Abstract
Under normal conditions, basal levels of wild-type p53 promote mitochondrial function through multiple mechanisms. Remarkably, some missense mutations of p53, in contrast to the null state, can result in the retention of its metabolic activities. These effects are particularly prominent in the mitochondria and demonstrate a functional role for mutant p53 in cancer metabolism. This review summarizes accumulating data on the mechanisms by which p53 missense mutations can regulate mitochondrial metabolism and promote the viability and survival of both normal and cancer cells, thus acting as a double edged sword for the host. Greater understanding of these mechanisms may provide insights for developing new treatment or preventive strategies against cancer.
Introduction
Cancer is a disease driven by genomic instability that causes the accumulation of DNA mutations, which then promote its aberrant growth [1]. p53 protein, encoded by the human TP53 gene, acts to suppress this process through various cellular mechanisms including DNA damage repair, cell cycle regulation, and cell death [2,3]. Based on the Cancer Gene Census mutation database, the pivotal role of TP53 in tumorigenesis has been further underscored by the high frequency (36.1%) of its somatic mutations in cancer patients across 20 tissues, the most of any known gene [4]. Only PIK3CA (14.3%) and BRAF (10%), the second and third most frequently mutated genes, were altered in 10% or more of patients out of the remaining 197 cancer genes. The somatic mutations of TP53 found in human cancers occur in domains of the gene that are most conserved amongst mammals, highlighting the importance of its wild-type activity in maintaining normal cells [5]. In contrast to other tumor suppressor genes, which frequently have frameshift or nonsense mutations, most mutations of TP53 are missense mutations suggesting that altered forms of p53 protein may play a role in tumorigenesis [6].
In 1969, Li and Fraumeni described a familial cancer syndrome, inherited in an autosomal-dominant manner, with a predisposition to multiple primary tumors at an early age [7]. Li-Fraumeni syndrome (LFS) was found to be caused by germline mutations of the TP53 gene [8]. Consistent with the higher frequency of missense TP53 mutations in tumors, the majority of LFS patients also carry missense mutations. In addition, patients with missense mutations develop cancer at an earlier age compared with those carrying deletion mutations [9]. Thus, even from a clinical perspective, the loss of wild-type activity by a missense mutation in the TP53 gene is not equivalent to a haploinsufficiency state caused by gene deletion. In the laboratory, mutant p53 has been shown to display gain-of-function activities through different mechanisms, including the interference of the transactivating activities of p63 and p73, and acquiring new transactivation target genes that promote cell proliferation such as EGFR, MYC and MAP2K3 [10–12].
Within the past decade, there has been a resurgence of interest in the role of metabolism in cancer biology [13–15]. Mitochondrial metabolism in particular appears to be critical for the selection and survival of cancer cells, with varying outcomes dependent on factors like tumor type and microenvironment [16,17]. For example, parallel studies utilizing different approaches have demonstrated the prominent role of the mitochondria in glioblastoma, lymphoma and melanoma [18–20]. Mitochondria are regulated through multiple pathways by wild-type p53, so these recent reports of the role of mitochondria in cancer biology provide impetus for investigating whether mutant p53, commonly overexpressed in human cancers, can also regulate the mitochondria [21]. Here we summarize some of the findings of a growing body of work that suggests that mutant p53 can regulate metabolism and mitochondrial function.
