Abstract
Targeting mutations and amplifications in the EGFR has been successful precision therapy for cancers of the lung, oral cavity and gastrointestinal track. However, a systemic immune reaction manifested by dose-limiting inflammation in the skin and gut has been a consistent adverse effect. To address the possibility that intra-tumoral immune changes contribute to the anti-cancer activity of EGFR inhibition, squamous cancers were produced by syngeneic orthografts of either EGFR null or wildtype mouse primary keratinocytes transduced with an oncogenic H-ras retrovirus. Flow cytometric, RNA and Bioplex immunoassay analyses of the tumor immune milieu were performed. Cancers forming from keratinocytes genetically depleted of EGFR were smaller than wildtype cancers and had fewer infiltrating FoxP3 Treg cells, lower Foxp3 RNA and a lower percentage of CD4 PD1 positive cells indicating a tumor cell autonomous regulation of its microenvironment. Hosts bearing wildtype cancers treated with gefitinib for one week showed a trend for smaller tumors. In this short term pharmacological model, there was also a trend to reduced FoxP3 cells and FoxP3 RNA in the tumors of treated mice as well as a substantial increase in the ratio of IL-1A/IL-1RA transcripts. These results suggest that relatively brief systemic inhibition of EGFR signaling alters the immune environment of the targeted cancer. Together these data imply that an EGFR dependent Treg function supports the growth of squamous cancers and is a target for the therapeutic activity of EGFR inhibition.
Keywords: EGFR, SCC, gefitinib, tumor immune-environment, T regulatory cells
Introduction
The Epidermal Growth Factor Receptor (EGFR) is a precision medicine cancer drug target to treat non-small cell lung cancer, colorectal cancer, squamous cell carcinoma of the head and neck and pancreatic cancer. Often overexpressed and/or mutated in these cancers, EGFR represents a desirable target in order to block cancer cell proliferation and survival, to reduce tumor neo-angiogenesis and cancer progression and to induce cell cycle arrest and apoptosis1. In the clinic, EGFR activity is blocked via the administration of tyrosine kinase inhibitors (gefitinib, erlotinib, lapatinib) or specific antibodies (cetuximab, panitumumab). Blockade of EGFR is of particular interest to the dermatology community because the typical adverse effect seen with EGFR inhibition therapy is a skin inflammatory response (papulo-pustular rash and pruritus) not unexpected since EGFR is also highly expressed in normal epithelial tissues2. Recent genetic approaches have indicated that macrophage and mast cell infiltration, responding to keratinocyte derived chemokines, initiate the skin rash3,4. This evidence raises the possibility that the tumor immune-environment can also be altered in the absence of a functional EGFR during pharmacological intervention. The recent introduction of immune checkpoint (CTLA4, PD1/PD-L1) inhibitors into the arena of cancer treatments proves that specific immune populations can determine the tumor fate in terms of growth or eradication5. To model the effects of EGFR ablation on the immune environment of cutaneous squamous cell carcinoma (SCC), we performed a series of exploratory experiments using genetic and pharmacologic approaches on FVB/N immunocompetent mice.
Materials and Methods
Cell cultures and tumor studies
In the genetic and in the pharmacologic models, the tumors were obtained by grafting FVB/N newborn mouse keratinocytes cultured as previously described6 with the following modifications: EGFRwt/wt and EGFRflx/flx keratinocytes were transformed with a constitutively active form of HRAS via infection for 5 days with the retrovirus7. At day 3 of HRAS infection, deletion of EGFR was carried out with a second infection with an adenovirus expressing Cre Recombinase8 that ablated EGFR only in EGFRflx/flx keratinocytes9 and not in the EGFRwt/wt keratinocytes. At grafting time, small pellets of cells were stored to perform a standard SDS PAGE immunoblot to document the effective deletion of EGFR (Antibodies: EGFR Millipore 06-847, HRAS Santa Cruz sc-520, B Actin Cell Signaling 4967).
