SUMMARY
Borrelia burgdorferi maintains a complex life cycle between tick and vertebrate hosts. Although some genes have been identified as contributing to bacterial adaptation in the different hosts, the list is incomplete. In this manuscript, we report the first use of transposon mutagenesis combined with high-throughput sequencing (Tn-seq) in B. burgdorferi. We utilize the technique to investigate mechanisms of carbohydrate utilization in B. burgdorferi and the role of carbohydrate metabolism during mouse infection. We performed genetic fitness analyses to identify genes encoding factors contributing to growth on glucose, maltose, mannose, trehalose, and N-acetyl-glucosamine. We obtained insight into the potential functions of proteins predicted to be involved in carbohydrate utilization and identified additional factors previously unrecognized as contributing to the metabolism of the tested carbohydrates. Strong phenotypes were observed for the putative carbohydrate phosphotransferase transporters BB0408 and BBB29 as well as the response regulator Rrp1. We further validated Tn-seq for use in mouse studies and were able to correctly identify known infectivity factors as well as additional transporters and genes on lp54 that may contribute to optimal mouse infection. As such, this study establishes Tn-seq as a powerful method for both in vitro and in vivo studies of B. burgdorferi.
INTRODUCTION
Borrelia burgdorferi, the causative agent of Lyme disease, maintains a complex life cycle between a specific arthropod host, the Ixodes tick, and a variety of vertebrates including small rodents and birds. The environments the bacteria encounter in these hosts vary extensively including, but not limited to, differences temperature, host immune responses, and available nutrients. A distinguishing feature of B. burgdorferi this is the ability of the bacteria to adapt to these diverse conditions and establish long-term infection. Previous work has shown that B. burgdorferi differentially regulates gene expression in response to changes in growth conditions (Iyer et al., 2015, Revel et al., 2002, Brooks et al., 2003, Angel et al., 2010, Hyde et al., 2007, Ojaimi et al., 2003, Tokarz et al., 2004, Bykowski et al., 2008, Miller et al., 2003, Pappas et al., 2011). However, compared to other pathogens, the mechanisms utilized by B. burgdorferi to adapt to shifts in environmental conditions are relatively unknown.
The ability of a bacterial pathogen to sense the available nutrients in the environment and alter metabolism accordingly is essential to survival and infectivity. The monosaccharide glucose is thought to be the preferred carbohydrate for B. burgdorferi, particularly in the vertebrate host where D-glucose is available (Corona & Schwartz, 2015). Glycolysis is the central metabolic pathway in B. burgdorferi as genes encoding all of the enzymes involved in this pathway are present in the genome whereas the genes encoding enzymes necessary for oxidative phosphorylation and the citric acid cycle are absent (Fraser et al., 1997).
The central pathway for glucose transport into the cell is the carbohydrate phosphoenolpyruvate-dependent phosphotransferase system (PTS). Through a series of phosphoryl transfer reactions, the PTS mediates the import and phosphorylation of carbohydrates from the environment. There are three basic components to the PTS. First, the cytoplasmic enzyme I (EI) is phosphorylated by phosphoenolpyruvate (PEP). EI then transfers the phosphate to the phosphocarrier protein Hpr that in turn phosphorylates the enzyme II complex (EII) localized to the plasma membrane. The EII complex phosphorylates the carbohydrate as it is transported into the cell. The phosphorylated carbohydrate is then processed for entry into glycolysis or alternate metabolic pathways. There are three subunits in the EII complex (A, B, and C) that can be encoded as three distinct polypeptides or within multidomain proteins. EII complexes often exhibit carbohydrate specificity although they are not necessarily limited to a single substrate (Deutscher et al., 2006). Ten genes in the B. burgdorferi genome are annotated as encoding EII components suggesting that the ability to utilize carbohydrates other than glucose is relevant to the B. burgdorferi life cycle. The only EII transporter with a confirmed function is the chitobiose transporter, ChbABC (Fraser et al., 1997, Tilly et al., 2001, Tilly et al., 2004, Rhodes et al., 2010).
The primary role of the PTS is in metabolism. However, in many bacteria, the PTS is multifunctional and can regulate general aspects of environmental adaptation. Either directly or through the activation of signaling pathways, PTS components have been shown to modulate virulence in multiple human pathogens (Hondorp et al., 2013, Poncet et al., 2009, Lim et al., 2007, Iyer et al., 2005, Antunes et al., 2012). For example, carbohydrate availability and the PTS can regulate the activity of the Listeria monocytogenes virulence gene transcription factor PrfA, Clostridium difficile toxin gene regulation, and Salmonella enterica expression of genes in the Salmonella pathogenicity island 1 (Ake et al., 2011, Antunes et al., 2011, Lim et al., 2007).
In B. burgdorferi, the ability to metabolize carbohydrates other than glucose is critical to survival in the tick. Glycerol acts as an anti-freeze enabling the tick to survive through the winter and thus is an available carbon and energy source for B. burgdorferi. Although B. burgdorferi mutants lacking the ability to metabolize glycerol are fully infectious in mice, they are less fit in the nymphal tick (Pappas et al., 2011, He et al., 2011). Chitin is an amino polysaccharide synthesized from units of N-acetyl-glucosamine (GlcNAc). In many insects, chitin is a constituent of the peritrophic matrices that form a barrier between the ingested food and the midgut epithelium during feeding (Merzendorfer & Zimoch, 2003). Chitobiose is a dimer of chitin. If chitin is present in the peritrophic membrane of Ixodes ticks, chitobiose could be released by the tick chitinase in the remodeling of the membrane during the blood meal. Although the B. burgdorferi chitobiose transporter, ChbABC is not required for tick colonization, it has been hypothesized that B. burgdorferi uses chitobiose shed from the peritrophic membrane as a source of GlcNAc (Tilly et al., 2004, Sze et al., 2013). GlcNAc is a required nutrient for B. burgdorferi survival and is thought to be available in limited quantities in the unfed tick (Tilly et al., 2001).
In contrast to the arthropod host, the utilization of specific carbohydrates during vertebrate infection is less explored. However, one EIIBC component of the PTS, BB0645 (PtsG), is known to be required for mouse infection (Khajanchi et al., 2015). In addition, expression of RpoS, the primary sigma factor regulating the expression of genes contributing to mammalian infection in B. burgdorferi, may be linked to carbohydrate availability through the transcription factor BadR. In in vitro conditions mimicking the unfed tick, BadR inhibits rpoS by direct binding to the promoter region. The presence of phosphorylated sugars, as may be encountered by the bacteria in the tick midgut during the blood meal, releases BadR from the rpoS promoter allowing for gene expression (Miller et al., 2013).
There have been no investigations into the mechanisms that govern the metabolism of carbohydrates other than glucose, glycerol, and chitobiose. B. burgdorferi is also capable of using mannose, trehalose, GlcNAc, and maltose as primary carbon sources (von Lackum & Stevenson, 2005, Hoon-Hanks et al., 2012). These alternate carbohydrates may be used by B. burgdorferi as it transverses the enzootic life cycle. B. burgdorferi may encounter mannose during dissemination in a vertebrate host as it is a component of all vertebrate sera tested including that of mice, dogs and humans (Alton et al., 1998). Trehalose may be available to B. burgdorferi in the tick as it is a precursor in the chitin biosynthesis pathway (Merzendorfer & Zimoch, 2003). GlcNAc is a ubiquitous metabolite, and although sources of GlcNAc may be limited in the unfed tick, it may be available to the bacteria in other relevant niches. It is more difficult to predict where B. burgdorferi may encounter maltose. However, given the limited metabolic capacities of B. burgdorferi, the ability of the bacteria to use maltose as a primary carbon source suggests it may play a biologically relevant role (von Lackum & Stevenson, 2005).
In this study, we examined the hypothesis that the utilization of carbohydrates other than glucose contributes to optimal B. burgdorferi growth and vertebrate infection using Tn-seq. Tn-seq is a high-throughput screening technique for performing genetic fitness analyses using transposon mutagenesis. In Tn-seq, bacterial fitness is determined by sequencing the genomic DNA flanking the transposon insertion sites in a mutant library en masse (van Opijnen et al., 2009). This is the first use of Tn-seq to study aspects of B. burgdorferi biology. We conducted in vitro screens to identify genes involved in the metabolism of different carbohydrates. We obtained insight into potential function for two annotated PTS transporters in the genome as well as an additional function for the response regulator Rrp1 in metabolism. To explore a potential role for these transporters and the PTS in vivo, we performed a second screen in mice. We were able to identify a role for PTS transporters in promoting bacterial survival during mouse infection as well as identify uncharacterized potential infectivity factors.
RESULTS AND DISCUSSION
Identification of Genes Involved in Carbohydrate Metabolism Using Tn-seq
In order to establish Tn-seq as a method to perform genetic screens in B. burgdorferi, we sought to identify genes involved in maltose, mannose, trehalose, and GlcNAc metabolism. We utilized a signature tagged Mariner himar1 transposon library (STM) in the infectious B. burgdorferi strain B31 5A18NP1 (Lin et al., 2012). The library is largely maintained as arrayed transposon mutants with mapped insertion sites. However, a single pool was created for the Tn-seq experiments that included mutants with both mapped and unmapped insertion sites. Multiple frozen stocks were made from the pooled library. In addition to the chromosome, the B. burgdorferi B31 5A18NP1 genome includes nine circular and ten linear plasmids (Kawabata et al., 2004). In order to reduce the chance that fitness defects observed in the screen were due to plasmid loss during repeated in vitro passage, a new stock was used for each replicate of the in vitro screen.
There is no minimal media available for B. burgdorferi. To identify B. burgdorferi genes contributing to growth in carbohydrates other than glucose, we grew the transposon library in BSK-Lite, a modified version of the standard B. burgdorferi growth medium, BSK-II (Barbour, 1984, von Lackum & Stevenson, 2005). BSK-Lite contains no added carbon sources although carbohydrates are present in essential components of the media such as rabbit serum and yeast extract. Bacteria grown in BSK-Lite reach maximum density after two days of growth. Bacteria cultured in BSK-Lite medium supplemented with a single carbohydrate reach a density that is approximately 10-fold higher than the BSK-Lite cultures after three days of growth (von Lackum & Stevenson, 2005, Hoon-Hanks et al., 2012).
For the screen, the bacteria were cultured in BSK-Lite supplemented with glucose, maltose, mannose, trehalose, or GlcNAc. It should be noted that the BSK-Lite base contains a small amount of GlcNAc in order to support cell wall synthesis. After the cultures reached late exponential phase three days post-inoculation, the bacteria were diluted into fresh medium containing the same carbon source. This served to further eliminate transposon mutants from the population that were unable to efficiently metabolize the available carbon source. The diluted cultures were grown for an additional three days. Tn-seq was performed on genomic DNA isolated from each culture.
