Abstract
We developed a chemically-induced oral cancer animal model and a computer aided method for tongue cancer diagnosis. The animal model allows us to monitor the progress of the lesions over time. Tongue tissue dissected from mice was sent for histological processing. Representative areas of hematoxylin and eosin (H&E) stained tissue from tongue sections were captured for classifying tumor and non-tumor tissue. The image set used in this paper consisted of 214 color images (114 tumor and 100 normal tissue samples). A total of 738 color, texture, morphometry and topology features were extracted from the histological images. The combination of image features from epithelium tissue and its constituent nuclei and cytoplasm has been demonstrated to improve the classification results. With ten iteration nested cross validation, the method achieved an average sensitivity of 96.5% and a specificity of 99% for tongue cancer detection. The next step of this research is to apply this approach to human tissue for computer aided diagnosis of tongue cancer.
Keywords: Tongue cancer diagnosis, squamous cell carcinoma, 4NQO-induced oral cancer, random forest, histological image classification, computer aided diagnosis
1. DESCRIPTION OF PURPOSE
More than half a million patients are diagnosed worldwide with squamous cell carcinoma (SCC) of the head and neck each year [1]. Only half of the people diagnosed with oral cancer live for 5 years. Current gold standard for cancer diagnosis is made by pathologists using visual examination of haematoxylin and eosin (H&E) stained sections under the microscope [2]. The diagnosis and grading of oral epithelial dysplasia is based on a combination of architectural and cytological changes. Specifically, the histological features for cancer diagnosis mainly include: 1) the proliferation of immature cells characterized by a loss of cellular organization or polarity, 2) variations in the size and shape of the nuclei, 3) increase in nuclear size relative to the cytoplasm, 4) increase in the nuclear chromatin with irregularity of distribution, and 5) increased mitoses, including atypical forms in all epithelial layers [3]. The locations of the architectural and cytomorphologic changes within the epithelium represent key diagnostic parameters. However, the effectiveness of cancer diagnosis is highly dependent on the attention and experience of pathologists. This process is time consuming, subjective, and inconsistent due to considerable inter- and intra-observer variations [4]. Therefore, a computer-aided image classification system with quantitative analysis of these histological features is highly desirable to provide rapid, consistent and quantitative cancer diagnosis.
The development of quantitative analysis methods for oral cancer diagnosis purposes is still in its early stage. Only a few studies have been conducted to quantify the oral tissue architectural and morphological changes. Landini et al. [5] measured statistical properties of the graph networks constructed based on the cell centroids for classifying normal, premalinant and malignant cells, and reported an accuracy of 67%, 100% and 80% to identify normal, premalignant and malignant cells, respectively. Krishnan et al. [6] explored the potential of texture features to grade the histopathological tissue sections into normal, oral sub-mucous fibrosis (OSF) without dysplasia, and OSF with dysplasia, and reported an accuracy of 95.7%, a sensitivity of 94.5%, and a specificity of 98.8%. Recently, Das et al [7] proposed an automated segmentation method for the identification of keratinization and keratin pearl from oral histological images and achieved a segmentation accuracy of 95.08% in comparison with the manual segmentation ground truth.
In this study, we developed a predictive model that combines multiple features, such as color, texture, morphormetry, and topology features from epithelium tissue and its constituent nuclei and cytoplasm, for the diagnosis of tongue cancer. We evaluated the effectiveness of the proposed features with multiple classifiers and performance metrics.
2. MATERIALS AND METHODS
2.1 Tongue Carcinogenesis Animal Model
Six-week-old female CBA/J mice were purchased from the Jackson Laboratory and were used for the studies. Animals were housed in the Animal Resource Facility of our institution under controlled conditions and fed sterilized special diet (Teklad global 10% protein rodent diet, Harlan) and autoclaved water. 4-NQO powder (Sigma Aldrich, St. Louis, USA) was diluted in the drinking water for mice. The water was changed once a week. Mice were allowed access to the drinking water at all times during the treatment. Mice were randomly divided into an experimental group and a control group. In the experimental group, the drinking water contained 4-NQO while no 4-NQO was added into the water in the control group. Mice in the experimental group were administered 4NQO (Sigma) at 100 μg/ml in their drinking water on a continuous basis for 16 weeks to induce epithelial carcinogenesis. The body weights of the mice were measured once a week to monitor the tumor burden. Mice were euthanized at different time points (weeks 12, 20, 24) to track different pathological grades.