Mutant p53 and metabolism
Hundreds of different somatic and germline mutations of TP53 have been reported, but only a minority have been biologically characterized (IARC TP53 Database) [6]. Therefore, it is important to stipulate that the properties of one specific somatic or LFS germline mutation cannot be generalized to all variants of p53. This makes drawing general conclusions from studies of mutant p53 challenging, but nonetheless, specific p53 mutations can still be informative for investigating the role of aberrant mitochondrial activity in cancer biology. Under normal conditions, wild-type p53 is short-lived and expressed at low levels, but mutant p53 protein can accumulate to high levels in cancer cells [22]. The loss of wild-type p53 transcriptional activation of MDM2 that destabilizes p53 protein can contribute to increased mutant p53 levels by eliminating a major negative feedback control. However, there also appear to be other factors involved in its abnormal accumulation [23]. From an evolutionary perspective of cancer development, the relatively late acquisition of TP53 mutations and frequent overexpression of mutant p53 protein in colon cancer suggest that it may result in cellular fitness required for malignant progression in this specific type of tumor [1,24,25]. Compared to p53 null mice, those harboring LFS hot-spot mutations of p53 (human p53 R175H and R273H missense mutations) display a distinct tumor spectrum with more aggressive metastatic potential [26,27]. Similarly, skin carcinomas in mice harboring the p53 R172H missense mutation (human R175H homolog) are more aggressive and metastatic compared to that from p53 null mice [28]. These observations begin to describe the gain-of-function properties exhibited by mutant p53 which are distinct from p53 null state.
Increased glycolysis is observed in many types of cancer cells, but this metabolic shift can also cause increased oxidative stress and DNA damage [16,29–31]. One of the earliest p53-regulated metabolic genes identified was TP53-induced Glycolysis and Apoptosis Regulator (TIGAR), which inhibits glycolysis by hydrolyzing an allosteric activator of the glycolytic enzyme 6-phosphofructo-1-kinase (PFK-1) [32]. This activity can indirectly increase antioxidant capacity by shunting glucose into the pentose phosphate pathway for glutathione biosynthesis. Therefore, TIGAR could be tumor suppressive by decreasing oxidative DNA damage [32]. On the other hand, a subsequent study has shown that the overexpression of TIGAR in human colon cancers can promote both tumorigenesis and the regeneration of normal tissue, demonstrating that cancer cells can also benefit from the metabolic activities of TIGAR [33]. p53 can also down-regulate other components of glycolysis through the destabilization of phosphoglycerate mutase and the repression of the glucose transporters 1, 3 and 4 [34,35]. In the absence of wild-type p53 activity, insulin receptor is overexpressed demonstrating another pathway by which p53 inhibits glycolysis [36].
The dissociation of wild-type p53 metabolic activities from classic cell cycle regulation has highlighted the importance of what was previously thought to be of lesser concern in tumorigenesis prevention. While cell cycle arrest, senescence and apoptosis are well-established tumor suppressive processes, a recent study elegantly demonstrated that even with the loss of these typical p53 cell cycle control pathways, its metabolic activities were sufficient to suppress thymic lymphoma development [37]. A p53 acetylation mutant mouse (p533KR/3KR) with the loss of three acetylation sites necessary for cell cycle arrest, senescence and apoptosis retains its capacity to inhibit glycolysis and reactive oxygen species (ROS) generation. Like wild-type p53, p533KR can still upregulate mitochondrial glutaminase 2 (GLS2), which promotes mitochondrial metabolism and redox homeostasis, and repress the expression of SLC7A11, a cystine-glutamate exchanger which promotes ferroptosis in response to oxidative stress [38,39]. The delineation of important cellular processes through specific structural modifications of p53 has helped to define the various roles of p53 mutants in tumor metabolism and cell survival.
Mutant p53 regulation of mitochondrial biogenesis and function
The promotion of mitochondrial respiration by p53 was first attributed to its transcriptional regulation of Synthesis of Cytochrome c Oxidase 2 (SCO2), an essential gene for the assembly of complex IV (cytochrome c oxidase) [40,41]. However, growing evidence indicates that p53 promotes mitochondrial respiration through multiple pathways that include both transcriptional and post-translational mechanisms involving mitochondrial biogenesis genes such as Mitochondrial Transcription Factor A (TFAM) (Figure 1) [21]. The marked decrease in swimming and running endurance of p53 null mice compared with p53 wild-type mice demonstrated the physiological importance of p53 in aerobic metabolism [41–43]. Notably, like p533KR, the LFS “hot-spot” TP53 R175H mutation retains the mitochondrial biogenesis activities of wild-type p53 and can amplify them due to its unregulated expression [37,44]. Cells and tissues of p53 R172H knockin mice (human R175H homolog) display a mutant allele dose-dependent increase in both mRNA and protein levels of TFAM and SCO2 with a concomitant loss in the expression of CDKN1A, the prototypical p53 target gene encoding p21 [44]. This dissociation between the cell cycle and metabolic activities of p53 also helps to conceptualize the important distinction between the p53 mutant and p53 null states. There is an opposite effect on exercise capacity between the homozygous states of p53 null (p53-/-) and p53 R172H mutation (p53H/H) mice compared to p53 wild-type mice (Figure 2) even though p21 transactivation is similarly lost in these two models [42–44]. In a translational study of patients with Li-Fraumeni syndrome, noninvasive P-31 magnetic resonance spectroscopy of skeletal muscle showed evidence of increased oxidative phosphorylation capacity as measured by phosphocreatine recovery kinetics after exercise [44]. These observations were further supported by mitochondrial studies using both blood and skeletal muscle needle biopsy specimens, therefore indicating that general observations in mouse LFS models may be applicable to humans in the clinics.
