Abstract
The process of memory consolidation requires transcription and translation to form long-term memories. Significant effort has been dedicated to understanding changes in hippocampal gene expression after contextual fear conditioning. However, alternative splicing by differential transcript regulation during this time period has received less attention. Here, we use RNA-seq to determine exon-level changes in expression after contextual fear conditioning and retrieval. Our work reveals that a short variant of Homer1, Ania-3, is regulated by contextual fear conditioning. The ribosome biogenesis regulator Las1l, small nucleolar RNA Snord14e, and the RNA-binding protein Rbm3 also change specific transcript usage after fear conditioning. The changes in Ania-3 and Las1l are specific to either the new context or the context-shock association, while the changes in Rbm3 occur after context or shock only. Our analysis revealed novel transcript regulation of previously undetected changes after learning, revealing the importance of high throughput sequencing approaches in the study of gene expression changes after learning.
Keywords: Transcription, Alternative splicing, RNA-seq, Fear conditioning, Homer1
1. Introduction
Contextual fear conditioning requires two waves of transcription and protein synthesis in the hippocampus to form long-term memory (Bourtchouladze et al., 1998; Igaz, Vianna, Medina, & Izquierdo, 2002). Our lab and others have focused on discovering the genes regulated during these transcriptional waves using both candidate gene and genome-wide approaches. Our microarray-based studies have indicated that the first wave of transcription induces the largest change in gene expression 30 min after contextual learning (Peixoto, Wimmer et al., 2015). However, gene regulation is a complex process that has multiple layers of control. Levels of particular mRNA isoforms can be regulated by alternative start sites, differential splicing including exon skipping and intron retention, and alternative poly(A) site selection (Leff, Rosenfeld, & Evans, 1986; Raj & Blencowe, 2015). Alternative splicing can lead to distinct protein function and interactions (Ellis et al., 2012) or regulate mRNA localization (Ehlers, Fung, O’Brien, & Huganir, 1998; Jaskolski et al., 2004; Papandrikopoulou, Doll, Tucker, Garner, & Matus, 1989), and thus is expected to be particularly important in neurons, which need to traffic mRNA to their long cellular processes.
Most previous research studying genome-wide gene expression in the hippocampus after contextual learning has relied on microarray technology (Barnes, Kirtley, & Thomas, 2012; Cavallaro, D’Agata, Manickam, Dufour, & Alkon, 2002; Keeley et al., 2006; Klur et al., 2009; Levenson et al., 2004; Mei et al., 2005; Peixoto, Wimmer et al., 2015). Although microarrays are a reliable tool to measure changes in gene expression, they are unable to distinguish exon-level effects that are indicative of alternative splicing. RNA-seq provides numerous advantages over microarrays (Peixoto, Risso et al., 2015), including the ability to study exon-level changes in gene expression. Isoform-specific gene expression changes are known to occur after fear conditioning, including upregulation of Bdnf IV, but not other Bdnf isoforms (Lubin, Roth, & Sweatt, 2008; Mizuno, Dempster, Mill, & Giese, 2012), and Homer1a, but not Homer1c (Mahan et al., 2012) in response to strong, three shock training protocols. The different Bdnf isoforms have distinct transcription start sites, while the expression of Homer1 isoforms is controlled by the splicing regulator SRp20 (Wang, Chikina, Pincas, & Sealfon, 2014), which is upregulated after learning (Antunes-Martins, Mizuno, Irvine, Lepicard, & Giese, 2007). These examples indicate that gene regulation after learning is more complex than gene-level differences and can be highly selective for particular isoforms of a gene.
Therefore, we used RNA-seq to study differential alternative splicing 30 min after contextual fear conditioning and 30 min after memory retrieval. Applying Remove Unwanted Variation (RUV), a recently designed normalization algorithm (Peixoto, Risso et al., 2015; Risso, Ngai, Speed, & Dudoit, 2014), to our data, we discovered 171 bins, corresponding to either an entire exon or any portion of a gene, across 138 genes that showed differential expression after learning independent of changes at the gene-level. After memory retrieval 450 differentially expressed bins corresponding to 311 unique genes were discovered. These bins include retained introns, unique start/end sites, or small RNA not yet spliced out of the polyadenylated mRNA. The differences include Snord14e, a small nucleolar RNA, which our lab has previously shown to be regulated at this time point (Peixoto, Wimmer et al., 2015). Sno-RNAs, which are commonly found within introns of genes, regulate RNA processing and have been implicated in memory consolidation (Rogelj, Hartmann, Yeo, Hunt, & Giese, 2003). In addition, Ania-3, an alternative short form of Homer1 that has not previously been linked to learning, ribosome biogenesis regulator Las1l, and the RNA-binding protein Rbm3 were also regulated by contextual fear conditioning. These findings demonstrate that alternative splicing is regulated by contextual learning on a genome-wide scale and also identify novel candidate isoforms that may be pertinent to memory consolidation.
2. Materials and methods
2.1. Subjects
C57Bl/6J mice were maintained under standard conditions with food and water available ad libitum. Adult male mice 2 months of age were kept on a 12-h light/12-h dark cycle with lights on at 7AM. All behavioral and biochemical experiments were performed during the light cycle with training starting at 10AM (ZT3). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania and were consistent with National Institutes of Health guidelines.
2.2. Behavior
Contextual fear conditioning was performed as previously described (Hawk et al., 2012; Vecsey et al., 2007) with handling for 3 days prior to conditioning. Briefly, the conditioning protocol entailed a single 2-s, 1.5 mA footshock terminating at 2.5 min after placement of the mouse in the chamber. Mice were left in the chamber for an additional 30 s and then returned to their homecage. One mouse per behavioral group (homecage, fear conditioned) was trained per day over 10 days to reduce unwanted variation caused by training and sacrifice times. One mouse was also tested the next day to ensure proper freezing levels (Peixoto, Wimmer et al., 2015).
2.3. RNA isolation
Hippocampi were dissected either from homecage mice or 30 min after training and placed into RNAlater (Qiagen Valencia, CA) and frozen on dry ice. Tissue was homogenized using a TissueLyser system and RNA was extracted using the RNAeasy Microarray Tissue kit (Qiagen) according to the manufacturer’s instructions. Samples were DNase treated using the RNase-Free DNase kit (Qiagen) off-column by incubating 5 μl DNase and 35 μl Buffer RDD for 25 min at RT with each sample. Samples were then ethanol precipitated and resuspended in water.
2.4. RNA-seq library preparation and sequencing
2 μg of RNA from n = 5 homecage and fear conditioned mice was used in the TruSeq RNA Sample Prep Kit (Illumina San Diego, CA) according to the manufacturer’s instructions with polyA selection. Completed libraries were size-selected on an agarose gel to remove any high basepair fragments, quantified by qPCR (KAPA Biosystems Boston, MA), and submitted to the PGFI sequencing core at the University of Pennsylvania. An Illumina HiSeq 2000 sequenced the libraries in paired-end 100 bp reads. 3 libraries were sequenced per lane on an Illumina HiSeq 2000, resulting in an average of 67,011,105 reads per sample in the homecage mice and 62,115,805 reads per sample after fear conditioning. Reads had good unique concordance (86.9% in homecage, 85.5% after fear conditioning) and mapping (90.7% of unique concordant reads in homecage and 93.1% after fear conditioning). RNA-seq data is available through GEO (GSE63412) (Peixoto, Risso et al., 2015).
2.5. Data analysis
Sequencing reads were aligned to the mouse mm9 genome using GSNAP (Wu & Nacu, 2010) (http://share.gene.com/gmap). An exon-level count table was produced by counting reads into unique, non-overlapping “bins” using Ensembl gene models and HTSeq (Anders, Reyes, & Huber, 2012) (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). A “bin” can either be any part of a gene or an entire exon depending on the uniqueness of the region. Bin counts were normalized using upper-quartile scaling implemented in edgeR (Robinson, McCarthy, & Smyth, 2010) followed by RUVs, which corrects for unwanted variation using replicate/negative control samples (Risso et al., 2014). Additionally, we used 8897 bins residing in 625 genes identified as unchanged from a previous microarray experiment as negative controls for RUV under the assumption that these bins are also not changing (Peixoto, Risso et al., 2015; Peixoto, Wimmer et al., 2015). We discovered that four factors of unwanted variation (k = 4) need to be adjusted for to resolve the differences caused by contextual fear, which was chosen using the method described by Peixoto, Risso et al. (2015). Differential splicing analysis was performed with the limma Bioconductor package, using the voom and diffSplice functions (Law, Chen, Shi, & Smyth, 2014; Ritchie et al., 2015). Functional annotation was performed through DAVID (Huang da, Sherman, & Lempicki, 2009a, 2009b) (http://david.abcc.ncifcrf.gov/). The annotation was limited to the following sources: GO Biological process, GO Molecular Function, KEGG pathways, and SwissProt and Protein Information Resource keywords and an EASE score restriction of 0.1.
2.6. qPCR analysis
RNA was isolated from a separate cohort of fear conditioned, immediate shock, or context only mice following the same behavioral paradigms described above. Immediate shock consisted of placing the mouse in the context with the footshock on and immediate removal, while context involved placing the mouse in the context for the same time as contextual conditioning with no shock. RNA was converted to cDNA using the RETROscript kit (Ambion) according to the manufacturer’s instructions. cDNA reactions were diluted to 200 μl and 2.25 μl was combined with 0.25 μl 5 μM primer mix and 2.5 μl SYBR Select Master Mix (Life Technologies Carlsbad, CA) and run on a Viia7 Real Time PCR system. The ΔΔCt method was used for analysis (Poplawski et al., 2014), with all primers showing >90% efficiency. The primers used were: Ania1F-AGTGGCTGGTTTTCTTGGACT, Ania1R-GGGAGGTGGATTGGTGACAA, Homer1Bin21F-CTGGAGTCCACTGCCAATGT, Home r1Bin21R-CTCTGCTTCCTCCTGGTACG, Las1lBin15F-TCAAAGTCAGAGGGGTCGGA, Las1Bin15R-AGACTTCGCTCTTGCTGCTT, Las1lBin17F-TGCTGGAGAAACACAGGCAT, Las1lBin17R-ACATTGTACACGTGGGGAAAGA, Rbm3Bin2F-ACCTGAGTTTTGGAGGCTGG, Rbm3Bi n2R-ACAACAGCGGACACCATAGG, Rbm3Bin7F-GGTGGCTATGACCGCTACTC, Rbm3Bin7R-TTTTGTGTGCATGCCCCATC, Rbm3Bin22F-TGCCCCTGGCAGACATAGAG, Rbm3Bin22R-GTCTGCCACTTTCTTCGTTCTTT. The comparison between three groups (homecage, immediate shock, context only) was analyzed using an ANOVA. The effect of bin (F(7, 160) = 11.90), condition (F(2, 160) = 7.835) and interaction (F(14, 160) = 3.719) were all significant, and Tukey tests were used to determine the significance of each bin.
