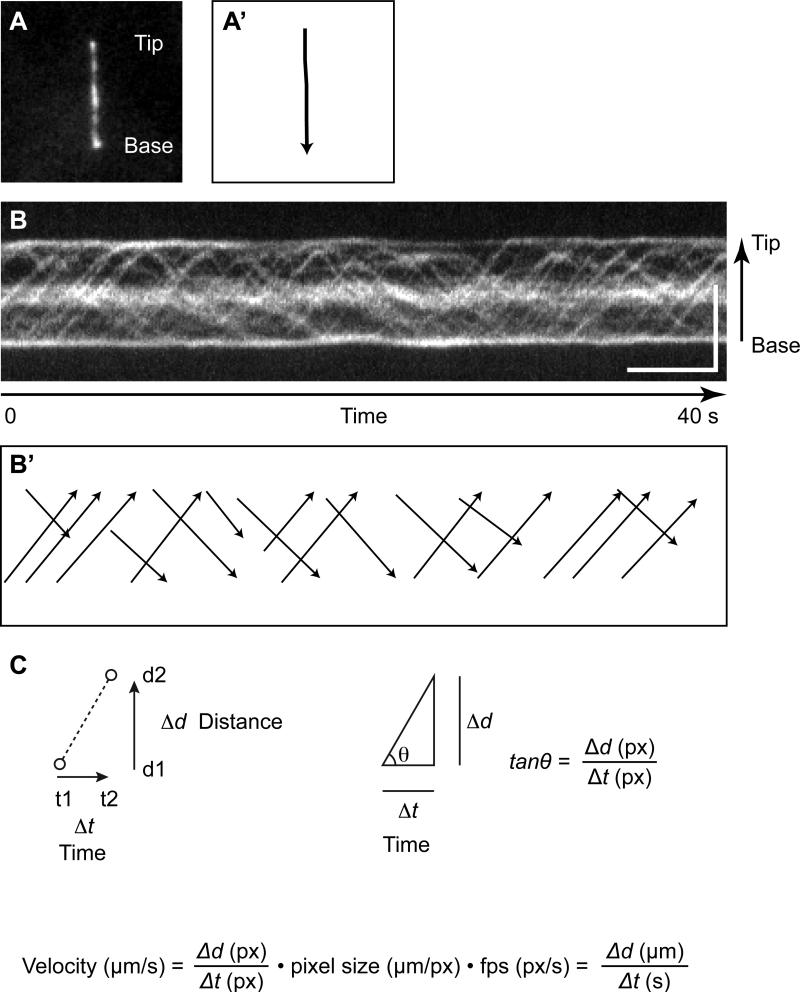
Open the image file with ImageJ. If the image sequence is composed of multiple files, open image files from the menu using File → Import → Image sequence... (Figure 3A).
Select the “Segmented Line” tool by right-clicking the Line tool in the Toolbar, start from the tip of the cilium and draw a segmented line along with the cilium (double click when finished) (Figure 3A’).
Adjust line width to fit with the thickness of the cilium by double clicking the Line tool or from the menu Edit → Option → Line Width...
Run the KymoResliceWide plugin to create a kymograph from Plugins → IFT Kymograph. Select “Maximum” from the pull down menu of Intensity value across width, check the box of “Rotate 90 degrees” and then click “OK”.
The Kymograph will then be shown in a new window. The horizontal axis of the kymograph shows time and the vertical axis shows the distance of IFT trains movement along with the cilium (Figure 3B).
Figure 3. Kymograph analysis for measuring IFT velocity.
(A) An image of IFT-88EGFP in the primary cilium of mIMCD-3 cells from Movie 1. (B) Kymographs of IFT-88EGFP moving in the cilium of mIMCD-3 cells. The kymographs was assembled from Movie 1. Bars: 5 s (horizontal), 5 μm (vertical). (A’ and B’) Draw lines along with the cilium or trajectories of IFT trains for making a kymograph and measuring the angle of the trajectory, respectively. (C) The IFT velocity is given from the angle of trajectory of IFT train.
3.2 Measure IFT velocity
Making a kymograph is just the first step in quantitative analysis. Once the kymograph is in hand, it can be analyzed to extract measures of IFT dynamics, for example speed, frequency, and size. Speed is the easiest parameter to measure and the one that we focus on here. An image sequence is converted to a kymograph to measure the velocity of IFT trains. In this case, the kymograph shows the locus of IFT-GFP along with the cilium over time (Figure 3B). When an IFT train moves distance Δd (from d1 to d2) during the time Δt (from t1 to t2), the speed of the IFT train is shown Δd/Δt (Figure 3C). Therefore, the speed of the IFT train is calculated by tanθ (θ is the angle of the trajectory of the IFT train against the horizontal axis). Because the unit of tanθ is given in pixel, the unit can be converted into μm/s by multiplying pixel size and fps (Figure 3D).
Select the “Straight Line” tool from the toolbar, and draw a line along with a straight IFT track in chronological order, i.e. left to right (Figure 3B’).
Measure angle from the menu using Analyze → Measure or press “M” key. You can get the angle against the horizontal axis in degree.
Angle degree is converted to radian by multiplying π/180.
Calculate tanθ to get the velocity of IFT train in unit of pixel.
To calculate the velocity of IFT train in actual unit, multiply tanθ by pixel size (μm/px) and fps (px/s).
