Abstract
While important strides have been made in cancer therapy by targeting certain oncogenes, KRAS, the most common among them, remains refractory to this approach. In recent years, a deeper understanding of the critical importance of inflammation in promoting KRAS-driven oncogenesis has emerged, and applies across the different contexts of lung, pancreatic, and colorectal tumorigenesis. Here we review why these tissue types are particularly prone to developing KRAS mutations, and how inflammation conspires with KRAS signaling to fuel carcinogenesis. We discuss multiple lines of evidence that have established NF-κB, STAT3, and certain cytokines as key transducers of these signals, and data to suggest that targeting these pathways has significant clinical potential. Furthermore, recent work has begun to uncover how inflammatory signaling interacts with other KRAS regulated survival pathways such as autophagy and MAPK signaling, and that co-targeting these multiple nodes may be required to achieve real benefit. In addition, the impact of KRAS associated inflammatory signaling on the greater tumor microenvironment has also become apparent, and taking advantage of this inflammation by incorporating approaches that harness T cell anti-tumor responses represents another promising therapeutic strategy. Finally, we highlight the likelihood that the genomic complexity of KRAS mutant tumors will ultimately require tailored application of these therapeutic approaches, and that targeting inflammation early in the course of tumor development could have the greatest impact on eradicating this deadly disease.
Keywords: KRAS, inflammation, NF-κB, STAT3, cytokines, autophagy
1. Introduction
The RAS family of oncogenes was one of the first to be identified as mutated in human cancer. But despite extensive investigation of the signaling networks that RAS activates to promote cellular transformation, effective therapy has yet to be discovered either against RAS itself or its multitude of downstream targets. Since mutations in KRAS are frequently observed in three of the leading causes of cancer deaths: lung adenocarcinoma (LUAC), pancreatic ductal adenocarcinoma (PDAC), and colorectal carcinoma (CRC), the ability to target KRAS effectively would almost certainly reduce the morbidity and mortality from these common cancers [1].
KRAS-driven carcinogenesis is tightly linked with tumor promoting inflammation, which is increasingly recognized as target for therapeutic development. Activating mutations in KRAS endow epithelial cells with the capacity to survive and expand in this setting, often fueled by the same cytokines that are attempting to recruit inflammatory cells and fend them off [2]. Indeed, the most common tumors in which KRAS mutations are observed arise from the epithelial linings of organs – lung, pancreas, and colon - that sustain both mutational and inflammatory insults over the course of time (Figure 1). In addition, KRAS-driven cancers are not uniform diseases. Tissue specificity shapes the microenvironment in which the cancer develops, co-mutation of different tumor suppressor genes can modify how inflammatory signals downstream of KRAS are elaborated, and a variety of other factors such can influence the unique immune cell ecosystem that each cancer is associated with. Thus, therapies that target tumor promoting inflammation downstream of KRAS will likely need to be tailored further based on certain genomic or immunologic contexts.
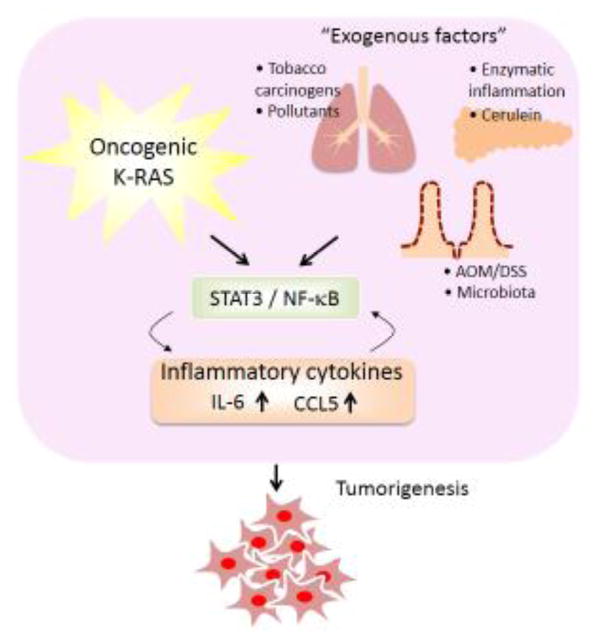
Figure 1. Cooperation of KRAS mutation and inflammatory signaling in lung, pancreatic and colorectal tumorigenesis.
Exogenous factors such as tobacco carcinogens or pollutants in the lung, enzymatic inflammation or cerulein in the pancreas, and microbiota or AOM/DSS (Azoxymethane/dextran sodium sulphate) in the colon trigger oncogenic KRAS mutation, NF-κB/STAT3, and inflammatory cytokine production, which promote tumorigenesis.
In this review we will focus specifically on the recent data that supports a critical role of inflammation in oncogenic KRAS mediated tumor development and maintenance, and how targeting inflammatory signaling pathways will likely be a necessary component of effective KRAS targeted therapy. First we will review the evidence for inflammatory signaling in promoting KRAS-associated lung, pancreatic, and colorectal carcinogenesis. Next, we discuss how cell intrinsic factors such as levels of basal autophagy may influence the relative engagement of immune signaling pathways, and how differences in the tumor immune microenvironment could influence immune targeted therapy. Finally, we end with a discussion of therapeutic considerations, and how developing combinations which incorporate drugs that counteract this tumor promoting inflammation may ultimately help to make KRAS-driven cancers a manageable disease.
2. Inflammation and KRAS-driven Lung Adenocarcinoma
Oncogenic mutations in KRAS are observed in approximately 1/3 of cases of LUAC and are strongly associated with smoking [3]. The effect of tobacco associated mutagens and chronic inflammation in fueling KRAS-driven lung cancer has been well documented in mouse models. For example, exposure to the tobacco carcinogen 3-methylcholanthrene (MCA), particularly when followed by induction of chronic inflammation with the non-carcinogenic pneumotoxin butylated hydroxytoluene (BHT), triggers a high frequency of murine lung cancers with Kras codon 12 mutations that mimic the spectrum observed in human LUAC [4]. More recent whole exome sequencing from mouse lung tumors arising from treatment with methyl-nitrosourea (MNU) also revealed common mutations in Kras codon 12, differing from other models that involve direct genetic activation of a KrasG12D allele, in that the former is dominated by high mutational load while the latter are associated with aneuploidy [5]. This study further examined mutational profiles following urethane treatment, another chemical carcinogen utilized in mouse lung cancer models that induces inflammation and NF-κB activation [6]. In contrast to MNU, urethane caused frequent KrasQ61L and KrasQ61R mutations, and a different mutational spectrum in general. Together, these studies highlight the fact that different mutagenic insults, coupled with inflammation, can promote distinct but common paths towards oncogenic KRAS activation in LUAC.