3. Results
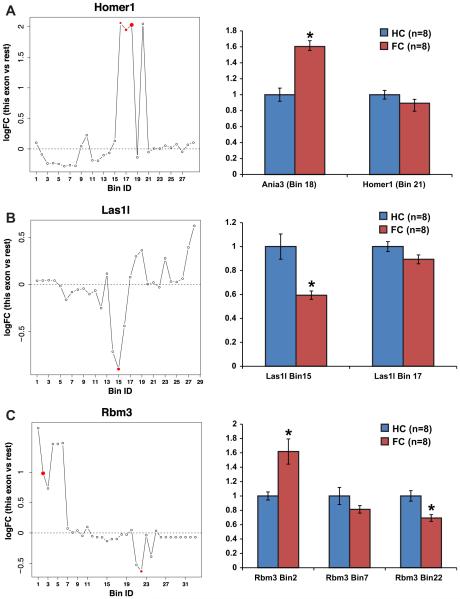
RNA-seq has the advantage of distinguishing exon-level reads that are difficult to identify by any other method, and therefore it is an ideal technique to study alternative splicing. We used RNA-seq to study gene expression in the hippocampus 30 min after contextual fear conditioning, a time point our lab has previously determined to show robust expression changes after fear conditioning (Peixoto, Wimmer et al., 2015). We used GSNAP (Wu & Nacu, 2010) to align reads to the mm9 mouse genome and HTSeq (Anders, Pyl, & Huber, 2015) to count reads into bins (Anders et al., 2012) using Ensembl gene models. Bins are separated based on overlap of Ensembl gene models, with any unique section of a transcriptional unit split into a separate bins. Therefore, a bin can represent either a whole exon or any other unique portion of the gene model. Thus, differential start sites, 3′ ends, or retained introns can be observed as unique bins if they are part of the Ensembl database. So as not to bias ourselves using gene models, we considered every bin as a potential site for alternative regulation. RUVs normalization performed as described (Risso et al., 2014), adjusting for four factors of unwanted variation (which can include biological and technical noise), was found to control for fear conditioning as the primary effector of variation between samples. Bioconductor package limma was then used to determine differential bin usage independent of gene-level changes (Ritchie et al., 2015). We identified 171 bins across 138 genes that displayed differential usage (FDR < 0.05) after contextual fear conditioning (Table 1). 129 of these bins were upregulated and 42 were downregulated, consistent with the general increase in gene expression after fear conditioning (Peixoto, Wimmer et al., 2015). We performed functional classification of genes showing at least 1 bin-specific change after fear conditioning. The SwissProt and Protein Information Resource keywords “phosphoprotein” and “alternative splicing” were enriched in our data set, indicating that our exon-level analysis discovers alternative splicing as expected. Clusters corresponding to protein catabolic processes and nucleotide binding were also enriched. The same analysis was performed on samples 30 min after memory retrieval (testing). In this analysis, we found 450 bins corresponding to 311 unique genes (Table 2). This list of genes contains 70 of the 138 genes observed to change after fear conditioning, highlighting the overlap between memory consolidation and retrieval (Peixoto, Wimmer et al., 2015).
Table 1.
List of bins showing differential expression after fear conditioning. Each differential bin contains the chromosome, start position, and end position for easy reference.
| GeneID | Gene name | Bin # |
Chr | Start | Stop | Strand | logFC | t | P value | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| ENSMUSG00000007617 | Homer1 | 018 | chr13 | 94136356 | 94137083 | + | 2.0 | 14.1 | 1.4E–33 | 3.7E–28 |
| ENSMUSG00000029657 | Hsph1 | 012 | chr5 | 150423259 | 150426039 | − | 1.1 | 10.3 | 2.4E–21 | 3.3E–16 |
| ENSMUSG00000039801 | 2410089E03Rik | 053 | chr15 | 8201065 | 8202148 | + | −1.1 | −9.2 | 9.6E–19 | 8.6E–14 |
| ENSMUSG00000031167 | Rbm3 | 002 | chrX | 7716104 | 7717909 | − | 1.0 | 9.0 | 3.5E–17 | 2.3E–12 |
| ENSMUSG00000007617 | Homer1 | 017 | chr13 | 94136233 | 94136355 | + | 1.9 | 8.4 | 3.5E–15 | 1.9E–10 |
| ENSMUSG00000020431 | Adcy1 | 024 | chr11 | 7072883 | 7078509 | + | −0.4 | −8.4 | 5.5E–15 | 2.4E–10 |
| ENSMUSG00000025372 | Baiap2 | 027 | chr11 | 119867673 | 119868096 | + | 0.5 | 7.7 | 3.5E–13 | 1.3E–08 |
| ENSMUSG00000034083 | C130022K22Rik | 009 | chr6 | 91835401 | 91838063 | + | 1.0 | 7.8 | 4.5E–13 | 1.5E–08 |
| ENSMUSG00000008153 | Clstn3 | 004 | chr6 | 124383426 | 124383521 | − | 1.2 | 7.4 | 1.5E–12 | 4.5E–08 |
| ENSMUSG00000043872 | Zmym1 | 001 | chr4 | 126724338 | 126724885 | − | 1.1 | 7.3 | 6.6E–12 | 1.8E–07 |
| ENSMUSG00000005089 | Slc1a2 | 036 | chr2 | 102621901 | 102630941 | + | −0.3 | −6.7 | 8.4E–11 | 2.0E–06 |
| ENSMUSG00000020287 | Mpg | 010 | chr11 | 32130054 | 32131244 | + | −0.6 | −7.1 | 1.5E–10 | 3.3E–06 |
| ENSMUSG00000024576 | Csnk1a1 | 028 | chr18 | 61745286 | 61746152 | + | 0.6 | 6.5 | 4.2E–10 | 8.5E–06 |
| ENSMUSG00000063077 | Kif1b | 001 | chr4 | 148550428 | 148552126 | − | −0.2 | −6.3 | 7.6E–10 | 1.5E–05 |
| ENSMUSG00000035206 | 3110056O03Rik | 016 | chr10 | 80329406 | 80330144 | + | 0.4 | 6.5 | 9.0E–10 | 1.6E–05 |
| ENSMUSG00000059495 | Arhgef12 | 002 | chr9 | 42771926 | 42776264 | − | −0.3 | −6.3 | 1.0E–09 | 1.7E–05 |
| ENSMUSG00000022710 | Usp7 | 020 | chr16 | 8697013 | 8697568 | − | 0.8 | 6.2 | 1.4E–09 | 2.1E–05 |
| ENSMUSG00000025372 | Baiap2 | 026 | chr11 | 119864352 | 119864399 | + | 0.6 | 6.3 | 1.6E–09 | 2.4E–05 |
| ENSMUSG00000024576 | Csnk1a1 | 027 | chr18 | 61744853 | 61745285 | + | 0.6 | 6.2 | 2.3E–09 | 3.2E–05 |
| ENSMUSG00000063077 | Kif1b | 002 | chr4 | 148552127 | 148552971 | − | −0.3 | −6.0 | 4.0E–09 | 5.3E–05 |
| ENSMUSG00000041879 | Ipo9 | 036 | chr1 | 137302594 | 137303043 | − | −0.7 | −5.9 | 5.5E–09 | 7.0E–05 |
| ENSMUSG00000057421 | Las1l | 015 | chrX | 93143543 | 93144773 | − | −0.9 | −6.0 | 6.0E–09 | 7.3E–05 |
| ENSMUSG00000034656 | Cacna1a | 068 | chr8 | 87163334 | 87163334 | + | 1.4 | 5.9 | 7.4E–09 | 8.6E–05 |
| ENSMUSG00000031878 | Nae1 | 015 | chr8 | 107042164 | 107043101 | − | 1.1 | 5.9 | 8.2E–09 | 9.1E–05 |
| ENSMUSG00000023033 | Scn8a | 030 | chr15 | 100869972 | 100876360 | + | −0.2 | −5.9 | 9.3E–09 | 1.0E–04 |
| ENSMUSG00000075876 + ENSMUSG00000064791 + ENSMUSG00000075924 + ENSMUSG00000015656 |
Snord14c/ Snord14e/ Snord14d/Hspa8 |
038 | chr9 | 40612831 | 40612920 | + | 1.4 | 5.9 | 9.7E–09 | 1.0E–04 |
| ENSMUSG00000027523 | Gnas | 027 | chr2 | 174155788 | 174155935 | + | 1.0 | 5.8 | 1.0E–08 | 1.0E–04 |
| ENSMUSG00000071984 | Fndc1 | 001 | chr17 | 7931434 | 7932195 | − | 0.4 | 5.8 | 2.0E–08 | 1.9E–04 |
| ENSMUSG00000038383 | Pigu | 004 | chr2 | 155104386 | 155108131 | − | 0.5 | 5.8 | 2.6E–08 | 2.4E–04 |
| ENSMUSG00000028053 | Ash1l | 002 | chr3 | 88785155 | 88789712 | + | 0.4 | 5.7 | 3.0E–08 | 2.6E–04 |
| ENSMUSG00000028826 | Tmem57 | 002 | chr4 | 134360480 | 134362431 | − | −0.4 | −5.7 | 3.9E–08 | 3.4E–04 |
| ENSMUSG00000075876 + ENSMUSG00000064791 + ENSMUSG00000075924 + ENSMUSG00000015656 |
Snord14c/ Snord14e/ Snord14d/Hspa8 |
037 | chr9 | 40612779 | 40612830 | + | 1.4 | 5.6 | 4.3E–08 | 3.6E–04 |
| ENSMUSG00000024576 | Csnk1a1 | 025 | chr18 | 61742498 | 61744058 | + | 0.5 | 5.6 | 4.5E–08 | 3.7E–04 |
| ENSMUSG00000023952 | Gtpbp2 | 035 | chr17 | 46303816 | 46303936 | + | 0.9 | 5.6 | 4.6E–08 | 3.7E–04 |
| ENSMUSG00000007617 | Homer1 | 016 | chr13 | 94136198 | 94136232 | + | 2.1 | 5.6 | 5.4E–08 | 4.1E–04 |
| ENSMUSG00000027429 | Sec23b | 030 | chr2 | 144405140 | 144406851 | + | 0.9 | 5.5 | 6.4E–08 | 4.8E–04 |
| ENSMUSG00000036052 | Dnajb5 | 011 | chr4 | 42963816 | 42965965 | + | 0.4 | 5.7 | 6.8E–08 | 4.9E–04 |
| ENSMUSG00000013033 | Lphn1 | 001 | chr8 | 86424004 | 86424471 | + | 0.7 | 5.5 | 8.0E–08 | 5.6E–04 |
