Abstract
DICER is the central enzyme that cleaves precursor microRNAs (miRNAs) into 21-25 nucleotide duplex in cell lineage differentiation, identity and survival. In the current study, we characterized the specific bone metabolism genes and corresponding miRNAs and found that DICER and Runt-related transcription factor 2 (Runx2) expressions increased simultaneously during osteogenic differentiation. Luciferase assay showed that Runx2 significantly increased the expression levels of DICER luciferase promoter reporter. Our analysis also revealed weaker DICER expression in embryos of Runx2 knock out mice (Runx2 −/−) compared with that of Runx2 +/− and Runx2 +/+ mice. We further established the calvarial bone critical-size defect (CSD) mouse model. The bone marrow stromal cells (BMSCs) transfected with siRNA targeting DICER were combined with silk scaffolds and transplanted into calvarial bone CSDs. Five weeks post-surgery, micro-CT analysis revealed impaired bone formation and repairing in calvarial defects with the siRNA targeting DICER group. In conclusion, our results suggest that DICER is specifically regulated by osteogenic master gene Runx2 that binds to the DICER promoter. Consequently, DICER cleaves precursors of miR-335-5p and miR-17-92 cluster to form mature miRNAs, which target and decrease the Dickkopf-related protein 1 (DKK1) and proapoptotic factor BIM levels, respectively, leading to an enhanced Wnt/β-catenin signaling pathway. These intriguing results reveal a central mechanism underlying lineage-specific regulation by a Runx2/DICER/miRNAs cascade during osteogenic differentiation and bone development. Our study also suggests a potential application of modulating DICER expression for bone tissue repair and regeneration.
Keywords: Bone, DICER, Runx2, miRNA, Osteogenesis
Introduction
MicroRNAs (miRNAs) are a family of noncoding, small RNAs that represent a sophisticated level of gene regulation that coordinates a broad spectrum of biological processes, such as cell differentiation, growth, and metabolism (Ambros, 2004; Bartel, 2004). miRNAs have a role in the regulation of bone remodeling by pairing with specific sequences in the 3’UTR of target mRNAs to induce post-transcriptional gene silencing. Functional mature miRNAs arise through several post-transcriptional processing steps including cleavage by Drosha/DGCR8 to pre-miRNA, export, and digestion by the RNaseIII endonuclease DICER, which also mediates loading onto RISC complexes (Bushati and Cohen, 2007; Filipowicz et al., 2008; Kim et al., 2009; Schickel et al., 2008; Winter et al., 2009). In the current literature, there is no report of DICER-independent miRNAs, suggesting that DICER is indispensable for miRNA biogenesis (Levy et al., 2010). DICER cleavage activity and mature miRNA expression in mammals are restricted to certain tissues and cell types, suggesting tissue-specific regulation of its activity (Obernosterer et al., 2006). Ablation of DICER in osteoprogenitors prevents their differentiation and compromises fetal survival at E15.5 while excision in differentiated osteoblasts increases adult bone mass in mice (Gaur et al., 2010). Runt-related transcription factor 2 (Runx2), is identified as a “master gene” required for osteoblastic differentiation process from mesenchymal precursors (Tu et al., 2007b; Vimalraj et al., 2015; Wysokinski et al., 2015). Our previous studies showed that haploinsufficiency of Runx2 results in decreased bone formation and changes in BSP expression patterns (Li et al., 2005a; Tu et al., 2009; Tu et al., 2007b; Tu et al., 2008). Our recent studies have shown a feedback link connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster (Hassan et al., 2010), and found that Runx2 negatively regulates expression of the miR cluster 23a~27a~24-2 that each miR directly targets the 3′-UTR of SATB2. However, transcriptional regulation of DICER expression by transcription factors particularly in bone development and regeneration remains incompletely understood.
We analyzed the DICER promoter region sequence and identified one consensus Runx-binding site (TGTGGT) (Dalle Carbonare et al., 2012; Wu et al., 2014) upstream of the previously characterized transcription start site. These findings prompted us to examine the regulation and functional activity of DICER in osteoblasts and in relation to Runx2, which is required for osteogenesis.
Here, we demonstrate that DICER and Runx2 expressions increased simultaneously during osteogenic differentiation. Significantly, embryos from Runx2 −/− mice showed weak DICER expression compared with embryos from Runx2 +/+ and Runx2 +/− mice. Additionally, we found that Runx2 directly increases the luciferase levels of DICER luciferase promoter reporter. BMSCs transfected with siRNA targeting DICER were then combined with silk scaffolds and transplanted into calvarial bone CSDs of mice expressed poor new bone formation compared with control groups. Our studies have identified a regulatory circuit involving Runx2, DICER, and the miRNAs, which has a critical and central role in controlling progression and attenuation of a specific cell phenotype.