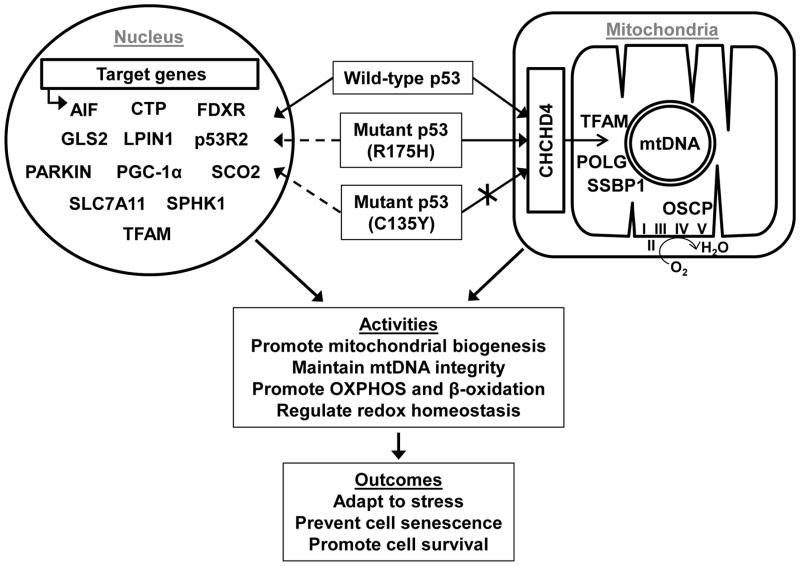
Figure 1. Transcriptional and post-translational regulation of the mitochondria by p53 is determined by multiple factors including its mutation status and translocation into the mitochondria.
Depicted are three possible genotype states: wild-type p53, mutant p53 (R175H) that can translocate into the mitochondria; and mutant p53 (C135Y) that cannot translocate into mitochondria (crossed arrow) due to disruption of its interaction with the disulfide relay protein import carrier CHCHD4. Proteins that have been reported to interact with p53 inside the mitochondria are shown along with schematic representations of circular mtDNA and respiratory complexes. p53 of all three indicated genotypes can translocate into the nucleus, but the ability to transactivate the indicated genes involved in mitochondrial function may be altered (dashed arrows) depending on its mutation status and the specific p53 target gene. Abbreviations: I, II, III, IV, and V, mitochondrial OXPHOS complexes; AIF, apoptosis-inducing factor; CTP, citrate transporter protein; FDXR, ferredoxin reductase; GLS2, mitochondrial glutaminase 2; LPIN1, lipin 1; p53R2, p53-inducible ribonucleotide reductase 2; PGC-1α, peroxisome-proliferator-activated receptor gamma co-activator-1α; SCO2, synthesis of cytochrome c oxidase 2; SLC7A11, solute carrier family 7 (cationic amino acid transporter, y+ system), member 11; SPHK1, sphingosine kinase 1; SSBP1, single-stranded DNA-binding protein; TFAM, transcription factor A, mitochondrial; OSCP, oligomycin sensitivity-conferring protein; OXPHOS, oxidative phosphorylation.
