Abstract
Hyperspectral imaging (HSI) is an emerging imaging modality for medical applications. HSI acquires two dimensional images at various wavelengths. The combination of both spectral and spatial information provides quantitative information for cancer detection and diagnosis. This paper proposes using superpixels, principal component analysis (PCA), and support vector machine (SVM) to distinguish regions of tumor from healthy tissue. The classification method uses 2 principal components decomposed from hyperspectral images and obtains an average sensitivity of 93% and an average specificity of 85% for 11 mice. The hyperspectral imaging technology and classification method can have various applications in cancer research and management.
Keywords: Hyperspectral Imaging, Head and neck cancer, Principal Component Analysis (PCA), Support vector machine (SVM), Superpixels, Feature Extraction, Image classification
1. INTRODUCTION
In the United States, there are more than 40,000 new cases of head and neck cancer and more than 8,000 deaths in 2015.1 This malignancy affects the cavities, glands, and sinuses in the head and neck area. Over 90% of head and neck cancer are diagnosed as squamous cell carcinoma.2 Early detection of these malignant tumors is important to improve survival and diagnosis. Traditional diagnosis of oral cancer screening includes a visual inspection using an incandescent light method.3 This method is followed by a biopsy of the suspicious lesion sites. However, the traditional diagnosis can be subjective. Optical spectroscopy has been used for diagnosis of head and neck cancer.4
Optical spectroscopy can detect malignant lesions using information from early biochemical and histological changes.4, 5 Spectroscopy, however, is only a point measurement method while hyperspectral imaging provides additional spatial information. Hyperspectral imaging acquires images at continuous spectral wavelengths and combines spectroscopy and imaging.
Another benefit of HSI is the ability to capture large areas of tissue non-invasively without an external contrast agent.6 Hyerspectral imaging produces large amounts of high dimensional data that becomes challenging to process. Pixel-wise classification considers each pixel independently without spatial relationship of neighboring pixels. Pixel-wise classification is also time consuming.
Superpixels have been used in computer vision and medical applications.7–10 Superpixels divide the image based on coherent regions of arbitrary shapes and sizes creating a local, coherent entity while maintaining overall structure.11 Superpixel based methods can speed up the processing of HSI big data. Furthermore, implementing superpixels with hyperspectral imaging reduces the complexity for classification.
2. EXPERIMENTS AND METHODS
2.1 Hyperspectral Imaging System and Experiments
Reflectance images were acquired using a CRI Maestro in-vivo imaging system (Caliper, Hopkinton, MA). The system consists of a 300 Watt Xenon light source, 12 bit high-resolution charge-coupled device (CCD), solid state liquid crystal filter, and spectrally optimized lens. The hypercube array collected a whole body image with a size of 1,040 × 1,392 and a 2-nm wavelength increment between 450 to 950 nm.12–14 Instead of using the whole body image, a 390 × 435 region of interest was selected around the tumor region, and wavelengths from 450 to 890 nm was used in this study.
A head and neck tumor xenograft model was used in this experiment. Head and neck cancer cells (M4E, 2∗106) with green fluorescence protein (GFP) were injected on the lower back of 12 female mice aged 4-6 weeks.12 Only 11 female mice were analyzed in this study. The animal experiments were approved by the Animal Care and Use Committee of Emory University. Hyperspectral scans were acquired three weeks after tumor cell injection. The mice were anesthetized with 2% isoflurane in oxygen during image acquisition. Reflectance images were acquired without moving the mouse. Autoexposure and reflectance mode were set on the hyperspectral imaging system for the image scans.12
Blue excitation light at 455 nm and blue emission filter 490 nm were used to acquire GFP fluorescence images. Tumor cells expressed green signals in the fluorescence images due to the GFP in the tumor cells. The GFP fluorescence images were utilized as the in-vivo gold standard for evaluation of cancer detection in this study.12 After cervical dislocation of the mice, histological slides were analyzed after image acquisitions to validate cancer diagnosis.