Materials and Methods
Cell culture
C3H10T-1/2 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. MC3T3-E1 cells were maintained in a modified essential medium (a-MEM) with 10% FBS and antibiotics. Additionally, calvarial cells from Runx2−/− embryos were cultured in α-MEM with 10% FBS and antibiotics. Osteogenesis was induced with 50 mg/mL of ascorbic acid for 1, 4, 7 and 10 days. BMSCs were cultured as described previously (Tu et al., 2007a). Briefly, BMSCs were obtained from femurs and tibias of 6- to 8-week-old male WT mice, and maintained in DMEM with 20% FBS, 1% penicillin-streptomycin.
Plasmids and transfection experiments
Plasmid pCMV-Runx2 was provided by Dr. Gerard Karsenty (Bayer College of Medicine, Houston, TX). Plasmid overexpression vector for DICER and siRNA targeting DICER was purchased from Addgene (Cambridge, MA). The empty vector was used as a control plasmid in transfection experiments. Transfection of plasmids was performed using Lipofectamine 2000 (Life Technologies) following the manufacturer’s recommendations.
Real-time RT-PCR for mRNA and miRNA analysis
Bone metabolism genes mRNA analysis was performed by quantitative real-time reverse-transcriptase PCR (qRT-PCR) assay using iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) on a Bio-Rad iQ5 thermal cycler (Bio-Rad Laboratories). The evaluation of PCR product amounts relative differences was carried out by the comparative cycle threshold method using GAPDH as a control. For miRNA analysis, total RNA was extracted using the miRNeasy Mini Kit (Qiagen), cDNA synthesis was performed with 1μg of total RNA, using an NCode miRNA First-Strand cDNA Synthesis Kit (Invitrogen) according to the manufacturer’s protocol. qRT- PCR was performed on a Bio-Rad iQ5 thermal cycler using an NCode Express SYBR GreenER miRNA qRT-PCR Kit (Invitrogen). Expression levels of PCR product amounts were evaluated by the comparative cycle threshold method using U6 snRNA as a control (Table 1).
Table 1.
The primers for real-time PCR.
| Primer of target | Sequence (5' to 3') |
|---|---|
| DICER | forward:5'- GTCAGCCGTCAGAACTCACTC-3' |
| reverse:5'-ACAGTCAAGGCGACATAGCAA-3' | |
| Runx2 | forward: 5'-CAGTCACCTCAGGCATGTCC-3' |
| reverse: 5'-GTGCTGCTGGTCTGGAAGG-3' | |
| SATB2 | forward : 5'-GAGATGAGTTGAAGAGGGCTAGTG–3' |
| reverse : 5'-CCCTGTGTGCGGTTGAAT -3' | |
| OSX | forward: 5'-ATGGCGTCCTCTCTGCTTG-3' |
| reverse: 5'-TGAAAGGTCAGCGTATGGCTT-3' | |
| DKK1 | forward:5’-CTCATCAATTCCAACGCGATCA-3’ |
| reverse:5’-GCCCTCATAGAGAACTCCCG-3’ | |
| GAPDH | forward: 5'-AGGTCGGTGTGAACGGATTTG-3' |
| reverse: 5'-TGTAGACCATGTAGTTGAGGTCA-3' | |
| miR-335-5p | 5'- GCGTCAAGAGCAATAACGAAAAATGT-3' |
| miR-342-3p | 5'-ACACAGAAATCGCACCCGT-3' |
| miR-17 | 5'-GCAAAGTGCTTACAGTGCAGGTAG-3' |
| miR-18a | 5'-CGCTAAGGTGCATCTAGTGCAGATAG-3' |
| miR-19a | 5'-CGCTGTGCAAATCTATGCAAAACTGA-3' |
| miR-20a | 5'-GCGTAAAGTGCTTATAGTGCAGGTAG-3' |
| miR-19b-1 | 5'-CGTGTGCAAATCCATGCAAAACTGA-3' |
| miR-92-1 | 5'-TATTGCACTTGTCCCGGCCTG-3' |
| U6 snRNA | 5'- CTTCGGCAGCACATATACTAAAATT-3' |
Western blot
Whole protein lysates for western blotting were prepared with RIPA lysis buffer (Santa Cruz Biotechnology, Inc.) supplemented with 1 mM PMSF. SDS-PAGE and Western blot analyses were then performed using NuPAGE 4–12% Bis-Tris gradient gels and 0.45μm polyvinylidene fluoride membranes (Invitrogen). Antibodies for DICER (1:400), RUNX2 (1:800), SATB2 (1:1000), Dickkopf-related protein 1 (DKK1) (1:1000), and β–actin (1:1000) were obtained from Santa Cruz Biotechnologies, Inc. (Santa Cruz, CA, USA). The secondary antibodies were horseradish peroxidase (HRP)–linked goat-anti rabbit IgG (Santa Cruz Biotechnologies, Inc.). Blots were visualized using ECL chemiluminescence reagents from Pierce Biotechnology (Rockford, IL, USA).
Luciferase assay
Cell was co-transfected with the DICER luciferase promoter reporter (from David E. Fisher, Cutaneous Biology Research Center, Mass. General Hospital, Harvard Medical School), pMIR-beta-gal and transcriptional factor cDNA plasmids (RUNX2, Osterix, or SATB2, respectively). Luciferase levels were determined by luminometer and normalized to β-galactosidase activity. The luciferase assay was performed using a Lumat LB9501 luminometer (Berthold Technologies) as described previously (Tu et al., 2008).