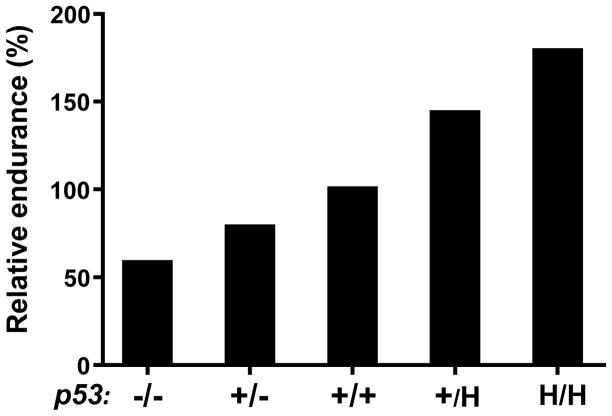
Figure 2. Effect of p53 genotype on relative exercise endurance as a marker of aerobic metabolic capacity.
This is a schematized composite graph based on the maximal treadmill exercise times of mice that are heterozygous or homozygous for p53 gene deletion or R172H knockin mutation (mouse homolog of the human LFS R175H mutation) compared to wild-type mice (set at 100% relative endurance) [43,44]. p53 wild-type (+/+), heterozygous knockout (+/−), homozygous knockout or null (−/−), heterozygous R172H knockin (+/H), homozygous R172H knockin (H/H).
Besides transactivating SCO2 expression, p53 is also known to transcriptionally regulate other mitochondrial biogenesis genes such as Apoptosis-Inducing Factor (AIF) gene, which is involved in complex 1 assembly, and Ferredoxin Reductase (FDXR), which is responsible for the maturation of Fe-S proteins essential in electron transfer reactions [41,45–48]. While the regulation of AIF and FDXR by mutant p53 has not been reported, the mitochondrial citrate transporter protein (CTP, encoded by SLC25A1) is induced by several mutant forms of p53, including the LFS TP53 R175H and R273H mutations, but not by the p53 null state [49]. CTP, localized in the inner membrane, facilitates the exchange of mitochondrial citrate for cytosolic malate, which stimulates respiration and helps maintain inner membrane integrity [50]. CTP is required for tumor proliferation, and it has been proposed to serve as a negative prognostic marker as its level and activity are elevated in human cancers [49,50]. p53 also regulates fatty acid metabolism through genes such as Sphingosine Kinase 1 (SPHK1) and Lipin 1 (LPIN1), which can promote cell growth and mediate nutritional and genotoxic stress signals [51,52]. In summary, p53 displays multiple transcriptional activities in promoting mitochondrial function, which is important for cell viability as well as human fitness. However, the retention and augmentation of these activities by the overexpressed mutant p53 in cancer cells ultimately compromises longer term organismal survival.
Mitochondrial genomic maintenance
The mitochondrial genome (mtDNA) encodes only 13 proteins, however, maintaining its integrity is paramount because each protein is essential for respiration and oxidative phosphorylation [53]. Like its role in maintaining the nuclear genome, there is increasing evidence that p53 is also involved in mtDNA homeostasis. mtDNA depletion is observed in both cultured cells and tissues with p53 deficiency, which has been attributed to p53-inducible ribonucleotide reductase 2 (p53R2) and TFAM down-regulation [43,54,55]. p53R2 functions to supply deoxyribonucleotides and repair nuclear DNA damage, but it is also necessary for mtDNA maintenance [56,57]. Paradoxically, high p53R2 expression was also observed in a number of different cancer types, and is associated with more malignant characteristics, thus raising speculation that mutated p53 may play a role in upregulating p53R2 expression [58].