2.2 Overview of the Classification Method
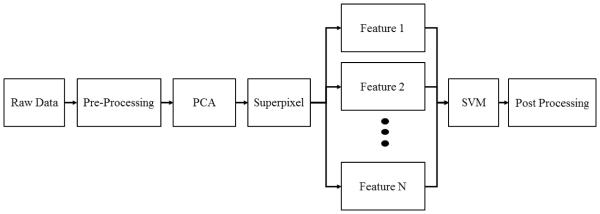
An overview of the algorithm is shown in Figure 1. Reflectance hyperspectral images were collected over multiple wavelengths. Pre-processing steps involved reflectance calibration and curvature correction.15 The pre-processed image cube underwent principal component analysis. In this study, 2 principal component images were used for further analysis. Superpixel segmentation was applied to the 2 principal component images. Within each principal component image, the pixels within each superpixel region were averaged. The average pixel values from principal component 1 image were used as Feature 1. Likewise, the averaged pixel values from principal component 2 image were used as Feature 2. SVM training and classification were applied on Feature 1 and Feature 2. After classification with SVM, the largest connected region classified as tumor was chosen to be the main tumor region. This final classified image was compared to the ground truth image to calculate the sensitivity and specificity.
Figure 1.
The Flow chart of the proposed superpixel-based classification algorithm.
2.3 Data Preprocessing
The entire processing was implemented using MATLAB 2015a (The Math Works Inc., Natick, Massachusetts, USA). A white reference image was acquired with a standard white reference board in the field of view. A dark reference image was acquired with the camera shutter closed. The dark current influence was removed by subtracting the dark image from the raw and white images. To remove the effects of non-uniform illumination, the raw image cube was normalized using the standard white reference.16
| (1) |
2.4 Principal Components
Principal component analysis (PCA) reduces the dimensionality of the hyperspectral image cube. This technique was applied to the pre-processed data before superpixel segmentation, as shown in Figure 1. PCA was applied to the entire image cube. Only the first few principal components that contained the largest variance of information was selected. Instead of analyzing all 220 bands, PCA reduces the data into the most important wavelengths that account for the largest variance in the dataset.17 Using eigenvalue decomposition of the covariance matrix of hyperspectral image cube, the image that corresponds to the largest eigenvalue was selected as the first principal component (Figure 2a). The image that corresponds to the second largest eigenvalue was chosen as the second principal component (Figure 2d). This study applied PCA to the dataset and selected 2 principal components. The number of principal components determines the number of features. One main benefit of PCA is to remove the step of manually choosing specific bands to analyze and perform classification.
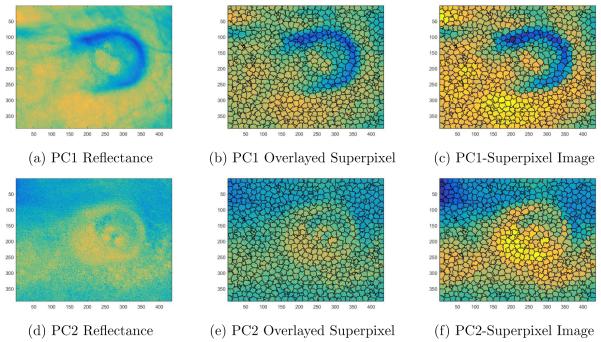
Figure 2.
Illustration of the SLIC superpixel segmentation for Mouse 7. 2a) Principal component 1 (PC1) image from the PCA technique. 2b) Principal component 1 image with superpixel boundary region overlayed. 2c) PC1-superpixel image: Setting each superpixel region in Figure 2b to the average value of each superpixel region. 2d) Principal component 2 (PC2) image from PCA. 2e) Principal component 2 (PC2) image with the same superpixel boundary region overlayed. 2f) PC2-Superpixel image: Setting each superpixel region in Figure 2e to the average value of each superpixel region.