Immunohistochemistry analysis
Runx2 +/+, Runx2 −/−, Runx2 +/− E.16 embryos from Runx2 mutant mice (a generous gift from Dr. Michael Owen, Imperial Cancer Research Fund, London, UK) were fixed in 10% formalin at 4°C overnight and demineralized in a 10% ethylenediaminetetraacetic acid (EDTA)-buffered solution (pH 7.0) for 14 days at room temperature. Samples were embedded in paraffin and serial sections of 5-μm thickness were obtained in a mesial-distal direction. The tissue slides were first deparaffinized and rehydrated, and then submerged in hydrogen peroxide for peroxidase quenching. Before using the primary antibodies, the slides were incubated with normal serum to block the nonspecific bindings. Antibody against DICER (Santa Cruz, CA, USA) was then used at a dilution of 1:200. After overnight incubation, the biotinylated secondary antibodies were applied to the slides. Finally, the substrate-chromogen 3-Amino-9-ethylcarbazole (AEC) was applied and the slides were counterstained with hematoxylin and mounted.
Silk scaffold preparation and cell seeding
The water based silk fibroin scaffolds (pore size 500-600 microns, disk-shaped, 4mm diameter and 2mm thick) were prepared as we previously described (Ye et al., 2011). For cell seeding, BMSCs transfected with siRNA targeting DICER after 3 days cultured in differentiation medium and were released from the culture substratum using trypsin/EDTA (0.25% w/v trypsin, 0.02% EDTA) and concentrated to 2×107cells/mL in serum-free medium. Then BMSCs were seeded onto the silk scaffold by pipetting the cell suspension onto the materials. The BMSCs/silk scaffold (SS) construct was incubated for an additional 4h to allow for cell attachment in vitro before implantation.
Mice and experimental calvarial bone CSDs model
This study was performed in accordance with the animal protocol approved by the Institutional Animal Care and Use Committee at Tufts University, as well as the Animal Research: Reporting In Vivo Experiments (ARRIVE) guidelines for animal research. 16 6-week-old male C57BL/6J mice (Stock Number 000664, The Jackson Laboratory, Bar Harbor, ME, USA) were anesthetized, and then a 4-mm-diameter calvarial bone CSD was created on each side of the calvarial bone using a dental bur attached to a slow-speed hand piece with minimal invasion of the dura mater. Following the CSDs surgery, mice were randomly divided into 4 groups: defects unfilled, defects filled with blank scaffolds, defects filled with scaffolds seeded with BMSCs, defects filled with scaffolds seeded with BMSCs transfected with siRNA targeting DICER (n=5).
Micro-CT measurement
Healing of calvarial defects was examined by micro-CT 4 weeks after surgery. The morphology of the reconstructed cranium was assessed using a micro-CT system (CT-40, Scanco Medical, Bassers-dorf, Switzerland). The CT settings were used as follows: pixel matrix, 1024*1024; slice thickness, 20mm. After scanning, the micro-CT images were segmented using a nominal threshold value of 225 as reported previously,(Yost et al., 1996) and a three dimensional (3D) histomorphometric analysis was performed automatically. The parameters of bone volume fraction (bone volume/total volume, BV/TV), volume of newly regenerated bone, and bone volume were used for comparison in this study.
Results
miRNA expression during osteogenesis
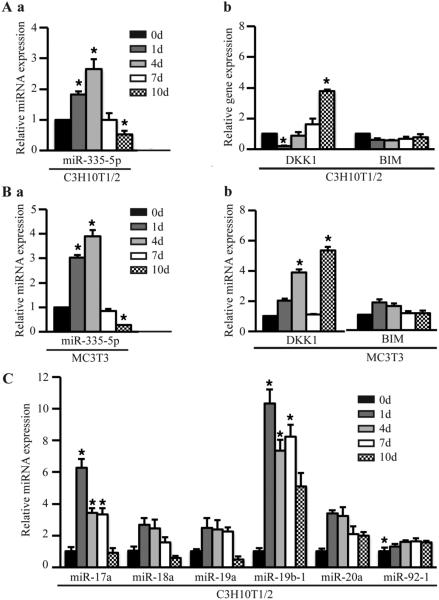
Our previous study has proved that miR-335-5p specifically targets DKK1 3’ UTR and controls DKK1 expression, canonical Wnt signaling activity, and osteogenic differentiation (Zhang et al., 2011a). Here miR-335-5p mRNA levels are increased initially when osteogenic differentiation is initiated in C3H10T1/2 cells, and the target gene DKK1 expression decreased (Fig. 1A). The same trend was observed during osteogenesis in MC3T3 and C3H10T1/2cells (Fig. 1B). The miR-17~92 cluster encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1), which are highly conserved in all vertebrates. Germline hemizygous deletions of miR-17~92 cluster were accounted for microcephaly, short stature and digital abnormalities(de Pontual et al., 2011; Zhou et al., 2014). In this study, miR-17~92 cluster increased during osteogenesis in C3H10T1/2cells (Fig. 1C), suggesting that they critically regulate osteoblast differentiation.