| ENSMUSG00000035640 | Dos | 014 | chr10 | 79598293 | 79598333 | − | 1.6 | 5.5 | 8.3E–08 | 5.7E–04 |
| ENSMUSG00000028488 | Sh3gl2 | 016 | chr4 | 85033579 | 85035284 | + | 0.2 | 5.5 | 1.1E–07 | 7.5E–04 |
| ENSMUSG00000027569 | 1600027N09Rik | 010 | chr2 | 180318228 | 180319110 | + | 0.4 | 5.5 | 1.5E–07 | 9.7E–04 |
| ENSMUSG00000008153 | Clstn3 | 005 | chr6 | 124386790 | 124386835 | − | 1.4 | 5.3 | 2.1E–07 | 1.3E–03 |
| ENSMUSG00000014873 | Surf2 | 009 | chr2 | 26773052 | 26774384 | + | 0.3 | 5.4 | 2.3E–07 | 1.5E–03 |
| ENSMUSG00000063160 + ENSMUSG00000003762 | Numbl/Adck4 | 037 | chr7 | 28047272 | 28049894 | + | 0.4 | 5.2 | 2.9E–07 | 1.8E–03 |
| ENSMUSG00000024777 | Ppp2r5b | 006 | chr19 | 6230276 | 6230385 | − | 0.5 | 5.3 | 3.1E–07 | 1.8E–03 |
| ENSMUSG00000031167 | Rbm3 | 022 | chrX | 7721600 | 7721698 | − | −0.6 | −5.2 | 3.1E–07 | 1.8E–03 |
| ENSMUSG00000053580 | Tanc2 | 043 | chr11 | 105786047 | 105790613 | + | −0.3 | −5.2 | 3.3E–07 | 1.9E–03 |
| ENSMUSG00000028161 | Ppp3ca | 030 | chr3 | 136598842 | 136598864 | + | 0.7 | 5.2 | 3.5E–07 | 1.9E–03 |
| ENSMUSG00000029765 | Plxna4 | 001 | chr6 | 32094565 | 32095925 | − | −0.3 | −5.2 | 3.7E–07 | 2.0E–03 |
| ENSMUSG00000075003 + ENSMUSG00000037876 | Jmjd1c/Jmjd1c | 041 | chr10 | 66707622 | 66708166 | + | 0.7 | 5.1 | 4.2E–07 | 2.3E–03 |
| ENSMUSG00000027799 | Nbea | 062 | chr3 | 55986894 | 55987623 | − | 0.6 | 5.1 | 4.4E–07 | 2.3E–03 |
| ENSMUSG00000023952 | Gtpbp2 | 031 | chr17 | 46302947 | 46303259 | + | 0.4 | 5.1 | 5.3E–07 | 2.7E–03 |
| ENSMUSG00000042605 | Atxn2 | 051 | chr5 | 122261639 | 122261939 | + | 0.6 | 5.1 | 5.6E–07 | 2.8E–03 |
| ENSMUSG00000003269 | Cyth2 | 023 | chr7 | 53068527 | 53069248 | − | 0.4 | 5.1 | 5.9E–07 | 2.9E–03 |
| ENSMUSG00000022451 | Twf1 | 001 | chr15 | 94408382 | 94410096 | − | 0.2 | 5.2 | 6.1E–07 | 3.0E–03 |
| ENSMUSG00000072647 + ENSMUSG00000029454 | Adam1a/ Mapkapk5 |
002 | chr5 | 121968622 | 121969392 | − | 0.4 | 5.1 | 6.9E–07 | 3.3E–03 |
| ENSMUSG00000038664 | Herc1 | 095 | chr9 | 66348328 | 66348982 | + | −0.6 | −5.0 | 7.3E–07 | 3.4E–03 |
| ENSMUSG00000030082 | Sec61a1 | 014 | chr6 | 88463896 | 88464200 | − | 0.8 | 5.1 | 7.6E–07 | 3.5E–03 |
| ENSMUSG00000032855 | Pkd1 | 017 | chr17 | 24709563 | 24711715 | + | 0.5 | 5.0 | 8.4E–07 | 3.8E–03 |
| ENSMUSG00000040929 | Rfx3 | 001 | chr19 | 27836211 | 27840930 | − | −0.3 | −5.0 | 9.7E–07 | 4.3E–03 |
| ENSMUSG00000038762 | Abcf1 | 037 | chr17 | 36105913 | 36106178 | − | 1.4 | 4.9 | 1.3E–06 | 5.6E–03 |
| ENSMUSG00000023952 | Gtpbp2 | 044 | chr17 | 46304840 | 46304970 | + | 0.4 | 4.9 | 1.3E–06 | 5.7E–03 |
| ENSMUSG00000006676 | Usp19 | 027 | chr9 | 108403525 | 108404028 | + | 0.2 | 5.0 | 1.4E–06 | 5.8E–03 |
| ENSMUSG00000078789 + ENSMUSG00000038268 | Dph1/Ovca2 | 001 | chr11 | 74989444 | 74991144 | − | 0.3 | 5.0 | 1.4E–06 | 5.8E–03 |
| ENSMUSG00000040896 | Kcnd3 | 009 | chr3 | 105468465 | 105469879 | + | 1.0 | 5.0 | 1.4E–06 | 5.9E–03 |
| ENSMUSG00000030207 | 8430419L09Rik | 017 | chr6 | 135182873 | 135183273 | + | −0.5 | −4.9 | 1.7E–06 | 6.9E–03 |
| ENSMUSG00000029587 | Zfp12 | 004 | chr5 | 143997458 | 143997932 | + | 1.0 | 5.0 | 1.7E–06 | 6.9E–03 |
| ENSMUSG00000021097 | Clmn | 001 | chr12 | 106001324 | 106010173 | − | −0.2 | −5.0 | 1.8E–06 | 7.0E–03 |
| ENSMUSG00000015536 | Mocs2 | 015 | chr13 | 115615731 | 115616365 | + | 0.6 | 4.9 | 2.0E–06 | 7.8E–03 |
| ENSMUSG00000045482 | Trrap | 027 | chr5 | 145557830 | 145558030 | + | 0.9 | 4.8 | 2.0E–06 | 7.8E–03 |
| ENSMUSG00000085832 | D430036J16Rik | 004 | chr9 | 81530442 | 81530544 | + | −1.8 | −5.1 | 2.1E–06 | 7.8E–03 |
| ENSMUSG00000020612 | Prkar1a | 011 | chr11 | 109522664 | 109523067 | + | 0.4 | 4.9 | 2.2E–06 | 8.2E–03 |
| ENSMUSG00000053470 | Kdm3a | 017 | chr6 | 71558999 | 71559041 | − | 2.5 | 4.8 | 2.6E–06 | 9.3E–03 |
| ENSMUSG00000042042 | Csgalnact2 | 007 | chr6 | 118074432 | 118076139 | − | 1.2 | 4.9 | 2.8E–06 | 1.0E–02 |
| ENSMUSG00000004070 | Hmox2 | 004 | chr16 | 4756845 | 4756902 | + | 1.1 | 4.9 | 2.8E–06 | 1.0E–02 |
| ENSMUSG00000040479 | Dgkz | 011 | chr2 | 91774090 | 91774212 | − | 0.5 | 4.7 | 3.1E–06 | 1.1E–02 |
| ENSMUSG00000021327 | Zkscan3 | 009 | chr13 | 21481162 | 21485100 | − | 0.3 | 4.8 | 3.1E–06 | 1.1E–02 |
| ENSMUSG00000055491 | Pprc1 | 039 | chr19 | 46146825 | 46147038 | + | 0.9 | 4.7 | 3.4E–06 | 1.2E–02 |
| ENSMUSG00000074247 | Dda1 | 013 | chr8 | 73996515 | 73996681 | + | 0.7 | 4.8 | 3.6E–06 | 1.2E–02 |
| ENSMUSG00000020654 | Adcy3 | 032 | chr12 | 4210892 | 4211481 | + | 0.5 | 4.7 | 3.6E–06 | 1.2E–02 |
| ENSMUSG00000022565 + ENSMUSG00000063268 | Plec/Parp10 | 003 | chr15 | 76001406 | 6005809 | − | −0.2 | −4.7 | 3.7E–06 | 1.2E–02 |
| ENSMUSG00000062296 | Trank1 | 011 | chr9 | 111267179 | 111270052 | + | 0.4 | 4.8 | 3.9E–06 | 1.3E–02 |
| ENSMUSG00000057897 | Camk2b | 049 | chr11 | 5965662 | 5965745 | − | 1.3 | 4.7 | 4.0E–06 | 1.3E–02 |
| ENSMUSG00000050357 | Rltpr | 040 | chr8 | 108219675 | 108220760 | + | 0.4 | 4.7 | 4.2E–06 | 1.3E–02 |
| ENSMUSG00000000416 | Cttnbp2 | 029 | chr6 | 18381940 | 18383819 | − | 0.4 | 4.7 | 4.2E–06 | 1.3E–02 |
| ENSMUSG00000028161 | Ppp3ca | 029 | chr3 | 136598744 | 136598841 | + | 0.6 | 4.7 | 4.4E–06 | 1.4E–02 |
| ENSMUSG00000024826 | Dpf2 | 011 | chr19 | 5902769 | 5903332 | − | 0.4 | 4.7 | 4.4E–06 | 1.4E–02 |
| ENSMUSG00000034171 | Faah | 014 | chr4 | 115672694 | 115673391 | − | 0.3 | 4.7 | 4.6E–06 | 1.4E–02 |
| ENSMUSG00000015869 | Prpsap1 | 009 | chr11 | 116338662 | 116339053 | − | 0.5 | 4.7 | 4.8E–06 | 1.4E–02 |
| ENSMUSG00000053483 | Usp21 | 037 | chr1 | 173215746 | 173216480 | − | 0.4 | 4.6 | 4.8E–06 | 1.4E–02 |
| ENSMUSG00000004947 | Dtx2 | 015 | chr5 | 136495428 | 136497624 | + | 0.9 | 4.7 | 5.0E–06 | 1.5E–02 |
| ENSMUSG00000025155 | Dus1l | 009 | chr11 | 120651195 | 120651761 | − | 0.4 | 4.6 | 5.1E–06 | 1.5E–02 |
| ENSMUSG00000006920 | Ezh1 | 022 | chr11 | 101068968 | 101069435 | − | 1.2 | 4.6 | 5.3E–06 | 1.5E–02 |
| ENSMUSG00000004110 | Cacna1e | 001 | chr1 | 156239649 | 156246065 | − | −0.3 | −4.6 | 5.5E–06 | 1.5E–02 |
| ENSMUSG00000019254 | Ppp1r12c | 052 | chr7 | 4453132 | 4453266 | − | 0.9 | 4.6 | 5.6E–06 | 1.5E–02 |
| ENSMUSG00000068221 + ENSMUSG00000022436 | Pdxp/Sh3bp1 | 031 | chr15 | 78744349 | 78744961 | + | 0.3 | 4.6 | 5.6E–06 | 1.5E–02 |
| ENSMUSG00000063077 | Kif1b | 004 | chr4 | 148552981 | 148554344 | − | −0.2 | −4.6 | 5.6E–06 | 1.5E–02 |
| ENSMUSG00000074247 | Dda1 | 012 | chr8 | 73996275 | 73996514 | + | 0.7 | 4.7 | 5.7E–06 | 1.5E–02 |
| ENSMUSG00000024012 | Mtch1 | 030 | chr17 | 29484705 | 29484849 | − | 0.7 | 4.6 | 5.7E–06 | 1.5E–02 |
| ENSMUSG00000053141 | Ptprt | 001 | chr2 | 161347726 | 161352092 | − | −0.3 | −4.6 | 5.7E–06 | 1.5E–02 |
| ENSMUSG00000039838 | Slc45a1 | 002 | chr4 | 150004156 | 150005026 | − | 0.4 | 4.8 | 5.7E–06 | 1.5E–02 |
| ENSMUSG00000000441 | Raf1 | 007 | chr6 | 115570346 | 115571833 | − | 0.2 | 4.6 | 5.7E–06 | 1.5E–02 |
| ENSMUSG00000054263 | Lifr | 025 | chr15 | 7141744 | 7147489 | + | −0.3 | −4.6 | 5.9E–06 | 1.5E–02 |
| ENSMUSG00000063160 + ENSMUSG00000003762 | Numbl/Adck4 | 035 | chr7 | 28046397 | 28047186 | + | 0.5 | 4.6 | 6.1E–06 | 1.6E–02 |
