Abstract
Cardiac mitochondria produce vast amounts of ATP through oxidative phosphorylation to maintain contractile function. They are also the primary source of reactive oxygen species, which contribute to mitochondrial dysfunction, cardiomyocyte death, and heart failure. To protect against mitochondrial damage, cardiomyocytes develop well-coordinated quality control mechanisms that maintain the overall mitochondrial health through mitochondrial biogenesis, mitochondrial dynamics, and mitochondrial autophagy (mitophagy). Mitophagy removes dysfunctional mitochondria in the heart not only under normal physiological conditions, but also in response to pathological stresses. Accumulating evidence suggests that mitophagy dysregulation can induce cardiomyocyte death and cardiomyopathy. In this review, we discuss what is currently known about mitophagic mechanisms, regulatory pathways, and function in the heart.
Keywords: mitochondria, autophagy, mitophagy, cardiomyopathy, heart failure
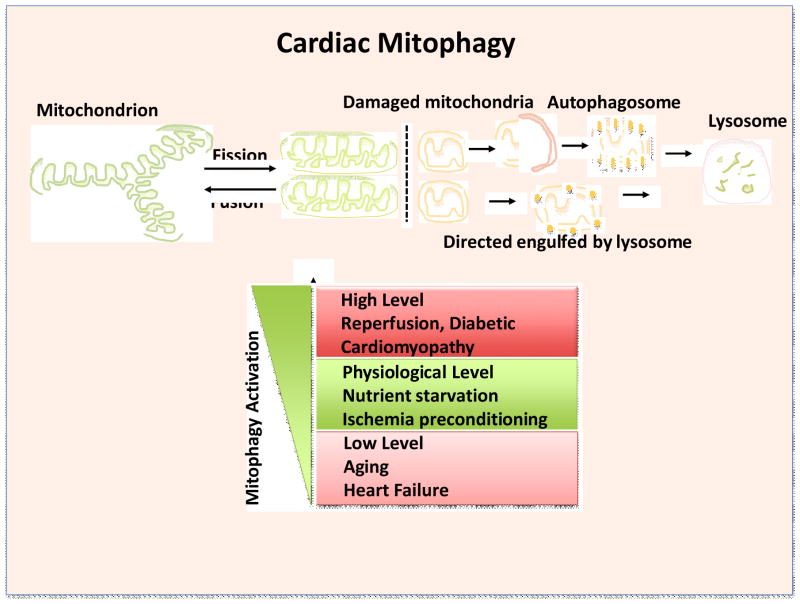
Maintenance of mitochondrial function and integrity is essential for normal cell physiology and survival. This is particularly true in cells with high-energy demand such as cardiomyocytes, where mitochondria comprise approximately 30% of the total cell volume and generate vast amounts of ATP through oxidative phosphorylation to maintain contractile function[1]. In addition, mitochondria regulate cell survival and death, including apoptosis and necrosis, by integrating cellular signals, including reactive oxygen species (ROS), Ca2+, and NAD+/NADH. To protect against stresses, cells have well-coordinated quality control mechanisms that maintain the overall health of mitochondria, including fusion, fission, mitochondrial autophagy[2] or mitophagy[3], and mitochondrial biogenesis[4,5] (Figure 1). It remains to be clarified whether autophagic removal of damaged mitochondria in cardiomyocytes is mediated by a truly mitochondria-specific form of autophagy, termed mitophagy, or by general autophagy. Morphologically, however, autophagosomes containing only mitochondria can be observed in electron microscopic analyses of heart sections, supporting the presence of mitophagy in cardiomyocytes[6]. Removal of damaged and dysfunctional mitochondria through autophagy and mitophagy appears particularly important in terminally differentiated cells such as cardiomyocytes, as evidenced by the high basal level of mitophagic activity[7] and the fact that accumulation of protein aggregates in dysfunctional cardiomyocytes is commonly accompanied by the suppression of autophagy[8]. Mitochondrial DNA is also degraded during the mitophagic process by DNaseII in lysosomes. Incomplete digestion of mitochondrial DNA induces inflammation in the heart and causes heart failure[9].
Figure 1. Outline of mitophagy in cardiomyopathy.
Mitophagy is essential for cardiac mitochondrial quality control. The regulation of mitophagy must be precise. Insufficient or exacerbated mitophagy results in cardiomyopathy. An appropriate level of mitophagy protects the heart by increasing mitochondrial health. However, mitophagy is suppressed below physiological levels in some cardiac conditions, such as aging and heart failure, which leads to mitochondrial dysfunction and cell death and exacerbates cardiac dysfunction. On the other hand, mitophagy may be stimulated excessively in some conditions, including ischemia/reperfusion and diabetic cardiomyopathy, and this may be harmful for the heart, although whether excessive mitophagy induces death of cardiomyocytes requires more investigation.