Figure 1.
Gene and miRNA expressions during osteogenesis. (A) miR-335-5p (a), Dkk1 and BIM (b) expression in C3H10T1/2 cells. (B) miR-335-5p (a), Dkk1 and BIM (b) expression in MC3T3-E1 cells. (C) miR-17~92 cluster expression in C3H10T1/2 cells. Cells were induced in osteogenic medium with 50 mg/mL of ascorbic acid for 1, 4, 7 and 10 days, respectively. mRNA and miRNA expression were detected by quantitative RT-PCR. These data are expressed as the mean ± SD (n=3). * p < 0.05, versus day 0 before induction.
DICER and Runx2 protein and mRNA levels are upregulated simultaneously during osteogenesis
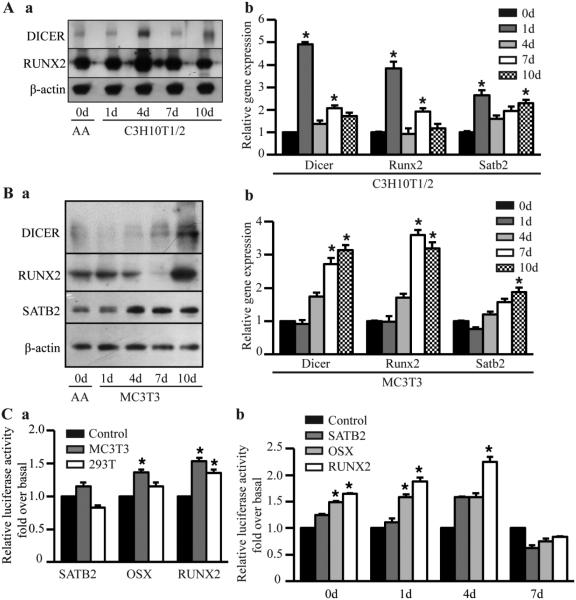
A striking increase was observed in DICER protein level in stimulated C3H10T1/2 cells, which occurred simultaneously with upregulation of Runx2 (Fig. 2Aa). In addition to elevated protein levels, DICER mRNA levels also increased upon osteoblast differentiation (Fig. 2Ab). In MC3T3-E1 cells, the Runx2 and DICER protein and mRNA expressions were parallel (Fig. 2B), suggesting that some direct relationship might exist between them. In the luciferase assay, DICER luciferase activity dramatically increased after cotransfection with Runx2 when compared with the empty vector control (Fig. 2C). Special AT-rich sequence-binding protein 2 (SATB2), a DNA-binding protein that regulates chromatin organization and gene expression including Runx2 expression (Alcamo et al., 2008; Zhang et al., 2011b), was also included as a control. The results suggested that if Runx2 can directly regulate DICER, it might bind the DICER transcription start site.
Figure 2.
Parallel upregulation of DICER and Runx2 protein and mRNA levels during osteogenesis. (A) Protein (a) and mRNA (b) levels in stimulated C3H10T1/2 cells. (B) Protein (a) and mRNA (b) levels in stimulated MC3T3 cells. Cells were induced in osteogenic medium for 1, 4, 7 and 10 days, respectively. Proteins were detected by Western blot, and mRNA and miRNA expressions checked by quantitative RT-PCR. (C) Luciferase assay. (a) MC3T3-E1 or T293 cells was co-transfected with the DICER promoter driven luciferase reporter, pMIR-beta-gal and transcriptional factor (TF) cDNA plasmids (RUNX2, Osterix, or SATB2, respectively). Empty vectors without harboring TF cDNAs were used as a control. Luciferase activity was detected 48 hours after transfection. (b) MC3T3 cells were induced in osteogenic medium for 1, 4, 7 and 10 days, respectively. The cells were cotransfected as described in Figure 2Ca one day before the induction was due. Luciferase activity was detected 48 hours after transfection. β-actin level was used for normalization of gene expression. These data are expressed as the mean ± SD (n=3). * p < 0.05, versus control.