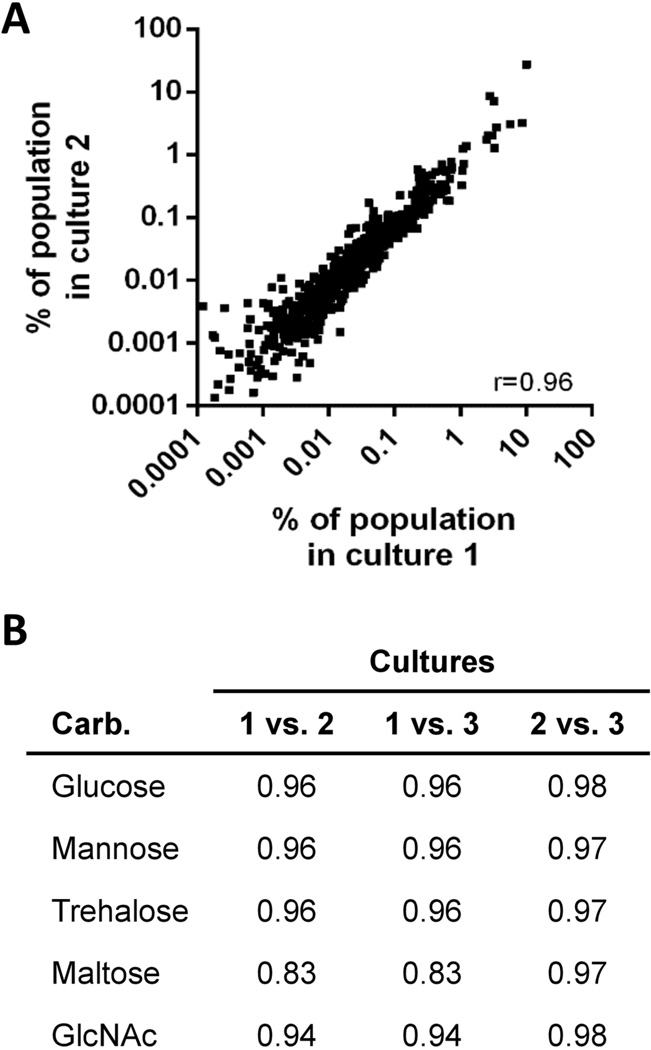
In Tn-seq, the relative abundance of each mutant in a library is determined by sequencing the genomic DNA flanking the transposon en masse. The frequency of a particular sequence is equivalent to the frequency of the transposon mutant containing a transposon insertion at that site within the bacterial population (van Opijnen et al., 2009). The Tn-seq identified 2,570 unique sequences with greater than 10 reads in all glucose samples. This represents transposon insertions in 39% of all annotated genes present in the genome of the parental strain of the transposon library, B31 5A18NP1 (677 of the 1,739 predicted open reading frames). Although this was not a saturated transposon library, to our knowledge it represents the most complete library currently available for B. burgdorferi. When comparing the results between replicates, reproducibility was high between all three cultures containing the same carbohydrate (Fig. 1).
Fig. 1.
Correlation between cultures of bacteria grown in specific carbohydrates. (A) Correlation between the mutant compositions of two cultures of the transposon library grown in glucose. Each data point represents the frequency of bacteria in the population that contained insertions in a specific gene. Only those genes for which there were greater than 10 sequences in all three glucose cultures are shown. Each axis represents a different biological replicate. (B) Spearman correlation coefficients between three bacterial cultures grown in media containing the indicated carbohydrate (Carb.). p-values for all comparisons were <0.001.
The relative fitness of a transposon mutant during growth in media containing a primary carbon source other than glucose was calculated by comparing growth on the alternate carbohydrate with growth on glucose. The frequency of insertions into a particular site or gene was calculated as a percentage of all reads in a given sample. A fitness value (FV) was obtained by dividing the average frequency of an insertion following growth in media containing an alternate carbohydrate by the average frequency following growth in media containing glucose. A fitness value of 1 indicates that the transposon mutant was able to grow equally well in media containing glucose and media containing the specified carbohydrate. A fitness value of less than 1 indicates that the transposon mutant decreased in frequency within the population following growth on the alternate carbohydrate relative to glucose. This implies that the disrupted gene or non-coding regulatory region contributes to optimal bacteria replication during growth on that carbon source.
Alternatively, the transposon mutant may have lost a plasmid necessary for optimal bacterial replication. It is not feasible to re-isolate and check the plasmid content of all individual mutants in the library following screening. However, the plasmid content of the majority of the arrayed mutant stocks was determined prior to inclusion in the library. The plasmids missing from the stocks of the individual transposon mutants highlighted in the analysis below are listed in Table S1 and Table S4. It is important to note that, if the arrayed stock contained more than one transposon mutant, these data would represent a composite of the plasmids present in all mutants in the stock.
Prior to performing confirmatory assays, we verified the purity and plasmid content of the mutants being tested (Table 6). Furthermore, when confirming the growth defects observed in the screen, we tested multiple mutants with insertions in the same gene. In all cases, mutants with insertions in the same gene displayed the same phenotype providing assurance that plasmid loss or unrecognized secondary site mutations are unlikely to be responsible for the phenotype (Fig. 2–5).
Table 6.
Transposon mutants used in single strain and competitive growth curves
| Strain Name Used In This Study |
Original Arrayed STM Stocka |
Description | Plasmids Missing |
Reference |
|---|---|---|---|---|
| bb0408Tn1 | T07TC473* | 5A18NP1 bb0408::Tn (GenR), Insertion Site 421164 | lp5 | This study |
| bb0408Tn2 | T04TC410 | 5A18NP1 bb0408::Tn (GenR), Insertion Site 420810 | lp5 | (Lin et al., 2012) |
| bb0408Tn3 | T11TC520 | 5A18NP1 bb0408::Tn (GenR), Insertion Site 420789 | lp5, lp21 | (Lin et al., 2012) |
| bb0408Tn4 | T08P01G11 | 5A18NP1 bb0408::Tn (GenR), Insertion Site 421348 | lp5 | (Lin et al., 2012) |
| bb0629Tn | T06TC053 | 5A18NP1 bb0629::Tn (GenR), Insertion Site 660192 | None | (Lin et al., 2012) |
| bb0645Tn | T10TC291 | 5A18NP1 bb0645::Tn (GenR), Insertion Site 684111 | lp5, lp28-2 | (Lin et al., 2012) |
| bbb29Tn1 | T04TC008 | 5A18NP1 bbb29::Tn (GenR), Insertion Site 25103 | None | (Lin et al., 2012) |
| bbb29Tn2 | T05TC538* | 5A18NP1 bbb29::Tn (GenR), Insertion Site 25453 | cp32-1 | This study |
| bbb29Tn3 | T06TC064* | 5A18NP1 bbb29::Tn (GenR), Insertion Site 25711 | lp5 | This study |
| rrp1Tn1 | T09TC402* | 5A18NP1 rrp1::Tn (GenR), Insertion Site 429701 | lp5, lp28-1 | This study |
| rrp1Tn2 | T10TC452* | 5A18NP1 rrp1::Tn (GenR), Insertion Site 430422 | lp5, lp28-1, cp32-1, cp32-6 |
This study |
| bb0408com1 | T04TC410/pKFFS1:: p0408::408407 (GenR, StrepR) | cp9, lp5 | This study | |
| bb0408com2 | T04TC410/pKFFS1:: p0408::408407 (GenR, StrepR) | cp9, lp5 | This study |
Strain name used in (Lin et al., 2012)
Arrayed mutant stock was contaminated based on PCR and mutant was re-purified for this study
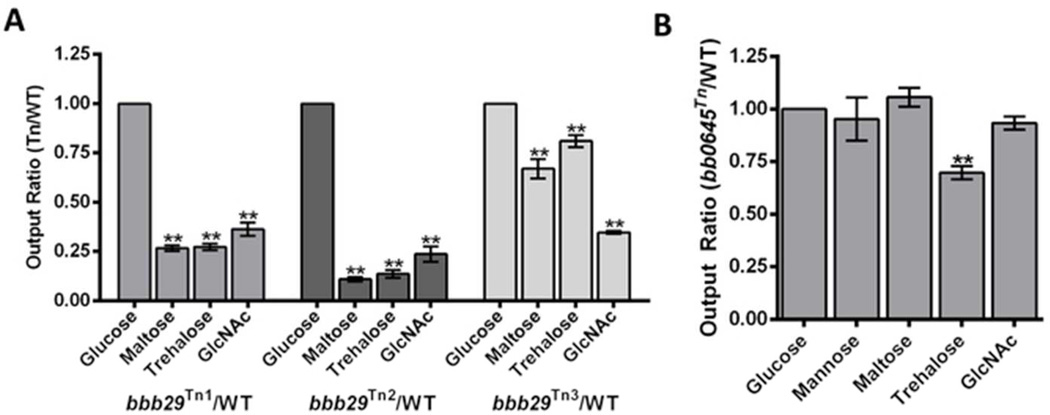
Fig. 2.
Differing contributions of genes encoding PTS Glc transporters to bacterial fitness. Competition assays between B. burgdorferi B31 5A18NP1 and transposon mutants in (A) bbb29 (malX2) and (B) bb0645 (ptsG). The transposon mutants were mixed 1:1 with B31 5A18NP1 and inoculated into BSK-Lite supplemented with the indicated carbohydrates at 1×105 cells/ml. After 5 days, the ratio of transposon mutant to B31 5A18NP1 in the culture as well as the inoculum was determined using qPCR with primers to recA and aacC1 as described in the Material and Methods. The output ratios of all carbohydrates were normalized to glucose. Values are the mean±SEM of three experiments. An asterisk indicates a statistically significant difference between the output ratio in the specific carbohydrate and glucose. Significance was determined using a one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test. *p<0.05; **p<0.01
A fitness value of greater than 1 indicates the transposon mutant increased in frequency within the population following growth in media containing an alternate carbohydrate relative to glucose. This implies that loss of gene function increased bacterial fitness. Alternatively, the transposon insertion could impair glucose metabolism while not affecting utilization of the alternate carbohydrate.
The data were analyzed by gene as well as individual insertion sites. In the per gene analysis, the total number of sequence reads from all insertions in a specific gene were combined to derive a single fitness value associated with that gene (Table S2). We prioritized genes with fitness values greater than 3 or less than 0.33, indicating a greater than 3-fold change in frequency, for further study (Table 1). The fitness values associated with individual insertions are reported in Table S3. The concordance between the results of the whole gene analysis and the analysis of individual insertions in that gene is included in Table 1. We prioritized genes based on fitness value rather than statistical significance because in our analysis we found that, in some cases, large decreases in mutant fitness lacked statistical significance due to variability in the magnitude of the fitness defects between replicates, whereas mutants with small changes in fitness but low variability were statistically significant (Table S2 and Table S3).
Table 1.