2.5 Superpixel Segmentation
Superpixels are becoming increasingly popular in computer vision applications. Superpixels algorithms combine pixels into meaningful regions. Superpixels capture image redundancy, provide a convenient way to capture image features, and reduce computational loads.18 Different superpixel algorithms exist with different advantages and drawbacks.18 The superpixel algorithm used in this study was the simple linear iterative clustering (SLIC) technique.18 This algorithm is based on k-means clustering approach. Instead of searching the entire image using the standard k-means clustering, the SLIC algorithm searches a limited region around the center of a superpixel. This method was chosen based on the simplicity to generate superpixels and provide better segmentation performance.18 An example of SLIC superpixel segmentation is shown in Figure 2 with the number of superpixels set to 1000. Based on the principal component analysis, the image with the most variance underwent superpixel segmentation. The same superpixel boundaries used for PC1 (Figure 2b) was applied for PC2 (Figure 2e). Within one superpixel region, the average pixel value was calculated and that entire superpixel region was set to the average pixel value as illustrated in Figure 2c and Figure 2f. Only the average value within each superpixel region was used as the data points for the superpivsed learning method in the following section. Because only the average value within each superpixel region was used, the number of data points was reduced to 1000. These principal component images that underwent superpixel segmentation will be called PCA-superpixel images (Figure 2c and Figure 2f).
2.6 Supervised Learning Method
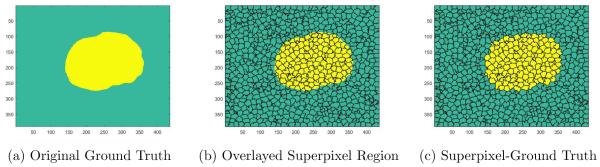
Support vector machine (SVM) classification was used as the supervised learning method. SVM was implemented using the library LIBSVM.19 A leave-one-out method was utilized to train and classify the PCA-superpixel images. For example, 1 mice was used as the testing set and the other 10 mice dataset was used as the training set. This was repeated for all 11 mice with the leave-one-out evaluation method. In order to perform SVM on the superpixel regions, a modified ground truth needed to be created, which will be called superpixel-ground truth. To maintain the same number of data points in both the testing set image and ground truth image, the ground truth image needed to have the same superpixel region. Figure 3a illustrates the original ground truth image segmented from the GFP fluorescence images. Figure 3b illustrates the boundaries of the 1000 superpixel regions. A modified ground truth image needed to be created to maintain the same number of data points in the SVM classifier. If the superpixel region contained values of 1 (tumor), the superpixel region was set as tumor. Likewise, if the superpixel region contained values of 0 (normal tissue), the superpixel region was set as normal tissue. Note that some superpixel regions contain both tumor and normal tissue. If more than half of that superpixel region consisted of tumor tissue, the superpixel-ground truth was labeled as tumor. If the superpixel region contained more normal tissue, the superpixel was labeled as normal tissue. The resulting superpixel-ground truth image is shown in Figure 3c. This ensures that the number of data points in the PCA-superpixel images matches the number of data points in the superpixel-ground truth image.
Figure 3.
Illustration of the modified ground truth for SVM classifier. 3a) Original ground truth image based on fluorescence data. 3b) Original ground truth image with superpixel regions overlayed. 3c) Superpixel-ground truth image for SVM classifier.
Furthermore, an SVM class weighting scheme was used for the choice of C parameter that controls the cost of misclassification. This allowed the SVM to reduce misclassification of the less frequent class because of the smaller number of tumor superpixels compared to normal superpixels. The class weights were determined by the number of superpixel ground truth in each class. The SVM produced a classified binary image of tumor and normal tissue.
2.7 Post-processing step
After classification with SVM, the largest connected region classified as tumor was chosen to be the main tumor region. The smaller areas labeled as tumor but not connected to the main tumor region was reclassified as normal tissue. Furthermore, if a tumor labeled region enclosed a smaller area labeled as normal tissue, the normal tissue was reclassified as tumor.
2.8 Evaluation
In this study, sensitivity and specificity were used as performance metrics for the classification.15, 20–22 Sensitivity measures the proportion of tumor pixels which are correctly classified as tumor. Specificity measures the proportion of normal pixels that are correctly classified as normal. The binary classified image after the post processing step was compared with the ground truth image to measure the sensitivity and specificity. The performance metrics was applied on all 11 mice.
3. RESULTS
The results of the PCA-superpixel-SVM based classifier is shown in Table 1 and Figure 4. For the result using two principal components and superpixels, the superpixel-PCA-SVM based classifier obtained the highest sensitivity and specificity when compared with PCA-SVM and only SVM classifier. The superpixel-PCA-SVM achieved a sensitivity of 93% and a sensitivity of 85%. Furthermore, a comparison of techniques with and without superpixel and PCA is shown in Table 1. The superpixel-PCA-SVM based method improved the average sensitivity by 35% and average specificity by 13% compared to the only SVM classifier. Furthermore, the addition of the superpixel step improved the average sensitivity by 10% and average specificity by 8% when compared to the PCA-SVM classifier.