Gain-and-loss of function analyses
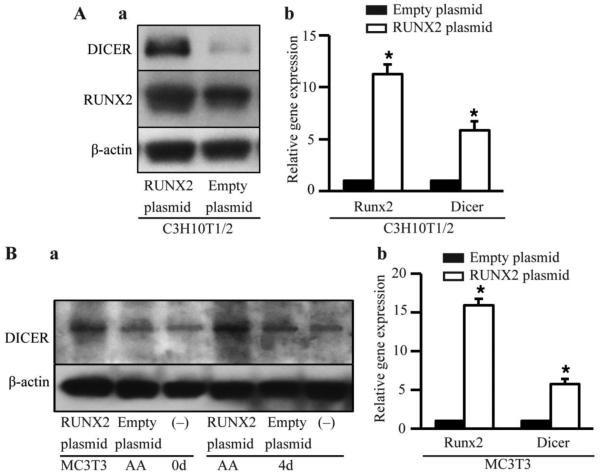
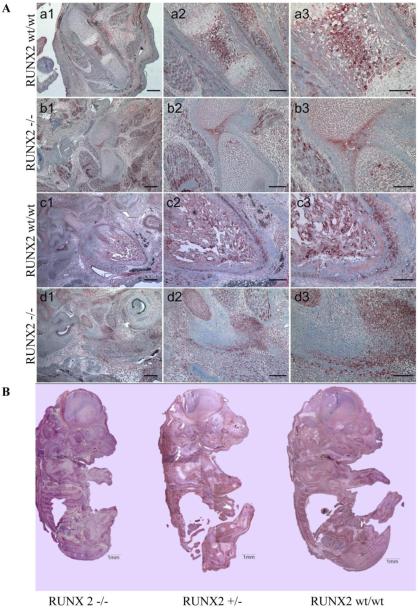
C3H10T1/2 and MC3T3 cells were transiently transfected with Runx2 overexpression vector or empty vector. Runx2 overexpression in C3H10T1/2 (Fig. 3A) and MC3T3 (Fig. 3B) cells led to 5- and 3- fold increases in DICER expression, respectively. In vivo excision of Runx2 impaired bone formation at E16.5 and induces embryonic lethality. Immunohistochemistry of femurs and mandible mesenchyme that surrounds Meckel's cartilage, and embryos showed severely delayed mineralization and less DICER expression in Runx2 −/− mice compared with Runx2 +/+ mice (Fig. 4A). The embryos from Runx2 −/− mice showed weak DICER expression compared with embryos from Runx2 +/+ and Runx2 +/− mice (Fig. 4B). Calvarial cells from Runx2-/− embryos were cultured in vitro, and were transiently transfected with DICER cDNAs or empty vector. The overexpression of DICER was confirmed by Immunohistochemical analysis (Fig. 5A). The expression of proapoptotic factor BIM that was previously found to be a direct target of the miR-17-92 cluster decreased by 2-3 folds while miR-17-92 cluster increased, suggesting that DICER overexpression increased the miR17-92 cluster expression, which may directly target BIM to rescue the osteogenesis in Runx2−/− cells (Fig. 5B).
Figure 3.
Upregulation of Dicer expression by Runx2 overexpression. (A) (a) Runx2 and DICER protein were detected by Western blot; (b) mRNA levels of Runx2 and DICER were detected by qRT-PCR in C3H10T1/2 cells. The cells were induced in ostegenic medium for 4 days, and collected 48 hours after Runx2 cDNA or empty vector transfection. (B) (a) DICER protein was detected by Western blot in MC3T3-E1 cells. The cells were induced in ostegenic medium for 0 and 4 days, respectively, and collected 48 hours after Runx2 cDNAs or empty vector transfection. (−), untransfected cells as a internal control. (b) mRNA levels of Runx2 and DICER were detected by qRT-PCR in MC3T3-E1 cells transfected with Runx2 cDNAs or empty vector. The cells were induced in ostegenic medium for 4 days. These data are expressed as the mean ± SD (n=3). * p < 0.05 versus empty vector group.
Figure 4.
Immunohistochemical analysis of Dicer (AEC) in Runx2 +/+, Runx2 +/− and Runx2 −/− mice (E16.5). (A) Representative pictures of femurs (a1-b3) and mandible mesenchyme that surrounds Meckel's cartilage (c1-d3) showed severely impaired mineralization and less DICER expression in Runx2 −/− mouse compared with Runx2 wt/wt mouse.(E16.5, scale bars, 20 µm). (B) The Specimens of Runx2 wt/wt, Runx2 +/− and Runx2 −/− mouse (E16.5, scale bars, 1 mm).
Figure 5.
miRNA and gene expression in calvarial cells from Runx2−/− embryos. after transfected with overexpression vector for Dicer or empty vector. (A) Runx2−/− cells were transfected with Dicer cDNA plasmids or empty vector. Immunohistochemical analysis was performed to confirm increased expression of DICER in Dicer cDNA group compared to with empty vector control (intense red signals, x200). (B) miRNA and gene expressions were detected by qRT-PCR in Runx2−/− cells transfected with DICER cDNAs or empty vector. These data are expressed as the mean ± SD (n=3). * p < 0.05, versus empty vector control.