Genetically engineered mouse models (GEMMs) of Kras-driven lung cancer have been instrumental in defining specific signaling components, including NF-κB, STAT3, and secreted cytokines, that fuel oncogenesis. Although they typically lack the high mutational load and more faithful evolution of carcinogen induced lung tumors, these models have nonetheless helped to define oncogenic KRAS biology in vivo and its collaboration with commonly mutated tumor suppressors. These models have also enabled testing of genetic requirements and novel therapeutics, given the relatively short latency of lung tumor development. Multiple studies have defined an important role for NF-κB signaling the aggressive lox-stop-lox (LSL) KrasG12D;p53Flox/Flox (KP) mouse model of lung cancer. Expression of the IκB super repressor gene inhibited tumorigenesis in this model [7], as did genetic deletion of the p65 subunit or chemical inhibition of IκB kinase (IKK) activity [8, 9]. In contrast, studies of STAT3 signaling have suggested a more complex role for this key downstream mediator of cytokines in KRAS-driven tumorigenesis. Lung epithelial specific knockout of Stat3 paradoxically increased inflammation and Kras-driven tumorigenesis following urethane treatment, and cooperated with oncogenic Kras in tumor initiation, but was required in both instances for sustained tumor growth [10]. Another study further demonstrated that Stat3 disruption enhanced lung tumorigenesis downstream of oncogenic Kras, though showed mechanistically that this resulted from dysregulated NF-κB signaling [11]. Specific deletion of IL6 in mouse lung epithelia has yielded similar paradoxical results. In two separate studies, IL6 deletion combined with KrasG12D mutation promoted lung tumorigenesis but impaired tumor growth [12, 13], although the effect on tumor growth required intact p53 [12]. However, a more defined role for IL6 in promoting lung cancer pathogenesis was recently demonstrated in the LSL-KrasG12D;Lkb1Flox/Flox (KL) model [14], an equally if not more aggressive model compared to KP mice [15]. In this setting, Lkb1 inactivation shifted the immune microenvironment towards the accumulation of neutrophils, which exhibited T cell suppressive activity through the release of multiple cytokines including IL-6. In this model tumor growth was inhibited by treatment with a neutralizing IL-6 antibody [14]. Together, these studies demonstrate a key role for these inflammatory signals in fueling KRAS-driven lung cancer, but highlight feedback compensation between STAT3 and NF-κB signaling and tumor suppressor background as important issues to consider.
2. KRAS signaling and Inflammation in Pancreatic Ductal Carcinogenesis
PDAC is associated with the highest frequency of KRAS mutations of any tumor type [16]. Similar to LUAC, inflammation plays a key role in its pathogenesis, as lifetime risk of developing PDAC in patients with familial pancreatitis syndromes approaches 69-fold that of the general population [17]. The most common causes of hereditary pancreatitis involve germline mutations in PRSS1, encoding trypsinogen, or SPINK1, a serine protease inhibitor that limits trypsin activity, suggesting that chronic exposure of the pancreas to its own damaging projects may be a major etiologic factor in pancreatic carcinogenesis [18]. In contrast to KRASG12C transversion mutations, which are associated with smoking and commonly found in LUAC, PDAC is commonly associated with KRASG12D or KRASG12V, revealing mutational predilection or selection for these particular amino acid changes [19].
Similar to LUAC, mouse models of pancreatitis and PDAC have helped define the specific role of inflammation in KRAS-driven dysplasia. A commonly employed model utilizes transient administration of supraphysiologic doses of the cholecystokinin analog cerulein, which hyperstimulates the pancreas and results in acute pancreatitis. Whereas pancreatitis resolves in a KRASWT background, activation of a doxycycline inducible KRASG12D allele (iKRAS* model) triggers prolonged inflammation that induces acinar to ductal metaplasia (ADM) after one week, followed by pancreatic intra-epithelial neoplasia (PanIN) formation and eventual PDAC [20]. This tissue pathology requires the persistent expression of mutant KRAS, since withdrawal of doxycycline could revert histologies back to normal. Cerulein-induced pancreatitis promotes RAS activation, which becomes fixed in the presence of an oncogenic KRAS allele [21]. Inhibition of NF-κB activity by deletion of IKK-β suppressed RAS-GTP, inflammation, and dysplasia, whereas transgenic over-expression of IKK-β collaborated with KRASG12D to fuel this circuit. Similarly, deletion of IL-6 in the iKRAS* model markedly interfered with prolonged pancreatic inflammation and dysplasia following cerulein treatment, downregulating both pSTAT3 and pERK levels [22]. Multiple other studies have shown a role for IKK signaling in promoting oncogenic KRAS induced inflammatory signaling and pancreatic carcinogenesis in mice [23, 24], as well as IL-6/STAT3 signaling [25–27]. Together, these data firmly demonstrate that oncogenic KRAS mutation cooperates with NF-κB, STAT3, and cytokine signaling in the pancreatic microenvironment to sustain a pathologic circuit that promotes the evolution of PDAC.
4. Colitis and KRAS-driven Colorectal Cancer
While inflammation also plays a key role in colorectal carcinogenesis, the colon has several unique features compared with the lung and pancreas, which shape how cancer develops in this setting. First, cells that arise deep in the intestinal crypt proliferate and migrate towards the surface as they differentiate, eventually sloughing off into the lumen. This high cell turnover rate differs from lung and pancreas, and helps serve to prevent the accumulation of deleterious mutations, as cells that sustain mutational insults are continuously shed. In this context, mutations in APC and dysregulation of WNT-β-catenin signaling enables fixation of cells and formation of polyps, which then become the nidus for further accumulation of mutations that fuel CRC [28]. The other key distinguishing feature of the colon, as compared with other tissues, is the presence of a microbiome. While the specific role of intestinal microbiota in promoting CRC continues to be elucidated [29], direct exposure to bacterial and other organisms, and the cellular adaptations that enable this commensal relationship, clearly shapes its development.