| ENSMUSG00000038406 | Scaf1 | 010 | chr7 | 52266722 | 52267492 | − | 0.7 | 4.6 | 6.5E–06 | 1.6E–02 |
| ENSMUSG00000020978 | Klhdc2 | 002 | chr12 | 70397709 | 70397741 | + | 0.8 | 4.6 | 6.8E–06 | 1.7E–02 |
| ENSMUSG00000019877 | Serinc1 | 001 | chr10 | 57235580 | 57237098 | − | 0.1 | 4.6 | 7.3E–06 | 1.8E–02 |
| ENSMUSG00000069045 | Ddx3y | 001 | chrY | 597158 | 599810 | − | 0.3 | 4.6 | 7.4E–06 | 1.8E–02 |
| ENSMUSG00000027893 | Ahcyl1 | 007 | chr3 | 107468310 | 107468433 | − | 0.8 | 4.6 | 7.4E–06 | 1.8E–02 |
| ENSMUSG00000021830 | Txndc16 | 017 | chr14 | 45787030 | 45787898 | − | 1.7 | 4.5 | 7.5E–06 | 1.8E–02 |
| ENSMUSG00000042726 | Trafd1 | 010 | chr5 | 121825256 | 121825804 | − | −0.4 | −4.5 | 8.4E–06 | 2.0E–02 |
| ENSMUSG00000052593 | Adam17 | 007 | chr12 | 21333841 | 21333900 | − | 2.5 | 4.5 | 9.0E–06 | 2.1E–02 |
| ENSMUSG00000022199 | Slc22a17 | 005 | chr14 | 55526468 | 55526722 | − | 0.4 | 4.6 | 9.2E–06 | 2.2E–02 |
| ENSMUSG00000060216 | Arrb2 | 011 | chr11 | 70249075 | 70249498 | + | 0.7 | 4.5 | 9.4E–06 | 2.2E–02 |
| ENSMUSG00000027185 | Nat10 | 027 | chr2 | 103574683 | 103575022 | − | 0.8 | 4.5 | 9.4E–06 | 2.2E–02 |
| ENSMUSG00000002280 | Narfl | 018 | chr17 | 25917898 | 25918137 | + | −0.5 | −4.5 | 9.4E–06 | 2.2E–02 |
| ENSMUSG00000032540 | Abhd5 | 002 | chr9 | 122260848 | 122260957 | + | −0.9 | −4.7 | 9.8E–06 | 2.2E–02 |
| ENSMUSG00000047342 | Zfp286 | 003 | chr11 | 62591891 | 62593106 | − | 0.4 | 4.5 | 9.9E–06 | 2.2E–02 |
| ENSMUSG00000038324 | Trpc4ap | 013 | chr2 | 155464719 | 155465208 | − | 0.5 | 4.5 | 1.0E–05 | 2.3E–02 |
| ENSMUSG00000063659 | Zfp238 | 002 | chr1 | 179375952 | 179377219 | + | 0.4 | 5.1 | 1.0E–05 | 2.3E–02 |
| ENSMUSG00000034739 + ENSMUSG00000079592 | Mfrp/C1qtnf5 | 030 | chr9 | 43915789 | 43916054 | + | −1.1 | −4.5 | 1.0E–05 | 2.3E–02 |
| ENSMUSG00000023087 | Ccrn4l | 003 | chr3 | 51044128 | 51051726 | + | 0.5 | 4.7 | 1.1E–05 | 2.3E–02 |
| ENSMUSG00000029765 | Plxna4 | 002 | chr6 | 32095926 | 32100584 | − | −0.2 | −4.5 | 1.1E–05 | 2.4E–02 |
| ENSMUSG00000040225 | Prrc2c | 029 | chr1 | 164640414 | 164640960 | − | 0.5 | 4.5 | 1.1E–05 | 2.4E–02 |
| ENSMUSG00000034675 | Dbn1 | 011 | chr13 | 55577678 | 55577992 | − | 0.3 | 4.5 | 1.1E–05 | 2.4E–02 |
| ENSMUSG00000028782 | Bai2 | 046 | chr4 | 129698499 | 129698806 | + | 0.3 | 4.4 | 1.3E–05 | 2.7E–02 |
| ENSMUSG00000039953 | Clstn1 | 001 | chr4 | 148960577 | 148960746 | + | 1.2 | 4.5 | 1.3E–05 | 2.7E–02 |
| ENSMUSG00000061751 | Kalrn | 052 | chr16 | 34152121 | 34152180 | − | 0.4 | 4.4 | 1.4E–05 | 2.9E–02 |
| ENSMUSG00000033059 | Pygb | 019 | chr2 | 150649343 | 150649711 | + | 0.9 | 4.4 | 1.4E–05 | 2.9E–02 |
| ENSMUSG00000035847 | Ids | 002 | chrX | 67596247 | 67599848 | − | 0.2 | 4.5 | 1.4E–05 | 2.9E–02 |
| ENSMUSG00000030603 | Psmc4 | 021 | chr7 | 28834222 | 28834719 | − | 0.5 | 4.5 | 1.4E–05 | 2.9E–02 |
| ENSMUSG00000021196 | Pfkp | 021 | chr13 | 6594283 | 6595229 | − | 0.6 | 4.4 | 1.5E–05 | 3.0E–02 |
| ENSMUSG00000040479 | Dgkz | 010 | chr2 | 91774000 | 91774089 | − | 0.5 | 4.4 | 1.5E–05 | 3.1E–02 |
| ENSMUSG00000029713 + ENSMUSG00000029711 | Gnb2/Epo | 041 | chr5 | 137972128 | 137972202 | − | 0.4 | 4.4 | 1.6E–05 | 3.1E–02 |
| ENSMUSG00000048148 | Nwd1 | 031 | chr8 | 75235492 | 75238645 | + | −0.3 | −4.4 | 1.6E–05 | 3.1E–02 |
| ENSMUSG00000022514 | Il1rap | 028 | chr16 | 26728315 | 26730203 | + | −0.3 | −4.4 | 1.7E–05 | 3.3E–02 |
| ENSMUSG00000044783 | Hjurp | 023 | chr1 | 90171673 | 90173793 | − | 0.5 | 4.4 | 1.7E–05 | 3.3E–02 |
| ENSMUSG00000045482 | Trrap | 013 | chr5 | 145545127 | 145545215 | + | 1.3 | 4.3 | 1.7E–05 | 3.3E–02 |
| ENSMUSG00000005378 | Wbscr22 | 030 | chr5 | 135537215 | 135537339 | − | 0.9 | 4.4 | 1.7E–05 | 3.3E–02 |
| ENSMUSG00000084896 + ENSMUSG00000020883 | Gm11632/Fbxl20 | 014 | chr11 | 97956818 | 97958242 | − | −0.6 | −4.4 | 1.7E–05 | 3.3E–02 |
| ENSMUSG00000052423 | B4galt3 | 014 | chr1 | 173201505 | 173201770 | + | −0.8 | −4.4 | 1.8E–05 | 3.4E–02 |
| ENSMUSG00000031878 | Nae1 | 009 | chr8 | 107040890 | 107040949 | − | 1.0 | 4.3 | 1.8E–05 | 3.4E–02 |
| ENSMUSG00000037996 | Slc24a2 | 002 | chr4 | 86629033 | 86637076 | − | −0.2 | −4.4 | 1.9E–05 | 3.5E–02 |
| ENSMUSG00000028703 | Lrrc41 | 005 | chr4 | 115751487 | 115751587 | + | 1.1 | 4.4 | 1.9E–05 | 3.5E–02 |
| ENSMUSG00000060206 | Zfp462 | 005 | chr4 | 55021187 | 55024237 | + | 0.4 | 4.4 | 1.9E–05 | 3.6E–02 |
| ENSMUSG00000037017 | Zscan21 | 015 | chr5 | 138575442 | 138575442 | + | 2.3 | 4.4 | 2.0E–05 | 3.6E–02 |
| ENSMUSG00000020716 | Nf1 | 029 | chr11 | 79258526 | 79258648 | + | 1.2 | 4.3 | 2.0E–05 | 3.6E–02 |
| ENSMUSG00000031389 + ENSMUSG00000031388 + ENSMUSG00000031391 |
Arhgap4/Naa10/ L1cam |
132 | chrX | 71163408 | 71164840 | − | 0.4 | 4.3 | 2.1E–05 | 3.7E–02 |
| ENSMUSG00000032589 | Bsn | 010 | chr9 | 108012745 | 108018857 | − | 0.4 | 4.4 | 2.1E–05 | 3.7E–02 |
| ENSMUSG00000091471 + ENSMUSG00000025204 + ENSMUSG00000051984 |
Gm20538/Ndufb8/ Sec31b |
015 | chr19 | 44599966 | 44600144 | − | −2.8 | −4.3 | 2.1E–05 | 3.7E–02 |
| ENSMUSG00000020894 | Vamp2 | 013 | chr11 | 68903553 | 68903678 | + | 0.5 | 4.4 | 2.1E–05 | 3.8E–02 |
| ENSMUSG00000001763 | Tspan33 | 001 | chr6 | 29644222 | 29644233 | + | 1.9 | 4.4 | 2.3E–05 | 4.0E–02 |
| ENSMUSG00000026596 | Abl2 | 018 | chr1 | 158572848 | 158579699 | + | −0.3 | −4.4 | 2.3E–05 | 4.0E–02 |
| ENSMUSG00000050875 | A730017C20Rik | 012 | chr18 | 59232072 | 59234318 | + | −0.6 | −4.4 | 2.3E–05 | 4.0E–02 |
| ENSMUSG00000030082 | Sec61a1 | 012 | chr6 | 88462600 | 88463804 | − | 0.5 | 4.3 | 2.4E–05 | 4.2E–02 |
| ENSMUSG00000040447 | Spns2 | 007 | chr11 | 72266618 | 72267055 | − | 0.5 | 4.3 | 2.4E–05 | 4.2E–02 |
| ENSMUSG00000048078 | Odz4 | 055 | chr7 | 104057065 | 104059603 | + | −0.2 | −4.3 | 2.5E–05 | 4.2E–02 |
| ENSMUSG00000057236 | Rbbp4 | 016 | chr4 | 129002068 | 129005831 | − | 0.4 | 4.3 | 2.5E–05 | 4.2E–02 |
| ENSMUSG00000044308 | Ubr3 | 054 | chr2 | 69858185 | 69858507 | + | −0.5 | −4.3 | 2.5E–05 | 4.2E–02 |
| ENSMUSG00000040209 | Zfp704 | 001 | chr3 | 9427011 | 9438898 | − | −0.4 | −4.4 | 2.6E–05 | 4.3E–02 |
| ENSMUSG00000023026 | Dip2b | 023 | chr15 | 100011740 | 100011867 | + | 0.6 | 4.2 | 2.8E–05 | 4.6E–02 |
| ENSMUSG00000056602 | Fry | 027 | chr5 | 151198318 | 151198442 | + | 0.5 | 4.2 | 2.8E–05 | 4.7E–02 |
| ENSMUSG00000051306 | Usp42 | 023 | chr5 | 144483224 | 144483814 | − | 1.8 | 4.3 | 2.9E–05 | 4.7E–02 |
| ENSMUSG00000035027 | Map2k2 | 009 | chr10 | 80581357 | 80581721 | + | −0.9 | −4.3 | 2.9E–05 | 4.7E–02 |
| ENSMUSG00000007850 | Hnrnph1 | 046 | chr11 | 50199824 | 50199891 | + | 0.6 | 4.2 | 2.9E–05 | 4.7E–02 |
| ENSMUSG00000029578 | Wipi2 | 016 | chr5 | 143140444 | 143140598 | + | 0.7 | 4.3 | 3.0E–05 | 4.8E–02 |
| ENSMUSG00000027797 | Dclk1 | 009 | chr3 | 55270495 | 55275239 | + | 0.3 | 4.3 | 3.0E–05 | 4.8E–02 |
| ENSMUSG00000028943 | Espn | 001 | chr4 | 151494440 | 151494444 | − | 0.8 | 4.2 | 3.0E–05 | 4.8E–02 |
| ENSMUSG00000042625 | Safb2 | 026 | chr17 | 56708446 | 56708857 | − | 0.4 | 4.2 | 3.1E–05 | 4.8E–02 |
| ENSMUSG00000058624 | Gda | 010 | chr19 | 21493215 | 21493863 | − | 1.6 | 4.3 | 3.1E–05 | 4.9E–02 |
Table 2.