2.2 Data Acquisition
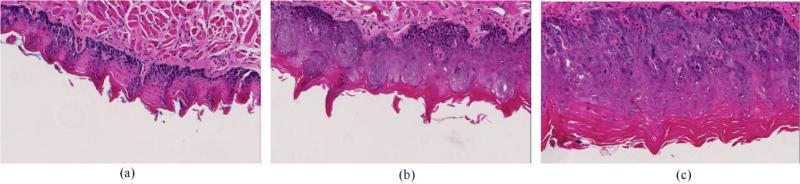
Immediately after the euthanasia of the mice, the mouse tongues were dissected and kept in 10% formalin overnight. Tongue tissue from both 4NQO-treated and control groups were embedded in paraffin, and sectioned vertically down the dorsal surface into 5 μm sections. Tissue samples were stained with hematoxylin and eosin (H&E). The H&E slides were then digitized and reviewed by an experienced pathologist specialized in head and neck cancer, who segmented the section into pathological regions of normal, dysplasia, carcinoma in situ, and squamous cell carcinoma. The images were captured at 20× magnification using the Aperio ImageScope software (Leica Biosystems) producing 1472 × 922-pixel color images. Some example images were shown in Figure 1.
Figure 1.
Example images of H&E stained pathological samples. (a) Healthy tissue. (b) Dysplastic tissue. (c) Carcinoma in situ.
2.3 Overview of the Computer Aided Diagnosis Approach
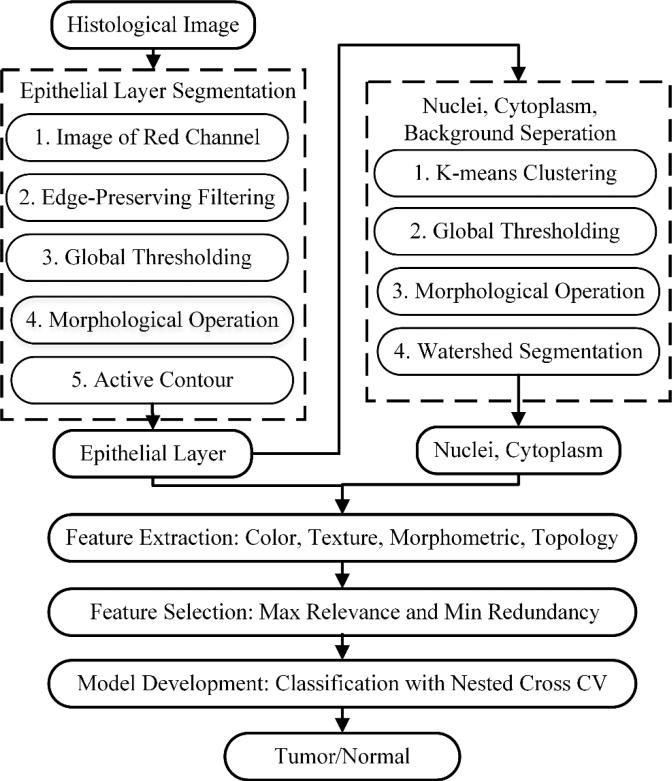
The flowchart of the proposed method is shown in Figure 2. First, the histological RGB image was segmented into epithelial layer, connective tissue and background. In clinical practices, the diagnosis of precursor lesions is based on the altered epithelium with an increased likelihood for progression to squamous cell carcinoma [8]. This step is to pre-process histological images for diagnosis. Second, the segmented nuclei image was further separated into nuclei, cytoplasm and background. Next, multiple features were extracted from both the whole epithelial image and its components (nuclei, cytoplasm). Finally, feature selection and supervised classification method with nested cross validation is conducted to build predictive models for cancer diagnosis.
Figure 2.
Flowchart of the proposed computer aided diagnosis method.