Mitophagy removes dysfunctional mitochondria in the heart under normal physiological conditions, and during nutrient starvation, ischemic preconditioning (IPC), myocardial infarction (MI), ischemia and reperfusion (I/R), and cardiac hypertrophy[10]. In order to determine the precise role of mitophagy in cardiomyocytes, reliable tools to accurately assess mitophagy are essential. Traditional methods have been successfully used to analyze mitophagy in cardiomyocytes[11], including electron microscopy, which recognizes mitochondrial remnants within autophagosomes, analysis of the mitochondrial protein turnover rate, and dual fluorescence labeling of mitochondria and autophagic markers such as LC3[12], or lysosome markers, such as Lamp1 and Lamp2[13]. In addition, recent innovative methods have been developed for the analysis of mitophagy, such as the novel pH-dependent fluorescent molecule, Keima [14,15]. When Keima is localized in cellular compartments with neutral pH it emits a green color, whereas when it is in lysosomesat acidic pH it emits a red color. Keima directed to mitochondria using a mitochondrial localization signal (Mito-Keima) allows for accurate evaluation of the presence of mitochondrial proteins in lysosomes (pH~4.5). The level of mitophagy can be quantified by the number of red pixels divided by the total number of all pixels[6,15–19]. In Mito-Keima transgenic mice, mitophagy is observed at high rates in the heart and the brain compared to other organs[7]. Another method potentially useful for monitoring mitophagy in the heart is MitoTimer, which is a mitochondria-targeted mutant of the red fluorescent protein DsRed (DsRed1-E5) that changes color during protein maturation; shortly after translation, it emits green light, whereas 12–18 hours after translation it emits red light [20,21]. Mice expressing MitoTimer in the heart are useful for investigating mitochondrial turnover in cardiomyocytes, where the accumulation of red-shifted mitochondria indicates low mitochondrial turnover as a result of low mitophagic flux[22].
Parkin-dependent mitophagy
The most well studied mechanism of mitophagy in cardiomyocytes is that mediated by the cytosolic E3 ubiquitin ligase Parkin[23] and the mitochondrial membrane kinase PTEN-induced putative kinase-1 (PINK1)[24]. When dysfunctional or damaged mitochondria occur, Parkin is recruited from the cytosol to damaged mitochondria[24] and ubiquitinates mitochondrial outer membrane proteins, such as mitofusin 1 (MFN1), mitofusin 2 (MFN2) and voltage dependent anion channel (VDAC)[24]. In healthy mitochondria, PINK1 is unstable and rapidly degraded, but in depolarized mitochondria, PINK1 is stabilized, allowing PINK1 accumulation only in depolarized mitochondria[25]. PINK1 phosphorylates MFN2, which promotes Parkin translocation to mitochondria[26]. PINK1 also phosphorylates ubiquitin and phosphorylated ubiquitin activates Parkin E3 ubiquitin ligase activity[27,28]. In cardiomyocytes, MFN2 deficiency prevents depolarization-induced recruitment of Parkin to damaged mitochondria, resulting in cardiomyopathy[26]. Pink1−/− mice develop left ventricular dysfunction and pathological cardiac hypertrophy[29], which is mediated by increased oxidative stress and dysfunctional mitochondria in cardiomyocytes, and PINK1 protein levels are significantly decreased in heart failure[29]. Interestingly, Parkin can still be recruited to damaged mitochondria after MI in the absence of PINK1 and compensates for PINK1 deficiency[30]. In addition, nitric oxide induces Parkin-dependent mitophagy even in the absence of PINK1 [31]. Conversely, PINK1 recruits autophagy receptors to induce mitophagy even without Parkin[32], indicating the presence of alternative pathways.
Parkin−/− mice exhibited normal myocardial function at baseline[33]. Autophagic clearance of damaged mitochondria is still possible in the absence of Parkin in the heart, most likely through activation of Parkin-independent mitophagy and upregulation of macroautophagy[34]. In fact, there is a compensatory increase of several Parkin-related E3 ubiquitin ligases of the RING families in Parkin−/− hearts[35]. TNF-receptor-associated factor 2 (TRAF2), an E3 ubiquitin ligase, is recruited to mitochondria and promotes the removal of ubiquitin-tagged damaged mitochondria during I/R[36]. TRAF2 co-localizes with ubiquitin, p62, and LC3, and deletion of TRAF2 induces accumulation of depolarized mitochondria in cardiomyocytes. Thus, TRAF2 functions independently of Parkin to mediate mitophagy in cardiomyocytes[36]. Smad-ubiquitin regulatory factor 1(SMURF1) is a HECT-domain ubiquitin ligase that is recruited to damaged mitochondria, where it promotes mitophagy[37]. SMURF1 is required for damaged mitochondria to be engulfed by autophagosomes. Cardiomyocytes in Smurf1−/− mice showed an accumulation of abnormal mitochondria that were swollen, fragmented and contained abnormal cristae, and an increased number of p62 aggregates[37], suggesting that SMURF1 is involved in cardiac mitochondrial quality control at baseline. It should be noted, however, that Parkin−/−hearts exhibit altered mitochondrial networks with small mitochondria and are more sensitive to MI[33]. Cardiac-specific Parkin−/− mice and MFN2 mutant mice lacking the PINK1 phosphorylation sites necessary for Parkin binding exhibited impaired metabolic maturation in the postnatal hearts, suggesting that Parkin-mediated mitophagy plays an important role in mediating developmental mitochondrial plasticity and metabolic remodeling in the heart [38]. In Parkin−/−Drosophila [35], mitochondria are depolarized and generate more ROS, and the heart is dilated even at baseline, most likely because Drosophila has only one Parkin gene. These results support the fundamental importance of Parkin in cardiac mitophagy throughout the evolutionary tree.