B. burgdorferi genes contributing to growth in specific carbohydrates
| Carbohydrate | Loc. | Locus | Name | Gene Fitness Valuea |
% of Insertion Sites In Agreementb |
Annotation |
|---|---|---|---|---|---|---|
| Mannose | ||||||
| chr | bb0407 | manA | 0.002±0.004* | 100 (1/1) | Mannose Isomerase | |
| chr | bb0408 | fruA1 | 0.022±0.015* | 100 (3/3) | PTS system transporter subunit IIABC | |
| lp54 | bba76 | thyX | 0.041±0.077 | 100 (1/1) | Thymidylate synthase | |
| chr | bb0789 | ftsH | 0.096±0.122 | 100 (1/1) | ATP-dependent zinc metalloprotease | |
| lp54 | bba03 | 0.194±0.286 | 100 (1/1) | Lipoprotein | ||
| chr | bb0825 | 0.280±0.388 | 50 (1/2) | Hypothetical protein | ||
| chr | bb0756 | 0.323±0.152 | 100 (1/1) | Hypothetical protein | ||
| cp26 | bbb29 | malX2 | 3.089±2.711 | 33.3 (6/18) | PTS system transporter subunit IIBC | |
| Maltose | ||||||
| chr | bb0155 | 0.301±0.274 | 100 (1/1) | Lipoprotein | ||
| lp54 | bba76 | thyX | 0.310±0.384 | 100 (1/1) | Thymidylate synthase | |
| cp26 | bbb29 | malX2 | 0.321±0.447 | 44 (8/18) | PTS system transporter subunit IIBC | |
| chr | bb0163 | 3.092±2.326 | 100 (1/1) | Hypothetical protein | ||
| lp54 | bba04 | 3.180±4.129 | 50 (1/2) | S2 lipoprotein antigen | ||
| cp32-9 | bbn39 | erpQ | 3.241±2.294 | 50 (1/2) | Outer surface lipoprotein | |
| lp28-2 | bbg30 | 3.776±5.402 | 100 (1/1) | Hypothetical protein | ||
| lp28-1 | bbf14 | 7.008±4.389 | 100 (1/1) | Hypothetical protein | ||
| chr | bb0726 | 9.225±13.320 | 100 (1/1) | ATP-binding protein | ||
| Trehalose | ||||||
| cp26 | bbb29 | malX2 | 0.097±0.085 | 94.4 (17/18) | PTS system transporter subunit IIBC | |
| lp54 | bba76 | thyX | 0.191±0.231 | 100 (1/1) | Thymidylate synthase | |
| lp54 | bba03 | 0.264±0.164* | 100 (1/1) | Surface lipoprotein | ||
| chr | bb0726 | 3.209±2.033 | 100 (1/1) | ATP-binding protein | ||
| GlcNAc | ||||||
| cp26 | bbb29 | malX2 | 0.067±0.079 | 100 (18/18) | PTS system transporter subunit IIBC | |
| cp26 | bbb16 | oppA4 | 0.142±0.118* | 83.3 (5/6) | Oligopeptide permease | |
| chr | bb0420 | hk1 | 0.195±0.190* | 90 (9/10) | Histine kinase 1 | |
| chr | bb0050 | 0.220±0.169 | 100 (4/4) | Hypothetical protein | ||
| chr | bb0224 | 0.267±0.072* | 75 (3/4) | Lipoprotein | ||
| chr | bb0619 | 0.283±0.157 | 100 (1/1) | Phosphoesterase | ||
| chr | bb0157 | 0.315±0.210* | 100 (2/2) | Hypothetical protein | ||
| cp26 | bbb01 | 0.324±0.343* | 100 (1/1) | Acylphosphatase | ||
| lp21 | bbu08 | 3.364±2.497 | 100 (1/1) | SUA5 subfamily putative translation factor | ||
| cp32-4 | bbr43 | 3.547±0.460* | 100 (1/1) | Hypothetical protein | ||
| lp25 | bbe22 | pncA | 3.626±1.230 | 50 (1/2) | Nicotinamidase | |
| lp21 | bbu06 | 3.861±3.551 | 100 (1/1) | Plasmid partition protein | ||
Loc, location of the gene in the B. burgdorferi genome; chr, Chromosome
average fitness value (% of total reads following growth in specified carbohydrate/% of total reads following growth in glucose) of the triplicate cultures±standard deviation
# of insertion sites with a fitness value of >3.00 or <0.33 in agreement with fitness values from the whole gene analysis/total # of insertion sites in different locations within a gene
T-test, p<0.05
B. burgdorferi genes contributing to optimal growth in specific carbohydrates
Overall, transposon mutants with insertions in 26 genes had fitness values ≤0.33 or ≥3.0 following growth in the different carbohydrates (Table 1). The fitness of the greatest number of transposon mutants was affected by growth in media containing GlcNAc as the primary carbon source (bbb29, bbb16, bb0420, bb0050, bb0224, bb0619, bb0157, bbb01, bbu08, bbr43, bbe22, bbu06). This is potentially due to the fact that in addition to being an energy source, GlcNAc is also a precursor in cell wall synthesis. In contrast, only four transposon mutants had increased or decreased fitness following growth in media containing trehalose (bbb29, bba76, bba03, bb0726), and all four of these genes were identified in multiple carbohydrates (Table 1). For example, transposon mutants in bbb29, a putative carbohydrate transporter, displayed decreased fitness in GlcNAc (FV 0.067±0.079), trehalose (FV 0.097±0.085), and maltose (FV 0.321±0.447) but increased fitness in mannose (FV 3.089±2.711). Transposon insertions in bba76 (thyX) had decreased fitness in mannose (FV 0.041±0.077), maltose (FV 0.310±0.384), and trehalose (FV 0.191±0.231). Transposon mutants with insertions in eight genes had fitness values of ≤0.33 or ≥3.0 following growth in media containing mannose (bb0407, bb0408, bba76, bb0789, bba03, bb0825, bb0756, bbb29), and transposon mutants in nine genes were affected following growth in media containing maltose (bb0155, bba76, bbb29, bb0163, bba04, bbn39, bbg30, bbf14, bb0726).
A majority of the genes identified as potentially contributing to carbohydrate metabolism were located on the chromosome, lp54, or cp26 (Table 1). This includes all of the genes potentially contributing to mannose or trehalose metabolism and a majority of the genes potentially contributing to GlcNAc and maltose metabolism. Lp54 and cp26 are two highly conserved plasmids in the B. burgdorferi species (Terekhova et al., 2006, Jewett et al., 2007). The location of the potential carbohydrate metabolism genes on conserved elements of the genome suggests that the ability to utilize multiple carbohydrates is important for optimal B. burgdorferi fitness. The fitness of multiple mutants with insertions in cp32 plasmids was increased during growth in GlcNAc (Table S3). Unfortunately, due to extensive sequence homology between the cp32 plasmids, it is not possible to identify the specific genes on these plasmids that contribute to growth using Tn-seq.
As expected, transposon mutants with insertions in genes encoding the chitobiose and glycerol transporters displayed less than a two-fold change in frequency in the population following growth in the tested conditions relative to the inoculum (Table 2). Transposon mutants with insertions in genes encoding the enzymes in glycolysis were not present in the library likely due to the essentiality of these genes for B. burgdorferi survival in BSK-II, the medium used during the transposon mutagenesis (Lin et al., 2012). There were also no transposon mutants with insertions in genes encoding the phosphotransfer components of the PTS.
Table 2.
Tn-seq analysis of metabolic genes during growth in specific carbohydrates
| Gene Category |
Location | Locus |
Name | Gene Fitness Valuea |
|||
|---|---|---|---|---|---|---|---|
| GlcNAc | Maltose | Trehalose | Mannose | ||||
| PTS | |||||||
| chr | bb0116 | malX1 | 0.79±0.27 | 0.75±0.51 | 1.42±0.92 | 0.81±0.46 | |
| chr | bb0408 | fruA1 | 1.28±0.49 | 1.15±0.74 | 0.77±0.31 | 0.02±0.02* | |
| chr | bb0629 | fruA2 | 0.97±0.18 | 1.38±0.83 | 0.89±0.10 | 0.60±0.12 | |
| chr | bb0645 | ptsG | 1.17±0.81 | 1.19±1.06 | 0.78±0.69 | 1.05±0.70 | |
| cp26 | bbb04 | chbC | 0.74±0.54 | 0.74±0.25 | 0.95±0.13 | 0.72±0.10* | |
| cp26 | bbb05 | chbA | 0.95±0.70 | 0.93±0.29 | 1.19±0.37 | 0.92±0.31 | |
| cp26 | bbb06 | chbB | 0.64±0.61 | 0.75±0.69 | 1.12±0.45* | 1.03±0.37 | |
| cp26 | bbb29 | malX2 | 0.07±0.08 | 0.32±0.45 | 0.10±0.09 | 3.09±2.71 | |
| General Metabolic | |||||||
| chr | bb0241 | glpK | 0.61±0.35 | 0.80±0.40 | 1.00±0.62 | 0.79±0.54 | |
| chr | bb0243 | glpD | 0.43±0.42 | 0.93±1.02 | 0.84±0.92 | 0.63±0.70 | |
| chr | bb0407 | manA | 1.85±0.99 | 1.87±0.43 | 2.00±0.32* | 2.40×10−3 ±4.16×10−3* | |
| Regulatory | |||||||
| chr | bb0419 | rrp1 | 0.00±0.00b | 0.09±0.18 | 0.91±1.16 | 0.40±0.43 | |
| chr | bb0420 | hk1 | 0.20±0.19* | 0.59±0.58* | 0.92±0.50 | 1.07±0.57 | |
chr, Chromosome
average fitness value (% of total reads following growth in specified carbohydrate/% total reads following growth in glucose)±standard deviation
No transposon mutants in rrp1 were detected in the population following growth in GlcNAc
T-test, p<0.05
Confirmation of the in vitro Tn-seq Results
To confirm the results of the Tn-seq screen we focused on genes predicted to encode PTS transporters. Given the small size of the B. burgdorferi genome, the presence of ten genes annotated to encode EII PTS transporter subunits (bb0116, bb0367, bb0408, bb0559, bb0629, bb0645, bbb04, bbb05, bbb06, bbb29) suggests that this method of importing carbohydrates plays a significant role in the enzootic cycle. Transposon mutants with insertions in eight of these genes were identified in the Tn-seq screen (bb0116, bb0408, bb0629, bb0645, bbb04, bbb05, bbb06, bbb29). In the Tn-seq screen, transposon mutants in bbb29 and bb0408 displayed some of the strongest phenotypes (Table 1). As such we chose to focus confirmatory assays on transposon mutants with insertions in these genes and obtained individual transposon mutants from the arrayed library. The stocks of the mutants used in these assays were tested for purity. If the stock contained more than one mutant, the desired mutant was isolated by plating for single colony and the plasmid content determined (Table 6).
A PTS transporter in the Glucose-Glucoside (Glc) family may import maltose, trehalose, and GlcNAc
The PTS Glucose-Glucoside (Glc) family includes transporters of glucose, glucosamine, GlcNAc, and a large variety of α- and β-glucosides. There are three genes annotated in the B. burgdorferi genome as encoding members of the PTS Glc family: bb0645 (ptsG), bb0116 (malX1) and bbb29 (malX2). All three genes are predicted to encode the EIIB and EIIC domains of a PTS transporter. It has been hypothesized that Crr (BB0559) is the EIIA component of these transporters. The fact that Crr may serve as the EIIA for multiple EIIBC transporters may explain why no transposon mutant in crr is present in the library.
Transposon mutants in bbb29 had decreased fitness during growth in GlcNAc, trehalose, and maltose and increased fitness in mannose (Table 1). In competition assays with wild-type bacteria, three transposon mutants with different insertions in bbb29 were outcompeted in media containing maltose, GlcNAc, and trehalose (Fig. 2A). This result suggests that BBB29 may be a PTS transporter capable of importing these carbohydrates. In support of this hypothesis, BBB29 has 39% identity (p=1e−119) to the maltose transporter MalX in E.coli, 30% identity (p=6e−74) to the maltose transporter MalP in Bacillus subtilis and 34% identity (p=5e−79) to the GlcNAc transporter NagP in B. subtilis
The frequency of transposon mutants with insertions in the genes encoding the other two annotated Glc-family PTS transporters, bb0645 and bb0116, remained relatively constant following growth in media containing each of the alternate carbohydrates tested relative to glucose, varying by less than 1.4-fold within the populations (Table 2). This was not unexpected as PTS transporters are not necessarily specific for particular carbohydrates and there may be redundancy, making it difficult to parse out the capabilities of the different transporters. Furthermore, we only investigated four carbohydrates. The bacterium likely encounters a larger variety of carbohydrates during the enzootic cycle. If bb0116 and bb0645 are annotated correctly and encode PTS transporters, their substrates may be different from the four carbohydrates tested in the Tn-seq screen.
To confirm the results of the Tn-seq, a competition assay was performed with wild-type bacteria and an individual transposon mutant in bb0645. Following five days of growth in media containing the different carbohydrates, the ratio between the two strains remained relatively unchanged (Fig. 2B). The transposon mutant was slightly outcompeted by the wild-type strain in trehalose. This trend mirrors what was observed in the Tn-seq screen. The fitness value for the bb0645 transposon mutant following growth on trehalose was 0.78±0.69 as opposed to between 1.05±0.70 and 1.19±1.06 for the other carbohydrates (Table 2).
A PTS transporter in the Fructose-Mannitol (Fru) family contributes to growth in mannose
There are two genes in the B. burgdorferi genome predicted to encode transporters in the PTS Fructose-Mannitol (Fru) family: bb0408 (fruA1) and bb0629 (fruA2). Both of these genes are predicted to encode all three EII domains of a PTS transporter. The PTS Fru family includes a number of fructose, mannose and mannitol transporters. BB0408 and BB0629 were initially annotated as fructose transporters. However, B. burgdorferi cannot grow in the presence of fructose as a primary carbon source, suggesting the genes serve an alternate function (von Lackum & Stevenson, 2005). Fitness values for bb0629 ranged from 0.60±0.12 to 1.38±0.83 suggesting that the gene product does not have a substantial impact on bacterial growth in any of the tested carbohydrates (Table 2). In contrast, mutants with insertions in bb0408 had severely decreased fitness during growth in mannose (FV 0.02±0.02) but not other sugars (Tables 1 and 2). A mutant with an insertion in the gene downstream of bb0408, bb0407 (manA), also had a severe growth defect in mannose (FV 0.002±0.004) (Tables 1 and 2). Although not a PTS transporter, BB0407 is homologous to mannose isomerase, which interconverts mannose-6-phosphate and fructose-6-phosphate, and thus would be predicted to be involved in mannose utilization downstream of entry into the cell.