2.4 Segmentation of Epithelial Layer
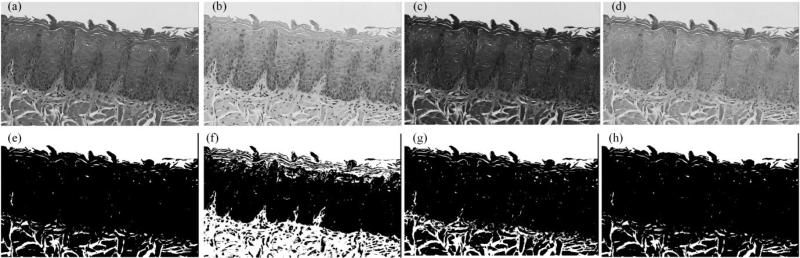
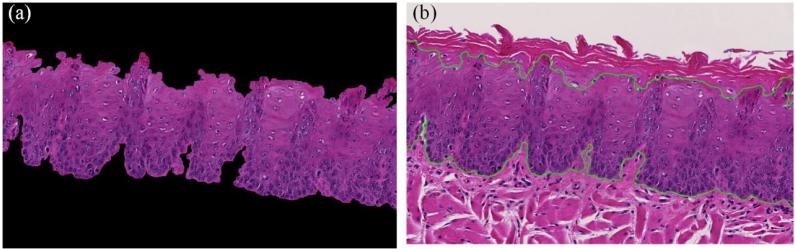
The first step in the analysis pipeline was the segmentation of the epithelium tissue from the connective tissue, muscle and image background. Figure 3 shows that image of the red channel exhibits the best contrast for separating epithelium from connective tissue. Figure 4 is the smoothed red image with edge-preserving decomposition [9], which generated an image with less noise and it is easy to binarize. Next, global thresholding was applied on the smoothed red image to obtain the mask of the epithelial layer. Green, blue and grayscale images have all been tested for thresholding, but none of them generated better results than the red image. The connected component with the largest area was kept on the image. In some cases, the epithelial layers may be disconnected after thresholding. So the first few connected components with the largest area were retained. On the initial nuclei mask, small spurious background regions were cleaned by morphological opening with a disk radius of 5. The generated mask was used as initialization for the Chan-Vese active contour algorithm, which smoothed the segmentation mask.
Figure 3.
Comparison of difference grayscale images for epithelial layer segmentation. (a)-(d) are the luminescence image from HSV color space, red channel, green channel and blue channel of RGB color space, respectively. (e)-(h) are the corresponding image to (a)-(d) after edge-preserving smoothing and global thresholding.
Figure 4.
Segmented epithelial layer. (a) Separated epithelium after the morphological processing of the mask in Figure 3 (f). (b) The boundary of the epithelium smoothed by active contour shown in green.
2.5 Nuclei and Cytoplasm Segmentation
Epithelial cell nucleus absorbs the haematoxylin which is a dark blue or violet stain, while eosin is absorbed by the cytoplasm which is a red or pink stain. So the colors consisting white, blue and pink in the histological image allows a clear distinguishing between different cellular components within the epithelium. To quantify these visual differences, RGB images were converted to the CIELab color space, which consists of a luminosity layer L, chromaticity-layer ‘a’ indicating where color falls along the red-green axis, and chromaticity-layer ‘b’ indicating where color falls along the blue-yellow axis. Since all of the color information is in the a and b layers, K-means clustering was applied on these two layers to segment the image into two or three clusters (nuclei, cytoplasm and background) using the Euclidean distance.
To better segment the nuclei out, global thresholding was performed on the red channel of the segmented nuclei image from clustering. Morphological opening with a disk radius of 1 was used to remove small spurious objects. Connected components with more than 25 pixels were also discarded. At this point, pure nuclei images were produced, but some nucleus were overlapping. So we identified the multi-nucleus with solidity lower than 0.85 and performed watershed segmentation to separate the touching cells. The single nuclei mask and separated multi-nucleus m were combined to form the final mask of the epithelial nuclei.
2.6 Feature Extraction
Multiple features were extracted from the epithelium, nuclei, and cytoplasm image. It has been shown in previous study that removing white pixels could improve the classification performance [10]. This is obtained by first transforming the image from the RGB color space into the YCbCr space and then applying the threshold to the luminance (Y) component. A threshold value 180 was used in this stage.
A key morphological change in dysplasia is the loss of stratification, due to a lack of normal maturation from the cells in basement membrane to the surface keratin. Cells located in the surface layer of the epithelium have the same immature appearance as those in the deep basal layers [11]. To reflect these abnormal changes, we extracted 64 color and 293 texture features from each epithelium image and each cytoplasm image respectively. Color features include 16-bin transformed RGB histogram and red-blue histogram difference. The RGB histogram itself is sensitive to photometric variations. However, the transformed RGB histogram has scale-invariance and shift-invariance with respect to light intensity [12]. Texture features include the Haralick feature [13], local binary pattern [14] and fractal textures [15].