p53 has been shown to interact with mitochondrial matrix proteins and participate in the repair of mtDNA damage primarily through base excision repair (BER) [21,59]. p53 interacts with DNA polymerase γ (POLG), TFAM, and single-stranded DNA-binding protein 1 (SSBP1) to repair and maintain mtDNA [60–62]. At least three different mechanisms have been proposed to explain how p53 translocates into mitochondria under non-apoptotic conditions [63–65]. One mechanism of interest is the import of p53 into the mitochondria via the coiled-coil-helix-coiled-coil-helix domain-containing protein 4 (CHCHD4), a carrier of the respiration-driven disulfide relay system [65]. Because electron transfer during active respiration may be associated with ROS generation, this import mechanism could sub-cellularly target p53 to the site of DNA damage where its repair activities are needed. Indeed, the accelerated repair of oxidative mtDNA damage has been shown to be dependent on the Cys-135 amino acid residue of p53 that is necessary for interaction with CHCHD4 and import into the mitochondria [65]. Given the multifaceted roles of p53, the CHCHD4 mediated translocation of p53 represents an ideal homeostatic mechanism by which its mtDNA repair activity can be targeted to the site of oxidative stress.
The CHCHD4-mediated repair of oxidative mtDNA damage is preserved in the p53 R175H mutant, both in vitro and in mouse tissues [65]. Remarkably, recent work has demonstrated decreased prevalence of mtDNA mutations in colon tumors, which frequently harbor TP53 mutations, relative to adjacent normal tissue [66]. Thus, it could be of interest to correlate such an observation with TP53 mutation status of the primary tumors. One prediction would be that colon cancers with p53 alterations that preserve CHCHD4 interaction and are sufficiently functional to exist as both somatic or germline mutation, such as p53 R175H, would maintain mtDNA integrity. On the other hand, tumors with severe p53 mutations that disrupt its translocation into mitochondria and are only encountered as somatic mutations, such as p53 C135Y, may show greater mtDNA instability [65].
Other mechanisms of mitochondrial regulation by mutant p53
There is further evidence that the translocation of p53 into the mitochondria impacts mitochondrial function and homeostasis. A mitochondrial pool of p53 has been reported to interact with the oligomycin sensitivity-conferring protein (OSCP) to promote complex V (F1F0-ATP synthase) assembly, increase respiration, and decrease ROS production [67]. Parkin, an E3 ubiquitin ligase responsible for initiating mitophagy of damaged mitochondria, can be both transcriptionally and post-translationally regulated by p53 [68,69]. Because cytosolic p53 is known to inhibit autophagy, the significant correlation between the high level of cytosolic mutant p53 expression and increased autophagy inhibition suggests that mutant p53 may also modulate parkin-mediated autophagy and mitophagy [70,71].
The role of p53 in regulating metabolism is becoming increasingly difficult to define and predict as there can be contradictory results in cultured cells and tissues dependent on the type of cellular stress, p53 expression levels and p53 mutation status. Under normal, relatively unstressed conditions, p53 promotes mitochondrial respiration that is essential for redox homeostasis, while highly induced p53 can be pro-oxidant and apoptotic [30,72]. This dual nature is exemplified by p53 modulation of the master mitochondrial biogenesis regulator, peroxisome-proliferator-activated receptor γ co-activator-1α (PGC-1α), expression levels [73,74]. Basal levels of wild-type p53 can stimulate PGC-1α expression and mitochondrial biogenesis in mice, while severe cellular stress associated with telomere dysfunction and increased p53 levels can repress both PGC-1α and PGC-1β expression [42,75]. DNA interaction studies show that p53 binds to the −2317 region on the mouse PGC-1α promoter and can increase PGC-1α expression, while its binding to elements at −564 and −954 suppresses its expression [75,76]. In addition to its dosage or activation state, p53 mutation status is also likely to be critical for determining how p53 interacts with the cis regulatory elements of PGC-1α.