The role of Drp1 in mitophagy in the heart
Dynamin-related protein 1 (Drp1) is a small GTPase involved in mitochondrial fission. Although Dnm1, a yeast homolog of Drp1, has been identified as an essential mediator of mitophagy in yeast[39], it has not yet been determined whether or not Drp1 also plays an essential role in mediating mitophagy in the mammalian heart. Drp1 is more strongly expressed in the brain and the heart than in other tissues or mouse embryonic fibroblast (MEF) cells[40]. Multiple reports have shown that Drp1 is essential for normal cardiac function at baseline and in response to stress[6,17,40–43]. Furthermore, postnatal cardiac-specific downregulation of Drp1 reproducibly induces dilated cardiomyopathy and rapid lethality in mice[6,17,40,42]. However, whether Drp1 plays an essential role in mediating mitophagy in the heart remains controversial. Song et al. reported that mitophagy is hyper-activated in Drp1-deficient mouse hearts. Since Parkin is upregulated in Drp1-deficient hearts, and since concomitant ablation of Parkin increases the survival of Drp1-deficient mice, Parkin-mediated hyper-activation of mitophagy may be detrimental for the heart[41]. On the other hand, using Mito-Keima, Ikeda et al.[6] reported that endogenous Drp1 is required to mediate mitophagy in cardiomyocytes in response to glucose deprivation. Furthermore, Drp1 downregulation in the adult heart inhibits mitochondrial autophagy and causes mitochondrial dysfunction and consequent cell death in the heart, both at baseline and under stress conditions. Kageyama et al.[17] reported that Drp1 mediates Parkin-independent mitophagy, whereas Parkin is critical for the maintenance of mitochondrial respiratory function in the absence of Drp1. Interestingly, ubiquitination of mitochondrial proteins was unaffected in Parkin/Drp1 knockout mice, indicating that other E3 ligases may mediate mitochondrial degradation in the absence of Parkin. Cahill et al.[43] reported that C452F mutation in Drp1 causes dilated cardiomyopathy with abnormal mitochondrial morphology and defective mitophagy. In a clinical study, a newborn girl with a Drp1 heterozygous mutation (A395D) displayed a defect in mitochondrial fission with persistently elevated levels of lactate and very-long chain fatty acids in the plasma[44]. In summary, further investigation is required to elucidate the role of Drp1 in mitophagy in the heart, as well as the precise molecular mechanism involved.
Other mechanisms mediating mitophagy in the heart
Damaged mitochondria are engulfed by autophagosomes through interaction between autophagy adaptors or receptors and LC3 or LC3-related proteins. Autophagy adaptors, including p62/SQSTM1, Tax1BP1, NBR1, NDP52, and optineurin, possess ubiquitin binding domains through which they recognize damaged mitochondria ubiquitinated through either Parkin-dependent [45] or –independent mechanisms [32,36], whereas mitochondrial receptors, such as NIX/BNIP3L, mitochondrial pro-apoptotic BH3-only domain protein (BNIP3) and FUNDC1, are located in the outer mitochondrial membrane and do not require ubiquitination of mitochondria to promote mitophagy. BNIP3 is upregulated in myocardia during hypoxia[46], while NIX is upregulated during pathological cardiac hypertrophy[47]. Both BNIP3 and NIX are mitophagy regulators in adult hearts[48]. FUNDC1 is an integral mitochondrial outer-membrane protein that interacts with LC3 to induce mitophagy during hypoxia in HeLa cells[49]. FUNDC1 is dephosphorylated at Serine 13 by phosphoglycerate mutase family member 5 (PGAM5), a Ser/Thr phosphatase, leading to activation of mitophagy[50]. PGAM5 has been shown to play a critical role in mediating PINK1-mediated mitophagy in the heart [51] but the connection between PINK1 and FUNDC1 is currently unknown. In addition, ULK1, a Ser/Thr kinase required for early autophagosome formation, is upregulated and translocates to mitochondria, where it interacts with FUNDC1 and phosphorylates it at Serine 17, thereby enhancing the interaction of FUNDC1 and LC3 in MEF cells[52]. However, it remains unclear whether a similar mechanism exists in the heart. Bcl2-like protein 13(Bcl2-L-13) has been identified as a mammalian mitophagy receptor[53] that is homologous to Atg32 in yeast[54,55]. Bcl2-L-13 was shown to bind to LC3 and induce mitochondrial fragmentation independently of either Drp1 or Parkin in HEK293A cells[53], providing a novel insight into the mechanism of mitophagy.