In a competitive growth assay with wild-type bacteria, the ratio of bb0629 transposon mutants to wild-type bacteria varied less than 1.2-fold between the initial mixed culture and each carbohydrate tested (Fig. 3A). Although some of the variation in the output ratios between carbohydrates was statistically significant, the small magnitude of the differences suggests that BB0629 plays a minor role in bacterial growth in the tested carbohydrates.
Fig. 3.
Differing contributions of genes encoding PTS Man transporters to bacterial fitness. (A) Competition assays between B. burgdorferi B31 5A18NP1 and transposon mutants in bb0629. Assays were performed as described in Fig. 2. (B) Growth of B. burgdorferi strain B31 5A18NP1 (WT) and four different transposon mutants in bb0408 in BSK-Lite or BSK-Lite supplemented with either glucose or mannose. Cultures were inoculated at 1×105 cells/ml and grown for 5 days at 37°C. Cell density was determined by dark field microscopy using a Petroff-Hauser counter. Values are the mean±SEM of three experiments. An asterisk indicates a statistically significant difference between growth in the specified carbohydrate and glucose. Significance was determined using a one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test. *p<0.05; **p<0.01
Four transposon mutants containing insertions in different sites within bb0408 were tested for growth in mannose as the primary carbon source and all showed decreased fitness. During growth in media containing glucose, the transposon mutants were able to grow to the same level as the parental wild-type strain. However, in media containing mannose as a primary carbohydrate source, the transposon mutants were unable to grow beyond the levels observed in BSK-Lite alone, while the WT strain was able to grow to a higher density (Fig. 3B). BB0408 has 43% identity to the mannose transporter in Bacillus subtilis, ManP (p=3e−119).
As fructose-6-phosphate is a glycolytic intermediate, BB0408 and BB0407 may function together to import and process mannose for use in glycolysis. The ability of a transposon mutant in bb0408 to grow in media containing mannose could be restored by expressing bb0408 and bb0407 in trans from a plasmid under the control of the bb0408 promoter (Fig. 4). This indicates that the transposon insertion is responsible for the phenotype observed in the Tn-seq screen and rules out the possibility that the phenotype is due to a second site mutation. However, we were unable to transform a complementation construct expressing only bb0408 into the bb0408 transposon mutant. Thus, we cannot exclude the possibility that the growth phenotype of the bb0408 mutant is due to polar effects on the expression of bb0407.
Fig. 4.
Complementation of the growth defect of a bb0408 transposon mutant in media containing mannose. B. burgdorferi B31 5A18NP1 (WT), bb0408Tn2 and two strains of bb0408Tn2 exogenously expressing bb0407 and bb0408 from the bb0408 promoter (bb0408com1 and bb0408com2) were grown in BSK-Lite or BSK-Lite supplemented with either glucose or mannose. Cultures were inoculated at 1×105 cells/ml and grown for 5 days at 37°C. Cell density was determined by dark field microscopy using a Petroff-Hauser counter. Values are the mean ±SEM of three experiments. *p<0.05; **p<0.01
Rrp1 may play a role in regulating maltose and GlcNAc metabolism through PTS components
The Hk1/Rrp1 two-component system is required for B. burgdorferi survival in the tick during acquisition and transmission of the bacteria between the tick and the vertebrate host. One critical function of this system may be to help the bacteria metabolically adapt to changing conditions. In in vitro assays, Rrp1 is required for growth on glycerol and chitobiose under conditions designed to mimic the tick environment (Sze et al., 2013, He et al., 2011). Furthermore, in microarray and RNA-seq analyses, Rrp1 has been implicated in the upregulation of carbohydrate metabolic genes including the glycerol transport genes, chbC, bb0116, bb0629, and bbb29 (Sze et al., 2013, Rogers et al., 2009, He et al., 2011, Caimano et al., 2015). The results from the Tn-seq screen suggest an additional role for Hk1/Rrp1 in maltose and GlcNAc metabolism. Multiple transposon mutants with insertions in hk1 (bb0420) displayed a fitness defect in media containing GlcNAc (FV 0.195±0.190) (Table 1). No individual transposon mutant in rrp1 had greater than ten sequence reads aligning to the insertion site in all three glucose cultures, a condition required for inclusion of the gene on Table 1. However, when the number of sequence reads aligning to each of the multiple mutants with insertions in rrp1 were added together in the whole gene analysis, there were greater than ten sequences in all three glucose cultures and the fitness values suggest that Rrp1 may contribute to bacterial fitness in media containing GlcNAc or maltose (FV 0.00±0.00 and 0.09±0.18, respectively) (Table 2).
To investigate the potential role of Rrp1 in regulating bacterial growth on these carbohydrates, we tested two transposon mutants with insertions in rrp1 in competition assays with the parental wild-type strain. The transposon mutants were out competed in media containing GlcNAc or maltose as predicted by the Tn-seq screen (Fig. 5A). As further confirmation, we performed competition assays using an independently constructed mutant lacking functional Rrp1 due to the insertion of a streptomycin resistance gene into rrp1 (rrp1−) and the parental wild-type strain. This mutant was used in a previous study to identify the role of Rrp1 in glycerol metabolism (He et al., 2011). In agreement with the growth phenotypes of the transposon mutants, rrp1− was outcompeted by the parental wild-type strain during growth in media containing maltose or GlcNAc as primary carbon sources. Complementation by restoration of expression of Rrp1 in the mutant under the control of the native promoter (rrp1com) allowed the strain to grow on GlcNAc (Fig. 5B). Of note, although rrp1− had fitness defects in GlcNAc, the strain out competed wild-type and rrp1com during growth in glucose, a phenotype that would not have been detected in the Tn-seq analysis.
Fig. 5.
Rrp1 contributes to B. burgdorferi growth in media containing maltose or GlcNAc as primary carbon sources. Competition assays between (A) two transposon mutants in rrp1 and WT (B31 5A18NP1) and (B) rrp1-and B. burgdorferi B31 5A4NP1 (WT) or rrp1com. Assays were performed as described in Fig. 2. An asterisk indicates a statistically significant difference between the output ratio in the specific carbohydrate and glucose. Significance was determined using a one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test. *p<0.05; **p<0.01
As a global gene regulator, the effect of Rrp1 on GlcNAc and maltose metabolism may be through transcriptional regulation of metabolic genes. Previous investigations have shown that expression of bbb29 is reduced in B. burgdorferi mutants lacking rrp1 (Rogers et al., 2009, Caimano et al., 2015). As transposon mutants with insertions in rrp1 and bbb29 have growth defects in GlcNAc and maltose, it is possible that the role of Rrp1 in GlcNAc and maltose metabolism may be through transcriptional regulation of bbb29.
Tn-seq determination of competitive fitness of B. burgdorferi transposon mutants in vivo
We next determined whether in addition to metabolism, PTS transporters play a role in bacterial fitness during vertebrate infection. In a previous investigation, we used Tn-seq to identify B. burgdorferi population bottlenecks in mice by monitoring the composition of a population of eight equally infectious B. burgdorferi transposon mutants during the establishment and maintenance of infection (Troy et al., 2013). As this study suggested Tn-seq could be a viable approach for transposon library screening in vivo, we conducted a Tn-seq screen in mice. However, due to the fact that the earlier study also discovered there was a significant population bottleneck at the site of infection, we performed a targeted screen rather than using the entire transposon library . We created a pool of 103 transposon mutants including mutants with insertions in genes annotated as involved in transport or genes on lp54 and two control strains, a fully infectious transposon mutant with an insertion in the oligopeptide permease peptide binding protein oppA1 (bb0328) and a non-infectious transposon mutant with an insertion in pncA (bbe22) (Purser et al., 2003, Troy et al., 2013). The transporter gene set contained mutants with insertions in predicted PTS transporters as well as other predicted carbohydrate and non-carbohydrate transporters. The set of mutants with insertions in genes on lp54 was chosen because, despite significant genomic variation between subspecies of B. burgdorferi, lp54 is present in all sequenced Lyme disease agent genomes, stable during in vitro propagation and encodes a number of genes known to be required for full infectivity in the mouse (Terekhova et al., 2006, Bestor et al., 2010).
Two groups of ten mice were infected with the mutant pool. The mice were inoculated with 1×105 bacteria representing approximately 1,000 copies of each mutant. A portion of the inoculum was diluted and grown in vitro in BSK-II as a control to confirm that changes in the composition of the population following infection were not due to general growth defects. Although B. burgdorferi are able to maintain long-term colonization in the mouse model, the mice were sacrificed at two weeks post-infection in order to maximize the number of mutants present and potentially reveal more intermediate infectivity phenotypes. The population bottleneck study conducted using the B. burgdorferi transposon mutants demonstrated that mutants present at lower frequencies in mice at two weeks post-infection were often lost by six weeks post-infection (Troy et al., 2013)
At two weeks post-infection, the tibiotarsal joints, knees, ear, and skin at the inoculation site of the infected mice were removed and individually cultured in BSK-II media. All of the samples were positive for B. burgdorferi. The tissues were cultured to expand the population of the bacteria, thereby increasing the limit of detection for minor members of the population during sequencing and reducing the amount of eukaryotic DNA in the sample. Genomic DNA was isolated from the cultured inoculum and organ cultures. In order to minimize the effects of the population bottleneck, the DNA obtained from the four mammalian tissues was pooled prior to genomic library preparation. Thus, each of the two mouse sequencing samples is comprised of bacteria isolated from four tissues from ten infected mice.
A fitness value for each mutant was obtained by dividing its frequency within the population following mouse infection by its frequency in the cultured inoculum. The results from the two experiments indicated that Tn-seq is an accurate and reproducible approach for performing B. burgdorferi transposon mutagenesis screens in the mouse model of infection. There was excellent correlation between the biological replicates of both the cultured inoculum populations and those recovered from the mice (r=0.93 and 0.81 respectively, Spearman correlation coefficient, p<0.0001). Positive and negative control strains behaved as expected. The mean fitness value (MFV) of the pncA insertion mutant was 5.75×10−5 and the MFV of the oppA1 mutant was 3.52 (Tables 3 and 4). The high correlation between experiments suggests that stochastic variability due to bottleneck issues did not inhibit our ability to identify mutants with true fitness changes using this library. The data set for all transporter and lp54 mutants pooled for the in vivo Tn-seq screen is presented in Table S4.
Table 3.