The variations in the size and shape of the cells and of the nuclei are characteristics of oral dysplastic changes. Cancer nuclei have an increased nuclear to cytoplasmic ratio compared to normal nuclei. To quantify these changes, we extracted 24 color, morphometric and topological features from each nuclei image. Color features include the mean, standard deviation, minimum, and maximum of the red, green, and blue channel. Morphometric features include the area, ratio of the major to minor axis, shape (solidity, eccentricity, and compactness), neighborhood radius, and the nucleus to cytoplasm ratio. Topology feature was extracted based on the centroids of connected components segmented in the nuclei image [16]. The number of nodes, number of edges, number of triangles, edge length, and cyclomatic number [17] of the Delaunay triangulation were selected as the topology features to characterize the distribution of individual nucleus. In summary, a total of 738 features were extracted from each epithelium image and its constituent nuclei and cytoplasm images.
2.7 Feature Selection
The goal of feature selection is to find a feature set S with n feature {λi}, that “optimally” characterize the difference between cancerous and normal tissue. To achieve the “optimal” condition, we used the maximal relevance and minimal redundancy (mRMR) [18] framework to maximize the dependency of each feature on the target class labels (tumor or normal), and minimize the redundancy among individual features simultaneously. Relevance is characterized by mutual information I(x; y), which measures the level of similarity between two random variables x and y:
| (1) |
where p(x, y) is the joint probability distribution function of x and y, and p(x) and p(y) are the marginal probability distribution functions of x and y respectively.
We represent the feature of each pixel with a vector λ = [λ1, λ2, ... , λi], i = 738, and the class label (tumor or normal) with c. Then the maximal relevance condition is:
| (2) |
The features selected by maximal relevance is likely to have redundancy, so the minimal redundancy condition is used to select mutually exclusive features:
| (3) |
So the simple combination (equation (5) and (6)) of these two conditions forms the criterion “minimal-redundancy-maximal-relevance” (mRMR).
| (4) |
i.e.
| (5) |
2.8 Model Development
The image set used for tumor/non-tumor classification consisted of a total of 214 images (114 tumor and 100 normal images). 738 features described in Section 2.6 were extracted from each image. To avoid the curse of dimensionality in supervised classification, we performed feature selection and built predictive models through nested cross validation (CV) consisting of 10 iterations of five-fold outer CV and 10 iterations of four-fold inner CV. The outer CV loop was used to estimate the classification performance; and the inner CV loop was used to tune the optimal parameters for the model development. For the stratified five-fold outer CV, the whole dataset was randomly split into five sets of roughly equal sizes. Splitting was performed such that the proportion of images per class was roughly equal across the subsets. Each run of the outer five-fold CV algorithm consisted of training models on four image subsets and testing on the remaining subset. A stratified four-fold inner CV was conducted to select the optimal feature numbers from three subsets of the training data and to validate the model using the remaining subset. Six different classifiers, including support vector machine (SVM) [19], random forest (RF) [20], Naive Bayes, linear discriminate analysis (LDA) [21], k-nearest neighbors (KNN) [22], and decision trees (DT) [23], were compared for classification.
2.9 Performance Evaluation
Following imaging experiments, tissue specimens were fixed in 10% formalin and then paraffin embedded. Each tongue specimen was sectioned into a series of 5 μm tissue sections with 100 μm intervals between sections. Each tissue section represented one longitudinal line parallel to the midline on the dorsal surface of the tongue. The H&E slide from each section was reviewed by an experienced head and neck pathologist, who segmented the section into regions of normal, dysplasia, carcinoma in situ and carcinoma as our gold standard. Receiver operating characteristic (ROC) curves, the area under the ROC curve (AUC), accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were used as metrics to assess the performance of supervised classification [24] [25] [26].
3. RESULTS AND DISCUSSIONS
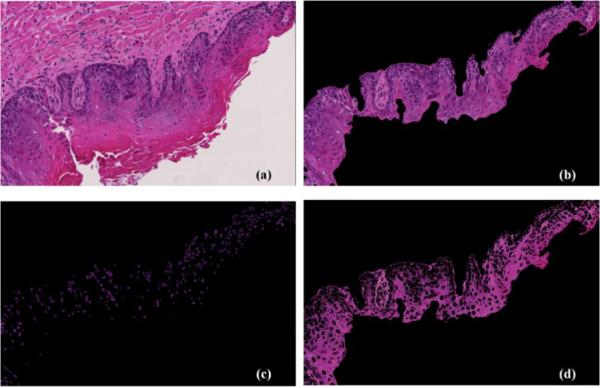
Figure 5 shows an example of the segmentation results. From visual assessment, the segmentation results of the epithelium, nuclei and cytoplasm were satisfactory.