Relevance to health and cancer
Generally, metabolic genes function to maintain the homeostasis of cells for growth and adaptation to their environment. It is clear that p53-regulated metabolism under normal conditions contributes to cellular homeostasis without which there are gross deficiencies in cellular and physiological function. Some examples include mtDNA depletion in p53 null cells and tissues and the poor intrinsic exercise capacity or adaptation to exercise training of p53 null mice, both of which could be critical for survival during evolution [41–43,54,55]. Some germline LFS TP53 mutations may retain or amplify p53 metabolic function, with resultant improvements in bioenergetic, thermogenic or antioxidant capacity that could be beneficial to both cellular and organismal survival under stressful conditions [44]. However, these advantages could be deleterious to the TP53 mutation carrier when normal cells transform into cancer cells which can use this mechanism for aggressive growth. In support of this notion, recent studies have shown that PGC-1α expression mediates increased mitochondrial respiration and resistance to oxidative stress in cancer cells and correlates with their metastatic potential [20,77]. A genetic analysis of the multi-cancer TCGA datasets comprised of >9000 primary and metastatic tumor samples have demonstrated the potential clinical significance of mutant p53-regulated SCO2 expression [78]. Both mutated TP53 state and high SCO2 mRNA expression, as well as amplification of SCO2 gene, significantly correlated with poor patient survival [78]. These observations of mutant p53 promotion of mitochondrial activity and correlation with poor clinical outcomes suggest that targeting the mitochondria may be one avenue for restraining tumorigenesis. Indeed, growing evidence from disparate investigations suggest that increased mitochondrial biogenesis and antioxidant capacity are conducive for cancer progression, while disrupting mitochondrial function inhibits it [79–84]. The effect of mutant p53 on metabolic regulation appears to be important for tumorigenisis and thus, as recently suggested for therapy-resistant cancers, may represent a selective vulnerability that can be used to inhibit their growth [85]. Cancer cells are adept at utilizing the double edged sword of mutant p53, but as with most advantages there are also disadvantages to which cancer is not immune.
Acknowledgments
We wish to thank Jie (Jerry) Li for assistance with the artwork and other members of our laboratory Ju-Gyeong Kang, Ji-Hoon Park, and Jie Zhuang for helpful discussions and critical review of the manuscript. This work was supported by the intramural program of National Heart, Lung and Blood Institute (NHLBI), NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
* of special interest
** of outstanding interest
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 5.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 6.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71:747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 8.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 9.Bougeard G, Sesboue R, Baert-Desurmont S, Vasseur S, Martin C, Tinat J, Brugieres L, Chompret A, de Paillerets BB, Stoppa-Lyonnet D, et al. Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J Med Genet. 2008;45:535–538. doi: 10.1136/jmg.2008.057570. [DOI] [PubMed] [Google Scholar]
- 10.Ludes-Meyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, Bigger JE, Brown DR, Deb SP, Deb S. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol. 1996;16:6009–6019. doi: 10.1128/mcb.16.11.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol. 1998;18:3735–3743. doi: 10.1128/mcb.18.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bossi G, Marampon F, Maor-Aloni R, Zani B, Rotter V, Oren M, Strano S, Blandino G, Sacchi A. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle. 2008;7:1870–1879. doi: 10.4161/cc.7.12.6161. [DOI] [PubMed] [Google Scholar]
- 13.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vander Heiden MG. Exploiting tumor metabolism: challenges for clinical translation. J Clin Invest. 2013;123:3648–3651. doi: 10.1172/JCI72391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 17.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, Polak K, Tondera D, Gounarides J, Yin H, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lago CU, Sung HJ, Ma W, Wang PY, Hwang PM. p53, aerobic metabolism, and cancer. Antioxid Redox Signal. 2011;15:1739–1748. doi: 10.1089/ars.2010.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozhok AI, DeGregori J. Toward an evolutionary model of cancer: Considering the mechanisms that govern the fate of somatic mutations. Proc Natl Acad Sci U S A. 2015;112:8914–8921. doi: 10.1073/pnas.1501713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Kinzler KW. The Path to Cancer --Three Strikes and You’re Out. N Engl J Med. 2015;373:1895–1898. doi: 10.1056/NEJMp1508811. [DOI] [PubMed] [Google Scholar]