Contribution of the B. burgdorferi transporter gene set to mouse infectivity
| Category | Loc. | Ins. Site | Locus | Gene Name |
Fitness Valuea |
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Mouse Group 1 |
Mouse Group 2 |
MFI in STM Screenb |
Annotation | |||||
| Metabolism | |||||||||
| chr | 113343 | bb0116 | malX1 | 9.33×10−5 | 1.87×10−4 | 0.00c | 38.95 | PTS, EIIABC component | |
| chr | 113729 | bb0116 | malX1 | 4.49×10−5 | 0.00c | 8.98×10−5 | 49.57 | PTS, EIIABC component | |
| chr | 114543 | bb0116 | malX1 | 8.91×10−5 | 1.42×10−4 | 3.61×10−5 | 188.36 | PTS, EIIABC component | |
| chr | 245916 | bb0240 | glpF | 15.26 | 10.48 | 20.04 | 290.11 | Glycerol uptake facilitator | |
| chr | 246891 | bb0241 | glpK | 15.40 | 15.56 | 15.23 | 3579.57 | Glycerol kinase | |
| chr | 249604 | bb0243 | glpD | 0.98 | 0.56 | 1.40 | 226.79 | Glycerol-3-phosphate dehydrogenase | |
| chr | 325023 | bb0318 | mglA | 1.87×10−4 | 9.54×10−5 | 2.78×10−4 | 14.39 | ABC-type sugar transporter | |
| chr | 421166 | bb0408 | fruA1 | 0.05 | 2.25×10−4 | 0.11 | 19.75 | PTS, EIIABC component | |
| chr | 630181 | bb0604 | lctP | 0.00 | 0.00 | 0.00 | 24.71 | L-lactate permease | |
| chr | 661151 | bb0629 | fruA2 | 0.00 | 0.00 | 0.00 | 34.95 | PTS, EIIABC component | |
| chr | 661155 | bb0629 | fruA2 | 1.41 | 0.31 | 2.51 | 5297.91 | PTS, EIIABC component | |
| chr | 684113 | bb0645 | ptsG | 0.00 | 0.00 | 0.00 | 25.18 | PTS, EIIBC component | |
| cp26 | 3500 | bbb04 | chbC | 0.05 | 0.09 | 1.40×10−3 | 1795.95 | PTS, chitibiose EIIC component | |
| cp26 | 3542 | bbb04 | chbC | 1.79 | 2.96 | 0.63 | 1067.25 | PTS, EIIC chitobiose | |
| cp26 | 4228 | bbb05 | chbA | 0.56 | 0.15 | 0.98 | 1921.04 | PTS, EIIA chitobiose | |
| cp26 | 19120 | bbb22 | pbuG1 | 9.93×10−5 | 1.81×10−4 | 1.72×10−5 | 28.14 | Purine permease | |
| cp26 | 25105 | bbb29 | malX2 | 0.01 | 0.03 | 3.39×10−5 | 787.09 | PTS, EIIBC | |
| ABC Transport | |||||||||
| chr | 335333 | bb0328 | oppA1 | 3.52 | 1.70 | 5.34 | N/Dd | Oligopeptide permease | |
| chr | 337852 | bb0329 | oppA2 | 8.03×10−5 | 1.61×10−4 | 0.00 | 186.39 | Oligopeptide permease | |
| chr | 586734 | bb0573 | 0.01 | 0.02 | 0.00 | 23.04 | ABC transporter, ATP-binding protein | ||
| lp54 | 22398 | bba34 | oppA5 | 0.25 | 0.51 | 1.25×10−4 | 20.68 | Oligopeptide permease | |
| lp38 | 19646 | bbj26 | 0.89 | 0.26 | 1.53 | 5550.18 | ABC transporter, ATP-binding protein | ||
| Ion Exchange /RND EffluxPump | |||||||||
| chr | 141502 | bb0140 | besB | 9.31 | 5.18 | 13.45 | 3038.00 | AcrB homolog | |
| chr | 142878 | bb0141 | besA | 1.95 | 0.94 | 2.95 | 46.96 | AcrA homolog | |
| chr | 143306 | bb0142 | besC | 0.00 | 0.00 | 0.00 | 499.71 | TolC homolog | |
| chr | 164996 | bb0164 | 4.99×10−5 | 9.97×10−5 | 0.00 | 53.96 | K+-dependent Na+/Ca+ antiporter | ||
| chr | 675846 | bb0637 | nhaC1 | 0.00 | 0.00 | 0.00 | 39.75 | Na+/H+ antiporter family | |
| chr | 676847 | bb0638 | nhaC2 | 3.63×10−5 | 0.00 | 7.27×10−5 | 28.86 | Na+/H+ antiporter family | |
| chr | 861758 | bb0814 | panF | 2.11 | 1.37 | 2.86 | 535.18 | Na+/symporter | |
| Regulation | |||||||||
| chr | 41270 | bb0042 | phoU | 0.00 | 0.00 | 0.00 | 24.79 | Transport regulatory protein | |
| chr | 144594 | bb0143 | hlyA | 0.09 | 0.18 | 1.06×10−4 | 64.93 | Membrane protein insertion factor | |
| chr | 775398 | bb0733 | plzA | 9.38×10−5 | 6.74×10−5 | 1.20×10−4 | 34.77 | Cyclic di-GMP receptor | |
Loc, location of the gene in the B. burgdorferi genome; chr, Chromosome; Ins. Site, Insertion site
% of total reads in population isolated from mice/% total reads in cultured inoculum
MFI is the mean fluorescence intensity of the mutant in the STM screen (Lin et al., 2012). Mutants with a MFI of less than 100 was considered non-infectious
A fitness value of 0.00 indicates the insertion sequence was not detected by the Tn-seq following infection
The mutant was not tested in the STM screen
Table 4.
Tn-seq analysis of genes on lp54 with previously established effects on mouse infectivity
| Infectivity | Ins. Site |
Locus | Gene Name |
Fitness Valuea |
MFI in STM Screenb |
Annotation | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Group 1 | Group 2 | |||||||
| Infectiousc | |||||||||
| 5049 | bba07 | chpA1 | 1.16 | 1.02 | 1.31 | 2440.71 | lipoprotein | (Xu et al., 2010a) | |
| 10837 | bba16 | ospB | 0.75 | 0.85 | 0.65 | 1575.82 | outer surface protein | (Yang et al., 2004, Neelakanta et al., 2007) |
|
| 4249 | bba05 | 0.19 | 0.37 | 6.15×10−4 | 478.43 | lipoprotein | (Xu et al., 2010b) | ||
| 4123 | bba05 | 0.07 | 2.21×10−4 | 0.15 | 108.46 | lipoprotein | (Xu et al., 2010b) | ||
| 42339 | bba62 | lp6.6 | 0.06 | 1.98E-04 | 0.12 | 59.21 | lipoprotein | (Promnares et al., 2009) | |
| 51718 | bba74 | osm28 | 2.38 | 1.58 | 3.18 | 27.43 | outer membrane porin | (Mulay et al., 2009) | |
| Non-infectious | |||||||||
| 16452 | bba24 | dbpA | 1.31 ×10−4 | 1.77 ×10−4 | 8.48×10−5 | 22.57 | decorin binding protein | (Shi et al., 2008a, Shi et al., 2008b, Weening et al., 2008) | |
| 16508 | bba24 | dbpA | 6.79×10−5 | 9.83×10−5 | 3.74×10−5 | 86.61 | decorin binding protein | ||
| 16973 | bba25 | dbpB | 1.31 ×10−4 | 1.42 ×10−4 | 1.20×10−4 | 130.86 | decorin binding protein | (Weening et al., 2008, Shi et al., 2008a, Shi et al., 2008b) | |
| 38515 | bba57 | 6.88 ×10−4 | 1.29 ×10−3 | 8.18 ×10−5 | 69.46 | outer surface lipoprotein | (Yang et al., 2013) | ||
| 15339 | bbe22 | pncA | 5.75×10−5 | 9.68×10−5 | 1.81×10−5 | 37.83 | nicotinamidase | (Purser et al., 2003) | |
Ins. Site, Insertion site
% of total reads in population isolated from mice/% total reads in cultured inoculum
MFI is the mean fluorescence intensity of the mutant in the STM screen (Lin et al., 2012). Mutants with a MFI of less than 100 was considered non-infectious
B. burgdorferi lacking the gene are able to infect mice following needle inoculation
Comparison of Tn-seq and signature tagged mutagenesis as methods for transposon library screening in B. burgdorferi
Many of the mutants tested in the in vivo Tn-seq screen have previously been tested in a STM screen. In the STM screen, 434 transposon mutants were tested in groups of 11 mutants. Each mutant in a group was associated with a unique fluorescent tag. Three mice infected with each group of transposon mutants were sacrificed at two weeks post-infection and the organs cultured to recover the bacteria. The STM screen identifies mutants that have decreased in frequency after infection by measuring changes in the mean fluorescence intensity (MFI) of the tag associated with each mutant in the input population versus the bacteria isolated from the organ culture (Lin et al., 2012). Table S4 compares the results of the in vivo Tn-seq and STM screens, including the average MFI and the percentage of the tissues that were positive for the transposon mutant. The results of the two methods are not directly comparable due to differences in how the results are reported. However, there was general agreement between the two screens. Of the 41 transposon mutants with a MFV below 0.10 in the Tn-seq (a greater than 10-fold decrease in frequency within the population), 31 had MFI values below the negative cutoff (MFI <100) and were deemed non-infectious in the STM screen (Table S4).
Although we combined 103 individual Tn mutants to generate the inoculum for our in vivo Tn-seq studies, we identified 150 mutants after sequencing. Twelve of the expected mutants were absent and there were 59 additional mutants that we had not intended to include. This is likely due to the presence of multiple Tn mutants in some of the arrayed stocks. The results for the unexpected mutants are included on Table S5. In general, there fewer sequences aligned to the insertion site in the unexpected mutants when compared to the expected mutants. It should be noted that mutants present at a low level in the inoculum (a small number of sequences aligning to the insertion site) are more likely to be lost from the population in the population bottleneck at the site of infection.
This presence of a large number of unexpected mutants highlights an important advantage of Tn-seq. The presence of additional mutants does not interfere with the identification of fitness defects among the expected mutants as all mutants are examined individually by the sequencing technology. In contrast, the presence of additional mutants in the arrayed stocks could have affected the results of the STM screen (Lin et al., 2012). If there were multiple mutants present in an arrayed stock, more than one mutant in a pool may have been labeled with the same fluorescent tag potentially leading to the mis-assignment of phenotypes to individual mutants.
Tn-seq as a method to identify B. burgdorferi transporters contributing to infectivity
Consistent with B. burgdorferi being a fastidious organism, most of the genes in the transporter gene set were identified as contributing to optimal bacterial growth during mouse infection, although there were some exceptions. The transposon mutant with an insertion in oppA1, previously shown to be dispensable for bacterial survival in a mouse, did not have a fitness defect during mouse infection (Table 3, MFV 3.52) (Troy et al., 2013, Wang et al., 2002). In contrast to oppA1, mutants with insertions in the alternate oligopeptide peptide binding protein, oppA2, did have a fitness defect during infection (Table 3, MFV 8.03×10−5). Expression of oppA2 but not oppA1 is regulated by RpoS, the sigma factor that regulates many genes important for infection in mammals. This supports the hypothesis that the substrates recognized by OppA2 are present in the mouse and that transport into the cell may contribute to B. burgdorferi infectivity (Medrano et al., 2007).
B. burgdorferi mutants with insertions in chitobiose and glycerol transport genes generally did not display decreased fitness during growth in the mouse (Table 3). This is consistent with findings suggesting their role is predominantly in the tick host (Pappas et al., 2011, He et al., 2011, Tilly et al., 2001, Sze et al., 2013). Of note, mutants with insertions in the glycerol uptake facilitator glpF (bb0240) and the glycerol kinase glpK (bb0241) had mean fitness values above 15 indicating a significant increase in frequency within the population following mouse infection (Table 3). It is possible that in the absence of glycerol, the expression of GlpK and/or GlpF could interfere with the optimal utilization of the other nutrients. The absence of these proteins during vertebrate infection may increase the efficiency of metabolism of the available nutrients, resulting in an increased growth rate. It is also possible that the apparent fitness advantage may be due to the fact that many of the other mutants displayed infectivity defects and were lost from the population resulting in an increase in frequency of the remaining transposon mutants. However, glpF and glpK increased out of proportion to other neutral genes suggesting that there is a true increase in fitness.
In vivo fitness of mutants with insertions in the carbohydrate PTS genes
The PTS transporters bb0116, bb0408, bb0645, and bbb29 may contribute to bacterial growth during infection as transposon mutants with insertions in these genes displayed severe fitness defects during mouse infection in the in vivo Tn-seq screen (Table 3, MFV<0.05). The transposon mutants in bb0116, bb0408, and bb0645 were similarly non-infectious in the STM screen while the transposon mutant in bbb29 displayed a more intermediate infectivity phenotype (Table 3). In contrast, the results suggest that the predicted PTS Fru transporter bb0629 does not contribute to bacterial fitness in the mouse. In both the Tn-seq and STM screens, one of the transposon mutants with an insertion in bb0629 displayed a neutral infectivity phenotype although another was non-infectious (Table 3, MFV 1.41 and 0.00, MFI 5297.91 and 34.95) (Lin et al., 2012). The same infectivity phenotypes were observed when the strains were tested individually in mice (Khajanchi et al., 2015). Thus, it is possible that the infectivity defect of the non-infectious mutant is unrelated to the transposon insertion.