Figure 5.
Example segmentation results from a dysplasia image. (a) Input H&E pathological image. (b) Segmented epithelial layer. (c) Segmented nuclei image. (d) Segmented cytoplasm image.
To compare image-level features with object-level features, we first performed a stratified five-fold cross validation on all the dataset with different feature types. Table 1 shows the classification results of six classifiers for cancer diagnosis with features extracted from the epithelium image alone. Table 2 shows the classification with features extracted from individual objects (nuclei and cytoplasm). Table 3 shows the classification with features extracted from both the whole image and individual objects (nuclei and cytoplasm). From these tables, we found that the combination of image-level and object-level features outperformed image-level features alone and object-level features alone. This indicated that image-level and object-level features contained complementary information for cancer diagnosis. SVM was the best performing classifier, with an average sensitivity of 96.5% and specificity of 99.0%.
Table 1.
Comparison of different classifiers for cancer diagnosis using image-level features alone
| Classifier | AUC | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| SVM | 0.955 | 86.9% | 89.5% | 89.0% | 85.1% | 89.0% |
| Random Forest | 0.924 | 85.1% | 84.3% | 91.0% | 79.9% | 91.0% |
| Naive Bayes | 0.852 | 72.8% | 81.5% | 81.0% | 62.6% | 84.0% |
| KNN | 0.818 | 81.8% | 81.7% | 82.0% | 81.7% | 82.0% |
| Decision Tree | 0.808 | 58.5% | 82.5% | 80.0% | 31.1% | 90.0% |
| LDA | 0.786 | 78.5% | 78.2% | 79.0% | 78.2% | 79.0% |
Table 2.
Comparison of different classifiers for cancer diagnosis using object-level features alone
| Classifier | AUC | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| SVM | 0.985 | 93.0% | 93.0% | 98.0% | 88.6% | 98.0% |
| Random Forest | 0.966 | 89.7% | 91.2% | 93.0% | 86.8% | 93.0% |
| KNN | 0.889 | 88.8% | 87.7% | 90.0% | 87.7% | 90.0% |
| LDA | 0.857 | 85.5% | 83.3% | 88.0% | 83.3% | 88.0% |
| Decision Tree | 0.850 | 69.2% | 85.1% | 86.0% | 49.4% | 92.0% |
| Naive Bayes | 0.835 | 79.4% | 88.6% | 74.0% | 84.2% | 74.0% |
Table 3.
Comparison of different classifiers for cancer diagnosis using both image and object features
| Classifier | AUC | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| SVM | 0.993 | 95.3% | 96.5% | 99.0% | 92.1% | 99.0% |
| Random Forest | 0.970 | 89.7% | 89.4% | 95.0% | 85.1% | 95.0% |
| KNN | 0.923 | 92.1% | 88.6% | 96.0% | 88.6% | 96.0% |
| LDA | 0.912 | 91.1% | 90.4% | 92.0% | 90.4% | 92.0% |
| Naive Bayes | 0.869 | 74.8% | 88.7% | 82.0% | 66.0% | 85.0% |
| Decision Tree | 0.834 | 73.9% | 82.5% | 82.0% | 61.7% | 88.0% |
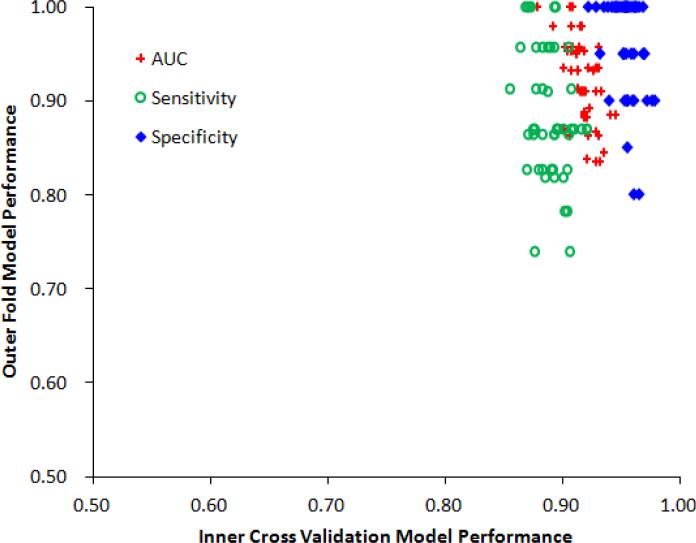
Figure 6 demonstrated the model performance (AUC, sensitivity, specificity) in both internal and external validation. X axis denotes the average cross validation model performance from four-fold inner cross validations, and Y axis denotes the corresponding outer fold model performance on testing dataset. It can be seen that the training models works well not only on training dataset but also in testing dataset, which indicated that the predictive models were robust and did not over-fit inner cross validation dataset.