Effect of DICER in bone regeneration of mouse CSDs
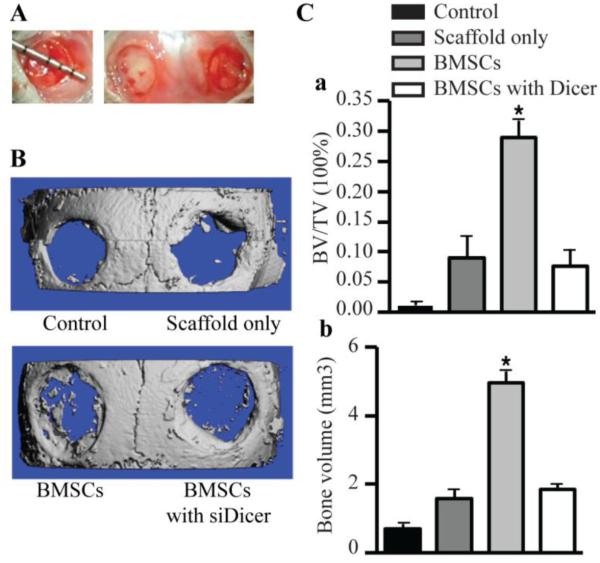
A non-healing full thickness defect of 4 mm in diameter was made on both sides of the cranial bone and defects were differently treated in all groups (defects unfilled, defects filled with scaffolds alone, defects filled with scaffolds seeded with BMSCs, defects filled with collagen seeded with BMSCs transfected with siRNA targeting DICER) (Fig. 6A). The morphology of the newly formed bone was reconstructed using micro-CT imaging, and the representative images from each group were shown in Fig. 6B. No obvious evidence of new bone growth was observed in the defects treated with BMSCs transfected with siRNA targeting DICER or scaffold alone or unfilled groups, only minimal amount of bone was visible in the periphery of the defect. The implantation of untransduced BMSCs culturing with scaffolds showed the formation of scattered new bone in the defect sites. To quantify the new bone regeneration within the calvarial defects, the ratio of bone volume to total volume (BV/TV), bone volume was measured. BV/TV was significantly higher for the untransduced BMSCs group when compared to BMSCs transfected with siRNA targeting DICER group (Fig. 6Ca). Significantly more bone volume was observed in the untransduced BMSCs group, and bone volume were also higher than the BMSCs transfected with siRNA targeting DICER group (Fig. 6Cb).
Figure 6.
Bone regeneration of calvarial bone critical-size defects (CSDs) in mice. (A) The non-healing full thickness defect of 4 mm in diameter was made in both sides of the cranial bone and filled with a silk scaffold seeded with gene-modified BMSCs. (B) Representative pictures of the morphology of the newly formed bone was reconstructed using micro-CT imaging in 4 groups (control, defects unfilled; scaffolds only, defects filled with scaffolds alone; BMSCs, defects filled with scaffolds seeded with BMSCs; BMSCs with siDicer, defects filled with scaffolds seeded with BMSCs transfected with siRNA targeting Dicer). (C) To quantify the new bone regeneration within the calvarial defects, the ratio of bone volume to total volume in region of interest (BV/TV) (a), bone volume (b) were measured in microCT scanned images 4 weeks after surgery. These data are expressed as the mean ± SD (n=4). * p < 0.05, versus control.
DISCUSSION
Expression of specific miRNAs may be transcriptionally regulated upon cell stimulation (Ozsolak et al., 2008; Sempere et al., 2003). However, like other RNAs, miRNA expression can be also regulated at the posttranscriptional level in either a tissue-specific or a developmentally regulated fashion (Winter et al., 2009). miRNAs are generated from long primary transcripts (pri-miRNAs) through multiple processing steps (Levy et al., 2010). pri-miRNAs are cleaved into small-hairpin pre-miRNAs by the microprocessor complex containing Drosha and DGCR8. Pre-miRNAs are exported into the cytoplasm, where the RNase III, DICER, removes the loop region of the hairpin. This step is essential for generation of mature miRNAs. It was previously shown that DICER cleavage activity is restricted to certain tissues and cell types, albeit by an unknown mechanism (Obernosterer et al., 2006). To explore the transcriptional regulation of DICER expression during osteogenesis, we detected DICER and Runx2 expressions and found that they were increased simultaneously during osteogenic differentiation.
To further determine the relationship between Runx2 and DICER, we identified one consensus Runx2-binding site (TGTGGT) within 2.5 kb upstream of the DICER transcription start site. We then conducted the luciferase assay and showed Runx2 overexpression could increase the luciferase activity of DICER promoter driven luciferase reporter.
Canonical Wnt signals are transmitted through stabilizing β-catenin protein by inhibiting GSK3b-mediated β-catenin phosphorylation (Balemans and Van Hul, 2007; Boyden et al., 2002; Galindo et al., 2005; Little et al., 2002; Lucero et al., 2013). DKK1 is essential to maintain skeletal homeostasis as an inhibitor of Wnt signaling and osteogenic differentiation (Glinka et al., 1998; Li et al., 2005b; Mukhopadhyay et al., 2001; Zhang et al., 2011a). MiR-335-5p has been proved to be a potential and useful targeting molecule for promoting bone formation and regeneration. MiR-335-5p activates Wnt signaling via directly binding to DKK1-3’UTR to downregulate DKK1 expression (Zhang et al., 2011a). In the current study, miR-335-5p levels are increased initially when osteogenic differentiation is initiated, while the target gene DKK1 expression decreased.