It should be noted that although transposon mutants in bb0116, bb0408, and bb0645 had severe infectivity defects using both Tn-seq and STM screening techniques, the transposon mutants in bb0116 and bb0408 were infectious when tested individually in single strain infection in mice (Khajanchi et al., 2015, Lin et al., 2012). This underscores an important difference in the information obtained by different techniques. As a large competition assay, the Tn-seq screen may reveal subtle phenotypes missed in traditional single strain infections. A similar phenomenon in mice has been reported for B. burgdorferi lacking luxS (Arnold et al., 2015). For many years, it was thought that luxS mutants were fully infectious (Hubner et al., 2003). However, an infectivity defect was uncovered in a recent publication in which luxS mutants were out competed by wild-type during mouse infection (Arnold et al., 2015).
There is experimental evidence suggesting that bb0116 and bb0408 may play a role in bacterial fitness during infection. Of the PTS transporter genes, bb0116 is the only one that has been identified as regulated by RpoS (Caimano et al., 2007). BB0408 was identified as an immunogenic protein in humans and mice implying it is expressed by B. burgdorferi during vertebrate infection (Barbour et al., 2008). If BB0408 is confirmed as a PTS transporter, the role of BB0408 during infection may be to import the mannose encountered in the vertebrate blood during dissemination.
Role of genes on lp54 in B. burgdorferi infectivity
Demonstrating the accuracy of the Tn-seq technique, mutants with transposon insertions in genes encoding proteins that do not play significant roles in the vertebrate portion of the life cycle (ospB and bba74) and an insertion in bba07, a gene not required for mouse infection following needle inoculation did not significantly decrease in frequency within the population following passage through a mouse (Table 4, MFV 0.75, 2.38, and 1.16, respectively) (Neelakanta et al., 2007, Yang et al., 2004, Xu et al., 2010a). In contrast, insertions in genes known to contribute to mouse infectivity (dbpA, dbpB, bba57, and pncA) were detected at a much lower frequency in the population following mouse infection (MFV <0.001) (Table 4) (Purser et al., 2003, Shi et al., 2008a, Shi et al., 2008b, Weening et al., 2008, Yang et al., 2013).
When tested individually in single strain infections, B. burgdorferi mutants lacking bba05 are fully infectious in all stages of the life cycle and mutants lacking bba62 (lp6.6) have no defect in mouse colonization following needle inoculation (Xu et al., 2010b, Promnares et al., 2009). However, the results of the Tn-seq screen suggest that transposon mutants in bba62 and bba05 have decreased fitness in mice (Table 4, MFV<0.02 ). These apparent infectivity defects were observed in the STM screen as well (Lin et al., 2012). The reconciliation for the different results may again be due to the fact that the screens are competition assays that can reveal subtle phenotypes. Although BBA05 and BBA62 may not be essential for infection, transposon mutants with insertions in bba05 or bba62 may be at a competitive disadvantage in a mixed population. It is also possible that the infectivity defects of the bba05 and bba62 transposon mutants are due to polar effects on downstream genes or non-coding regulatory regions. However, there is some evidence that these proteins have the potential to contribute to bacterial fitness during infection. B. burgdorferi lacking bba62 have decreased infectivity in mice following tick inoculation and BBA62 has been identified as an immunogen in mice and humans (Promnares et al., 2009, Barbour et al., 2008). BBA05 was initially identified as an antigen eliciting an antibody response in Lyme disease patients, is upregulated by the Rrp2-RpoN-RpoS pathway, and is expressed by B. burgdorferi in fed nymphs and in dialysis membrane chambers (DMCs) implanted in the peritoneal cavity of rats (Feng et al., 1995, Iyer et al., 2015, Caimano et al., 2007). Further mechanistic studies are required to fully reconcile the conflicting results and determine the specific function of BBA05 and BBA62 during infection..
Our studies identified 9 mutants that had insertions in genes with uncharacterized infectivity phenotypes and mean fitness values representing greater than a five-fold change in frequency in the population following mouse infection (Table 5). Three of the mutants (bb48, bba50, and bba71) were infectious when tested in the STM screen. This may be due to differences in technique or the arrayed stocks of these mutants may be contaminated with infectious mutants that masked the infectivity defect. Experimentally, antibodies recognizing BBA50 and BBA51 have been found in the cerebral spinal fluid and sera of patients with neurologic Lyme disease suggesting that they may be present on the bacterial surface at some point during vertebrate infection (Fikrig et al., 2004). Together with bba48, bba50 and bba51 may constitute an operon contributing to bacterial fitness during infection. Particularly intriguing is bba71, which is regulated by RpoS and is expressed by B. burgdorferi residing in DMCs (Iyer et al., 2015, Caimano et al., 2007). The genes identified do not have significant homology to any known genes or proteins and are candidates for novel B. burgdorferi infectivity factors warranting further study.
Table 5.
Potential mouse infectivity factors on lp54
| Insertion Site |
Locus | Fitness Valuea |
MFI in STM Screenb |
||
|---|---|---|---|---|---|
| Mean | Group 1 | Group 2 | |||
| 8951 | bba14 | 0.01 | 0.01 | 8.14×10−5 | 19.93 |
| 18257 | bba30 | 8.39×10−5 | 1.22 ×10−4 | 4.62×10−5 | 29.75 |
| 27741 | bba40 | 0.05 | 0.00c | 0.10 | 29.39 |
| 32713 | bba48 | 4.38 ×10−5 | 8.76 ×10−5 | 0.00 | 491.96 |
| 33872 | bba50 | 0.00 | 0.00 | 0.00 | 2089.79 |
| 34774 | bba51 | 0.12 | 0.03 | 0.21 | N/Dd |
| 39966 | bba59 | 0.00 | 0.00 | 0.00 | 27.82 |
| 49809 | bba71 | 3.49 ×10−4 | 3.02 ×10−4 | 3.96 ×10−4 | 584.32 |
| 8485 | bba13 | 5.39 | 3.71 | 7.07 | 1138.54 |
% of total reads in population isolated from mice/% total reads in cultured inoculum
MFI is the mean fluorescence intensity of the mutant in the STM screen (Lin et al., 2012). Mutants with a MFI of less than 100 was considered non-infectious
A fitness value of 0.00 indicates the insertion sequence was not detected by the Tn-seq following infection
The mutant was not tested in the STM screen
CONCLUSIONS
In this manuscript, we have shown for the first time that Tn-seq can be used to conduct both in vitro and in vivo high-throughput genetic fitness screens in B. burgdorferi. We first conducted an in vitro screen to identify genes contributing to B. burgdorferi growth in different carbohydrates. Although this topic has been extensively studied in other pathogens, our understanding of B. burgdorferi metabolism is largely dependent on inferences made by identifying homologs to known genes in other species. While multiple genes in the B. burgdorferi genome were annotated as encoding transporters in the carbohydrate PTS system, their function and relative contribution to bacterial fitness in different growth conditions remained largely untested. The Tn-seq screen revealed potential functions for two predicted PTS transporters: BB0408 and BBB29. Furthermore, we identified a potential role for Rrp1 in the regulation of maltose and GlcNAc metabolism.
In this study, we also validated Tn-seq as a method to identify B. burgdorferi infectivity factors in a vertebrate model of infection. Using a selected pool of transposon mutants, we were able to correctly identify mutants with known phenotypes. We identified several genes that potentially contribute to mouse infectivity that can be explored in future investigations. Taken together, this study opens the door for Tn-seq to be used to rapidly screen and identify genes involved in B. burgdorferi fitness under a large variety of both in vivo and in vitro conditions.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The transformable and infectious B. burgdorferi B31 clone 5A18NP1 is the parental strain for the transposon library used in this study. The signature tagged Mariner himar1 transposon mutagenesis library was constructed using modified versions of the B. burgdorferi transposon vector pGKT. pGKT is derived from pMarGent and has a kanamycin resistance cassette (flaB::aph1) inserted outside the himar1-based transposable element and a gentamicin resistance cassette (flgB::aacC1) inserted within the transposable element. The bacteria were grown in Barbour-Stoenner-Kelly II (BSK-II) media while constructing the library. A detailed explanation of transposon library construction is provided in (Lin et al., 2012). The transposon mutants used in individual and competitive growth assays to confirm screen results are listed in Table 6. The rrp1 insertion mutant (rrp1−) and complemented strain (rrp1com) were a kind gift of Dr. Frank Yang and were derived from the infectious strain of B. burgdorferi B31 5A4. To construct rrp1−, the wild-type rrp1 gene was disrupted with a streptomycin resistance cassette (flaB::aadA). In the complemented strain rrp1com, the mutated rrp1 gene was replaced with a wild-type copy of rrp1 linked to an ermC marker (He et al., 2011).
With the exception of the carbohydrate Tn-seq screen and confirmatory growth assays, the liquid cultures of B. burgdorferi were grown in BSK-II in sealed test tubes at 37°C. When plating the B. burgdorferi transposon mutants for the isolation of single clones, the bacteria were plated in semi-solid BSK-II media and incubated in sealed containers at 32°C and 1% carbon dioxide. Cultures of B31 5A18NP1 were maintained in kanamycin (200 µg/ml). Individual transposon mutants and library pools were maintained in kanamycin (200 µg/ml) and gentamicin (40 µg/ml). B31 5A4NP1 was maintained in kanamycin (200 µg/ml), rrp1− was maintained in streptomycin (100 µg/ml) and rrp1com was maintained in erythromycin (50 µg/ml). No antibiotics were present in the media during the individual and competitive growth assays.
For the carbohydrate Tn-seq screen and confirmatory growth assays, the bacteria were grown in BSK-Lite, a modified BSK-II that contained glucose-free CMRL (United Stated Biologicals, Swampscott, MA) and no added free glucose. BSK-Lite was supplemented with the carbohydrates being tested to a final concentration of 0.4% w/v as described previously (von Lackum & Stevenson, 2005). Concentrated carbohydrate solutions used to supplement the BSK-Lite media were prepared at 20% w/v in distilled water and sterilized by passage through a 0.20 µM filter. All carbohydrates were obtained from Sigma-Aldridge (St. Louis, MO).
Confirmation of the purity of mutant stocks and plasmid typing
Individual transposon mutants were grown in BSK-II. When the cultures reached late exponential phase, the bacteria were spun down and genomic DNA was isolated from the cells using alkaline lysis as described previously (Priem et al., 1997). Briefly, the cultures were spun down at 12,000 rcf for 5 minutes and re-suspended in 50 µl 50 mM NaOH. After a 15-minute incubation at 95°C, 12 µl 1M Tris-HCl pH 7.3 was added to neutralize the solution. A PCR was performed using primers that flanked the insertion site of the transposon mutant expected to be in the frozen stock (Table S6, Primers bb408_For and bbo408_Rev for insertions in bb0408, primers bbb29_For and bbb29_Rev for insertions in bbb29, and primers rrp1_For and rrp1_Rev for insertions in rrp1). The PCR products were then run on a 1% agarose gel. If the mutant stock contained only the desired mutant, the PCR would amplify a single product the size of the native gene and the transposon. The presence of a smaller amplicon the size of the native gene alone indicated the mutant stock was potentially contained with more than one transposon mutant.
If amplicons the size of the native gene were present, a liquid culture of the mixed transposon mutant stock was plated on semisolid BSK-II agarose at a density that would result in single colonies. Ten well-isolated colonies were removed with sterile pipet tips and inoculated into BSK-II. When the cultures reached late exponential phase, a frozen stock was made with half of the culture and the rest was spun down at 12,000 rcf for 5 minutes. Bacterial DNA was isolated from the pellet using alkaline lysis. PCR was performed again using the primers flanking the desired insertion site. If the PCR produced a single product of the correct size, the frozen stock made from the original colony was cultured in BSK and frozen stocks made from the expanded culture. Transposon mutants that had to be re-isolated from the original arrayed stocks are marked with an asterisk on Table 6.