Other mechanisms of mitochondrial degradation
Several recent studies in yeast have shown that mitochondria can be degraded by micromitophagy, where the lysosome directly engulfs mitochondria[13,56–58]. Similarly, in the heart, damaged mitochondria can be taken up directly by lysosomes during I/R[13]. This micromitophagy is independent of macroautophagy, which involves the formation of double membranes around organelles targeted for destruction that then fuse with lysosomes, and is regulated by δPKC-mediated phosphorylation of GAPDH[13]. Pharmacological upregulation of GAPDH-driven mitophagy promotes the clearance of damaged mitochondria and inhibits cell death during I/R[13].
Another form of micromitophagy is mediated by mitochondria-derived vesicles (MDVs) that bud off from mitochondria and are engulfed by lysosomes[59,60]. During oxidative stress, MDVs act as an early response, delivering their cargo of dysfunctional proteins and lipids from mitochondria to lysosomes independently of the mitochondrial fission machinery and without requiring mitochondrial depolarization. Although MDV formation and delivery to lysosomes are independent of the macroautophagic machinery, including ATG5 and LC3, the vesicles do require PINK1 and Parkin. However, whether MDVs exist in the heart is currently unknown.
Upstream signaling mechanisms of mitophagy
ULK1, a mammalian homolog of yeast Atg1, is an mTOR substrate. Phosphorylation of ULK1 at Ser757/758 (mouse/human) by mTOR inhibits its activity and autophagy[61,62]. AMP-activated protein kinase (AMPK) is a sensor for the metabolic state activated during starvation. Activation of AMPK inhibits the TORC1 pathway, activates ULK1, and positively regulates mitophagy [63–65].
Hexokinase-II (HK-II), a kinase in the first step of glycolysis, is a predominant isoform in the heart, adipose tissue and skeletal muscle. Reduction in HK-II levels results in decreases in cardiac function during IPC, I/R and cardiac hypertrophy[66–69]. In a recent study, HK-II was shown to positively regulate autophagy in response to glucose starvation in cardiomyocytes through interaction with and inhibition of mTOR[70]. HK-II overexpression enhances glucose deprivation-induced dephosphorylation of downstream targets of TORC1, whereas HK-II deficiency inhibits this response. Furthermore, HK-II overexpression leads to reduced phosphorylation of Ser757 in ULK1 and increased AMPK-mediated phosphorylation of Ser555, thereby stimulating autophagy[70].
Mitogen-activated protein kinase 1 (MAPK1) and MAPK14 and their upstream signaling pathways are essential for mitophagy[19]. Using Mito-Keima, it was demonstrated that only a small portion of mitochondria was degraded through mitophagy during starvation and hypoxia in HeLa cells. This process is ATG5-ATG7-independent, but requires ULK1, Beclin1, and class III PI3K. Knockdown of MAPK1 and MAPK14 efficiently inhibited mitophagy, without affecting macroautophagy. Phosphorylation of MAPK1 and MAPK14 is significantly increased during starvation and hypoxia, indicating that MAPK1 and MAPK14 are upstream of and regulate the activation of mitophagy, which is mediated through non-conventional mechanisms of autophagy[19].
Lysosomes also regulate mitophagy. TFEB, a transcription factor involved in lysosome biogenesis[71], facilitates the removal of damaged mitochondria and attenuates cardiomyocyte death during I/R[72]. Downregulation of TFEB results in impairment of autophagic flux, accumulation of damaged mitochondria, and increases in myocardial oxidative stress[73], suggesting that endogenous TFEB-mediated stimulation of autophagic flux protects cardiomyocytes against hypoxia-reoxygenation injury in cardiomyocytes[73].
Nutrient- and stress-dependent modification of mitochondrial proteins by acetylation/deacetylation plays a fundamental role in mitochondrial function. Both acetyltransferase and deacetylase enzymes regulate macroautophagy[74,75]. In particular, GCN5L1, a component of the mitochondrial acetyltransferase machinery, has been shown to regulate mitophagy[76]. GCN5L1 deficiency not only increases deacetylation of mitochondrial proteins and initiates mitophagy[76] but also activates the expression of TFEB, with a coordinated increase in mitochondrial biogenesis through increased PGC-1α in MEFs. Thus, GCN5L1 serves as a negative regulator of both mitochondrial biogenesis and mitophagy[5].