Thus, in contrast to other tumor types, activation of β-catenin is the primary initiating event fueled by dysregulated inflammation. For example, IL-10 deficient mice, which develop colitis and spontaneous colorectal tumors, exhibit nuclear β-catenin through engagement of multiple inflammatory signaling pathways involving NF-κB and cytokine signal transduction [30]. Similarly, mesalamine, a commonly used anti-inflammatory agent for ulcerative colitis, inhibits dysplasia in IL-10 knockout mice with suppressed β-catenin signaling [31]. Recent whole exome sequencing studies of CRC from patients with underlying chronic inflammatory bowel disease are consistent with this idea–while fewer APC and KRAS mutations were identified in these tumors as compared with sporadic CRC, alternative mechanisms to activate WNT and RAS pathway signaling were identified [32].
Another classic animal model of colon carcinogenesis involves treatment of mice or rats with azoxymethane (AOM), which induces colon cancer along the typical adenoma-carcinoma sequence observed in common human sporadic tumors. In this model, KRAS mutations are frequently identified, and in a proportion similar to human CRC [33], as are both APC and β-catenin mutations [34, 35]. Perhaps the strongest evidence that inflammation fuels both β-catenin and KRAS-driven carcinogenesis in this context comes from the fact that co-administration of the inflammatory agent dextran sodium sulphate (DSS), markedly shortens the latency of tumor formation in this model [36]. Thus, in the context of an appropriate mutagenic insult that provides a direct source for genomic alterations in APC/β-catenin and KRAS, inflammation accelerates the adenoma-carcinoma sequence and drives tumorigenesis, similar to LUAC and PDAC. In this model, important roles for NF-κB and JAK/STAT pathway activation have also been implicated as key signals that support KRAS-associated tumorigenesis. Genetic deletion of IKK-β in enterocytes decreased tumor incidence, but was associated with increased DSS induced inflammation [37]. In contrast, myeloid-specific deletion of IKK-β suppressed inflammation and inhibited the expression of multiple cytokines including IL-6, also impairing tumorigenesis. These findings confirm the role of both tumor cell autonomous and non-cell autonomous NF-κB signaling in fueling oncogenesis in this model. In another study, intestinal epithelial cell specific deletion of suppressor of cytokine signaling-3 (SOCS3), a negative regulator of JAK-STAT signaling, enhanced AOM/DSS induced tumorigenesis, also associated with dysregulation of IL-6 and NF-κB activation [38]. Interestingly, TLR4-deficient mice (germline deletion) are also protected from AOM/DSS induced colorectal tumorigenesis [39]. These findings suggest a unique role of the intestinal microbiota, a rich source for TLR ligands, in promoting this inflammatory state that supports colon tumorigenesis.
5. Modulation of KRAS-driven inflammation and tumorigenesis by autophagy
Macroautophagy (herein referred to as autophagy), which degrades large macromolecular complexes and removes damaged organelles such as dysfunctional mitochondria, is a tumor suppressive process, yet also can promote oncogenesis in established KRAS dependent cancers [40]. Multiple studies have shown that the metabolic consequences of autophagy inhibition in aggressive KRAS-driven malignancies due to the accumulation of damaged mitochondria, in addition to elevated levels of reactive oxygen species (ROS), limits the ability of these tumors to survive and expand [41]. But disruption of autophagy also promotes incipient KRAS dysplasia, even in aggressive PDAC models where established tumors respond to autophagy inhibition [42]. Although the basis for this phenomenon has remained enigmatic, recent studies have shown that autophagy inhibition restrains tumor promoting inflammation downstream of oncogenic KRAS. For example, levels of key autophagy adaptor proteins such as p62 (encoded by SQSTM1) are increased when autophagy is inhibited [43] and p62 promotes Ras driven NF-κB activation [44]. Multiple additional studies have also implicated mitochondrial quality control in limiting inflammation; in particular, release of mitochondrial DNA (mtDNA) activates the cytoplasmic double stranded DNA sensing protein STING, which fuels activation of canonical and non-canonical IκB kinases, especially TBK1 [45]. Thus, the accumulation of damaged mitochondria in response to defective autophagy could also enhance this inflammatory response, and indeed TBK1 may play a key role in this feedback downstream of KRAS [46, 47]. Following exposure to poly-IC, which favors IRF3 activation, autophagy deficiency induced TBK1 signaling to produce IFN-β, tipping the balance towards cell death [46]. In contrast, priming cells with IL-1β, which activates NF-κB, increased TBK1 regulated pro-survival chemokines and cytokines including CCL5 and IL-6, promoting both T cell and pro-tumorigenic neutrophil infiltration in vivo and fostering KrasG12D-induced dysplasia [47] (Figure 2). These results further correlate with the IL-6 mediated tumor promotion observed in murine KL lung cancer [14] which lack autophagy [48]. Notably, ATG5 deficiency in the KP lung cancer model also enhanced tumorigenesis, in this case by promoting the influx of FOXP3+ regulatory T cells as a means of impairing anti-tumor immunosurveillance [49].
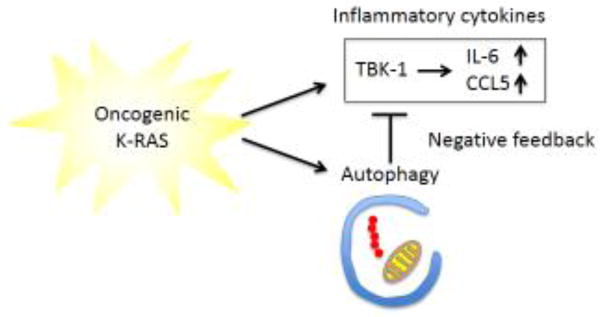
Figure 2. Interrelationship of autophagy and inflammation.
Oncogenic KRAS activates both autophagy and inflammation during tumorigenesis. While autophagy counterbalances cellular stress, it also acts as a negative feedback mechanism to suppress KRAS-induced inflammation.
Autophagy deficiency has also been strongly linked to the development of inflammatory bowel disease, as risk alleles in NOD2 and ATG16L1 have linked functionally to autophagy of bacterial peptidoglycans [50]. Impairment of autophagy by colonic epithelial cell deletion of ATG7 [51] or disruption of the NLRP6 inflammasome [52] disrupts mucus secretion and maintenance of the normal gut flora, resulting in colitis. Though as mentioned above, KRAS mutations themselves are less common in CRC resulting from inflammatory bowel disease, these findings nonetheless highlight the fact that autophagy plays a key role in regulating the unique microenvironment of the colon that also shapes sporadic CRC development.