The plasmid typing was done in two separate PCRs using the DNA obtained from the alkaline lysis. The first contained primers to the circular plasmids in B. burgdorferi B31 (cp9, cp26, cp32-1, cp32-3, cp32-4, cp32-6, cp32-7, cp32-8, and cp32-9). The second contained primers to the linear plasmids of B. burgdorferi B31 (lp5, lp17, lp21, lp25, lp28-1, lp28-2, lp28-3, lp28-4, lp36, lp38, lp54, and lp56). Primer sequences were obtained from (Bunikis et al., 2011).
Genetic complementation of the bb0408 transposon mutant
The complementation construct for the bb0408 transposon mutant was generated by first amplifying the contiguous bb0408 and bb0407 gene cluster with the native bb0408 promoter from the B. burgdorferi chromosome by PCR using primers p408(Kpn1)-F and bb407(XbaI)-R, which contained engineered restriction sites to facilitate downstream genetic manipulation (Table S6). The p0408::408407 construct was cloned into the PCR2.1 vector according to the manufacturer’s protocol (Life Technologies, Grand Island, NY). The expression construct was then excised from PCR2.1 using the KpnI and XbaI restriction sites and subcloned into the B. burgdorferi shuttle vector pKFSS1 resulting in the complementation plasmid gl408407 (Frank et al., 2003). The content of gl408407 was confirmed by PCR, restriction digest, and DNA sequencing. The gl408407 plasmid was transformed into the bb0408 transposon mutant bb408Tn2(T04TC410) using electroporation, resulting in the transformants bb0408com1 and bb0408com2 (Table 6). Electroporation was performed as described previously (Lin et al., 2012, Lin et al., 2009). Transformants were confirmed by PCR and sequencing.
In vitro carbohydrate Tn-seq screen
To construct a pool of the entire arrayed transposon library, 70–80 individual transposon mutants were used to inoculate a single culture and grown to a final density of approximately 3×107–5×107 bacteria/ml. Subsequently, aliquots of nine or ten cultures were mixed in equal amounts to create pools of up to 700 mutants. Aliquots of these pools were combined into one sample to generate the whole library. Frozen stocks were made from the pools of 70–80 mutants, 700 mutants and the entire library.
When performing the Tn-seq screen, a frozen stock of the entire transposon library was grown overnight in BSK-II media. The culture were centrifuged and re-suspended in BSK-Lite to a concentration of 6×106 B. burgdorferi/ml. The bacteria were then diluted 1:10 in 6 ml of BSK-Lite supplemented with one of the carbohydrates tested. After 3 days of growth, 6×105 B. burgdorferi were removed and added to 6 ml of BSK-Lite supplemented with the same carbohydrate as the original culture. Following three days of additional growth, the bacteria were centrifuged and the pellet frozen for genomic DNA isolation. The experiment was performed in triplicate.
In vivo mouse Tn-seq screen
The pool of transposon mutants used in the mouse screen was created by growing up individual cultures of 38 transporter and 62 lp54 mutants to a density of 3×107–5×107 B. burgdorferi/ml. The cultures were combined into groups of ten mutants and frozen stocks were made. Prior to mouse inoculation, overnight cultures of each group of 10 mutants as well as individual cultures of transposon mutants in pncA (bbE22), oppA1 (bb0328), and bb0051 were grown in BSK-II medium at 37°C. Cell density of the 13 cultures was determined by dark-field microscopy. The cultures were mixed such that the inoculum contained approximately equal numbers of all transposon mutants.
Adult C57BL/6 mice were used in all experiments. The mice were originally obtained from Jackson Laboratory (Bar Harbor, ME) and bred at Tufts University School of Medicine. All mice were housed in microisolator cages and provided with antibiotic-free food and water. Mice were injected subcutaneously at the base of the tail with 1×105 B. burgdorferi. As a control to prevent in vitro growth defects from affecting the results of the in vivo screen, 1×105 organisms were cultured in 5 ml BSK-II broth supplemented with kanamycin and gentamicin. When bacteria in the culture reached late exponential phase (5×107/ml-1×108/ml) the bacteria were centrifuged for 20 minutes at 3,000 rcf and the pellet frozen at −80°C.
The mice were sacrificed two weeks post-infection. Skin samples from the injection site and the right ear as well as the knee and tibiotarsal joints from each mouse were removed under aseptic conditions and cultured individually in 5 ml of BSK-II broth supplemented with kanamycin and gentamicin. The cultures were checked daily for growth. When the density of the cultures reached late exponential phase, the bacteria were centrifuged and the pellet frozen as described above. All organ cultures from a particular tissue were combined prior to centrifugation. The experiment was performed in duplicate.
Construction and sequencing of the DNA libraries
Genomic DNA was obtained from the frozen bacteria pellets using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) as per the manufacturer’s instructions. Libraries for sequencing were constructed as described previously (Troy et al., 2013). Briefly, an aliquot of the genomic DNA was placed in a 2 mL microfuge tube and sheared by sonication. Cytosine tails (C-tails) were added to 1 µg sheared DNA using terminal deoxynucleotidyl transferase (TdT, Promega, Madison, WI). Transposon containing fragments were amplified in a PCR containing DNA from the TdT reaction as template and primers specific to the ColE1 site on the 5’ end of the transposon, pMargent1 and the C-tail, olj376 (Table S6). To prepare the DNA for sequencing and further amplify the transposon-genomic DNA junction, a nested PCR reaction was performed using DNA from the first PCR as a template, a primer specific to the transposon end, pMargent2, and an indexing primer containing the specific sequences required for sequencing on an Illumina platform and where NNNNNN represents a six-base-pair barcode sequence allowing samples to be multiplexed in a single sequencing lane (Table S6). Within an experiment, a unique indexing primer was used for each individual B. burgdorferi sample. A majority of the PCR products were between 200 and 600 bp. For the in vivo samples, separate sequencing libraries were constructed from individual tissues. The libraries made from each tissues were pooled at equal concentrations prior to sequencing. The pooled libraries were sequenced on an Illumina HiSeq 2500 at the Tufts University Core Facility as 50 bp single-end reads using the custom sequencing primer pMargent3 and the standard Illumina index primer (Table S6).
Sequencing data analysis
Sequenced reads were demultiplexed using the index sequence on the primers. A separate fastq file was generated for for each sample. All downstream data analysis was done using the Galaxy platform (Blankenberg & Hillman-Jackson, 2014, Doerks et al., 2002, Goecks et al., 2010). The C-tail was removed from the sequence reads, and reads shorter than 30 bp were discarded. The remaining reads were filtered for quality. Reads for which 90% of the cycles did not have a quality score of greater than 15 were discarded. The remaining reads were aligned to the B. burgdorferi B31 genome using the short read aligner Bowtie. A custom script was then used to compile the resulting SAM files into a Microsoft Excel spreadsheet with the number of reads aligned to each site or annotated gene listed.
In the in vitro carbohydrate screen, the number of sequence reads obtained for each carbohydrate sample ranged from 7.6×106 to 9.9×106 with an average of 8.9×106. The data was analyzed in two ways. First by combining the number of reads from all insertions in a gene together to obtain a single fitness value for the gene. Second by examining the number of reads associated with each insertion site separately. The reads associated with a particular site in the genome represent an individual mutant in the transposon library. The frequency of transposon mutants with insertions in a particular site or gene in the population was assessed by determing the number of sequence reads aligned to that mutant or gene as a percentage of all reads in a given sample. A fitness value was obtained by dividing the average frequency of transposon mutants with insertions in the particular site or gene following growth in one of the alternate carbohydrates by the frequency in glucose. For inclusion in the analysis, each of the glucose cultures had to have at least 10 reads in all three glucose cultures. There were reads that aligned to multiple locations in the B. burgdorferi genome. Bowtie divided these reads between the all genome locations containing that sequence. This resulted in some sequences being aligned to lp28-4 and lp-56, plasmids not present in the parental strain of the transposon library, B31 5A18NPI. Sequences that aligned to multiple locations in the genome were excluded from Table 2. All of the sequencing results are presented in Table S2 and Table S3.
In the in vivo mouse screen, the number of sequence reads obtained for the mouse samples ranged from 2.92×106 to 5.36×106 with an average of 4.60×106. Reads that could be aligned to multiple locations in the genome were excluded from the analysis. The sequencing data was analyzed solely by insertion site. For inclusion in the analysis, both of the inoculum cultures had to have at least 10 reads. A fitness value was obtained by dividing the frequency of a particular sequence in the population following passage through a mouse by the frequency of that sequence in the cultured inoculum.
Single strain in vitro growth assays
Individual cultures of B31 5A18NP1 and the indicated transposon mutants were grown overnight in BSK-II. The following day, the bacteria were spun down at 8,000 rcf for 5 min. and re-suspended in BSK-Lite to a concentration of 1×106 bacteria/ml. The bacteria were added to BSK-Lite or BSK-Lite supplemented with the carbohydrate being tested in a 1.5 mL reaction tube for a final concentration of 105 bacteria/ml. After five days of growth, cell density was determined by dark-field microscopy.
Competitive Growth Assays
Individual cultures of indicated mutants and the corresponding wild-type B. burgdorferi (B31 5A18NP1 or B31 5A4) were grown overnight. The following day, an equal number of wild-type and mutant bacteria were mixed in BSK-Lite. The mixture was added to BSK-Lite or BSK-Lite supplemented with the carbohydrate being tested in a 1.5 mL reaction tube to a final concentration of 1×105 bacteria/ml. To determine the ratio of mutant bacteria to wild-type in the initial inoculum, 107 bacteria were spun down and the pellet frozen at −80°C. At five days post-inoculation, genomic DNA was isolated from the mixed cultures as well as the pellet of the starting mixture using alkaline lysis as described above (Priem et al., 1997). DNA was stored at −80°C.
The ratio of mutant bacteria to wild-type in the original inoculum and following five days of growth was determined using quantitative PCR (qPCR). qPCR was carried out using SYBR® Green Mastermix (Bio-Rad, Hercules, CA). Equal volumes of the cell lysates from each sample were analyzed with primers to B. burgdorferi recA to determine the total number of B. burgdorferi in the sample and primers specific to the gentamicin resistance gene on the pMarGent transposon, the streptomycin resistance gene in rrp1− or the erythromycin gene in rrp1com to determine the number of mutant bacteria in the sample (Table S6). Genomic DNA isolated from 5×107 B. burgdorferi was used to generate a standard curve in all amplification reactions. In the data analysis, the ratio of transposon mutant to wild-type B. burgdorferi recovered following growth in media containing the alternate carbohydrates was normalized to the ratio of mutant to wild-type B. burgdorferi grown in media containing glucose (set to 1).
Correlation between the frequencies of each insertion in the population between samples was determined by calculating the Spearman correlation coefficient. T-tests were used to assess the statistical significance of changes in the frequency of transposon mutants in the population following growth in the alternate carbohydrates relative to growth in glucose. One-way analysis of variance (ANOVA) with Dunnett’s Multiple Comparison Test was used to assess the significance of the differences between strains in the single strain and competitive growth assays. Significance was defined by a p-value of ≤0.05.
Supplementary Material
Acknowledgments
The authors thank Dr. X. Frank Yang for generously sharing the Rrp1 genetic mutants. We are grateful for the scientific advice and insight provided by Drs. Meghan Ramsey and Xin Li. This project was funded by the following grants from the U.S. National Institutes of Allergy and Infectious Diseases: R21AI111317 (L.T.H and T.L), R21AI103905 (L.T.H. and T.L) and F32AI098388 (E.B.T.)