In non-cardiomyocytes, starvation induces autophagic degradation of mitochondria later than autophagic and proteasomal degradation of cytosolic proteins, possibly because re-synthesis of intracellular organelles is energetically more costly[77]. During the initial hour of starvation, mitochondria begin to elongate, are spared from autophagic degradation, and try to maintain ATP production[78]. As starvation continues, mitophagy follows. However, since cardiomyocytes have a high energy demand and one third of their volume consists of mitochondria, whether a similar mechanism exists in cardiomyocytes is yet to be determined.
p53/TIGAR (TP53-induced glycolysis and apoptosis regulator)-mediated inhibition of mitophagy causes damage to mitochondrial integrity and apoptosis in cardiomyocytes[79]. Deletion of p53 or its downstream mediator, TIGAR, promotes mitophagy, thereby decreasing abnormal mitochondria and promoting resistance to ischemic injury[79] in the heart. In addition, cytosolic p53 inhibits Parkin-dependent mitophagy through interaction with Parkin[80]. Likewise, ALCAT1, a lysocardiolipin acyltransferase, was potently upregulated by hyperthyroid cardiomyopathy, leading to oxidative stress and mitochondrial dysfunction[81], whereas loss of ALCAT1 increased mitophagy and prevented cardiomyopathy. In contrast, PGAM5 promotes the clearance of dysfunctional mitochondria to maintain mitochondrial homeostasis through mitophagy[82].
Clinical significance.
Mitophagy is essential for cardiac mitochondrial quality control[10]. The regulation of mitophagy is diverse but must be precise. Either insufficient or exacerbated mitophagy can result in cardiomyopathy. For instance, in type 1 diabetes, diminished autophagy is a beneficial adaptive response to protect against diabetic cardiac injury, most likely by upregulating alternative pathways of autophagy and mitophagy[83]. Mainly, however, mitophagy exerts a protective role in cardiac function. IPC induces Parkin translocation to mitochondria and promotes mitophagy. Parkin knockout attenuates IPC-induced p62 translocation to mitochondria and abolishes the cardioprotective effects of IPC[45]. Mitophagy is also important for the clearance of damaged mitochondria during aging. Parkin deficiency results in the accumulation of damaged mitochondria in cardiomyocytes with age[84]. During myogenesis, mitophagy is upregulated by a process mediated by Drp1 and p62/SQSTM1[85]. Differentiation of primitive myoblasts into mature myotubes requires a metabolic switch from glycolysis to oxidative phosphorylation in skeletal muscle. The process requires dramatic remodeling of the mitochondrial network through coordinated actions of both mitochondrial biogenesis and clearance[85]. Whether the same mechanism exists in the heart remains to be shown. Addressing the in vivo importance of mitophagy is challenging, since some mediators of mitophagy, such as Parkin, have multiple functions besides mitophagy[86]. In addition, since multiple layers of back-up mechanisms exist for mitochondrial degradation, loss of function of one mechanism of mitophagy is often compensated for by other mechanisms[87]. Given that mitophagy is essential in mitochondrial quality control but can be harmful when excessively activated, it is of particular interest to investigate the participation of mitophagy in cardiovascular disease caused by distinct mechanisms in a context-dependent manner. In summary, if mitophagy is to emerge as a promising therapeutic target, it will be critical to determine when and how manipulating mitophagy activity can protect patients from cardiovascular diseases.