Although functions of canonical Wnt signaling in Runx2 expression and activation are still controversial, it is evident that Runx2 is the first transcription factor required for determination of the osteoblast lineage, while canonical Wnt signaling further directs the fate of mesenchymal cells to osteoblasts (Behrens et al., 1996; Moon et al., 2002; Nelson and Nusse, 2004; Takahashi-Yanaga and Sasaguri, 2007; Yost et al., 1996). In the current study, Runx2 deficient mice (Runx2−/−) is associated to incomplete bone mineralization. In Runx2−/− mice, there is a complete lack of mineralized bone and the weak DICER expression, which is consistent with an early block in osteogenic development in these mice.
The miR-17~92 cluster encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1), which are highly conserved in all vertebrates (Northcott et al., 2009; Tagawa and Seto, 2005; Uziel et al., 2009; Ventura et al., 2008). Germline hemizygous deletions of miR-17~92 clusters were accounted for microcephaly, short stature and digital abnormalities (de Pontual et al., 2011). In this study, we found that miR-17~92 cluster increased during osteogenesis, suggesting that they may critically regulate osteoblast differentiation. Furthermore, DICER overexpression significantly upregulated miR-17~92 cluster expression and downregulated the proapoptotic factor BIM which was previously found to be a direct target of the miR-17-92 cluster (Fontana et al., 2008; Koralov et al., 2008; Ventura et al., 2008).
In our in vivo study, the calvarial bone CSD mouse model was established and the BMSCs transfected with or without siRNA targeting DICER were combined with silk scaffolds and transplanted into calvarial bone CSDs. Four weeks post-surgery, micro-CT analysis revealed impaired new bone formation in calvarial defects in siRNA targeting DICER group.
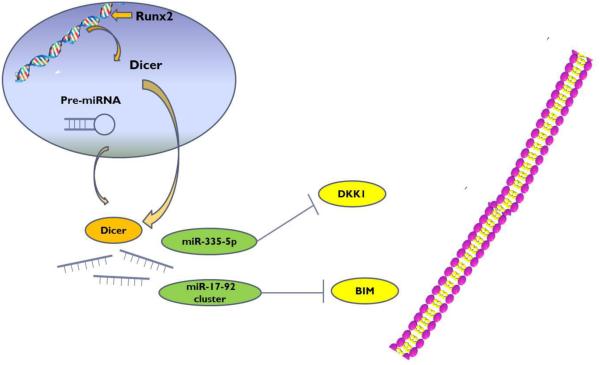
In conclusion, our results suggest that DICER processed miRNAs control commitment and differentiation of osteoblasts during bone development, which is mechanistically dependent upon transcriptional regulation of DICER by Runx2 (Fig. 7). Runx2 is able to transcriptionally regulate DICER potentially by binding to DICER promoter. Consequently, DICER cleaves precursors of miR-335-5p and miR-17-92 cluster to form mature miRNAs, which target and decrease the DKK1 and BIM levels, respectively. Therefore, Wnt-β-catenin signaling pathway is enhanced. These observations reveal a central mechanism underlying lineage-specific regulation by miRNAs during osteogenic differentiation and bone development. This cell- and development-specific regulation is essential and mandatory for the initiation and progression of osteogenic differentiation. Furthermore, our study also suggests a potential application of specifically regulating DICER for bone tissue repair and regeneration.
Figure 7.
Abridged general view for the interplay among Runx2/Dicer/miRNA pathway in osteogenesis. MiR-335-5p acts as a suppressor by targeting DKK1, which activates Wnt signaling and promotes osteogenic differentiation. MiRNA 17-92 cluster directly targets the proapoptotic factor BIM to downregulate BIM expression.
Acknowledgements
Dr. Jake Chen is supported by NIH Grant DE21464, IADR/AO 2012 Innovation in Implant Sciences Award, IADR/GSK 2014 Innovation in Oral Care Award, and an International Team for Implantoloty (ITI) Award. Dr. Leilei Zheng is supported by Natural Science Foundation of China (81470772).