Mouse studies were performed under a protocol approved by the National Cancer Institute and the NIH Animal Care and Use Committee. 3.5×106 transformed keratinocytes plus 5×106 primary fibroblasts (support cells) were grafted on the back of syngeneic mice as described6. Tumor growth was observed for a month and tumor volumes recorded every week by measuring them with a caliper. In the pharmacologic model at day 21 post grafting, the mice carrying tumors were randomized into two groups. For one week, one group received gefitinib 100 mg/kg in 10% DMSO and water once daily by gavage and the other group received only the vehicle. The genetic model experiment was repeated in three independent sets with a total number of initial grafted mice of n=23 EGFRwt/wt and n=19 EGFRflx/flx. The pharmacologic model was repeated twice with a total number of initial grafted mice of n=11 10% DMSO and n=11 gefitinib. Depending on the size of the tumors, the samples were processed (partial tumor used or pooled 2 or 3 tumors of the same genetic group to obtain enough material for flow cytometry analysis of the sample) to have at least 300mg to digest for flow cytometry analysis, a portion for the histological evaluation and a portion to process for total mRNA and/or protein extraction. In some cases the tumor material was not enough to be processed for all the uses described above. This accounts for the differences in number of mice plotted in the various experiments. Inhibition of EGFR phosphorylation after a week of gefitinib treatment was tested by western blot analysis on total tumor lysates (P-EGFR CST 3777, EGFR Millipore 06-847, HSP BD 610418).
Flow Cytometry
Freshly harvested tumors were dissected (or pooled depending on the size) to achieve a mass of 300 mg, then minced with razor blades and digested with Collagenase I (5 ml/tumor 2000 U dissolved in DMEM medium with no serum) for 45 min while stirring at 37 degrees in a glass beaker. Samples were placed on ice while 10 additional ml of DMEM 10% serum (complete DMEM) was added by pipetting up and down. The cell prep was then filtered with a 100 μm cell strainer and centrifuged at 4 degree /1200 rpm for 5 min. The pellet was resuspended and incubated with 3ml cold ACK buffer (Thermo Fisher) on ice for 5 min. ACK lysis was stopped by adding 20 ml of complete DMEM on ice then filtered with a 70 μm strainer and finally centrifuged at 4 degree/ 1200 rpm for 5 min. Cells were counted and for the staining procedure 1 ×106 cells were aliquoted in 12×75 mm tubes. Cells were washed with 2 ml of cold PBS, centrifuged, decanted and then incubated with 50 μl cold PBS with 1 μl Aqua or Blue (pre dilution 1:100) viability dyes (Life Technologies) for 15 min at room temperature. Then Fc block (1:100) was added to the same mix and incubated for additional 15 min at room temperature. Next the cells were washed with 2 ml cold PBS and centrifuged and resuspended with 50 μl of PBS to which premixed antibodies (Supplementary Table 1 and Table 2) were added according to manufacturer's protocols and incubated for 30 min at 4 degrees. Cells were washed with 2 ml of cold PBS, centrifuged, decanted and alternatively fixed (2% PFA for 20 min on ice) if only surface markers were stained or permeabilized if staining also included nuclear markers. To this aim, cells were treated with 1 ml of FOXP3 Fix/Perm solution (eBiosciences) for 15 min at room temperature then washed with 2 ml and incubated with 50 μl of FOXP3 Perm Buffer in the presence of nuclear antibodies for 30 min at room temperature. Finally cells were washed with 2 ml of FOXP3 Perm Buffer, centrifuged and resuspended in PBS. Samples were acquired the next day after filtering on BD tubes (BD352235). Only one FACS experiment set for the genetic model and one set for the pharmacologic model gave reliable readings for the FOXP3 populations (optimal permeabilization of the cells) while the surface PD1 staining data are the pool of the three genetic and the two pharmacologic experiments. For the pharmacologic model, samples were analyzed on BDLSRII , and for the genetic model, samples were analyzed on a BD LSR Fortessa instrument from BD Biosciences. Data were analyzed with FlowJo 9.8 software (TreeStar).
RNA isolation and Real Time RT-PCR
Total RNA was extracted from snap frozen portions of tumors that were cold processed at 2000 rpm for 2 min in a Mikro-Dismembrator S from Sartorius and then dissolved in TRIzol Reagent (Life Technologies), according to the manufacturer's instructions. For cDNA synthesis, 1 μg of total RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Life Technologies). PCR was performed in a volume of 20 μl using iQ SYBR Green Supermix from (Biorad) and 1:100 dilution of cDNA. Primers used for this analysis were from Quantitech Qiagen validated primer sets and Real Time analysis was performed using a Bio-Rad iCycler iQ. Results are presented as the mean of the relative quantity (specific gene levels normalized by Gapdh levels) ± SD.