6. Role of the tumor immune microenvironment
Given the extraordinary complexity of the immune microenvironment, and the fact that multiple variables beyond KRAS signaling alone can influence the specific fate of an individual tumor, it remains a challenge to generalize the impact of oncogenic KRAS signaling on infiltrating immune cells. Several principles have emerged, however, in addition to autophagy, which can modify how KRAS-driven tumors interact with the immune microenvironment, including the type of mutated tumor suppressor, mutational load, local immune compartments, and the influence of KRAS antigen presentation itself.
In LUAC, several additional recent studies examining gene expression patterns of human KRAS mutated tumors have highlighted key differences between cancers with TP53 or STK11/LKB1 co-mutation. In one study, unsupervised clustering of RNA-Sequencing data from the Cancer Genome Atlas (TCGA) [53] revealed segregation into three major subtypes, which coincided with TP53 mutation (KP), STK11/LKB1 mutation (KL), or lack thereof [54]. Gene-set enrichment analysis (GSEA) of the KP cluster highlighted upregulation of anti-tumor immunity and immune tolerance, including increased levels of PD-L1, PD1, and CTLA4 as compared with the KL subtype, and consistent with the preferential IL-6 activation in the KL mouse model [14]. In another study, gene expression profiling was performed on 442 LUAC samples, and the impact of TP53 and STK11/LKB1 co-mutations with KRAS was determined on features of tumor proliferative response and immune surveillance [55]. In this dataset, STK11/LKB1 co-mutation was associated with activation of an NF-κB signature previously linked to intra-tumoral T cell enrichment [56]. Though relative expression of PD-L1 or IL-6 as immune suppressive mechanisms were not examined in this study, these data further confirm that TP53 or STK11/LKB1 status alters how KRAS interacts with the tumor immune microenvironment.
Another confounder high mutational burden frequently associated with smoking and KRAS mutation status in LUAC. Elevated mutational load has been correlated with response to PD1 immune checkpoint activation in lung cancer [57], and neoantigen clonal diversity may be the predominant influence that regulates immune checkpoint activation in these tumors [58]. While TP53 inactivation enables cells to tolerate genotoxic stress and accumulate these mutational loads, multiple other factors influence mutational burden, such as alterations in repair pathway genes and smoking exposure. Indeed, levels of PD-L1 expression by immunohistochemistry were significantly correlated with smoking status in KRAS-mutant lung cancer and highest among current smokers, as compared with former or never smokers [59]. Conversely, KRAS mutant PDAC and CRC have been more refractory to anti-PD1/PDL1 therapies, except when mutational load is increased by microsatellite instability (MSI) status in CRC, for example [60].
Accessory immune compartments have also been implicated KRAS-driven tumorigenesis, especially in the presence of strong tumor antigens. Using a KP mouse model in which Cre mediated Kras activation and Trp53 inactivation is coupled with the expression of ovalbumin antigens, a recent study demonstrated the critical importance of Tregs and tertiary lymphoid structures (TLS) in enabling tumor immune evasion [61]. Deletion of Tregs by Foxp3 regulated expression of the diphtheria toxin receptor resulted in potent T cell infiltration and destruction of KP tumors. Rather than localizing within KP tumors themselves, Tregs accumulated in perivascular immune patches that exhibited features of tumor associated TLS, also observed in human cancers. These sites were responsible for immune cell proliferation and activation in response to Treg depletion, and importantly immune responses were also observed in KP mice even without ovalbumin antigen exposure. Similar immune destruction has also been identified in the pancreatic KrasG12D/+;Trp53R172H/+;Pdx1-Cre (KPC) mouse PDAC model following Listeria vaccine exposure and depletion of Tregs [62]. In this model Treg depletion also localized to pancreatic lymph nodes; however, response was limited to early stage PanINs and not late stage PanINs, suggesting a predominant role during tumor initiation. Extratumoral Ly6Clow F4/80+ macrophages have also been implicated as immunosuppressive in the KPC model [63]. Depletion of these cell resulted in the specific influx of CD8+ T cells and regression of established tumors following combined treatment with gemcitabine and a CD40 agonist.
Finally, oncogenic KRAS itself is a tumor specific neoantigen. Recent proof of principle studies in mice showed that HLA-A*11:01 can present KRAS mutant variant peptides including G12V and G12D and be recognized by specific T cell receptors (TCRs) [64]. Adoptive transfer of these T cells into NOD scid gamma (NSG) mice bearing a KRAS G12D mutated PDAC line resulted in tumor growth impairment. Next generation sequencing and high-throughput immunologic screening of human GI cancers also identified neoantigen specific tumor infiltrating lymphocytes, including an HLA-C*08:02 restricted TCR from CD8+ T cells targeting KRASG12D that could potentially be harnessed for personalized immunotherapy [65]. Further study of how commonly KRAS restricted T cells are found across LUAC, PDAC, and CRC patients, is an important area of future research, both to identify the factors that determine whether they have undergone immunoediting during tumor development, and to determine how they might potentially be deployed as tumor specific immunotherapy.
7. Therapeutic targeting of KRAS-associated immune pathways
Given the integral role of inflammation in promoting KRAS-driven tumorigenesis across a variety of different tumor types, targeting immune signaling pathways will almost certainly represent an integral component of achieving disease control (Figure 3). One attractive strategy is to target the NF-κB and JAK-STAT pathways, given their prominent roles in tumor development and the reported preclinical efficacy of inhibiting these pathways across multiple KRAS mouse tumor models. Although selective NF-κB inhibitors have yet to enter the clinic, multiple JAK kinase inhibitors have been developed based on the common activating JAK2V617F mutation in myeloproliferative neoplasms [66]. Ruxolitinib, a potent and selective inhibitor of JAK1/JAK2 kinases, has been FDA approved for this indication, and is currently being tested in clinical trials across a variety of different solid tumors, including LUAC, PDAC, and CRC. One study in metastatic refractory PDAC, combining ruxolitinib with the chemotherapeutic agent capecitabine, showed a trend towards improvement in progression free and overall survival, with significant improvement in a subgroup of patients with elevated levels of C-reactive protein (CRP) as a marker of systemic inflammation [67], although these results await further confirmation in larger randomized controlled studies. Momelotinib (previously known as CYT387), is another JAK1/2 inhibitor that is currently undergoing a randomized phase 3 trial with ruxolitinib in myelofibrosis, based on promising efficacy in phase 2 studies that not only demonstrated reduction in splenomegaly and constitutional symptoms, but also showed improvement in transfusion dependence [68]. In contrast to ruxolitinib, momelotinib is a multitargeted kinase that also inhibits both JAK1/2 and the IKK-related kinases TBK1 and IKKε, which results not only in inhibition of JAK/STAT signaling downstream of IFNγ and IL-6, but also TBK1/IKKε signaling downstream of LPS and IL-1 [69]. This broad suppression of STAT, NF-κB, and IRF3 transcription results in a potent anti-cytokine effect, and preclinical efficacy in murine Kras lung cancer models. Similar results in preventing PanIN formation in the iKRAS* pancreatic model were also recently reported with momelotinib, which was further shown to inhibit autophagy as well as feedback cytokine activation [47]. Together, these data suggest particular benefit of inhibiting both JAK/STAT and TBK1/IKKε activity in suppressing KRAS induced dysplasia, and are consistent with the findings that inhibiting only STAT3, for example, can result in feedback NF-κB activation [11]. While treatment with momelotinib itself represents one strategy to achieve multitargeted pathway inhibition, combination of potent and selective JAK and TBK1/IKKε inhibitors may be a more appealing strategy, to avoid alternate off-target effects.