Figure 6.
Model performances in internal and external validations. Red points represent AUC values, green points represent sensitivity values, and blue points represent specificity values.
Table 4 shows the most frequently selected features in the top 10 ranks in the cross validation. These top features included features from epithelium, cytoplasm as well as nuclei, which indicated the complementary value of image and object level features for cancer diagnosis and was consistent with the observations in Table 1-3. Local binary pattern capturing the texture of epithelium and cytoplasm may be related to the abnormal changes in epithelial layer as normal tissue progresses into tumor. Red and blue channel histogram differences of cytoplasm and epithelium are likely associated with the increased nuclei to cytoplasm ratio in tumor tissue. Enlarged nuclei sizes in tumors are also highly discriminating.
Table 4.
Most frequently selected top 10 ranking features
| Ranking | Feature name | Feature type | Location | Frequency |
|---|---|---|---|---|
| 1 | Local binary pattern | Texture | Epithelium | 38% |
| 2 | Red-blue difference | Color | Cytoplasm | 34% |
| 3 | Red-blue difference | Color | Epithelium | 54% |
| 4 | Mean nuclei size | Morphology | Nuclei | 28% |
| 5 | Local binary pattern | Texture | Cytoplasm | 28% |
| 6 | Fractal Dimension | Texture | Epithelium | 16% |
| 7 | Local binary pattern | Texture | Cytoplasm | 14% |
| 8 | Transformed RGB Histogram | Color | Epithelium | 16% |
| 9 | Mean nuclei solidity | Morphology | Nuclei | 12% |
| 10 | Mean nuclei solidity | Morphology | Nuclei | 10% |
To the best of our knowledge, this is the first time that multi-level (image and object) features, which include color, texture, morphometry and topology features, were extracted and combined for the improved classification of pathological images of tongue cancer. The animal experiment was specially designed to acquire images with progressing pathological grades. The proposed method was effective for the distinction of tumor and normal tissue.
4. CONCLUSIONS
In this paper, we developed a chemically-induced oral cancer animal model and designed a computer aided diagnosis method for the detection of the tongue cancer. We evaluated various image features from histological images for quantitative cancer evaluation. We found that the diagnostic performances of image-level features combined with object-level features outperformed image features or object features alone, which indicated their complementary effects. Texture feature describing epithelium structures was the most discriminating feature for cancer detection. The computer aided diagnosis models could provide objective and reproducible diagnosis for tongue cancer. The image features and quantitative analysis method can also be applied to human images for computer-aided diagnosis of human tongue cancer.
ACKNOWLEDGEMENTS
This research is supported in part by NIH grants (CA176684 and CA156775) and the Emory Center for Systems Imaging (CSI) of Emory University School of Medicine. We thank Ms. Jennifer Shelton from the Pathology Core Lab at Winship Cancer Institute of Emory University for her help in histologic processing.