Bioplex assays and ELISA of tumor lysates
Total cell lysates were prepared from the powderized tumors as for the RNA extraction but lysed in cold RIPA buffer containing anti-protease cocktail Complete Roche, 1mM sodium orthovanadate, 1mM sodium fluoride. Tumor lysates were vortexed and incubated on ice for 20 min and then centrifuged at 14000 rpm for 15 min. Supernatants were quantified with Bradford method and 10 μg of protein lysates were run for the R&D luminex magnetic bead assay with a custom configuration of 19 cytokines and chemokines (listed on the website as: HGF, TNFA, IL1B, IL1A, IFNG, GCSF, MCSF, EPO, LIX, MDC, KC, MCP-1, RANTES, IL6, Lipocalin2, MIP1A, MIP1B, IGF1, Eotaxin). Quantitation of IL1RA was performed using the ELISA Duo set kit from R&D Systems. Data are expressed as picograms per ml ± SD.
Immunohistochemistry
Paraffin embedded sections of biopsies fixed with 4% paraformaldehyde, were incubated with citrate buffer pH 6 and microwaved for 10 min at high power. Slides were allowed to cool down for 20 min at room temperature, treated with 3% hydrogen peroxide, and then incubated for 1 hour with 10% rabbit serum. After overnight incubation at 4 degrees with 1:600 anti PD-L1 primary antibody (AF1019 R&D) diluted in 5% rabbit serum, slides were washed in TBST and treated for 1 hour at room temperature with 1:200 anti-goat biotinylated secondary antibody. Immunoreactivity was revealed using avidin-biotin-peroxidase system and DAB as chromogen from Vector Laboratories. Sections were counterstained with Mayer's hematoxylin.
Statistical analysis
The Mann Whitney U test (GraphPad Prism software) was applied to compare differences between groups of data. Significance was assumed at a P value of 0.05 or less and indicated in the graphs as *.
Results and Discussion
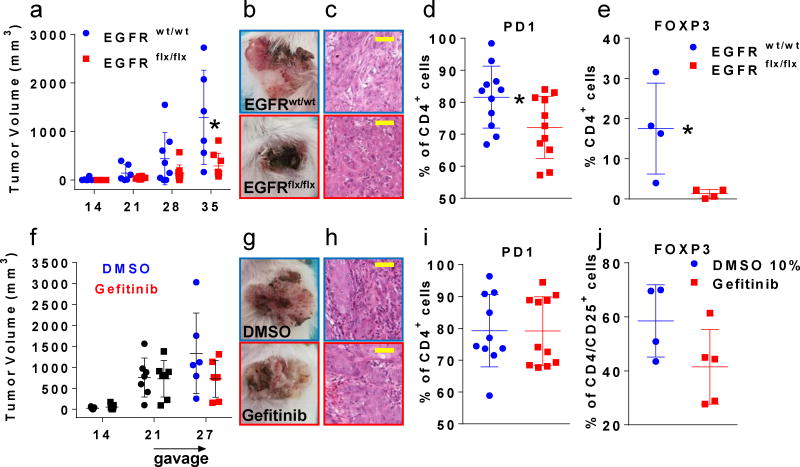
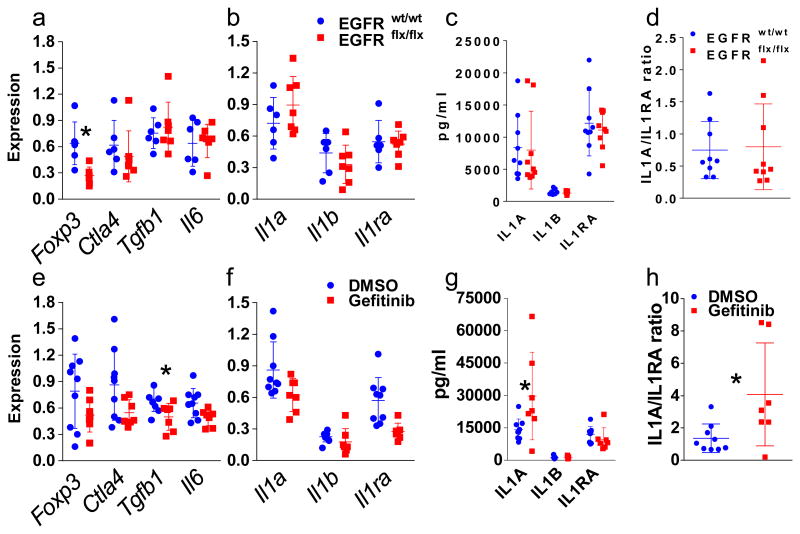
To study tumor autonomous immune alterations, we grafted the cells onto the back of syngeneic mice and followed tumor growth for one month (Fig. 1a). In the genetic model (Fig. 1 panels a to e and Fig. 2 panels a to d) we deleted EGFR with an adenovirus expressing Cre Recombinase in cultured primary FVB/N EGFRflx/flx keratinocytes (Suppl. Fig.1a) and treated EGFRwt/wt keratinocytes identically. Both groups were transformed with a constitutively active form of HRAS that is expressed equally in both genotypes (Suppl Fig.1a). Signaling downstream from oncogenic RAS is dependent on an intact EGFR in mouse keratinocytes10. Accordingly, in the genetic model, tumor growth was consistently impaired by the absence of EGFR in tumor epithelium (Fig. 1a). For the pharmacologic model (Fig. 1 panels f to j and Fig. 2 panels e to h), wild-type FVB/N keratinocytes were transformed with a constitutively active form of HRAS in culture, orthografted to syngeneic FVB/N immunocompetent mice where tumors developed over three weeks and then half of the mice were treated for one week with gefitinib and the other half with vehicle11. This model tested the primary changes in the immune environment as an immediate consequence of systemic EGFR inhibition. We chose to study early events occurring after systemic EGFR blockade to avoid major changes in tumor biology (cell death or necrosis) as confounding factors in the evaluation of the immune populations and in the analysis of the