4. Discussion
The imaging of individual IFT components fused to a fluorescent protein has been performed in a variety of ciliated organisms, such as Chlamydomonas, Caenorhabditis elegans, Trypanosoma brucei, Xenopus and mammals (Absalon et al., 2008; Brooks & Wallingford, 2012; Follit, Tuft, Fogarty, & Pazour, 2006; Orozco et al., 1999; Qin, Wang, Diener, & Rosenbaum, 2007). Interestingly, the velocity of IFT varies with the variety of model organisms. For example, the anterograde IFT velocity is 2.5 μm/s in Chlamydomonas whereas C. elegans show anterograde IFT trains moving at 0.7 μm/s in the middle segment of sensory cilia (Engel et al., 2009; Snow et al., 2004). These differences might just depend on the ability of the motor protein that drives IFT trains. However, it seems that there is a correlation between the length of cilia and speed of IFT.
IFT trains in mammalian cells sometimes stop and accumulate in the middle of the cilium (Figure 3B). This kind of IFT behavior is not seen in Chlamydomonas flagella.
The imaging of IFT in mammalian primary cilia presented here can be combined with a genetic analysis, such as siRNA or targeted gene knockout using CRISPR. We expect that this combination of molecular genetics with live cell image analysis will ultimately shed new light on the regulation and mechanism of IFT mediated cargo transport. Analyzing IFT in mutants or knockdown lines defective in ciliopathy disease genes holds the promise of revealing key mechanisms in the pathogenesis of human ciliopathies.
Much of the work on IFT in mammalian cells to date has concentrated on verifying and following up discoveries first made in other model organisms, such as Chlamydomonas or C. elegans. However, the availability of large collections of eGFP tagged constructs and siRNA libraries for mammalian cells means that we can harness the existing infrastructure for mammalian cell biology to probe IFT mechanisms in a systematic manner. In particular, the ease with which new fluorescent tagged protein expressing lines can be generated in mammalian cells compared to Chlamydomonas reinhardtii, in which stable expression is still difficult to achieve, is a particular advantage of the mammalian cell culture system, provided care is taken to optimize imaging conditions as we have outlined in this chapter.
Supplementary Material
Acknowledgement
We thank Kurt Thorn, DeLaine Larsen, and the UCSF Nikon Imaging Center for assistance with TIRF microscope imaging. We also thank Susanne Rafelski and Ben Engel for technical advice on ImageJ macro and calculation of IFT velocity, respectively. This work was supported by the National Institutes of Health grants R01 GM097017 and GM077004 (W.F.M).
References
- Absalon S, Blisnick T, Kohl L, Toutirais G, Doré G, Julkowska D, Tavenet A, Bastin P. Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Molecular Biology of the Cell. 2008;19(3):929–944. doi: 10.1091/mbc.E07-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Marshall WF. Stages of ciliogenesis and regulation of ciliary length. Differentiation. 2012;83(2):S30–42. doi: 10.1016/j.diff.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Engel BD, Lorentzen E. Intraflagellar transport complex structure and cargo interactions. Cilia. 2013a;2:10. doi: 10.1186/2046-2530-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamia S, Bastin P, Nigg EA, Lorentzen E. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013b;341(6149):1009–12. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB. Control of vertebrate intraflagellar transport by the planar cell polarity effector Fuz. The Journal of Cell Biology. 2012;198(1):37–45. doi: 10.1083/jcb.201204072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141(4):993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic. 2003;4(7):435–42. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11(20):1586–90. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Engel BD, Ludington WB, Marshall WF. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. The Journal of Cell Biology. 2009;187(1):81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Molecular Biology of the Cell. 2006;17(9):3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Thein M, Brust-Mascher I, Civelekoglu-Scholey G, Lu Y, Acar S, Prevo B, Shaham S, Scholey JM. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat. Cell Biol. 2011;13(7):790–8. doi: 10.1038/ncb2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Kapoor TM, Anderson KV. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nature Cell Biology. 2014;16(7):663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 2001;153(1):13–24. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nature Rev. Cell Mol. Biol. 2011;(12):222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ide T, Yagi T, Jiang X, Hirono M, Sasaki H, Yanagisawa H, Wemmer KA, et al. TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. eLife. 2014;3:e01566. doi: 10.7554/eLife.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U.S.A. 1993;90(12):5519–23. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludington WB, Wemmer KA, Lechtreck KF, Witman GB, Marshall WF. Avalanche-like behavior in ciliary import. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(10):3925–3930. doi: 10.1073/pnas.1217354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Orozco JT, Wedaman KP, Signor D, Brown H, Rose L, Scholey JM. Movement of motor and cargo along cilia. Nature. 1999;398(6729):674. doi: 10.1038/19448. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption, and signaling. Curr. Top. Dev. Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Pigino G, Geimer S, Lanzavecchia S, Paccagnini E, Cantele F, Diener DR, Rosenbaum JL, Lupetti P. Electron tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 2009;187(1):135–48. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova OV, Pugacheva EN, Golemis EA. Primary cilia and the cell cycle. Methods in Cell Biology. 2009;94:137–160. doi: 10.1016/S0091-679X(08)94007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellear precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164(2):255–66. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Current Biology: CB. 2007;17(3):193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H. Regulation of intraflagellar transport and ciliogenesis by small G proteins. Int. Rev. Cell Mol. Biol. 2012;293:149–68. doi: 10.1016/B978-0-12-394304-0.00010-5. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3(11):813–25. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–43. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MRS, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nature Cell Biology. 2004;6(11):1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17(3):527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- Wemmer KA, Marshall WF. Flagellar length control in Chlamydomonas – a paradigm for organelle size regulation. Int. Rev. Cytol. 2007;260:175–212. doi: 10.1016/S0074-7696(06)60004-1. [DOI] [PubMed] [Google Scholar]
- Wheatley DN. Cilia in cell-cultured fibroblasts. 3. Relationship between mitotic activity and cilium frequency in mouse 3T6 fibroblasts. Journal of Anatomy. 1971;110(Pt 3):367–382. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.