- 26.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, Lozano G, Roop DR. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest. 2007;117:1893–1901. doi: 10.1172/JCI31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 30.Sung HJ, Ma W, Wang P-y, Hynes J, O’Riordan TC, Combs CA, McCoy JP, Bunz F, Kang J-G, Hwang PM. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:1–8. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang SJ, Gu W. To be, or not to be: functional dilemma of p53 metabolic regulation. Curr Opin Oncol. 2014;26:78–85. doi: 10.1097/CCO.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 33**.Cheung EC, Athineos D, Lee P, Ridgway RA, Lambie W, Nixon C, Strathdee D, Blyth K, Sansom OJ, Vousden KH. TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev Cell. 2013;25:463–477. doi: 10.1016/j.devcel.2013.05.001. p53-inducible TIGAR was initially reported to suppress tumor growth. However, this study shows that TIGAR deficiency can reduce tumor burden and increase survival in a mouse intestinal adenoma model and that increased expression of TIGAR may promote human colon cancer progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 35.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 36.Webster NJ, Resnik JL, Reichart DB, Strauss B, Haas M, Seely BL. Repression of the insulin receptor promoter by the tumor suppressor gene product p53: a possible mechanism for receptor overexpression in breast cancer. Cancer Res. 1996;56:2781–2788. [PubMed] [Google Scholar]
- 37**.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. Tumor Suppression in the Absence of p53-Mediated Cell-Cycle Arrest, Apoptosis, and Senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. This study demonstrates that retention of p53 regulated metabolic activity with loss of cell-cycle arrest, apoptosis and senescence by an acetylation mutant of p53 (p533KR) is sufficient to suppress lymphomagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blum R, Kloog Y. Metabolism addiction in pancreatic cancer. Cell Death Dis. 2014;5:e1065. doi: 10.1038/cddis.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 41.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 42.Saleem A, Adhihetty PJ, Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 43.Park JY, Wang PY, Matsumoto T, Sung HJ, Ma W, Choi JW, Anderson SA, Leary SC, Balaban RS, Kang JG, et al. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ Res. 2009;105:705–712. 711–712. doi: 10.1161/CIRCRESAHA.109.205310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Wang PY, Ma W, Park JY, Celi FS, Arena R, Choi JW, Ali QA, Tripodi DJ, Zhuang J, Lago CU, et al. Increased oxidative metabolism in the Li-Fraumeni syndrome. N Engl J Med. 2013;368:1027–1032. doi: 10.1056/NEJMoa1214091. This translational study of Li-Fraumeni syndrome (LFS) patients and a LFS mouse model shows that germline mutations of p53 can increase mitochondrial function and exercise capacity. It demonstrates LFS mutations of p53 can retain their mitochondrial activities while losing cell cycle control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, Vogelstein B. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, et al. AIF deficiency compromises oxidative phosphorylation. Embo J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stambolsky P, Weisz L, Shats I, Klein Y, Goldfinger N, Oren M, Rotter V. Regulation of AIF expression by p53. Cell Death Differ. 2006;13:2140–2149. doi: 10.1038/sj.cdd.4401965. [DOI] [PubMed] [Google Scholar]
- 48.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 49*.Kolukula VK, Sahu G, Wellstein A, Rodriguez OC, Preet A, Iacobazzi V, D’Orazi G, Albanese C, Palmieri F, Avantaggiati ML. SLC25A1, or CIC, is a novel transcriptional target of mutant p53 and a negative tumor prognostic marker. Oncotarget. 2014;5:1212–1225. doi: 10.18632/oncotarget.1831. This study shows that the mitochondrial citrate transporter is a direct transcriptional target of multiple p53 mutants and can promote mitochondrial respiration and tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catalina-Rodriguez O, Kolukula VK, Tomita Y, Preet A, Palmieri F, Wellstein A, Byers S, Giaccia AJ, Glasgow E, Albanese C, et al. The mitochondrial citrate transporter, CIC, is essential for mitochondrial homeostasis. Oncotarget. 2012;3:1220–1235. doi: 10.18632/oncotarget.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Assaily W, Rubinger DA, Wheaton K, Lin Y, Ma W, Xuan W, Brown-Endres L, Tsuchihara K, Mak TW, Benchimol S. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 52.Heffernan-Stroud LA, Helke KL, Jenkins RW, De Costa AM, Hannun YA, Obeid LM. Defining a role for sphingosine kinase 1 in p53-dependent tumors. Oncogene. 2012;31:1166–1175. doi: 10.1038/onc.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787:328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 57.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 58.Yousefi B, Rahmati M, Ahmadi Y. The roles of p53R2 in cancer progression based on the new function of mutant p53 and cytoplasmic p21. Life Sci. 2014;99:14–17. doi: 10.1016/j.lfs.2014.01.063. [DOI] [PubMed] [Google Scholar]