List of bins showing differential expression 30 min after memory retrieval. Each differential bin contains the chromosome, start position, and end position for easy reference.
| GeneID | Gene name | Bin # |
Chr | Start | Stop | Strand | logFC | t | P value | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| ENSMUSG00000029657 | Hsph1 | 012 | chr5 | 150423259 | 150426039 | − | 1.2 | 12.8 | 1.7E–29 | 4.7E–24 |
| ENSMUSG00000031167 | Rbm3 | 002 | chrX | 7716104 | 7717909 | − | 1.2 | 11.8 | 3.5E–26 | 4.6E–21 |
| ENSMUSG00000007617 | Homer1 | 018 | chr13 | 94136356 | 94137083 | + | 1.5 | 10.5 | 1.6E–21 | 1.4E–16 |
| ENSMUSG00000039801 | 2410089E03Rik | 053 | chr15 | 8201065 | 8202148 | + | −1.0 | −9.2 | 1.1E–18 | 7.3E–14 |
| ENSMUSG00000041879 | Ipo9 | 036 | chr1 | 137302594 | 137303043 | − | −1.1 | −9.0 | 6.4E–18 | 3.4E–13 |
| ENSMUSG00000034083 | C130022K22Rik | 009 | chr6 | 91835401 | 91838063 | + | 1.1 | 9.1 | 1.2E–16 | 5.5E–12 |
| ENSMUSG00000027523 | Gnas | 027 | chr2 | 174155788 | 174155935 | + | 1.3 | 8.2 | 3.8E–15 | 1.4E–10 |
| ENSMUSG00000034656 | Cacna1a | 068 | chr8 | 87163334 | 87163334 | + | 1.7 | 7.9 | 1.2E–14 | 4.0E–10 |
| ENSMUSG00000057897 | Camk2b | 049 | chr11 | 5965662 | 5965745 | − | 2.2 | 7.6 | 1.9E–13 | 5.6E–09 |
| ENSMUSG00000035206 | 3110056O03Rik | 016 | chr10 | 80329406 | 80330144 | + | 0.5 | 8.0 | 2.4E–13 | 6.5E–09 |
| ENSMUSG00000035640 | Dos | 014 | chr10 | 79598293 | 79598333 | − | 2.1 | 7.6 | 2.8E–13 | 6.8E–09 |
| ENSMUSG00000039953 | Clstn1 | 001 | chr4 | 148960577 | 148960746 | + | 1.9 | 7.5 | 1.6E–12 | 3.6E–08 |
| ENSMUSG00000008153 | Clstn3 | 004 | chr6 | 124383426 | 124383521 | − | 1.1 | 7.4 | 2.0E–12 | 4.0E–08 |
| ENSMUSG00000075876 + ENSMUSG00000064791 + ENSMUSG00000075924 + ENSMUSG00000015656 |
Snord14c/ Snord14e/ Snord14d/Hspa8 |
038 | chr9 | 40612831 | 40612920 | + | 1.6 | 7.1 | 6.7E–12 | 1.3E–07 |
| ENSMUSG00000031167 | Rbm3 | 022 | chrX | 7721600 | 7721698 | − | −0.8 | −7.1 | 8.5E–12 | 1.5E–07 |
| ENSMUSG00000057421 | Las1l | 015 | chrX | 93143543 | 93144773 | − | −1.0 | −7.1 | 1.1E–11 | 1.8E–07 |
| ENSMUSG00000036052 | Dnajb5 | 011 | chr4 | 42963816 | 42965965 | + | 0.4 | 7.3 | 1.1E–11 | 1.8E–07 |
| ENSMUSG00000031878 | Nae1 | 015 | chr8 | 107042164 | 107043101 | − | 1.2 | 6.9 | 1.7E–11 | 2.5E–07 |
| ENSMUSG00000022199 | Slc22a17 | 005 | chr14 | 55526468 | 55526722 | − | 0.5 | 7.1 | 4.1E–11 | 5.8E–07 |
| ENSMUSG00000075876 + ENSMUSG00000064791 + ENSMUSG00000075924 + ENSMUSG00000015656 |
Snord14c/ Snord14e/ Snord14d/Hspa8 |
037 | chr9 | 40612779 | 40612830 | + | 1.4 | 6.7 | 9.5E–11 | 1.3E–06 |
| ENSMUSG00000075003 + ENSMUSG00000037876 | Jmjd1c/Jmjd1c | 041 | chr10 | 66707622 | 66708166 | + | 0.9 | 6.6 | 1.1E–10 | 1.3E–06 |
| ENSMUSG00000031167 | Rbm3 | 003 | chrX | 7717910 | 7718334 | − | 1.1 | 6.7 | 1.5E–10 | 1.9E–06 |
| ENSMUSG00000023965 | Fbxl17 | 017 | chr17 | 63848374 | 63849366 | − | 0.5 | 6.8 | 1.9E–10 | 2.2E–06 |
| ENSMUSG00000028826 | Tmem57 | 002 | chr4 | 134360480 | 134362431 | − | −0.5 | −6.6 | 3.8E–10 | 4.2E–06 |
| ENSMUSG00000038664 | Herc1 | 093 | chr9 | 66346564 | 66348060 | + | −0.7 | −6.3 | 4.3E–10 | 4.6E–06 |
| ENSMUSG00000019254 | Ppp1r12c | 052 | chr7 | 4453132 | 4453266 | − | 1.1 | 6.4 | 5.3E–10 | 5.4E–06 |
| ENSMUSG00000028797 + ENSMUSG00000086797 | 2510006D16Rik/ Gm12965 |
034 | chr4 | 129284424 | 129284574 | + | 1.1 | 6.3 | 7.6E–10 | 7.5E–06 |
| ENSMUSG00000028782 | Bai2 | 001 | chr4 | 129662114 | 129662407 | + | 0.6 | 6.3 | 8.8E–10 | 8.4E–06 |
| ENSMUSG00000024012 | Mtch1 | 030 | chr17 | 29484705 | 29484849 | − | 0.8 | 6.3 | 1.2E–09 | 1.1E–05 |
| ENSMUSG00000024576 | Csnk1a1 | 028 | chr18 | 61745286 | 61746152 | + | 0.5 | 6.3 | 1.4E–09 | 1.2E–05 |
| ENSMUSG00000027569 | 1600027N09Rik | 010 | chr2 | 180318228 | 180319110 | + | 0.5 | 6.5 | 1.4E–09 | 1.2E–05 |
| ENSMUSG00000031660 | Brd7 | 032 | chr8 | 90885914 | 90886093 | − | 1.1 | 6.3 | 1.5E–09 | 1.3E–05 |
| ENSMUSG00000046667 | Rbm12b | 012 | chr4 | 12072176 | 12073847 | + | 0.8 | 6.5 | 1.6E–09 | 1.3E–05 |
| ENSMUSG00000024392 | Bag6 | 032 | chr17 | 35277897 | 35278103 | + | 0.8 | 6.1 | 2.0E–09 | 1.5E–05 |
| ENSMUSG00000052423 | B4galt3 | 014 | chr1 | 173201505 | 173201770 | + | −1.0 | −6.2 | 2.0E–09 | 1.5E–05 |
| ENSMUSG00000019854 | Reps1 | 001 | chr10 | 17775667 | 17775711 | + | 1.1 | 6.1 | 2.5E–09 | 1.8E–05 |
| ENSMUSG00000061887 | Ssbp3 | 003 | chr4 | 106584116 | 106584137 | + | 1.8 | 6.1 | 2.7E–09 | 2.0E–05 |
| ENSMUSG00000074247 | Dda1 | 012 | chr8 | 73996275 | 73996514 | + | 0.9 | 6.3 | 2.9E–09 | 2.0E–05 |
| ENSMUSG00000056413 | Adap1 | 022 | chr5 | 139801419 | 139801576 | − | 1.8 | 6.2 | 3.2E–09 | 2.2E–05 |
| ENSMUSG00000026090 | 2010300C02Rik | 018 | chr1 | 37776641 | 37776930 | − | 1.5 | 6.3 | 3.4E–09 | 2.3E–05 |
| ENSMUSG00000007617 | Homer1 | 017 | chr13 | 94136233 | 94136355 | + | 1.4 | 6.0 | 5.7E–09 | 3.7E–05 |
| ENSMUSG00000037266 | D4Wsu53e | 022 | chr4 | 134481737 | 134481940 | + | −0.4 | −6.0 | 5.9E–09 | 3.8E–05 |
| ENSMUSG00000037266 | D4Wsu53e | 024 | chr4 | 134482066 | 134482649 | + | −0.4 | −6.0 | 6.1E–09 | 3.8E–05 |
| ENSMUSG00000037098 | Rab11fip3 | 027 | chr17 | 26206181 | 26206354 | − | 0.9 | 6.0 | 6.7E–09 | 4.1E–05 |
| ENSMUSG00000043872 | Zmym1 | 001 | chr4 | 126724338 | 126724885 | − | 0.9 | 6.1 | 7.1E–09 | 4.2E–05 |
| ENSMUSG00000024826 | Dpf2 | 011 | chr19 | 5902769 | 5903332 | − | 0.5 | 6.0 | 8.4E–09 | 4.9E–05 |
| ENSMUSG00000025372 | Baiap2 | 027 | chr11 | 119867673 | 119868096 | + | 0.3 | 6.0 | 8.6E–09 | 4.9E–05 |
| ENSMUSG00000037266 | D4Wsu53e | 021 | chr4 | 134481701 | 134481736 | + | −0.5 | −5.9 | 1.1E–08 | 6.3E–05 |
| ENSMUSG00000013033 | Lphn1 | 001 | chr8 | 86424004 | 86424471 | + | 0.8 | 5.8 | 1.3E–08 | 7.3E–05 |
| ENSMUSG00000040479 | Dgkz | 011 | chr2 | 91774090 | 91774212 | − | 0.6 | 5.8 | 1.4E–08 | 7.5E–05 |
| ENSMUSG00000027893 | Ahcyl1 | 007 | chr3 | 107468310 | 107468433 | − | 1.0 | 5.8 | 1.7E–08 | 9.0E–05 |
| ENSMUSG00000001729 | Akt1 | 034 | chr12 | 113912418 | 113912487 | − | 1.4 | 5.8 | 1.8E–08 | 9.3E–05 |
| ENSMUSG00000037266 | D4Wsu53e | 020 | chr4 | 134481692 | 134481700 | + | −0.5 | −5.8 | 2.0E–08 | 1.0E–04 |
| ENSMUSG00000031167 | Rbm3 | 021 | chrX | 7721572 | 7721599 | − | −0.8 | −5.8 | 2.1E–08 | 1.0E–04 |
| ENSMUSG00000055491 | Pprc1 | 039 | chr19 | 46146825 | 46147038 | + | 1.0 | 5.7 | 2.2E–08 | 1.1E–04 |