REFERENCES
- 1.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359(11):1143–54. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Speight PM. Update on oral epithelial dysplasia and progression to cancer. Head Neck Pathol. 2007;1(1):61–6. doi: 10.1007/s12105-007-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenig BM. Squamous Cell Carcinoma of the Upper Aerodigestive Tract: Precursors and Problematic Variants. Mod Pathol. 2002;15(3):229–254. doi: 10.1038/modpathol.3880520. [DOI] [PubMed] [Google Scholar]
- 4.Ismail SM, Colclough AB, Dinnen JS, et al. Observer variation in histopathological diagnosis and grading of cervical intraepithelial neoplasia. BMJ. 1989;298(6675):707–10. doi: 10.1136/bmj.298.6675.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landini G, Othman IE. Architectural analysis of oral cancer, dysplastic, and normal epithelia. Cytometry Part A. 2004;61A(1):45–55. doi: 10.1002/cyto.a.20082. [DOI] [PubMed] [Google Scholar]
- 6.Krishnan MMR, Venkatraghavan V, Acharya UR, et al. Automated oral cancer identification using histopathological images: A hybrid feature extraction paradigm. Micron. 2012;43(2–3):352–364. doi: 10.1016/j.micron.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Das DK, Chakraborty C, Sawaimoon S, et al. Automated identification of keratinization and keratin pearl area from in situ oral histological images. Tissue and Cell. 2015;47(4):349–358. doi: 10.1016/j.tice.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Barnes L, Eveson J, Sidransky D. Pathology and genetics of head and neck tumours. IARC Press; Lyon: 2005. [Google Scholar]
- 9.Bo G, Wujing L, Minyun Z, et al. Local Edge-Preserving Multiscale Decomposition for High Dynamic Range Image Tone Mapping. IEEE Transactions on Image Processing. 2013;22(1):70–79. doi: 10.1109/TIP.2012.2214047. [DOI] [PubMed] [Google Scholar]
- 10.Tabesh A, Teverovskiy M, Ho-Yuen P, et al. Multifeature Prostate Cancer Diagnosis and Gleason Grading of Histological Images. IEEE Transactions on Medical Imaging. 2007;26(10):1366–1378. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- 11.Speight PM, Farthing PM, Bouquot JE. The pathology of oral cancer and precancer. Current Diagnostic Pathology. 1996;3(3):165–176. [Google Scholar]
- 12.Lu G, Yan G, Wang Z. Bleeding detection in wireless capsule endoscopy images based on color invariants and spatial pyramids using support vector machines. IEEE Engineering in Medicine and Biology Society. 2011:6643–6646. doi: 10.1109/IEMBS.2011.6091638. [DOI] [PubMed] [Google Scholar]
- 13.Qin X, Lu G, Sechopoulos I, et al. Breast tissue classification in digital tomosynthesis images based on global gradient minimization and texture features. Proc. SPIE. 2014:9034, 90341V–90341V-8. doi: 10.1117/12.2043828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2002;24(7):971–987. [Google Scholar]
- 15.Costa AF, Humpire-Mamani G, Traina AJM. An Efficient Algorithm for Fractal Analysis of Textures. Graphics, Patterns and Images (SIBGRAPI), 2012 25th SIBGRAPI Conference on. 2012;39-46 [Google Scholar]
- 16.Champion A, Lu G, Walker M, et al. Semantic interpretation of robust imaging features for Fuhrman grading of renal carcinoma. IEEE Engineering in Medicine and Biology Society (EMBC) 2014:6446–6449. doi: 10.1109/EMBC.2014.6945104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurcan MN, Boucheron LE, Can A, et al. Histopathological Image Analysis: A Review. Biomedical Engineering, IEEE Reviews in. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng H, Fulmi L, Ding C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 19.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2(3):1–27. [Google Scholar]
- 20.Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 21.Guo Y, Hastie T, Tibshirani R. Regularized linear discriminant analysis and its application in microarrays. Biostatistics. 2007;8(1):86–100. doi: 10.1093/biostatistics/kxj035. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell TM. Machine Learning. McGraw-Hill, Inc.; 1997. [Google Scholar]
- 23.Breiman L, Friedman J, Stone CJ, et al. Classification and regression trees. CRC press; 1984. [Google Scholar]
- 24.Lu G, Halig L, Wang D, et al. Spectral-spatial classification for noninvasive cancer detection using hyperspectral imaging. J Biomed Opt. 2014;19(10):106004. doi: 10.1117/1.JBO.19.10.106004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu G, Wang D, Qin X, et al. Framework for Hyperspectral Image Processing and Quantification for Cancer Detection during Animal Tumor Surgery. J Biomed Opt. 2015;20(12):126012. doi: 10.1117/1.JBO.20.12.126012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pike R, Lu G, Wang D, et al. A Minimum Spanning Forest Based Method for Noninvasive Cancer Detection with Hyperspectral Imaging. IEEE Transactions on Biomedical Engineering. 2015;PP(99):1–1. doi: 10.1109/TBME.2015.2468578. [DOI] [PMC free article] [PubMed] [Google Scholar]