Acknowledgments
The authors thank Daniela Zablocki and Christopher D. Brady for critical reading of the manuscript. This work was supported in part by U.S. Public Health Service Grants HL67724, HL91469, HL102738, HL112330 and AG23039 and by the Leducq Foundation Transatlantic Network of Excellence.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Highlight at least 10% of references as being papers of special interest(•) or outstanding interest(••)
- 1.Vasquez-Trincado C, Garcia-Carvajal I, Pennanen C, Parra V, Hill JA, Rothermel BA, Lavandero S. Mitochondrial dynamics, mitophagy and cardiovascular disease. J Physiol. 2016;594:509–525. doi: 10.1113/JP271301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 4.Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- 5.Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Ikeda Y, Shirakabe A, Maejima Y, Zhai P, Sciarretta S, Toli J, Nomura M, Mihara K, Egashira K, Ohishi M, et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res. 2015;116:264–278. doi: 10.1161/CIRCRESAHA.116.303356. This study, together with [17••], reveals that Drp1-dependent mitophagy is essential in cardiac function and is independent of Parkin-mediated mitophagy. [DOI] [PubMed] [Google Scholar]
- 7••.Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, et al. Measuring in vivo mitophagy. Mol Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. This study shows an effective method for monitoring mitophagy in vivo using transgenic mice expressing Mito-Keima. This study also reveals the high activation rate of mitophagy in the heart. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109:151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito T, Sadoshima J. The molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circ Res. 2015;116:1477–1490. doi: 10.1161/CIRCRESAHA.116.303790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Kim TY, Wang D, Kim AK, Lau E, Lin AJ, Liem DA, Zhang J, Zong NC, Lam MP, Ping P. Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics. 2012;11:1586–1594. doi: 10.1074/mcp.M112.021162. This study reveals a novel type of micromitophagy, which is inactive-GAPDH-derived, and during which the lysosome directly engulfs mitochondria without the process of autophagosome formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolman NJ, Chambers KM, Mandavilli B, Batchelor RH, Janes MS. Tools and techniques to measure mitophagy using fluorescence microscopy. Autophagy. 2013;9:1653–1662. doi: 10.4161/auto.24001. [DOI] [PubMed] [Google Scholar]
- 13.Yogalingam G, Hwang S, Ferreira JC, Mochly-Rosen D. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) phosphorylation by protein kinase Cdelta (PKCdelta) inhibits mitochondria elimination by lysosomal-like structures following ischemia and reoxygenation-induced injury. J Biol Chem. 2013;288:18947–18960. doi: 10.1074/jbc.M113.466870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosado CJ, Mijaljica D, Hatzinisiriou I, Prescott M, Devenish RJ. Rosella a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy. 2008;4:205–213. doi: 10.4161/auto.5331. [DOI] [PubMed] [Google Scholar]
- 15.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 17••.Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. This study, together with [6••], reveals that Drp1-dependent mitophagy is essential in cardiac function and is independent of Parkin-mediated mitophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, Kitada T, Glass K, Owen CA, Mahmood A, Washko GR, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest. 2014;124:3987–4003. doi: 10.1172/JCI74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Hirota Y, Yamashita S, Kurihara Y, Jin X, Aihara M, Saigusa T, Kang D, Kanki T. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 2015;11:332–343. doi: 10.1080/15548627.2015.1023047. This research demonstrates that MAPKs are upstream regulators that are essential and specific to regulation of the activation of ULK1- and Beclin1-mediated mitophagy, but not ATG5- and ATG7-dependent macroautophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottlieb RA, Stotland A. MitoTimer a novel protein for monitoring mitochondrial turnover in the heart. J Mol Med (Berl) 2015;93:271–278. doi: 10.1007/s00109-014-1230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trudeau KM, Gottlieb RA, Shirihai OS. Measurement of mitochondrial turnover and life cycle using MitoTimer. Methods Enzymol. 2014;547:21–38. doi: 10.1016/B978-0-12-801415-8.00002-3. [DOI] [PubMed] [Google Scholar]
- 22.Stotland A, Gottlieb RA. alpha-MHC MitoTimer mouse. In vivo mitochondrial turnover model reveals remarkable mitochondrial heterogeneity in the heart. J Mol Cell Cardiol. 2015;90:53–58. doi: 10.1016/j.yjmcc.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 24.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. This research, together with [29••], reveals the mechanism of the PINK1/Parkin pathway in the quality control of cardiac mitochondria. PINK1 phosphorylates MFN2 and promotes Parkin translocation to mitochondria in cardiomyocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 28.