| ENSMUSG00000021327 | Zkscan3 | 009 | chr13 | 21481162 | 21485100 | − | 0.4 | 5.8 | 2.2E–08 | 1.1E–04 |
| ENSMUSG00000021262 | Evl | 001 | chr12 | 109792930 | 109793423 | + | 0.5 | 5.9 | 2.3E–08 | 1.1E–04 |
| ENSMUSG00000015869 | Prpsap1 | 009 | chr11 | 116338662 | 116339053 | − | 0.6 | 5.8 | 2.5E–08 | 1.1E–04 |
| ENSMUSG00000014873 | Surf2 | 009 | chr2 | 26773052 | 26774384 | + | 0.3 | 5.8 | 2.5E–08 | 1.1E–04 |
| ENSMUSG00000038383 | Pigu | 004 | chr2 | 155104386 | 155108131 | − | 0.4 | 5.8 | 2.8E–08 | 1.3E–04 |
| ENSMUSG00000022514 | Il1rap | 028 | chr16 | 26728315 | 26730203 | + | −0.4 | −5.7 | 3.0E–08 | 1.3E–04 |
| ENSMUSG00000004929 | Thop1 | 013 | chr10 | 80541613 | 80542202 | + | 0.6 | 5.8 | 3.2E–08 | 1.4E–04 |
| ENSMUSG00000021040 | 1810035L17Rik | 009 | chr12 | 88790351 | 88790753 | + | 0.6 | 5.9 | 3.3E–08 | 1.4E–04 |
| ENSMUSG00000023353 | Agap3 | 002 | chr5 | 23958025 | 23958073 | + | 1.8 | 5.7 | 3.3E–08 | 1.4E–04 |
| ENSMUSG00000050875 | A730017C20Rik | 012 | chr18 | 59232072 | 59234318 | + | −0.7 | −5.8 | 3.4E–08 | 1.4E–04 |
| ENSMUSG00000001729 | Akt1 | 033 | chr12 | 113912211 | 113912417 | − | 0.7 | 5.7 | 3.6E–08 | 1.4E–04 |
| ENSMUSG00000093290 + ENSMUSG00000035632 | Mir3572/Cnot3 | 026 | chr7 | 3610347 | 3610426 | + | 0.6 | 5.7 | 3.7E–08 | 1.5E–04 |
| ENSMUSG00000009073 | Nf2 | 014 | chr11 | 4684567 | 4685110 | − | 0.9 | 5.6 | 3.8E–08 | 1.5E–04 |
| ENSMUSG00000084708 + ENSMUSG00000065862 + ENSMUSG00000059796 |
//Eif4a1 | 040 | chr11 | 69485107 | 69485232 | − | 0.8 | 5.6 | 4.0E–08 | 1.5E–04 |
| ENSMUSG00000037266 | D4Wsu53e | 019 | chr4 | 134481361 | 134481691 | + | −0.4 | −5.7 | 4.0E–08 | 1.5E–04 |
| ENSMUSG00000015536 | Mocs2 | 015 | chr13 | 115615731 | 115616365 | + | 0.6 | 5.7 | 4.7E–08 | 1.8E–04 |
| ENSMUSG00000034675 | Dbn1 | 011 | chr13 | 55577678 | 55577992 | − | 0.3 | 5.6 | 4.8E–08 | 1.8E–04 |
| ENSMUSG00000003269 | Cyth2 | 023 | chr7 | 53068527 | 53069248 | − | 0.4 | 5.6 | 5.0E–08 | 1.8E–04 |
| ENSMUSG00000031065 | Cdk16 | 027 | chrX | 20274091 | 20274245 | + | 0.5 | 5.6 | 5.6E–08 | 2.0E–04 |
| ENSMUSG00000025499 | Hras1 | 021 | chr7 | 148379860 | 148379893 | − | 3.4 | 5.6 | 5.9E–08 | 2.1E–04 |
| ENSMUSG00000080683 + ENSMUSG00000087376 + ENSMUSG00000080352 + ENSMUSG00000045411 |
/Gm15517// 2410002F23Rik |
030 | chr7 | 51503682 | 51504911 | + | 0.4 | 5.5 | 7.4E–08 | 2.6E–04 |
| ENSMUSG00000009549 | Srp14 | 005 | chr2 | 118304301 | 118304567 | − | 0.8 | 5.7 | 8.4E–08 | 2.9E–04 |
| ENSMUSG00000031392 + ENSMUSG00000076127 + ENSMUSG00000092907 |
Irak1/Mir718/ Mir5132 |
056 | chrX | 71269148 | 71269165 | − | 1.7 | 5.4 | 8.6E–08 | 2.9E–04 |
| ENSMUSG00000078789 + ENSMUSG00000038268 | Dph1/Ovca2 | 001 | chr11 | 74989444 | 74991144 | − | 0.3 | 5.5 | 1.1E–07 | 3.6E–04 |
| ENSMUSG00000031660 | Brd7 | 031 | chr8 | 90885805 | 90885913 | − | 0.7 | 5.4 | 1.2E–07 | 3.9E–04 |
| ENSMUSG00000020923 | Ubtf | 012 | chr11 | 102169844 | 102170012 | − | 1.3 | 5.4 | 1.2E–07 | 4.1E–04 |
| ENSMUSG00000032047 | Acat1 | 004 | chr9 | 53391700 | 53391862 | − | 0.7 | 5.6 | 1.3E–07 | 4.2E–04 |
| ENSMUSG00000003345 | Csnk1g2 | 001 | chr10 | 80085525 | 80085695 | + | 0.6 | 5.5 | 1.3E–07 | 4.3E–04 |
| ENSMUSG00000072770 + ENSMUSG00000030330 | Acrbp/Ing4 | 042 | chr6 | 125003508 | 125003819 | + | 0.5 | 5.4 | 1.4E–07 | 4.4E–04 |
| ENSMUSG00000028796 | Phc2 | 020 | chr4 | 128404926 | 128404953 | + | 2.3 | 5.4 | 1.4E–07 | 4.4E–04 |
| ENSMUSG00000002393 | Nr2f6 | 023 | chr8 | 73905806 | 73905859 | − | 1.7 | 5.5 | 1.5E–07 | 4.6E–04 |
| ENSMUSG00000037266 | D4Wsu53e | 023 | chr4 | 134481941 | 134482065 | + | −0.4 | −5.4 | 1.5E–07 | 4.6E–04 |
| ENSMUSG00000006575 | Rundc3a | 019 | chr11 | 102261951 | 102261974 | + | 0.4 | 5.4 | 1.6E–07 | 4.9E–04 |
| ENSMUSG00000092679 + ENSMUSG00000026872 | Mir5129/Zeb2 | 001 | chr2 | 44839154 | 44842640 | − | −0.2 | −5.4 | 1.8E–07 | 5.3E–04 |
| ENSMUSG00000001211 | Agpat3 | 029 | chr10 | 77814958 | 77815187 | − | 0.5 | 5.4 | 1.8E–07 | 5.4E–04 |
| ENSMUSG00000016933 | Plcg1 | 004 | chr2 | 160557454 | 160557468 | + | 0.9 | 5.3 | 2.0E–07 | 5.9E–04 |
| ENSMUSG00000042625 | Safb2 | 026 | chr17 | 56708446 | 56708857 | − | 0.4 | 5.3 | 2.1E–07 | 6.2E–04 |
| ENSMUSG00000027546 | Atp9a | 040 | chr2 | 168567301 | 168567462 | − | 0.7 | 5.3 | 2.6E–07 | 7.4E–04 |
| ENSMUSG00000027367 | Stard7 | 014 | chr2 | 127115811 | 127116507 | + | 0.5 | 5.3 | 2.8E–07 | 7.9E–04 |
| ENSMUSG00000038644 | Pold1 | 018 | chr7 | 51789560 | 51789754 | − | 1.7 | 5.2 | 2.8E–07 | 7.9E–04 |
| ENSMUSG00000002812 | Flii | 019 | chr11 | 60532729 | 60533194 | − | 0.5 | 5.2 | 3.1E–07 | 8.5E–04 |
| ENSMUSG00000042605 | Atxn2 | 051 | chr5 | 122261639 | 122261939 | + | 0.6 | 5.2 | 3.1E–07 | 8.6E–04 |
| ENSMUSG00000040479 | Dgkz | 054 | chr2 | 91803443 | 91803814 | − | 0.3 | 5.2 | 3.2E–07 | 8.8E–04 |
| ENSMUSG00000059995 | Atxn7l3 | 020 | chr11 | 102157717 | 102157943 | − | 0.7 | 5.3 | 3.3E–07 | 8.9E–04 |
| ENSMUSG00000092870 + ENSMUSG00000020349 | Mir3061/Ppp2ca | 001 | chr11 | 51912183 | 51912652 | + | 0.9 | 5.5 | 3.3E–07 | 8.9E–04 |
| ENSMUSG00000027303 | Ptpra | 001 | chr2 | 130276014 | 130276279 | + | 0.6 | 5.3 | 3.5E–07 | 9.2E–04 |
| ENSMUSG00000023353 | Agap3 | 001 | chr5 | 23957995 | 23958024 | + | 1.6 | 5.3 | 3.6E–07 | 9.4E–04 |
| ENSMUSG00000028484 | Psip1 | 026 | chr4 | 83132179 | 83132357 | − | 0.5 | 5.2 | 3.9E–07 | 1.0E–03 |
| ENSMUSG00000033423 | Eri3 | 004 | chr4 | 117223367 | 117225214 | + | −0.5 | −5.2 | 4.3E–07 | 1.1E–03 |
| ENSMUSG00000026277 | Stk25 | 009 | chr1 | 95522184 | 95522357 | − | 0.3 | 5.2 | 4.7E–07 | 1.2E–03 |
| ENSMUSG00000037907 | Ankrd13b | 036 | chr11 | 77303169 | 77303180 | − | 2.3 | 5.1 | 4.8E–07 | 1.2E–03 |
| ENSMUSG00000038664 | Herc1 | 095 | chr9 | 66348328 | 66348982 | + | −0.5 | −5.0 | 5.6E–07 | 1.4E–03 |
| ENSMUSG00000034254 | Agpat1 | 012 | chr17 | 34747951 | 34748122 | + | 0.4 | 5.1 | 5.8E–07 | 1.4E–03 |
| ENSMUSG00000040479 | Dgkz | 010 | chr2 | 91774000 | 91774089 | − | 0.5 | 5.1 | 5.8E–07 | 1.4E–03 |
| ENSMUSG00000024858 | Adrbk1 | 046 | chr19 | 4306215 | 4306222 | − | 1.6 | 5.1 | 5.9E–07 | 1.4E–03 |
| ENSMUSG00000004929 | Thop1 | 012 | chr10 | 80541376 | 80541500 | + | 0.8 | 5.2 | 5.9E–07 | 1.4E–03 |
| ENSMUSG00000084708 + ENSMUSG00000065862 + ENSMUSG00000059796 |
//Eif4a1 | 037 | chr11 | 69484479 | 69484603 | − | 0.5 | 5.1 | 6.1E–07 | 1.5E–03 |