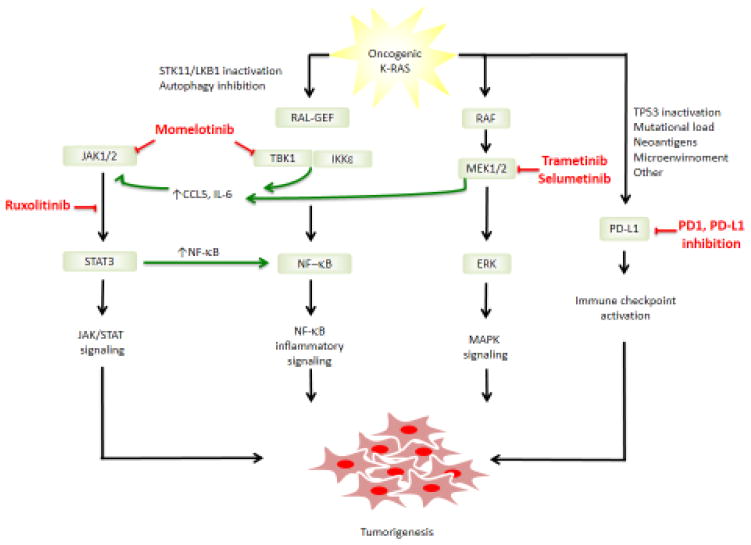
Figure 3. Schematic of therapeutic strategies targeting KRAS associated inflammation.
Shown are approaches to inhibit JAK/STAT, NF-κB, MAPK, and immune checkpoint signaling pathways, which are being evaluated as alone or as combination therapies. Also shown are potential factors that could influence susceptibility to protumorigenic cytokine inhibition (e.g. STK11/LKB1 inactivation) or immune checkpoint blockade (e.g. TP53 inactivation).
Downstream MAPK pathway inhibition has also emerged as a critical factor in targeting KRAS-driven cancers, and is the backbone of multiple clinical trials [70]. Important cross-talk also exists between IL-6 and the MEK/ERK signaling cascade [22]. Indeed, multiple studies have also shown that pharmacologic or genetic TBK1 inhibition results in feedback ERK activation, suggesting that JAK/TBK1/IKKε inhibition may not be sufficient on its own to fully impair tumor growth, and may require concomitant MEK inhibition [69, 71]. Conversely, observations across multiple KRAS-driven lung and colorectal cell lines have revealed that treatment with the MEK inhibitor selumetinib results in feedback activation of IL-6 and STAT3, as a mechanism of adaptive resistance [72]. Consistent with these findings, combination of momelotinib with the MEK inhibitor selumetinib was required to prevent this feedback and fully suppress both pathways, which was effective in potently inhibiting IL-6 production and limiting tumorigenesis in the aggressive KP model [69]. Further evidence for developing targeted therapeutic strategies that ablate IL-6 in combination with MAPK signaling comes from the iKRAS* model. Whereas deletion of IL-6 alone disrupted the inflammatory microenvironment and limited PanIN progression [22], similar to treatment with momelotinib [47], inhibition of MEK alone using the kinase inhibitor PD325901was able to suppress PanIN growth, but did not prevent pancreatitis [73]. Together, these findings suggest more broadly that therapeutic strategies designed to target NF-κB and JAK-STAT signaling will be more efficacious in combination with MEK inhibitors. As discussed above, immune checkpoint blockade with anti-PD1/PD-L1 antibodies is another emerging therapeutic strategy for KRAS-driven cancers. Multiple attempts to increase the response rate of these agents in lung and other malignancies are underway, both through increasing the mutational load (i.e. tumor radiation exposure) and by combination with therapies that modulate the immune microenvironment and T cell priming. In addition to producing IL-6 feedback activation, MEK inhibitor treatment of tumors in syngeneic mouse models also was shown to increase the percentage of CD8+ T cell infiltration in tumors, including T-bethi populations, which is induced following naïve T cell priming and is required for effector cytokine production [74]. While MEK inhibition (MEKi) was actually found to limit naïve T cell priming and expansion, it impaired the TCR triggered death of chronically stimulated CD8+ T cells, and synergized with anti-PD-L1 to enhance anti-tumor efficacy in vivo in the CT26 CRC tumor model. Another study showed similar efficacy of combining trametinib with anti-PD1 in this syngeneic model, accompanied by the accumulation of tumor infiltrating CD8+ T cells [75]. These results have spurred several clinical trials combining a variety of different MEK inhibitors with anti-PD1 or anti-PD-L1 inhibitors across different tumor types, including KRAS mutant LUAC, PDAC, and CRC. However, multiple issues remain such as the continuous versus pulse dosing of MEKi given the potential negative effects on T cell activation, and the fact that neoantigen specific T cells need to be present in the first place for them to be primed by MEKi. Regardless, the results from these trials and the particular efficacy in KRAS-driven cancers are eagerly awaited.
Finally, although initial clinical trials of these novel experimental drug combinations are being pursued in advanced treatment refractory patients, it is likely that targeting inflammatory signaling pathways will be even more effective during tumor development. Perhaps the strongest clinical evidence for this approach comes from the wealth of data showing that COX2 inhibition with aspirin or other non-steroidal anti-inflammatory agents can prevent colorectal cancer development [76]. Although the risk/benefit ratio of these particular agents remains controversial, and their potency limited, these findings suggest that disrupting tumor promoting inflammation with newer therapies, alone or in combination, will be particularly effective during KRAS-driven cancer formation.