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jcp.25406]
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57(3):364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. The genetics of low-density lipoprotein receptor-related protein 5 in bone: a story of extremes. Endocrinology. 2007;148(6):2622–2629. doi: 10.1210/en.2006-1352. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382(6592):638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. The New England journal of medicine. 2002;346(20):1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annual review of cell and developmental biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Dalle Carbonare L, Innamorati G, Valenti MT. Transcription factor Runx2 and its application to bone tissue engineering. Stem cell reviews. 2012;8(3):891–897. doi: 10.1007/s12015-011-9337-4. [DOI] [PubMed] [Google Scholar]
- de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Genevieve D, Goldenberg A, Oufadem M, Manouvrier S, Munnich A, Vidigal JA, Vekemans M, Lyonnet S, Henrion-Caude A, Ventura A, Amiel J. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nature genetics. 2011;43(10):1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews Genetics. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, Peschle C, Fruci D. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PloS one. 2008;3(5):e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. The Journal of biological chemistry. 2005;280(21):20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur T, Hussain S, Mudhasani R, Parulkar I, Colby JL, Frederick D, Kream BE, van Wijnen AJ, Stein JL, Stein GS, Jones SN, Lian JB. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340(1):10–21. doi: 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391(6665):357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107(46):19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Robinson KC, Veguilla RA, Chen PH, Yokoyama S, Makino E, Lu J, Larue L, Beermann F, Chin L, Bosenberg M, Song JS, Fisher DE. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141(6):994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhu J, Tu Q, Yamauchi M, Sodek J, Karsenty G, Tang J, Chen J. An in vivo model to study osteogenic gene regulation: targeting an avian retroviral receptor (TVA) to bone with the bone sialoprotein (BSP) promoter. J Bone Miner Res. 2005a;20(8):1403–1413. doi: 10.1359/JBMR.050316. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. The Journal of biological chemistry. 2005b;280(20):19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- Little RD, Recker RR, Johnson ML. High bone density due to a mutation in LDL-receptor-related protein 5. The New England journal of medicine. 2002;347(12):943–944. doi: 10.1056/NEJM200209193471216. author reply 943-944. [DOI] [PubMed] [Google Scholar]
- Lucero CM, Vega OA, Osorio MM, Tapia JC, Antonelli M, Stein GS, van Wijnen AJ, Galindo MA. The cancer-related transcription factor Runx2 modulates cell proliferation in human osteosarcoma cell lines. Journal of cellular physiology. 2013;228(4):714–723. doi: 10.1002/jcp.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, Bowerman B, Boutros M, Perrimon N. The promise and perils of Wnt signaling through beta-catenin. Science. 2002;296(5573):1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, Niehrs C, Izpisua Belmonte JC, Westphal H. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Developmental cell. 2001;1(3):423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer research. 2009;69(8):3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. Rna. 2006;12(7):1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes & development. 2008;22(22):3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27(45):5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Developmental biology. 2003;259(1):9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19(11):2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- Takahashi-Yanaga F, Sasaguri T. The Wnt/beta-catenin signaling pathway as a target in drug discovery. Journal of pharmacological sciences. 2007;104(4):293–302. doi: 10.1254/jphs.cr0070024. [DOI] [PubMed] [Google Scholar]
- Tu Q, Valverde P, Li S, Zhang J, Yang P, Chen J. Osterix Overexpression in Mesenchymal Stem Cells Stimulates Healing of Critical-Sized Defects in Murine Calvarial Bone. Tissue Engineering. 2007a;13(10):2431–2440. doi: 10.1089/ten.2006.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Zhang J, Fix A, Brewer E, Li YP, Zhang ZY, Chen J. Targeted overexpression of BSP in osteoclasts promotes bone metastasis of breast cancer cells. J Cell Physiol. 2009;218(1):135–145. doi: 10.1002/jcp.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Zhang J, James L, Dickson J, Tang J, Yang P, Chen J. Cbfa1/Runx2-deficiency delays bone wound healing and locally delivered Cbfa1/Runx2 promotes bone repair in animal models. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2007b;15(3):404–412. doi: 10.1111/j.1524-475X.2007.00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Zhang J, Paz J, Wade K, Yang P, Chen J. Haploinsufficiency of Runx2 results in bone formation decrease and different BSP expression pattern changes in two transgenic mouse models. J Cell Physiol. 2008;217(1):40–47. doi: 10.1002/jcp.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimalraj S, Arumugam B, Miranda PJ, Selvamurugan N. Runx2: Structure, function, and phosphorylation in osteoblast differentiation. International journal of biological macromolecules. 2015;78:202–208. doi: 10.1016/j.ijbiomac.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature cell biology. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome biology. 2014;15(3):R52. doi: 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokinski D, Pawlowska E, Blasiak J. RUNX2: A Master Bone Growth Regulator That May Be Involved in the DNA Damage Response. DNA and cell biology. 2015;34(5):305–315. doi: 10.1089/dna.2014.2688. [DOI] [PubMed] [Google Scholar]
- Ye JH, Xu YJ, Gao J, Yan SG, Zhao J, Tu Q, Zhang J, Duan XJ, Sommer CA, Mostoslavsky G, Kaplan DL, Wu YN, Zhang CP, Wang L, Chen J. Critical-size calvarial bone defects healing in a mouse model with silk scaffolds and SATB2-modified iPSCs. Biomaterials. 2011;32(22):5065–5076. doi: 10.1016/j.biomaterials.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes & development. 1996;10(12):1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, Chen J. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011a;26(8):1953–1963. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tu Q, Grosschedl R, Kim MS, Griffin T, Drissi H, Yang P, Chen J. Roles of SATB2 in osteogenic differentiation and bone regeneration. Tissue Eng Part A. 2011b;17(13-14):1767–1776. doi: 10.1089/ten.tea.2010.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Ma J, Chen S, Chen X, Yu X. MicroRNA-17-92 cluster regulates osteoblast proliferation and differentiation. Endocrine. 2014;45(2):302–310. doi: 10.1007/s12020-013-9986-y. [DOI] [PubMed] [Google Scholar]