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. This research, together with [26••], reveals the mechanism of the PINK1/Parkin pathway in the quality control of cardiac mitochondria. PINK1 is essential for heart function. PINK1 deficiency leads to development of early left ventricular dysfunction and pathological cardiac hypertrophy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson AB. PINK1 Is Dispensable for Mitochondrial Recruitment of Parkin and Activation of Mitophagy in Cardiac Myocytes. PLoS One. 2015;10:e0130707. doi: 10.1371/journal.pone.0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han JY, Kang MJ, Kim KH, Han PL, Kim HS, Ha JY, Son JH. Nitric oxide induction of Parkin translocation in PTEN-induced putative kinase 1 (PINK1) deficiency. functional role of neuronal nitric oxide synthase during mitophagy. J Biol Chem. 2015;290:10325–10335. doi: 10.1074/jbc.M114.624767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, Jimenez R, Petrosyan S, Murphy AN, Gustafsson AB. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piquereau J, Godin R, Deschenes S, Bessi VL, Mofarrahi M, Hussain SN, Burelle Y. Protective role of PARK2/Parkin in sepsis-induced cardiac contractile and mitochondrial dysfunction. Autophagy. 2013;9:1837–1851. doi: 10.4161/auto.26502. [DOI] [PubMed] [Google Scholar]
- 35.Bhandari P, Song M, Chen Y, Burelle Y, Dorn GW., 2nd Mitochondrial contagion induced by Parkin deficiency in Drosophila hearts and its containment by suppressing mitofusin. Circ Res. 2014;114:257–265. doi: 10.1161/CIRCRESAHA.114.302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Yang KC, Ma X, Liu H, Murphy J, Barger PM, Mann DL, Diwan A. Tumor necrosis factor receptor-associated factor 2 mediates mitochondrial autophagy. Circ Heart Fail. 2015;8:175–187. doi: 10.1161/CIRCHEARTFAILURE.114.001635. This study elucidates that TRAF2 functions independently of Parkin in mitophagy. Deletion of TRAF2 induces accumulation of depolarized mitochondria in cardiomyocytes, indicating that there is a Parkin-alternative pathway involved in cardiac mitophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ, Dorn GW., 2nd Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, et al. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol Cell Biol. 2015;35:211–223. doi: 10.1128/MCB.01054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song M, Gong G, Burelle Y, Gustafsson AB, Kitsis RN, Matkovich SJ, Dorn GW., 2nd Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. Circ Res. 2015;117:346–351. doi: 10.1161/CIRCRESAHA.117.306859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Song M, Mihara K, Chen Y, Scorrano L, Dorn GW., 2nd Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–285. doi: 10.1016/j.cmet.2014.12.011. This study elucidates how mitochondrial fission and fusion affect mitochondrial quality control. This study reveals distinct effects of mitochondrial fission and fusion on mitophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cahill TJ, Leo V, Kelly M, Stockenhuber A, Kennedy NW, Bao L, Cereghetti G, Harper AR, Czibik G, Lao C, et al. Resistance of Dynamin-related Protein 1 Oligomers to Disassembly Impairs Mitophagy, Resulting in Myocardial Inflammation and Heart Failure. J Biol Chem. 2015;290:25907–25919. doi: 10.1074/jbc.M115.665695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 45.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Regula KM, Ens K, Kirshenbaum LA. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91:226–231. doi: 10.1161/01.res.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- 47.Galvez AS, Brunskill EW, Marreez Y, Benner BJ, Regula KM, Kirschenbaum LA, Dorn GW., 2nd Distinct pathways regulate proapoptotic Nix and BNip3 in cardiac stress. J Biol Chem. 2006;281:1442–1448. doi: 10.1074/jbc.M509056200. [DOI] [PubMed] [Google Scholar]
- 48.Dorn GW., 2nd Mitochondrial pruning by Nix and BNip3 an essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3:374–383. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia- induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 50.Chen G, Han Z, Feng D, Chen Y, Chen L, Wu H, Huang L, Zhou C, Cai X, Fu C, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 51.Lu W, Sun J, Yoon JS, Zhang Y, Zheng L, Murphy E, Mattson MP, Lenardo MJ. Mitochondrial Protein PGAM5 Regulates Mitophagic Protection against Cell Necroptosis. PLoS One. 2016;11:e0147792. doi: 10.1371/journal.pone.0147792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, Yasui H, Ueda H, Akazawa Y, Nakayama H, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 56.Li WW, Li J, Bao JK. Microautophagy. lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. See the annotation below Ref. [60••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells. revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. See the annotation below Ref. [59••] [DOI] [PubMed] [Google Scholar]
- 58.Hwang S, Disatnik MH, Mochly-Rosen D. Impaired GAPDH-induced mitophagy contributes to the pathology of Huntington’s disease. EMBO Mol Med. 2015;7:1307–1326. doi: 10.15252/emmm.201505256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59••.Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, McBride HM. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. This study, together with [57••], reveals another mechanism of micromitophagy, during which MDVs bud off from mitochondria and are engulfed by lysosomes during oxidative stress. [DOI] [PubMed] [Google Scholar]
- 60•.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. This study, together with [56••], reveals another mechanism of micromitophagy, during which MDVs bud off from mitochondria and are engulfed by lysosomes during oxidative stress. The vesicles require PINK1 and Parkin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takagi H, Matsui Y, Hirotani S, Sakoda H, Asano T, Sadoshima J. AMPK mediates autophagy during myocardial ischemia in vivo. Autophagy. 2007;3:405–407. doi: 10.4161/auto.4281. [DOI] [PubMed] [Google Scholar]