Table 1.
Comparison of sensitivity and specificity between only SVM classifier, Superpixel-PCA-SVM classifier, and PCA-SVM classifier for the case with 2 principal components.
| Mouse No. | Superpixel-PCA-SVM | SVM | PCA-SVM | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| 1 | 0.84 | 0.91 | 0.41 | 0.73 | 0.72 | 0.82 |
| 2 | 0.99 | 0.84 | 0.77 | 0.71 | 0.90 | 0.77 |
| 3 | 0.96 | 0.88 | 0.47 | 0.73 | 0.80 | 0.78 |
| 4 | 0.69 | 0.89 | 0.30 | 0.64 | 0.65 | 0.71 |
| 5 | 0.99 | 0.77 | 0.55 | 0.72 | 0.88 | 0.72 |
| 6 | 0.99 | 0.84 | 0.79 | 0.80 | 0.95 | 0.79 |
| 7 | 1.00 | 0.91 | 0.88 | 0.75 | 0.89 | 0.75 |
| 8 | 1.00 | 0.84 | 0.64 | 0.70 | 0.94 | 0.74 |
| 9 | 0.98 | 0.72 | 0.45 | 0.64 | 0.76 | 0.69 |
| 10 | 0.80 | 0.92 | 0.60 | 0.77 | 0.78 | 0.88 |
| 11 | 0.93 | 0.84 | 0.49 | 0.71 | 0.81 | 0.78 |
|
| ||||||
| Average | 0.93 | 0.85 | 0.58 | 0.72 | 0.83 | 0.77 |
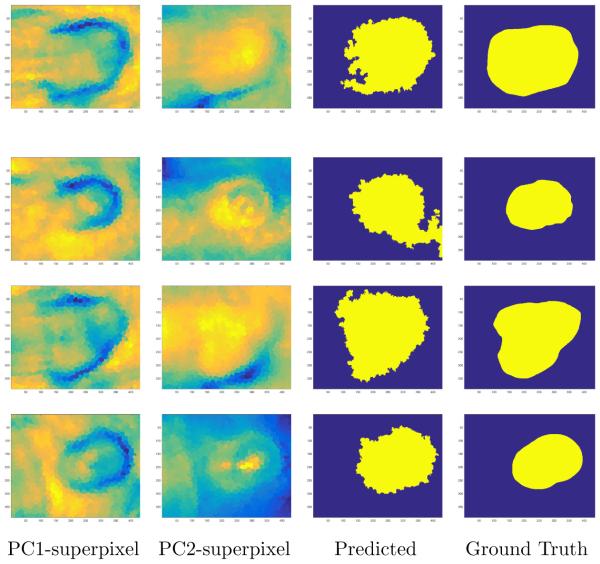
Figure 4.
An illustration of the superpixel segmented regions, classified tumor and normal tissue images, and original ground truth images for three different mice. Row 1: Mouse No. 1; Row 2: Mouse No. 2; Row 3: Mouse No. 3, Row 4: Mouse No. 7. Column 1: Superpixel segmented principal component 1 image using the SLIC superpixel algorithm. Principal component 1 image accounted for the most variance of the data. Column 2: Superpixel segmented principal component 2 image using the same superpixel segmented regions from superpixel PC1. Column 3: SVM Classified Image. Column 4: Original Ground Truth Image.
4. DISCUSSION
The results in Table 1 show the improvements of using both PCA and superpixels to detect cancer regions using hyperspectral images. The PCA achieved dimensionality reduction of wavelengths and superpixels combined groups of similar pixels.
4.1 Feature Selection
In previous trials, selecting different bands for feature selection was based on manual selection. The bands of interest usually consisted of wavelengths with the greatest reflectance intensity difference between the tumor and normal tissue. However, the problem with a manual selection of wavelength is that the wavelength with the greatest reflectance intensity difference between the tumor and normal tissue varies from mouse to mouse. To improve the band selection, this study utilizes principal component analysis to select the different features based on exploiting the statistical correlation of different bands. Principal component can be a powerful tool for dimensionality reduction and band selection.