Footnotes
Table S1. Plasmid content of transposon mutants on Tables 1 and 2
Table S2. Fitness values of all genes identified by the Tn-seq in the carbohydrate screen
Table S3. Fitness values of all transposon insertions identified by the Tn-seq in the carbohydrate screen
Table S4. Tn-seq and STM results for transport and lp54 insertion mutants
Table S5. Tn-seq results for mutants not expected in mouse Tn-seq screen
Table S6: Primer sequences used in this study
REFERENCES
- Ake FM, Joyet P, Deutscher J, Milohanic E. Mutational analysis of glucose transport regulation and glucose-mediated virulence gene repression in Listeria monocytogenes. Mol Microbiol. 2011;81:274–293. doi: 10.1111/j.1365-2958.2011.07692.x. [DOI] [PubMed] [Google Scholar]
- Alton G, Hasilik M, Niehues R, Panneerselvam K, Etchison JR, Fana F, Freeze HH. Direct utilization of mannose for mammalian glycoprotein biosynthesis. Glycobiology. 1998;8:285–295. doi: 10.1093/glycob/8.3.285. [DOI] [PubMed] [Google Scholar]
- Angel TE, Luft BJ, Yang X, Nicora CD, Camp DG, 2nd, Jacobs JM, Smith RD. Proteome analysis of Borrelia burgdorferi response to environmental change. PloS one. 2010;5:e13800. doi: 10.1371/journal.pone.0013800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile . Nucleic acids research. 2012;40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A, Martin-Verstraete I, Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol. 2011;79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- Arnold WK, Savage CR, Antonicello AD, Stevenson B. Apparent role for Borrelia burgdorferi LuxS during mammalian infection. Infection and immunity. 2015;83:1347–1353. doi: 10.1128/IAI.00032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG. Isolation and cultivation of Lyme Disease spirochetes. The Yale Journal of Biology and Medicine. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi . Infection and immunity. 2008;76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor A, Stewart PE, Jewett MW, Sarkar A, Tilly K, Rosa PA. Use of the Cre-lox recombination system to investigate the lp54 gene requirement in the infectious cycle of Borrelia burgdorferi . Infection and immunity. 2010;78:2397–2407. doi: 10.1128/IAI.01059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Hillman-Jackson J. Analysis of next-generation sequencing data using Galaxy. Methods in molecular biology. 2014;1150:21–43. doi: 10.1007/978-1-4939-0512-6_2. [DOI] [PubMed] [Google Scholar]
- Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global Analysis of Borrelia burgdorferi Genes Regulated by Mammalian Host-Specific Signals. Infection and immunity. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis I, Kutschan-Bunikis S, Bonde M, Bergstrom S. Multiplex PCR as a tool for validating plasmid content of Borrelia burgdorferi . J Microbiol Methods. 2011;86:243–247. doi: 10.1016/j.mimet.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Bykowski T, Woodman ME, Cooley AE, Brissette CA, Wallich R, Brade V, Kraiczy P, Stevenson B. Borrelia burgdorferi complement regulator-acquiring surface proteins (BbCRASPs): Expression patterns during the mammal-tick infection cycle. International journal of medical microbiology : IJMM. 2008;(298 Suppl 1):249–256. doi: 10.1016/j.ijmm.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Dunham-Ems S, Allard AM, Cassera MB, Kenedy M, Radolf JD. Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission. Infection and immunity. 2015;83:3043–3060. doi: 10.1128/IAI.00315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Molecular microbiology. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona A, Schwartz I. Borrelia burgdorferi: Carbon Metabolism and the Tick-Mammal Enzootic Cycle. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MBP-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome research. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Das S, Lam T, Flavell RA, Fikrig E. A 55-kilodalton antigen encoded by a gene on a Borrelia burgdorferi 49-kilobase plasmid is recognized by antibodies in sera from patients with Lyme disease. Infection and immunity. 1995;63:3459–3466. doi: 10.1128/iai.63.9.3459-3466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikrig E, Coyle PK, Schutzer SE, Chen M, Deng Z, Flavell RA. Preferential presence of decorin-binding protein B (BBA25) and BBA50 antibodies in cerebrospinal fluid of patients with neurologic Lyme disease. J Clin Microbiol. 2004;42:1243–1246. doi: 10.1128/JCM.42.3.1243-1246.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA Confers Streptomycin Resistance in Borrelia burgdorferi . Journal of bacteriology. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi . Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Ouyang Z, Troxell B, Xu H, Moh A, Piesman J, Norgard MV, Gomelsky M, Yang XF. Cyclic di-GMP is essential for the survival of the lyme disease spirochete in ticks. PLoS Pathog. 2011;7:e1002133. doi: 10.1371/journal.ppat.1002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol Microbiol. 2013;88:1176–1193. doi: 10.1111/mmi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon-Hanks LL, Morton EA, Lybecker MC, Battisti JM, Samuels DS, Drecktrah D. Borrelia burgdorferi malQ mutants utilize disaccharides and traverse the enzootic cycle. FEMS immunology and medical microbiology. 2012;66:157–165. doi: 10.1111/j.1574-695X.2012.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Revel AT, Nolen DM, Hagman KE, Norgard MV. Expression of a luxS Gene Is Not Required for Borrelia burgdorferi Infection of Mice via Needle Inoculation. Infection and immunity. 2003;71:2892–2896. doi: 10.1128/IAI.71.5.2892-2896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Trzeciakowski JP, Skare JT. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. Journal of bacteriology. 2007;189:437–445. doi: 10.1128/JB.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae . Journal of bacteriology. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Caimano MJ, Luthra A, Axline D, Jr, Corona A, Iacobas DA, Radolf JD, Schwartz I. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Molecular microbiology. 2015;95:509–538. doi: 10.1111/mmi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MW, Byram R, Bestor A, Tilly K, Lawrence K, Burtnick MN, Gherardini F, Rosa PA. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi . Molecular microbiology. 2007;66:975–990. doi: 10.1111/j.1365-2958.2007.05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata H, Norris SJ, Watanabe H. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infection and immunity. 2004;72:7147–7154. doi: 10.1128/IAI.72.12.7147-7154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajanchi BK, Odeh E, Gao L, Jacobs MB, Philipp MT, Lin T, Norris SJ. Phosphoenolpyruvate Phosphotransferase System Components Modulate Gene Transcription and Virulence of Borrelia burgdorferi . Infection and immunity. 2015 doi: 10.1128/IAI.00917-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Yun J, Yoon H, Park C, Kim B, Jeon B, Kim D, Ryu S. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic acids research. 2007;35:1822–1832. doi: 10.1093/nar/gkm060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, Norris SJ. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi . PLoS pathogens. 2009;5:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT, Norris SJ. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS One. 2012;7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano MS, Ding Y, Wang XG, Lu P, Coburn J, Hu LT. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi . J Bacteriol. 2007;189:2653–2659. doi: 10.1128/JB.01760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzendorfer H, Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. The Journal of experimental biology. 2003;206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- Miller CL, Karna SL, Seshu J. Borrelia host adaptation Regulator (BadR) regulates rpoS to odulate host adaptation and virulence factors in Borrelia burgdorferi . Molecular microbiology. 2013;88:105–124. doi: 10.1111/mmi.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B. Temporal Analysis of Borrelia burgdorferi Erp Protein Expression throughout the Mammal-Tick Infectious Cycle. Infection and immunity. 2003;71:6943–6952. doi: 10.1128/IAI.71.12.6943-6952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay VB, Caimano MJ, Iyer R, Dunham-Ems S, Liveris D, Petzke MM, Schwartz I, Radolf JD. Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. J Bacteriol. 2009;191:2783–2794. doi: 10.1128/JB.01802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelakanta G, Li X, Pal U, Liu X, Beck DS, DePonte K, Fish D, Kantor FS, Fikrig E. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS pathogens. 2007;3:e33. doi: 10.1371/journal.ppat.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. Profiling of Temperature-Induced Changes in Borrelia burgdorferi Gene Expression by Using Whole Genome Arrays. Infection and immunity. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas CJ, Iyer R, Petzke MM, Caimano MJ, Radolf JD, Schwartz I. Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS pathogens. 2011;7:e1002102. doi: 10.1371/journal.ppat.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet S, Milohanic E, Mazé A, Nait Abdallah J, Aké F, Larribe M, Deghmane AE, Taha MK, Dozot M, De Bolle X, Letesson JJ, Deutscher J. Correlations between carbon metabolism and virulence in bacteria. Contrib Microbiol. 2009;16:1–15. doi: 10.1159/000219374. [DOI] [PubMed] [Google Scholar]
- Priem S, Rittig MG, Kamradt T, Burmester GR, Krause A. An optimized PCR leads to rapid and highly sensitive detection of Borrelia burgdorferi in patients with Lyme borreliosis. J Clin Microbiol. 1997;35:685–690. doi: 10.1128/jcm.35.3.685-690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promnares K, Kumar M, Shroder DY, Zhang X, Anderson JF, Pal U. Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Molecular microbiology. 2009;74:112–125. doi: 10.1111/j.1365-2958.2009.06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Molecular microbiology. 2003;48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes RG, Atoyan JA, Nelson DR. The chitobiose transporter, chbC, is required for chitin utilization in Borrelia burgdorferi . BMC microbiology. 2010;10:21. doi: 10.1186/1471-2180-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, Marconi RT. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Molecular microbiology. 2009;71:1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Xu Q, McShan K, Liang FT. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi . Infection and immunity. 2008a;76:1239–1246. doi: 10.1128/IAI.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Xu Q, Seemanaplli SV, McShan K, Liang FT. Common and unique contributions of decorin-binding proteins A and B to the overall virulence of Borrelia burgdorferi . PloS one. 2008b;3:e3340. doi: 10.1371/journal.pone.0003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Smith A, Choi YH, Yang X, Pal U, Yu A, Li C. Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi . Infection and immunity. 2013;81:1775–1787. doi: 10.1128/IAI.00050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terekhova D, Iyer R, Wormser GP, Schwartz I. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. Journal of bacteriology. 2006;188:6124–6134. doi: 10.1128/JB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Elias AF, Errett J, Fischer E, Iyer R, Schwartz I, Bono JL, Rosa P. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi . Journal of bacteriology. 2001;183:5544–5553. doi: 10.1128/JB.183.19.5544-5553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Grimm D, Bueschel DM, Krum JG, Rosa P. Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector borne and zoonotic diseases. 2004;4:159–168. doi: 10.1089/1530366041210738. [DOI] [PubMed] [Google Scholar]
- Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infection and immunity. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy EB, Lin T, Gao L, Lazinski DW, Camilli A, Norris SJ, Hu LT. Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infection and immunity. 2013;81:2347–2357. doi: 10.1128/IAI.00266-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lackum K, Stevenson B. Carbohydrate utilization by the Lyme borreliosis spirochete, Borrelia burgdorferi . FEMS microbiology letters. 2005;243:173–179. doi: 10.1016/j.femsle.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Wang XG, Lin B, Kidder JM, Telford S, Hu LT. Effects of Environmental Changes on Expression of the Oligopeptide Permease (opp) Genes of Borrelia burgdorferi . Journal of bacteriology. 2002;184:6198–6206. doi: 10.1128/JB.184.22.6198-6206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Hook M, Skare JT. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infection and immunity. 2008;76:5694–5705. doi: 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He M, He JJ, Yang XF. Role of the surface lipoprotein BBA07 in the enzootic cycle of Borrelia burgdorferi . Infection and immunity. 2010a;78:2910–2918. doi: 10.1128/IAI.00372-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He M, Pang X, Xu ZC, Piesman J, Yang XF. Characterization of the highly regulated antigen BBA05 in the enzootic cycle of Borrelia burgdorferi. Infection and immunity. 2010b;78:100–107. doi: 10.1128/IAI.01008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Qin J, Promnares K, Kariu T, Anderson JF, Pal U. Novel microbial virulence factor triggers murine lyme arthritis. The Journal of infectious diseases. 2013;207:907–918. doi: 10.1093/infdis/jis930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. The Journal of experimental medicine. 2004;199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.