inflammatory milieu. After one week of treatment, the gefitinib group showed a trend to smaller tumor size (Fig. 1f mean of 1333 mm3 versus mean of 724 mm3) that did not reach statistical significance. Nevertheless, the drug hit its target (Suppl. Fig. 1b). All tumors in both models were SCC (Fig. 1c and h) with some adjacent areas of squamous papilloma of various sizes in the different mice. We analyzed several CD45+ expressing populations from enzymatically digested tumors by flow cytometry (Suppl. Table 1 and 2) and characterized mRNA and protein from flash frozen tumors for immune markers by Real Time PCR and Bioplex immunoassays. Although only assayed in a subgroup of tumors in the genetic model, we detected a significantly reduced percentage of FoxP3 expressing cells in the CD4 population by flow cytometry (Fig. 1e) and reduced FoxP3 mRNA (Fig. 2a) indicating the importance of EGFR dependent keratinocyte derived factors in modulating the immune environment of SCCs. This CD4 cell population also displayed a significantly lower percentage of PD1 expressing cells (Fig. 1d) although relative numbers of PD1 expressing CD8 cells were not altered (data not shown). Given the high tumor heterogeneity and the short term treatment with gefinitib, we could observe very few significant differences in the pharmacologic model. Fig. 1j and Fig. 2e showed trends to a reduction both in the CD4+/CD25+/FOXP3+ regulatory T cell frequency and in the expression of FoxP3 mRNA in those tumors. Also the percentage of PD1 expressing cells was not reduced in the CD4 population after an acute one week treatment with gefitinib (Fig. 1i). Gefitinib treatment is going to affect not only EGFR expressing transformed keratinocytes, but also all the stromal cells that express this receptor. Interestingly, an EGFR ligand, amphiregulin, is known to enhance regulatory T cells suppressive function12. This suggests both tumor cell autonomous and systemic routes through which EGFR inhibitors could change regulatory T cell activity within the tumor mass, potentially contributing to an immune effector mediated inhibition of tumor growth. Nevertheless, PD-L1 detected by immunohistochemistry was abundant in tumor and stromal compartments of both models and unchanged by EGFR blockade (Suppl. Fig. 2). In contrast, in both models Ctla4 mRNA trended to decrease while Tgfb1 mRNA decreased significantly and Il6 decreased modestly after gefitinib treatment only (Fig. 2a and 2e). While multiple other mediators showed only trends to increase or decrease in both the genetic model and the pharmacologic model at the mRNA (Suppl. Fig. 3) and protein levels (Suppl. Fig. 4), we found an interesting significant increase in IL1A protein levels and as ratio of IL1A over IL1RA upon gefitinib administration (Fig. 2g and h). An IL1A-IL-1R autocrine loop is a key element in oncogenic RAS signaling in transformed keratinocytes that is EGFR dependent in vitro13. In contrast, pharmacological blockade of EGFR activity in immunocompetent mice bearing malignant tumors caused a significant increase in IL1A at the protein level. Genetic deletion of EGFR in tumor cells only was not sufficient to increase IL-1A in the tumor suggesting the IL-1A increase came from other cells in the tumor microenvironment affected by gefitinib. If the reduction of FOXP3 regulatory T cells is a