4.2 Superpixel
PCA-Superpixel-SVM technique performed well in classifying the tumor region as shown in the third column of Figure 4. Incorporating superpixels into the algorithm improved sensitivity and specificity of the classification results as shown in Table 1. The PCA-superpixel-SVM technique did overestimated the tumor region. One possible reason is that more superpixels with both tumor and normal tissue were classified as tumor than normal tissue. Reducing the superpixel regions by increasing the number of superpixels could overcome the overestimation of the tumor region.
4.3 Reduction in Computation Time
Both of these reduction methods helped improve not only the classification accuracy but also computation time. The SVM pixelwise method needed to train millions of pixels for the classifier because the dimension of the image is 390 × 435. Especially when the images have large sizes as the ones used in this study, millions pixels needed to be trained for the classifier. The large hypercube caused the computation to take a full day (i3-5020U, 2.20GHz, 8GB) for feature extraction, training, and testing for SVM pixel-wise classification. On the other hand, segmenting the images into superpixel regions allowed significant computational improvement. The superpixel-PCA-SVM based classification took ≤ 15 minutes for feature extraction, training and testing. As briefly stated before, one of the main reason for the significant difference is the number of data points in the SVM classifier. For the superpixel-PCA-SVM based classification, the training data consisted of a few bands and 1000 superpixels for each mouse. Thus, the superpixel PCA method performs significantly faster than the pixel-wise method. With such a significant computational improvement, real-time analysis of hyperspectral imaging for cancer detection in image-guided surgery may be possible in the future.
5. CONCLUSION
This paper introduced a superpixel-based classification method for cancer detection on hyperpsectral images in a head and neck cancer animal model. The method uses principle component to reduce the data dimension and uses support vector machine to perform supervise classification of tumor and normal tissue. The method has been tested in a head and neck cancer mouse model. The hyperspectral imaging and classification technology may provide a noninvasive tool for the detection of head and neck cancer.
6. FUTURE RESEARCH
For future studies, another area of interest would be understanding the superpixel size constraint and its influence on the segmentation and classification. One would predict that as the number of superpixels size decreases, the computational time gradually increases. This is because smaller superpixel size means more superpixels to train and classify.
ACKNOWLEDGMENTS
This research is supported in part by NIH grants (CA176684 and CA156775). The authors would also like to thank Radhakrishna Achanta for making the SLIC source code available online. The work was conducted in the Quantitative BioImaging Laboratory in the Emory Center for Systems Imaging (CSI) of Emory University School of Medicine.
REFERENCES
- [1].Siegal MKJA. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. R. [DOI] [PubMed] [Google Scholar]
- [2].Walden M, Aygun N. Head and neck cancer. Seminars in Roentgenology. 2013;48(1):75–86. doi: 10.1053/j.ro.2012.09.002. [DOI] [PubMed] [Google Scholar]
- [3].Singh S, Ibrahim O, Byrne HJ, Mikkonen JW, Koistinen AP, Kullaa AM, Lyng FM. Recent advances in optical diagnosis of oral cancers: Review and future perspectives. Head and Neck. 2015:1–9. doi: 10.1002/hed.24293. [DOI] [PubMed] [Google Scholar]
- [4].Swinson B, Jerjes W, El-Maaytah M, Norris P, Hopper C. Optical techniques in diagnosis of head and neck malignancy. Oral Oncology. 2006;42(3):221–228. doi: 10.1016/j.oraloncology.2005.05.001. [DOI] [PubMed] [Google Scholar]