8. Conclusions
In summary, although much remains to be learned, our understanding of how inflammation contributes to KRAS-driven oncogenesis has improved immensely over the past several years. At the same time, drugs that target the signal transduction pathways that propagate these signals have entered the clinic. Understanding how to combine these therapies safely and achieve effective target inhibition in patients will hopefully result in meaningful clinical responses for patients suffering with advanced KRAS-driven malignancies. In addition, defining which patients might benefit from suppressing tumor promoting inflammation, versus harnessing its activity with immune checkpoint blockade, will be of major importance, and informed by analysis of the features beyond KRAS mutation that correlate with response. Finally, as effective regimens are identified and targeted to the appropriate patients, moving this therapy earlier in the course of disease is likely to have a greater impact, and bring us closer to the goal of finally controlling these refractory cancers.
Acknowledgments
This work was supported by the strategic young researcher overseas visits program for accelerating brain circulation (S.K.), NCI-R01 CA190394-01 (D.B.), NCI-K08 CA138918-01A1 (D.B.), and the Gloria T. Maheu Fund for Lung Cancer Research (D.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shunsuke Kitajima, Email: shunsuke_kitajima@dfci.harvard.edu.
Rohit Thummalapalli, Email: rohit_thummalapalli@hms.harvard.edu.
David A. Barbie, Email: dbarbie@partners.org.
References
- 1.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–81. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Golay HG, Barbie DA. Targeting cytokine networks in KRAS-driven tumorigenesis. Expert Rev Anticancer Ther. 2014;14(8):869–71. doi: 10.1586/14737140.2014.928596. [DOI] [PubMed] [Google Scholar]
- 3.Guibert N, Ilie M, Long E, Hofman V, Bouhlel L, Brest P, Mograbi B, Marquette CH, Didier A, Mazieres J, Hofman P. KRAS Mutations in Lung Adenocarcinoma: Molecular and Epidemiological Characteristics, Methods for Detection, and Therapeutic Strategy Perspectives. Curr Mol Med. 2015;15(5):418–32. doi: 10.2174/1566524015666150505161412. [DOI] [PubMed] [Google Scholar]
- 4.Fritz JM, Dwyer-Nield LD, Russell BM, Malkinson AM. The Kras mutational spectra of chemically induced lung tumors in different inbred mice mimics the spectra of KRAS mutations in adenocarcinomas in smokers versus nonsmokers. J Thorac Oncol. 2010;5(2):254–7. doi: 10.1097/JTO.0b013e3181c8ce04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, Delrosario R, Jen KY, Gurley KE, Kemp CJ, Fredlund E, Quigley DA, Adams DJ, Balmain A. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517(7535):489–92. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stathopoulos GT, Sherrill TP, Cheng DS, Scoggins RM, Han W, Polosukhin VV, Connelly L, Yull FE, Fingleton B, Blackwell TS. Epithelial NF-kappaB activation promotes urethane-induced lung carcinogenesis. Proc Natl Acad Sci U S A. 2007;104(47):18514–9. doi: 10.1073/pnas.0705316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–7. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basseres DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;70(9):3537–46. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basseres DS, Ebbs A, Cogswell PC, Baldwin AS. IKK is a therapeutic target in KRAS-Induced lung cancer with disrupted p53 activity. Genes Cancer. 2014;5(1–2):41–55. doi: 10.18632/genesandcancer.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Qu Z, Yan S, Sun F, Whitsett JA, Shapiro SD, Xiao G. Differential roles of STAT3 in the initiation and growth of lung cancer. Oncogene. 2015;34(29):3804–14. doi: 10.1038/onc.2014.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabner B, Schramek D, Mueller KM, Moll HP, Svinka J, Hoffmann T, Bauer E, Blaas L, Hruschka N, Zboray K, Stiedl P, Nivarthi H, Bogner E, Gruber W, Mohr T, Zwick RH, Kenner L, Poli V, Aberger F, Stoiber D, Egger G, Esterbauer H, Zuber J, Moriggl R, Eferl R, Gyorffy B, Penninger JM, Popper H, Casanova E. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat Commun. 2015;6:6285. doi: 10.1038/ncomms7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan X, Carretero J, Chen Z, Zhang J, Wang Y, Chen J, Li X, Ye H, Tang C, Cheng X, Hou N, Yang X, Wong KK. Loss of p53 attenuates the contribution of IL-6 deletion on suppressed tumor progression and extended survival in Kras-driven murine lung cancer. PLoS One. 2013;8(11):e80885. doi: 10.1371/journal.pone.0080885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu Z, Sun F, Zhou J, Li L, Shapiro SD, Xiao G. Interleukin-6 Prevents the Initiation but Enhances the Progression of Lung Cancer. Cancer Res. 2015;75(16):3209–15. doi: 10.1158/0008-5472.CAN-14-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, Buczkowski KA, Liu Y, Awad MM, Denning WL, Diao L, Wang J, Parra Cuentas ER, Wistuba, Soucheray M, Thai TC, Asahina H, Kitajima S, Altabef A, Cavanaugh JD, Rhee K, Gao P, Zhang H, Fecci PE, Shimamura T, Hellmann M, Heymach JV, Hodi FS, Freeman GJ, Barbie DA, Dranoff G, Hammerman PS, Wong KK. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T cell activity in the lung tumor microenvironment. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, Liang MC, Cai D, Naumov GN, Bao L, Contreras CM, Li D, Chen L, Krishnamurthy J, Koivunen J, Chirieac LR, Padera RF, Bronson RT, Lindeman NI, Christiani DC, Lin X, Shapiro GI, Janne PA, Johnson BE, Meyerson M, Kwiatkowski DJ, Castrillon DH, Bardeesy N, Sharpless NE, Wong KK. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 16.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grutzmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM I. Australian Pancreatic Cancer Genome. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 17.Becker AE, Hernandez YG, Frucht H, Lucas AL. Pancreatic ductal adenocarcinoma: risk factors, screening, and early detection. World J Gastroenterol. 2014;20(32):11182–98. doi: 10.3748/wjg.v20.i32.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaRusch J, Whitcomb DC. Genetics of pancreatitis. Curr Opin Gastroenterol. 