| ENSMUSG00000038324 | Trpc4ap | 013 | chr2 | 155464719 | 155465208 | − | 0.5 | 5.1 | 6.6E–07 | 1.6E–03 |
| ENSMUSG00000038546 | Ranbp9 | 018 | chr13 | 43576298 | 43576342 | − | 1.6 | 5.2 | 7.1E–07 | 1.7E–03 |
| ENSMUSG00000025499 | Hras1 | 020 | chr7 | 148379782 | 148379859 | − | 1.0 | 5.1 | 7.3E–07 | 1.7E–03 |
| ENSMUSG00000034730 + ENSMUSG00000093340 | Bai1/ | 020 | chr15 | 74394251 | 74394282 | + | 0.8 | 5.0 | 7.6E–07 | 1.7E–03 |
| ENSMUSG00000068267 + ENSMUSG00000027329 | Cenpb/Spef1 | 018 | chr2 | 131005613 | 131005803 | − | 0.9 | 5.1 | 7.6E–07 | 1.7E–03 |
| ENSMUSG00000029571 | Tmem106b | 020 | chr6 | 13034188 | 13039269 | + | 0.2 | 5.1 | 7.7E–07 | 1.7E–03 |
| ENSMUSG00000025964 | Adam23 | 005 | chr1 | 63492509 | 63492986 | + | 0.5 | 5.0 | 8.2E–07 | 1.8E–03 |
| ENSMUSG00000025372 | Baiap2 | 026 | chr11 | 119864352 | 119864399 | + | 0.4 | 5.1 | 8.2E–07 | 1.8E–03 |
| ENSMUSG00000020894 | Vamp2 | 013 | chr11 | 68903553 | 68903678 | + | 0.5 | 5.1 | 8.6E–07 | 1.9E–03 |
| ENSMUSG00000031065 | Cdk16 | 003 | chrX | 20265544 | 20265932 | + | 0.2 | 5.0 | 8.6E–07 | 1.9E–03 |
| ENSMUSG00000036555 | Iqce | 001 | chr5 | 141137781 | 141139458 | − | 0.5 | 5.0 | 9.1E–07 | 2.0E–03 |
| ENSMUSG00000016503 | Gtf3a | 004 | chr5 | 147761583 | 147762108 | + | 0.5 | 5.1 | 9.1E–07 | 2.0E–03 |
| ENSMUSG00000001366 | Fbxo9 | 006 | chr9 | 77933505 | 77933713 | − | 0.7 | 5.1 | 9.3E–07 | 2.0E–03 |
| ENSMUSG00000037098 | Rab11fip3 | 026 | chr17 | 26206123 | 26206180 | − | 0.7 | 5.0 | 9.3E–07 | 2.0E–03 |
| ENSMUSG00000004071 | 5730403B10Rik | 008 | chr16 | 4769309 | 4769639 | − | −0.4 | −5.1 | 9.4E–07 | 2.0E–03 |
| ENSMUSG00000020612 | Prkar1a | 011 | chr11 | 109522664 | 109523067 | + | 0.3 | 5.1 | 9.8E–07 | 2.0E–03 |
| ENSMUSG00000018861 | Fdxr | 018 | chr11 | 115137497 | 115138038 | − | 0.6 | 5.1 | 9.9E–07 | 2.1E–03 |
| ENSMUSG00000034730 + ENSMUSG00000093340 | Bai1/ | 021 | chr15 | 74394283 | 74394341 | + | 0.7 | 5.0 | 1.0E–06 | 2.1E–03 |
| ENSMUSG00000024533 | Spire1 | 022 | chr18 | 67770059 | 67770443 | − | 0.7 | 5.1 | 1.0E–06 | 2.1E–03 |
| ENSMUSG00000024576 | Csnk1a1 | 027 | chr18 | 61744853 | 61745285 | + | 0.5 | 5.0 | 1.1E–06 | 2.1E–03 |
| ENSMUSG00000028412 | Slc44a1 | 001 | chr4 | 53453285 | 53453542 | + | 0.9 | 5.0 | 1.1E–06 | 2.3E–03 |
| ENSMUSG00000026074 | Map4k4 | 001 | chr1 | 39957758 | 39958024 | + | 0.7 | 5.0 | 1.1E–06 | 2.3E–03 |
| ENSMUSG00000026977 | March7 | 022 | chr2 | 60083318 | 60085399 | + | 0.4 | 5.0 | 1.1E–06 | 2.3E–03 |
| ENSMUSG00000002949 | Timm44 | 019 | chr8 | 4267402 | 4267540 | − | −0.6 | −5.0 | 1.2E–06 | 2.4E–03 |
| ENSMUSG00000027634 | Ndrg3 | 005 | chr2 | 156756888 | 156756951 | − | 1.0 | 4.9 | 1.2E–06 | 2.4E–03 |
| ENSMUSG00000074247 | Dda1 | 011 | chr8 | 73996146 | 73996274 | + | 0.8 | 5.1 | 1.3E–06 | 2.5E–03 |
| ENSMUSG00000024392 | Bag6 | 033 | chr17 | 35278104 | 35278338 | + | 0.7 | 4.9 | 1.4E–06 | 2.7E–03 |
| ENSMUSG00000057522 | Spop | 019 | chr11 | 95346773 | 95346850 | + | −1.2 | −5.0 | 1.4E–06 | 2.7E–03 |
| ENSMUSG00000022771 | Ppil2 | 003 | chr16 | 17087409 | 17088082 | − | 0.4 | 4.9 | 1.6E–06 | 2.9E–03 |
| ENSMUSG00000035202 | Lars2 | 021 | chr9 | 123370617 | 123371782 | + | −0.6 | −5.0 | 1.6E–06 | 2.9E–03 |
| ENSMUSG00000052423 | B4galt3 | 015 | chr1 | 173201771 | 173201773 | + | −1.3 | −4.9 | 1.6E–06 | 3.0E–03 |
| ENSMUSG00000016346 | Kcnq2 | 047 | chr2 | 180869912 | 180869948 | − | 1.4 | 4.9 | 1.8E–06 | 3.3E–03 |
| ENSMUSG00000020402 | Vdac1 | 013 | chr11 | 52199869 | 52199973 | + | −0.6 | −4.9 | 1.8E–06 | 3.3E–03 |
| ENSMUSG00000003279 | Dlgap1 | 004 | chr17 | 70318713 | 70318741 | + | 0.7 | 4.8 | 1.8E–06 | 3.4E–03 |
| ENSMUSG00000018040 | Rrp7a | 009 | chr15 | 82948605 | 82948776 | − | 0.7 | 5.0 | 1.9E–06 | 3.4E–03 |
| ENSMUSG00000042042 | Csgalnact2 | 007 | chr6 | 118074432 | 118076139 | − | 1.2 | 5.0 | 2.0E–06 | 3.5E–03 |
| ENSMUSG00000030189 | Csda | 018 | chr6 | 131338232 | 131338468 | − | 1.2 | 4.9 | 2.0E–06 | 3.6E–03 |
| ENSMUSG00000026918 | Brd3 | 013 | chr2 | 27308971 | 27309832 | − | −1.2 | −4.8 | 2.0E–06 | 3.6E–03 |
| ENSMUSG00000021830 | Txndc16 | 017 | chr14 | 45787030 | 45787898 | − | 1.7 | 4.8 | 2.1E–06 | 3.7E–03 |
| ENSMUSG00000065452 + ENSMUSG00000028410 | Mir207/Dnaja1 | 027 | chr4 | 40678833 | 40679108 | + | 0.5 | 4.9 | 2.1E–06 | 3.7E–03 |
| ENSMUSG00000059552 | Trp53 | 018 | chr11 | 69403365 | 69404007 | + | 0.7 | 4.9 | 2.2E–06 | 3.8E–03 |
| ENSMUSG00000037058 | Paip2 | 010 | chr18 | 35769938 | 35770524 | + | 0.3 | 4.9 | 2.2E–06 | 3.8E–03 |
| ENSMUSG00000068221 + ENSMUSG00000022436 | Pdxp/Sh3bp1 | 031 | chr15 | 78744349 | 78744961 | + | 0.3 | 4.8 | 2.2E–06 | 3.8E–03 |
| ENSMUSG00000028796 | Phc2 | 019 | chr4 | 128404829 | 128404925 | + | 2.0 | 4.8 | 2.3E–06 | 3.9E–03 |
| ENSMUSG00000039219 | Arid4b | 042 | chr13 | 14283524 | 14284215 | + | −0.8 | −4.8 | 2.3E–06 | 3.9E–03 |
| ENSMUSG00000027655 | Dhx35 | 035 | chr2 | 158676365 | 158676477 | + | 1.4 | 4.8 | 2.3E–06 | 3.9E–03 |
| ENSMUSG00000024163 + ENSMUSG00000073436 | Mapk8ip3/Eme2 | 062 | chr17 | 25038763 | 25039222 | − | 0.3 | 4.7 | 2.4E–06 | 4.0E–03 |
| ENSMUSG00000039470 | Zdhhc2 | 004 | chr8 | 41509194 | 41509212 | + | 1.5 | 4.8 | 2.4E–06 | 4.0E–03 |
| ENSMUSG00000068921 | Dap3 | 001 | chr3 | 88724725 | 88727556 | − | −0.3 | −4.8 | 2.4E–06 | 4.0E–03 |
| ENSMUSG00000042605 | Atxn2 | 002 | chr5 | 122161618 | 122162285 | + | 0.7 | 4.8 | 2.4E–06 | 4.0E–03 |
| ENSMUSG00000006024 | Napa | 016 | chr7 | 16698993 | 16699469 | + | 0.3 | 4.8 | 2.5E–06 | 4.1E–03 |
| ENSMUSG00000066900 | Suds3 | 014 | chr5 | 117565681 | 117566002 | − | 0.3 | 4.9 | 2.6E–06 | 4.2E–03 |
| ENSMUSG00000050530 | Fam171a1 | 002 | chr2 | 3035654 | 3035684 | + | 2.0 | 4.9 | 2.6E–06 | 4.2E–03 |
| ENSMUSG00000038291 | Snx25 | 029 | chr8 | 47237136 | 47237511 | − | 1.9 | 4.8 | 2.8E–06 | 4.5E–03 |
| ENSMUSG00000039108 | Lsm14b | 001 | chr2 | 179759692 | 179760017 | + | 0.5 | 4.9 | 2.8E–06 | 4.5E–03 |
| ENSMUSG00000084896 + ENSMUSG00000020883 | Gm11632/Fbxl20 | 014 | chr11 | 97956818 | 97958242 | − | −0.6 | −4.8 | 2.9E–06 | 4.7E–03 |
| ENSMUSG00000017412 | Cacnb4 | 001 | chr2 | 52283845 | 52290269 | − | 0.3 | 4.8 | 3.0E–06 | 4.8E–03 |
| ENSMUSG00000002984 | Tomm40 | 008 | chr7 | 20288492 | 20288617 | − | 0.6 | 4.8 | 3.2E–06 | 5.0E–03 |
| ENSMUSG00000039759 | Thap3 | 004 | chr4 | 151359568 | 151359777 | − | −0.5 | −5.0 | 3.3E–06 | 5.2E–03 |