desirable effect for inhibiting tumor growth, the increase of IL1A is not. Indeed the neutralization of IL1A is seen as an amenable strategy not only in patients with chronic inflammatory conditions but also in cancer patients14. Moreover, IL1A expression modifies the response to EGFR inhibitors and negatively correlates with survival of patients with head and neck cancer suggesting a wider association with SCC, anti-EGFR therapy and IL-1A15. Our results indicate that systemic EGFR blockade alters the immune tumor microenvironment rather rapidly through a decrease in regulatory T cells and increase in the IL1A/IL1-RA ratio. The study also indicates that ablation of EGFR in tumor cells contributes to the differential infiltration of regulatory T cells.
Fig.1.
Tumor growth curve (a,f), tumor appearance and histology (b,c and g,h) and FACS analysis (d,e and i,j) of tumors originated from HRAS transformed FVB/N primary keratinocytes grafted on the back of immunocompetent syngeneic mice. The first row of panels (a to e) depicts data from the genetic approach where deletion of EGFR is achieved via Cre/lox system in EGFRflx/flx keratinocytes that formed the malignant squamous tumors. The second row panels (f to j) shows data collected after pharmacological blockade of EGFR by systemic administration of gefitinib or vehicle administration for 1 week on established wildtype malignant squamous tumors. The genetic or pharmacological blockade of EGFR is indicated in red while active EGFR is indicated in blue in all series of panels. Tumor growth was followed over time and plotted at the indicated days (x axis) in panels a and f. Representative tumor appearance (panels b and g and histology (panels c and h, yellow magnification bar = 50 μm, objective 100×). FACS analysis of tumor cell suspensions shows the percentage of CD4+ cells expressing PD1 (d) or FOXP3 (e) in the genetic model and PD1 in the pharmacologic model (i). The percentage of CD4+/CD25+ cells expressing FOXP3 (j) is shown in the pharmacologic model. * = p < 0.05
Fig. 2.
Real time RT-PCR analysis of mRNA (a and b for the genetic model and e and f for the pharmacologic model) and protein expression analysis by Bioplex assays and ELISA (c and d in the genetic model and g and h in the pharmacologic model) of a subset of inflammatory mediators. Ratios of IL1A over IL1RA protein levels for the genetic (d) and the pharmacologic (h) models. Genetic and pharmacological blockade of EGFR is indicated in red while active EGFR is indicated in blue. * = p < 0.05
These findings support the need for further exploration in the immune mediated consequences of EGFR blockade in the tumor microenvironment with closer observation on the T regulatory cells populations and IL1A levels.
Supplementary Material
Relevance statement.
Precision therapy targeting EGFR is effective for cancer of the lung, oral cavity and gastrointestinal track but causes systemic inflammation. Could immune effectors contribute to the anti-cancer response? We produced squamous cancers genetically deleted of EGFR or squamous cancers subjected to systemic treatment with gefitinib in mice. Tumor cell autonomous (genetic) or systemic (pharmacologic) inhibition of EGFR signaling reduces tumor growth and Treg infiltration in the microenvironment. Thus EGFR targeted cancer therapy may involve immunomodulation.