- [5].Dong L, Kudrimoti M, Cheng R, Shang Y, Johnson E, Stevens S, Shelton B, Yu G. Noninvasive diffuse optical monitoring of head and neck tumor blood flow and oxygenation during radiation delivery. Biomedical Optics Express. 2012;3(2):259–272. doi: 10.1364/BOE.3.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Akbari H, Halig L, Zhang H, Wang D, Chen Z, Fei B. Detection of cancer metastasis using a novel macroscopic hyperspectral method. Medical Imaging 2012: Biomedical Applications in Molecular, Structural, and Functional Imaging. 2012;8317:831711. doi: 10.1117/12.912026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tian Z, Liu L, Zhang Z, Fei B. Superpixel-based segmentation for 3d prostate mr images. IEEE Transactions on Medical Imaging. 2015;(99):1–1. doi: 10.1109/TMI.2015.2496296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yu N, Wu J, Weinstein SP, Gaonkar B, Keller BM, Ashraf AB, Jiang Y, Davatzikos C, Conant EF, Kontos D. A superpixel-based framework for automatic tumor segmentation on breast dce-mri. Proc. SPIE. 2015;9414 94140O-94140O-7. [Google Scholar]
- [9].Lu G, Fei B. Medical hyperspectral imaging: a review. Journal of Biomedical Optics. 2014;19(1) doi: 10.1117/1.JBO.19.1.010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Calin M, Parasca S, Savastru D, Manea D. Hyperspectral imaging in the medical field: Present and future. Applied Spectroscopy Reviews. 2014;49(6):435–447. [Google Scholar]
- [11].Mori G, Ren X, Efros A, Malik J, I. C. Society Recovering human body configurations: Combining segmentation and recognition. Proceedings of the 2004 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2004;2:326–333. [Google Scholar]
- [12].Lu G, Halig L, Wang D, Chen Z, Fei B. Hyperspectral imaging for cancer surgical margin delineation: Registration of hyperspectral and histological images. SPIE Medical Imaging 2014: Image-Guided Procedures, Robotic Interventions, and Modeling. 2014;9036:90360S. doi: 10.1117/12.2043805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lu G, Halig L, Wang D, Chen Z, Fei B. Spectral-spatial classification using tensor modeling for cancer detection with hyperspectral imaging. Medical Imaging 2014: Image Processing. 2014;9034:903413. doi: 10.1117/12.2043796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lu G, Halig L, Wang D, Qin X, Chen ZG, Fei B. Spectral-spatial classification for noninvasive cancer detection using hyperspectral imaging. Journal of Biomedical Optics. 2014;19(10):106004. doi: 10.1117/1.JBO.19.10.106004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lu G, Wang D, Qin X, Halig L, Muller S, Zhang H, Chen A, Pogue BW, Chen ZG, Fei B. Framework for hyperspectral image processing and quantification for cancer detection during animal tumor surgery. Journal of Biomedical Optics. 2015;20(12):126012. doi: 10.1117/1.JBO.20.12.126012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Akbari H, Halig L, Schuster D, Osunkoya A, Master V, Nieh P, Chen G, Fei B. Hyperspectral imaging and quantitative analysis for prostate cancer detection. Journal of Biomedical Optics. 2012;17(7) doi: 10.1117/1.JBO.17.7.076005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xia J, Chanussot J, Du P, He X. (semi-) supervised probabilistic principal component analysis for hyperspectral remote sensing image classification. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing. 2014;7(6):224–2236. [Google Scholar]
- [18].Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Susstrunk S. Slic superpixels compared to state-of-the-art superpixel methods. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2012;34(11):2274–2281. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- [19].Chang C, Lin C. Libsvm: A library for support vector machines. Acm Transactions on Intelligent Systems and Technology. 2011;2(3) [Google Scholar]
- [20].Lu G, Qin X, Wang D, Chen ZG, Fei B. Quantitative wavelength analysis and image classification for intraoperative cancer diagnosis with hyperspectral imaging. Proc. SPIE. 2015;9415 doi: 10.1117/12.2082284. 94151B-94151B-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pike R, Lu G, Wang D, Chen Z, Fei B. A minimum spanning forest based method for noninvasive cancer detection with hyperspectral imaging. IEEE Transactions on Biomedical Engineering. 2015;(99):1–1. doi: 10.1109/TBME.2015.2468578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pike R, Patton SK, Lu G, Halig LV, Wang D, Chen ZG, Fei B. A minimum spanning forest based hyperspectral image classification method for cancerous tissue detection. Proc. SPIE. 2014;9034 doi: 10.1117/12.2043848. 90341W-90341W-8. [DOI] [PMC free article] [PubMed] [Google Scholar]