- 65.Tian W, Li W, Chen Y, Yan Z, Huang X, Zhuang H, Zhong W, Chen Y, Wu W, Lin C, et al. Phosphorylation of ULK1 by AMPK regulates translocation of ULK1 to mitochondria and mitophagy. FEBS Lett. 2015;589:1847–1854. doi: 10.1016/j.febslet.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 66•.Wu R, Wyatt E, Chawla K, Tran M, Ghanefar M, Laakso M, Epting CL, Ardehali H. Hexokinase II knockdown results in exaggerated cardiac hypertrophy via increased ROS production. EMBO Mol Med. 2012;4:633–646. doi: 10.1002/emmm.201200240. Studies [66•–70•] reveal that HK-II, a predominant isoform in the heart, is essential in normal cardiac function. Reduction of HK-II decreases cardiac function in IPC, I/R, and hypertrophy. HK-II regulates ULK1-mediated mitophagy through inhibition of mTOR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu R, Smeele KM, Wyatt E, Ichikawa Y, Eerbeek O, Sun L, Chawla K, Hollmann MW, Nagpal V, Heikkinen S, et al. Reduction in hexokinase II levels results in decreased cardiac function and altered remodeling after ischemia/reperfusion injury. Circ Res. 2011;108:60–69. doi: 10.1161/CIRCRESAHA.110.223115. See the annotation below Ref. [66•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smeele KM, Southworth R, Wu R, Xie C, Nederlof R, Warley A, Nelson JK, van Horssen P, van den Wijngaard JP, Heikkinen S, et al. Disruption of hexokinase II-mitochondrial binding blocks ischemic preconditioning and causes rapid cardiac necrosis. Circ Res. 2011;108:1165–1169. doi: 10.1161/CIRCRESAHA.111.244962. See the annotation below Ref. [66•] [DOI] [PubMed] [Google Scholar]
- 69.Roberts DJ, Tan-Sah VP, Smith JM, Miyamoto S. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. J Biol Chem. 2013;288:23798–23806. doi: 10.1074/jbc.M113.482026. See the annotation below Ref. [66•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roberts DJ, Tan-Sah VP, Ding EY, Smith JM, Miyamoto S. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol Cell. 2014;53:521–533. doi: 10.1016/j.molcel.2013.12.019. See the annotation below Ref. [66•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Ma X, Liu H, Murphy JT, Foyil SR, Godar RJ, Abuirqeba H, Weinheimer CJ, Barger PM, Diwan A. Regulation of the transcription factor EB-PGC1alpha axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Mol Cell Biol. 2015;35:956–976. doi: 10.1128/MCB.01091-14. Studies [72*–73*] elucidate how lysosomes regulate mitophagy through TFEB, a transcription factor involved in lysosome biogenesis. TFEB facilitates the removal of damaged mitochondria and attenuates cardiomyocyte death during I/R. TFEB stimulates autophagic flux and protects against cardiac injury during I/R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Godar RJ, Ma X, Liu H, Murphy JT, Weinheimer CJ, Kovacs A, Crosby SD, Saftig P, Diwan A. Repetitive stimulation of autophagy-lysosome machinery by intermittent fasting preconditions the myocardium to ischemia-reperfusion injury. Autophagy. 2015;11:1537–1560. doi: 10.1080/15548627.2015.1063768. See the annotation below Ref. [72•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 75.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013;126:4843–4849. doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kristensen AR, Schandorff S, Hoyer-Hansen M, Nielsen MO, Jaattela M, Dengjel J, Andersen JS. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics. 2008;7:2419–2428. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- 78.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, Matoba S. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 80.Hoshino A, Ariyoshi M, Okawa Y, Kaimoto S, Uchihashi M, Fukai K, Iwai-Kanai E, Ikeda K, Ueyama T, Ogata T, et al. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic beta-cell function in diabetes. Proc Natl Acad Sci U S A. 2014;111:3116–3121. doi: 10.1073/pnas.1318951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu X, Ye B, Miller S, Yuan H, Zhang H, Tian L, Nie J, Imae R, Arai H, Li Y, et al. Ablation of ALCAT1 mitigates hypertrophic cardiomyopathy through effects on oxidative stress and mitophagy. Mol Cell Biol. 2012;32:4493–4504. doi: 10.1128/MCB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu W, Karuppagounder SS, Springer DA, Allen MD, Zheng L, Chao B, Zhang Y, Dawson VL, Dawson TM, Lenardo M. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson’s-like movement disorder. Nat Commun. 2014;5:4930. doi: 10.1038/ncomms5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu X, Kobayashi S, Chen K, Timm D, Volden P, Huang Y, Gulick J, Yue Z, Robbins J, Epstein PN, et al. Diminished autophagy limits cardiac injury in mouse models of type 1 diabetes. J Biol Chem. 2013;288:18077–18092. doi: 10.1074/jbc.M113.474650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kubli DA, Quinsay MN, Gustafsson AB. Parkin deficiency results in accumulation of abnormal mitochondria in aging myocytes. Commun Integr Biol. 2013;6:e24511. doi: 10.4161/cib.24511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sin J, Andres AM, Taylor DJ, Weston T, Hiraumi Y, Stotland A, Kim BJ, Huang C, Doran KS, Gottlieb RA. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy. 2015 doi: 10.1080/15548627.2015.1115172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL, et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest. 2011;121:3701–3712. doi: 10.1172/JCI44736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]