2011;27(5):467–74. doi: 10.1097/MOG.0b013e328349e2f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrow SL, Simon E, Prinz E, Bick T, Shentzer T, Nagawkar SS, Sabo E, Ben-Izhak O, Hershberg R, Hershkovitz D. Variation in KRAS driver substitution distributions between tumor types is determined by both mutation and natural selection. Sci Rep. 2016;6:21927. doi: 10.1038/srep21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–53. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniluk J, Liu Y, Deng D, Chu J, Huang H, Gaiser S, Cruz-Monserrate Z, Wang H, Ji B, Logsdon CD. An NF-kappaB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J Clin Invest. 2012;122(4):1519–28. doi: 10.1172/JCI59743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Yan W, Collins MA, Bednar F, Rakshit S, Zetter BR, Stanger BZ, Chung I, Rhim AD, di Magliano MP. Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res. 2013;73(20):6359–74. doi: 10.1158/0008-5472.CAN-13-1558-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier HJ, Wagner M, Schips TG, Salem HH, Baumann B, Wirth T. Requirement of NEMO/IKKgamma for effective expansion of KRAS-induced precancerous lesions in the pancreas. Oncogene. 2013;32(21):2690–5. doi: 10.1038/onc.2012.272. [DOI] [PubMed] [Google Scholar]
- 24.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H, Liu J, Lemischka IR, Hung MC, Chiao PJ. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(1):105–20. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, Yoshimura A, Reindl W, Sipos B, Akira S, Schmid RM, Algul H. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19(4):456–69. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, Hebrok M. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19(4):441–55. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, Levy DE, Settleman J, Engelman JA, Bardeesy N. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71(14):5020–9. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 29.Nistal E, Fernandez-Fernandez N, Vivas S, Olcoz JL. Factors Determining Colorectal Cancer: The Role of the Intestinal Microbiota. Front Oncol. 2015;5:220. doi: 10.3389/fonc.2015.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol. 2013;35(2):229–44. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown JB, Lee G, Managlia E, Grimm GR, Dirisina R, Goretsky T, Cheresh P, Blatner NR, Khazaie K, Yang GY, Li L, Barrett TA. Mesalamine inhibits epithelial beta-catenin activation in chronic ulcerative colitis. Gastroenterology. 2010;138(2):595–605. 605 e1–3. doi: 10.1053/j.gastro.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robles AI, Traverso G, Zhang M, Roberts NJ, Khan MA, Joseph C, Lauwers GY, Selaru FM, Popoli M, Pittman ME, Ke X, Hruban RH, Meltzer SJ, Kinzler KW, Vogelstein B, Harris CC, Papadopoulos N. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology. 2016;150(4):931–43. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vivona AA, Shpitz B, Medline A, Bruce WR, Hay K, Ward MA, Stern HS, Gallinger S. K-ras mutations in aberrant crypt foci, adenomas and adenocarcinomas during azoxymethane-induced colon carcinogenesis. Carcinogenesis. 1993;14(9):1777–81. doi: 10.1093/carcin/14.9.1777. [DOI] [PubMed] [Google Scholar]
- 34.Maltzman T, Whittington J, Driggers L, Stephens J, Ahnen D. AOM-induced mouse colon tumors do not express full-length APC protein. Carcinogenesis. 1997;18(12):2435–9. doi: 10.1093/carcin/18.12.2435. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi M, Nakatsugi S, Sugimura T, Wakabayashi K. Frequent mutations of the beta-catenin gene in mouse colon tumors induced by azoxymethane. Carcinogenesis. 2000;21(6):1117–20. [PubMed] [Google Scholar]
- 36.De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, Signori E, Fazio VM. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Rigby RJ, Simmons JG, Greenhalgh CJ, Alexander WS, Lund PK. Suppressor of cytokine signaling 3 (SOCS3) limits damage-induced crypt hyper-proliferation and inflammation-associated tumorigenesis in the colon. Oncogene. 2007;26(33):4833–41. doi: 10.1038/sj.onc.1210286. [DOI] [PubMed] [Google Scholar]
- 39.Fukata M, Chen A, Vamadevan AS, Cohen J, Breglio K, Krishnareddy S, Hsu D, Xu R, Harpaz N, Dannenberg AJ, Subbaramaiah K, Cooper HS, Itzkowitz SH, Abreu MT. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133(6):1869–81. doi: 10.1053/j.gastro.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25(19):1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White E, Mehnert JM, Chan CS. Autophagy, Metabolism, and Cancer. Clin Cancer Res. 2015;21(22):5037–46. doi: 10.1158/1078-0432.CCR-15-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4(8):905–13. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–9. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13(4):343–54. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Feng M, Guan W. Mitochondrial DNA sensing by STING signaling participates in inflammation, cancer and beyond. Int J Cancer. 2016 doi: 10.1002/ijc.30074. [DOI] [PubMed] [Google Scholar]
- 46.Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol Cell. 2014;55(6):916–30. doi: 10.1016/j.molcel.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang S, Imamura Y, Jenkins R, Canadas I, Kitajima S, Aref AR, Brannon AL, Oki E, Castoreno A, Zhu Z, Thai TC, Reibel J, Qian Z, Ogino S, Wong KK, Baba H, Kimmelman AC, Pasca di Magliano M, Barbie DA. Autophagy inhibition dysregulates TBK1 signaling and promotes pancreatic inflammation. Cancer Immunol Res. 2016 doi: 10.1158/2326-6066.CIR-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, Shaw RJ. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23(2):143–58. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, Sykacek P, Frank L, Schramek D, Komnenovic V, Sigl V, Aumayr K, Schmauss G, Fellner N, Handschuh S, Glosmann M, Pasierbek P, Schlederer M, Resch GP, Ma Y, Yang H, Popper H, Kenner L, Kroemer G, Penninger JM. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 50.de Lange KM, Barrett JC. Understanding inflammatory bowel disease via immunogenetics. J Autoimmun. 2015;64:91–100. doi: 10.1016/j.jaut.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 51.Tsuboi K, Nishitani M, Takakura A, Imai Y, Komatsu M, Kawashima H. Autophagy Protects against Colitis by the Maintenance of Normal Gut Microflora and Secretion of Mucus. J Biol Chem. 2015;290(33):20511–26. doi: 10.1074/jbc.M114.632257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, Elinav E, Finlay BB, Flavell RA. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156(5):1045–59. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.N. Cancer Genome Atlas Research. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, Zhang J, Giri U, Gudikote J, Cortez MA, Yang C, Fan Y, Peyton M, Girard L, Coombes KR, Toniatti C, Heffernan TP, Choi M, Frampton GM, Miller V, Weinstein JN, Herbst RS, Wong KK, Zhang J, Sharma P, Mills GB, Hong WK, Minna JD, Allison JP, Futreal A, Wang J, Wistuba, Heymach JV. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860–77. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, Engel BE, Xie M, Berglund AE, Creelan BC, Antonia SJ, Gray JE, Eschrich SA, Chen DT, Cress WD, Haura EB, Beg AA. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2015 doi: 10.1038/onc.2015.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hopewell EL, Zhao W, Fulp WJ, Bronk CC, Lopez AS, Massengill M, Antonia S, Celis E, Haura EB, Enkemann SA, Chen DT, Beg AA. Lung tumor NF-kappaB signaling promotes T cell-mediated immune surveillance. J Clin Invest. 2013;123(6):2509–22. doi: 10.1172/JCI67250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–9. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calles A, Liao X, Sholl LM, Rodig SJ, Freeman GJ, Butaney M, Lydon C, Dahlberg SE, Hodi FS, Oxnard GR, Jackman DM, Janne PA. Expression of PD-1 and Its Ligands, PD-L1 and PD-L2, in Smokers and Never Smokers with KRAS-Mutant Lung Cancer. J Thorac Oncol. 2015;10(12):1726–35. doi: 10.1097/JTO.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 60.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res. 2016;22(4):813–20. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 61.Joshi NS, Akama-Garren EH, Lu Y, Lee DY, Chang GP, Li A, DuPage M, Tammela T, Kerper NR, Farago AF, Robbins R, Crowley DM, Bronson RT, Jacks T. Regulatory T Cells in Tumor-Associated Tertiary Lymphoid Structures Suppress Anti-tumor T Cell Responses. Immunity. 2015;43(3):579–90. doi: 10.1016/j.immuni.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keenan BP, Saenger Y, Kafrouni MI, Leubner A, Lauer P, Maitra A, Rucki AA, Gunderson AJ, Coussens LM, Brockstedt DG, Dubensky TW, Jr, Hassan R, Armstrong TD, Jaffee EM. A Listeria vaccine and depletion of T-regulatory cells activate immunity against early stage pancreatic intraepithelial neoplasms and prolong survival of mice. Gastroenterology. 2014;146(7):1784–94 e6. doi: 10.1053/j.gastro.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beatty GL, Winograd R, Evans RA, Long KB, Luque SL, Lee JW, Clendenin C, Gladney WL, Knoblock DM, Guirnalda PD, Vonderheide RH. Exclusion of T Cells From Pancreatic Carcinomas in Mice Is Regulated by Ly6C(low) F4/80(+) Extratumoral Macrophages. Gastroenterology. 2015;149(1):201–10. doi: 10.1053/j.gastro.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang QJ, Yu Z, Griffith K, Hanada K, Restifo NP, Yang JC. Identification of T-cell Receptors Targeting KRAS-Mutated Human Tumors. Cancer Immunol Res. 2016;4(3):204–14. doi: 10.1158/2326-6066.CIR-15-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, Wunderlich JR, Somerville RP, Rosenberg SA. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–90. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenthal A, Mesa RA. Janus kinase inhibitors for the treatment of myeloproliferative neoplasms. Expert Opin Pharmacother. 2014;15(9):1265–76. doi: 10.1517/14656566.2014.913024. [DOI] [PubMed] [Google Scholar]
- 67.Hurwitz HI, Uppal N, Wagner SA, Bendell JC, Beck JT, Wade SM, Nemunaitis JJ, Stella PJ, Pipas JM, Wainberg ZA, Manges R, Garrett WM, Hunter DS, Clark J, Leopold L, Sandor V, Levy RS. Randomized, Double-Blind, Phase II Study of Ruxolitinib or Placebo in Combination With Capecitabine in Patients with Metastastic Pancreatic Cancer for Whom Therapy with Gemcitabine Has Failed. J Clin Oncol. 2015 doi: 10.1200/JCO.2015.61.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pardanani A, Laborde RR, Lasho TL, Finke C, Begna K, Al-Kali A, Hogan WJ, Litzow MR, Leontovich A, Kowalski M, Tefferi A. Safety and efficacy of CYT387, a JAK1 and JAK2 inhibitor, in myelofibrosis. Leukemia. 2013;27(6):1322–7. doi: 10.1038/leu.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, Moody SE, Shen RR, Schinzel AC, Thai TC, Reibel JB, Tamayo P, Godfrey JT, Qian ZR, Page AN, Maciag K, Chan EM, Silkworth W, Labowsky MT, Rozhansky L, Mesirov JP, Gillanders WE, Ogino S, Hacohen N, Gaudet S, Eck MJ, Engelman JA, Corcoran RB, Wong KK, Hahn WC, Barbie DA. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014;4(4):452–65. doi: 10.1158/2159-8290.CD-13-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu Z, Golay HG, Barbie DA. Targeting pathways downstream of KRAS in lung adenocarcinoma. Pharmacogenomics. 2014;15(11):1507–18. doi: 10.2217/pgs.14.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim JY, Welsh EA, Oguz U, Fang B, Bai Y, Kinose F, Bronk C, Remsing Rix LL, Beg AA, Rix U, Eschrich SA, Koomen JM, Haura EB. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc Natl Acad Sci U S A. 2013;110(30):12414–9. doi: 10.1073/pnas.1220674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26(2):207–21. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 73.Collins MA, Yan W, Sebolt-Leopold JS, Pasca di Magliano M. MAPK signaling is required for dedifferentiation of acinar cells and development of pancreatic intraepithelial neoplasia in mice. Gastroenterology. 2014;146(3):822–834 e7. doi: 10.1053/j.gastro.2013.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ebert PJ, Cheung J, Yang Y, McNamara E, Hong R, Moskalenko M, Gould SE, Maecker H, Irving BA, Kim JM, Belvin M, Mellman I. MAP Kinase Inhibition Promotes T Cell and Anti-tumor Activity in Combination with PD-L1 Checkpoint Blockade. Immunity. 2016;44(3):609–21. doi: 10.1016/j.immuni.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 75.Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, Yang J, Seestaller-Wehr L, Zhang SY, Hopson C, Tsvetkov L, Jing J, Zhang S, Smothers J, Hoos A. The BRAF and MEK Inhibitors Dabrafenib and Trametinib: Effects on Immune Function and in Combination with Immunomodulatory Antibodies Targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res. 2015;21(7):1639–51. doi: 10.1158/1078-0432.CCR-14-2339. [DOI] [PubMed] [Google Scholar]
- 76.Ranger GS. Current concepts in colorectal cancer prevention with cyclooxygenase inhibitors. Anticancer Res. 2014;34(11):6277–82. [PubMed] [Google Scholar]