- 59.de Souza-Pinto NC, Harris CC, Bohr VA. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene. 2004;23:6559–6568. doi: 10.1038/sj.onc.1207874. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 61.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. Embo J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong TS, Rajagopalan S, Townsley FM, Freund SM, Petrovich M, Loakes D, Fersht AR. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic Acids Res. 2009;37:568–581. doi: 10.1093/nar/gkn974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boopathi E, Srinivasan S, Fang JK, Avadhani NG. Bimodal protein targeting through activation of cryptic mitochondrial targeting signals by an inducible cytosolic endoprotease. Mol Cell. 2008;32:32–42. doi: 10.1016/j.molcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De S, Kumari J, Mudgal R, Modi P, Gupta S, Futami K, Goto H, Lindor NM, Furuichi Y, Mohanty D, et al. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J Cell Sci. 2012;125:2509–2522. doi: 10.1242/jcs.101501. [DOI] [PubMed] [Google Scholar]
- 65.Zhuang J, Wang PY, Huang X, Chen X, Kang JG, Hwang PM. Mitochondrial disulfide relay mediates translocation of p53 and partitions its subcellular activity. Proc Natl Acad Sci U S A. 2013;110:17356–17361. doi: 10.1073/pnas.1310908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O’Sullivan J, Salk JJ, Bielas JH. Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PLoS Genet. 2012;8:e1002689. doi: 10.1371/journal.pgen.1002689. Cancer cells have been thought to have increased mtDNA mutations, but this study reports the surprising finding of decreased frequencies of mtDNA mutations in colorectal cancer. The authors suggest that preservation of mtDNA integrity could promote cancer cell survival and disease progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergeaud M, Mathieu L, Guillaume A, Moll UM, Mignotte B, Le Floch N, Vayssiere JL, Rincheval V. Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F(1)F0-ATP synthase. Cell Cycle. 2013;12:2781–2793. doi: 10.4161/cc.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, Matoba S. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 70.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 71.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhuang J, Ma W, Lago CU, Hwang PM. Metabolic regulation of oxygen and redox homeostasis by p53: lessons from evolutionary biology? Free Radic Biol Med. 2012;53:1279–1285. doi: 10.1016/j.freeradbiomed.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–890. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Austin S, St-Pierre J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 75.Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aquilano K, Baldelli S, Pagliei B, Cannata SM, Rotilio G, Ciriolo MR. p53 orchestrates the PGC-1alpha-mediated antioxidant response upon mild redox and metabolic imbalance. Antioxid Redox Signal. 2013;18:386–399. doi: 10.1089/ars.2012.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77*.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. 1001–1015. doi: 10.1038/ncb3039. The mitochondrial biogenesis master regulator PGC-1α can be coopted in cancers cells to improve mitochondrial function for proliferation and metastasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nath A, Chan C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci Rep. 2016;6:18669. doi: 10.1038/srep18669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Salem AF, Tsirigos A, Lamb R, Sneddon S, Hulit J, Howell A, Lisanti MP. Mitochondria “fuel” breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle. 2012;11:4390–4401. doi: 10.4161/cc.22777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Sci Transl Med. 2014;6:221ra215. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- 81.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Fryknas M, Hernlund E, Fayad W, De Milito A, Olofsson MH, Gogvadze V, Dang L, Pahlman S, Schughart LA, et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat Commun. 2014;5:3295. doi: 10.1038/ncomms4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83*.Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. This paper demonstrates the essential role of mtDNA for cancer cell growth. [DOI] [PubMed] [Google Scholar]
- 84.Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolf DA. Is reliance on mitochondrial respiration a “chink in the armor” of therapy-resistant cancer? Cancer Cell. 2014;26:788–795. doi: 10.1016/j.ccell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]