Acknowledgments
The authors thank Marta Custer and Steve Jay for their technical assistance with the mouse colony. This work was supported by funds from Intramural Research Program of the NIH, NCI, Center for Cancer Research, Project BC005445-30.
Abbreviations
- EGFR
epidermal growth factor receptor
Footnotes
Conflict of Interest: The authors declare no conflict of interest with any aspect of this work.
References
- 1.Hynes NE, Lane HA. ERBB Receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.Lacouture M. Mechanisms of cutaneous toxicities to EGFR inhibitors. Nat Rev Cancer. 2006;6:803–812. doi: 10.1038/nrc1970. [DOI] [PubMed] [Google Scholar]
- 3.Mascia F, Lam G, Keith C, Garber C, Steinberg SM, Kohn E, Yuspa SH. Genetic ablation of epidermal EGFR reveals the dynamic origin of adverse effects of anti-EGFR therapy. Sci Transl Med. 2013;5:199RA110. doi: 10.1126/scitranslmed.3005773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtenberger BM, Gerber PA, Holcmann M, Buhren BA, Amberg N, Smolle V, Schrumpf H, Boelke E, Ansari P, Mackenzie C, Wollenberg A, Kislat A, Fischer JW, Röck K, Harder J, Schröder JM, Homey B, Sibilia M. Epidermal EGFR controls cutaneous host defense and prevents inflammation. Sci Transl Med. 2013;5:199RA111. doi: 10.1126/scitranslmed.3005886. [DOI] [PubMed] [Google Scholar]
- 5.Pardoll P. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protocols. 2008;3:799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roop DR, Lowy DR, Tambourin PE, Strickland J, Harper JR, Balaschak M, Spangler EF, Yuspa SH. Inactivated Harvey ras oncogene produces benign tumours on mouse epidermal tissue. Nature. 1986;323:822–824. doi: 10.1038/323822a0. [DOI] [PubMed] [Google Scholar]
- 8.Lee YH, Sauer B, Johnson PF, Gonzalez FJ. Disruption of the c/ebpa gene in adult mouse liver. Mol Cell Biol. 1997;17:6014–6022. doi: 10.1128/mcb.17.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TC, Threadgill DW. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis. 2009;47:85–92. doi: 10.1002/dvg.20464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cataisson C, Michalowski AM, Shibuya K, Ryscavage A, Klosterman M, Wright L, Dubois W, Liu F, Zhuang A, Rodrigues KB, Hoover S, Dwyer J, Simpson MR, Merlino G, Yuspa SH. MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis. Sci Signal. 2016;9:ra62. doi: 10.1126/scisignal.aaf5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y, Lu Y, Wang M, Vikis H, Yao R, Wang Y, Lubet RA, You M. Effect of an epidermal growth factor receptor inhibitor in mouse models of lung cancer. Mol Cancer Res. 2006;4:971–981. doi: 10.1158/1541-7786.MCR-06-0086. [DOI] [PubMed] [Google Scholar]
- 12.Zaiss DM, van Loosdregt J, Gorlani A, Bekker CP, Gröne A, Sibilia M, van Bergen en Henegouwen PM, Roovers RC, Coffer PJ, Sijts AJ. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity. 2013;21:275–284. doi: 10.1016/j.immuni.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cataisson C, Salcedo R, Hakim S, Moffit AB, Wright L, Yi M, Stephens R, Dai RM, Lyakh L, Schenten D, Yuspa SH, Trinchieri G. IL-1R–MyD88 signaling in keratinocyte transformation and carcinogenesis. J Exp Med. 2012;209:1689–1702. doi: 10.1084/jem.20101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinarello CA. Neutralization of IL1A in patients with cancer. The Lancet. 2014;5:552–553. doi: 10.1016/S1470-2045(14)70164-0. [DOI] [PubMed] [Google Scholar]
- 15.Koch AT, Love-Homan L, Espinosa-Cotton M, Stanam A, Simons AL. MyD88-dependent signaling decreases the antitumor efficacy of epidermal growth factor receptor inhibition in head and neck cancer cells. Cancer Res. 2015;75:1657–1667. doi: 10.1158/0008-5472.CAN-14-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.