| ENSMUSG00000038822 | Hace1 | 010 | chr10 | 45325391 | 45325393 | + | 2.1 | 4.7 | 3.4E–06 | 5.3E–03 |
| ENSMUSG00000052373 | Mpp3 | 012 | chr11 | 101870999 | 101871520 | − | 0.7 | 4.7 | 3.4E–06 | 5.3E–03 |
| ENSMUSG00000084708 + ENSMUSG00000065862 + ENSMUSG00000059796 |
//Eif4a1 | 038 | chr11 | 69484604 | 69484724 | − | 0.5 | 4.7 | 3.4E–06 | 5.3E–03 |
| ENSMUSG00000025155 | Dus1l | 009 | chr11 | 120651195 | 120651761 | − | 0.4 | 4.7 | 3.5E–06 | 5.3E–03 |
| ENSMUSG00000036545 | Adamts2 | 028 | chr11 | 50617071 | 50621075 | + | −0.8 | −4.8 | 3.5E–06 | 5.3E–03 |
| ENSMUSG00000038429 | Usp5 | 025 | chr6 | 124772861 | 124773037 | − | 0.7 | 4.7 | 3.5E–06 | 5.3E–03 |
| ENSMUSG00000061887 | Ssbp3 | 002 | chr4 | 106584075 | 106584115 | + | 1.9 | 4.7 | 3.7E–06 | 5.6E–03 |
| ENSMUSG00000084708 + ENSMUSG00000065862 + ENSMUSG00000059796 |
//Eif4a1 | 039 | chr11 | 69484725 | 69484952 | − | 0.5 | 4.7 | 3.8E–06 | 5.6E–03 |
| ENSMUSG00000021087 | Rtn1 | 003 | chr12 | 73313276 | 73313276 | − | −0.2 | −4.8 | 3.8E–06 | 5.6E–03 |
| ENSMUSG00000024012 | Mtch1 | 029 | chr17 | 29484412 | 29484704 | − | 0.3 | 4.7 | 3.8E–06 | 5.6E–03 |
| ENSMUSG00000026885 | Ttll11 | 024 | chr2 | 35835145 | 35835286 | − | 0.6 | 4.7 | 3.9E–06 | 5.6E–03 |
| ENSMUSG00000040896 | Kcnd3 | 009 | chr3 | 105468465 | 105469879 | + | 0.9 | 4.8 | 3.9E–06 | 5.6E–03 |
| ENSMUSG00000040859 | Bsdc1 | 017 | chr4 | 129146293 | 129147418 | + | 0.7 | 4.7 | 3.9E–06 | 5.7E–03 |
| ENSMUSG00000028063 | Lmna | 005 | chr3 | 88286535 | 88286786 | − | 0.6 | 4.7 | 4.0E–06 | 5.7E–03 |
| ENSMUSG00000053046 + ENSMUSG00000092652 | Brsk2/Mir3104 | 002 | chr7 | 149135751 | 149135911 | + | 0.9 | 4.7 | 4.0E–06 | 5.7E–03 |
| ENSMUSG00000032997 + ENSMUSG00000026211 | Chpf/Obsl1 | 060 | chr1 | 75499791 | 75499941 | − | −2.6 | −4.7 | 4.0E–06 | 5.8E–03 |
| ENSMUSG00000023952 | Gtpbp2 | 035 | chr17 | 46303816 | 46303936 | + | 0.7 | 4.7 | 4.2E–06 | 6.0E–03 |
| ENSMUSG00000050989 | Sepn1 | 019 | chr4 | 134107852 | 134108081 | − | 1.2 | 4.8 | 4.3E–06 | 6.1E–03 |
| ENSMUSG00000027223 | Mapk8ip1 | 018 | chr2 | 92241186 | 92241420 | − | 0.4 | 4.8 | 4.3E–06 | 6.1E–03 |
| ENSMUSG00000031167 | Rbm3 | 005 | chrX | 7719487 | 7719493 | − | 1.7 | 4.7 | 4.4E–06 | 6.1E–03 |
| ENSMUSG00000047617 | BC029214 | 024 | chr2 | 25316142 | 25316174 | − | −0.7 | −4.7 | 4.4E–06 | 6.1E–03 |
| ENSMUSG00000030447 | Cyfip1 | 051 | chr7 | 63185842 | 63185868 | + | −0.8 | −4.6 | 4.5E–06 | 6.3E–03 |
| ENSMUSG00000013593 | Ndufs2 | 015 | chr1 | 173170159 | 173170186 | − | 0.8 | 4.7 | 4.7E–06 | 6.4E–03 |
| ENSMUSG00000000441 | Raf1 | 007 | chr6 | 115570346 | 115571833 | − | 0.2 | 4.7 | 4.7E–06 | 6.5E–03 |
| ENSMUSG00000027001 + ENSMUSG00000026999 | Dusp19/Nup35 | 032 | chr2 | 80496235 | 80497345 | + | −0.9 | −4.7 | 5.0E–06 | 6.7E–03 |
| ENSMUSG00000038822 | Hace1 | 034 | chr10 | 45420831 | 45429686 | + | −0.3 | −4.6 | 5.0E–06 | 6.7E–03 |
| ENSMUSG00000073174 + ENSMUSG00000040003 | Magi2/Magi2 | 030 | chr5 | 20208194 | 20208297 | + | 0.8 | 4.7 | 5.0E–06 | 6.7E–03 |
| ENSMUSG00000025487 | Psmd13 | 024 | chr7 | 148076311 | 148076393 | + | 0.8 | 4.6 | 5.0E–06 | 6.7E–03 |
| ENSMUSG00000036067 | Slc2a6 | 009 | chr2 | 26879856 | 26880104 | − | −0.6 | −4.7 | 5.1E–06 | 6.8E–03 |
| ENSMUSG00000057236 | Rbbp4 | 016 | chr4 | 129002068 | 129005831 | − | 0.5 | 4.7 | 5.1E–06 | 6.8E–03 |
| ENSMUSG00000031511 | Arhgef7 | 028 | chr8 | 11830238 | 11831492 | + | 0.3 | 4.7 | 5.1E–06 | 6.8E–03 |
| ENSMUSG00000030207 | 8430419L09Rik | 017 | chr6 | 135182873 | 135183273 | + | −0.4 | −4.7 | 5.2E–06 | 6.8E–03 |
| ENSMUSG00000026977 | March7 | 025 | chr2 | 60085570 | 60085946 | + | 0.6 | 4.7 | 5.3E–06 | 6.9E–03 |
| ENSMUSG00000028782 | Bai2 | 046 | chr4 | 129698499 | 129698806 | + | 0.3 | 4.6 | 5.4E–06 | 7.1E–03 |
| ENSMUSG00000021772 | Nkiras1 | 014 | chr14 | 19109961 | 19110079 | + | 0.7 | 4.7 | 5.5E–06 | 7.1E–03 |
| ENSMUSG00000027674 | Pex5l | 032 | chr3 | 33042005 | 33042169 | − | 0.7 | 4.6 | 5.5E–06 | 7.1E–03 |
| ENSMUSG00000074886 | Grk6 | 001 | chr13 | 55546695 | 55546921 | + | 0.5 | 4.7 | 5.5E–06 | 7.1E–03 |
| ENSMUSG00000031392 + ENSMUSG00000076127 + ENSMUSG00000092907 |
Irak1/Mir718/ Mir5132 |
057 | chrX | 71269166 | 71269170 | − | 1.9 | 4.6 | 5.6E–06 | 7.2E–03 |
| ENSMUSG00000025134 | Alyref | 011 | chr11 | 120459546 | 120459679 | − | 1.4 | 4.8 | 5.6E–06 | 7.2E–03 |
| ENSMUSG00000023952 | Gtpbp2 | 036 | chr17 | 46303937 | 46304171 | + | 0.5 | 4.6 | 5.7E–06 | 7.2E–03 |
| ENSMUSG00000024392 | Bag6 | 034 | chr17 | 35278339 | 35278370 | + | 0.8 | 4.6 | 5.7E–06 | 7.2E–03 |
| ENSMUSG00000005469 | Prkaca | 001 | chr8 | 86496877 | 86497131 | + | 0.5 | 4.8 | 5.9E–06 | 7.3E–03 |
| ENSMUSG00000031167 | Rbm3 | 004 | chrX | 7719485 | 7719486 | − | 1.7 | 4.6 | 5.9E–06 | 7.3E–03 |
| ENSMUSG00000031878 | Nae1 | 008 | chr8 | 107040635 | 107040889 | − | 0.7 | 4.6 | 6.1E–06 | 7.6E–03 |
| ENSMUSG00000008348 | Ubc | 003 | chr5 | 125866669 | 125866896 | − | −0.3 | −4.8 | 6.1E–06 | 7.6E–03 |
| ENSMUSG00000022514 | Il1rap | 019 | chr16 | 26722520 | 26723017 | + | 0.3 | 4.6 | 6.1E–06 | 7.6E–03 |
| ENSMUSG00000001847 | Rac1 | 011 | chr5 | 144288631 | 144288861 | − | 0.5 | 4.7 | 6.3E–06 | 7.7E–03 |
| ENSMUSG00000038244 | Mical2 | 037 | chr7 | 119497490 | 119498460 | + | 0.2 | 4.6 | 6.4E–06 | 7.8E–03 |
| ENSMUSG00000027429 | Sec23b | 030 | chr2 | 144405140 | 144406851 | + | 0.7 | 4.6 | 6.5E–06 | 7.9E–03 |
| ENSMUSG00000042726 | Trafd1 | 024 | chr5 | 121835049 | 121835317 | − | 0.6 | 4.6 | 6.7E–06 | 8.1E–03 |
| ENSMUSG00000092367 + ENSMUSG00000011751 + ENSMUSG00000089832 |
Gm20479/ Spnb4/Shkbp1 |
007 | chr7 | 28127780 | 28127940 | − | 0.4 | 4.5 | 6.8E–06 | 8.1E–03 |
| ENSMUSG00000003808 | Farsa | 027 | chr8 | 87391986 | 87392122 | + | 0.4 | 4.6 | 6.8E–06 | 8.1E–03 |
| ENSMUSG00000049327 | Setd8 | 006 | chr5 | 124895592 | 124895672 | + | 1.4 | 4.6 | 6.8E–06 | 8.1E–03 |
| ENSMUSG00000006392 | Med8 | 007 | chr4 | 118082953 | 118083489 | + | 0.6 | 4.6 | 6.8E–06 | 8.1E–03 |
| ENSMUSG00000031167 | Rbm3 | 006 | chrX | 7719494 | 7719666 | − | 1.7 | 4.6 | 7.6E–06 | 8.9E–03 |
| ENSMUSG00000033184 + ENSMUSG00000056130 | Tmed7/Ticam2 | 012 | chr18 | 46756969 | 46757170 | − | 0.6 | 4.7 | 7.9E–06 | 9.2E–03 |
| ENSMUSG00000032301 | Psma4 | 007 | chr9 | 54799151 | 54799235 | + | −0.8 | −4.6 | 7.9E–06 | 9.3E–03 |
| ENSMUSG00000007670 | Khsrp | 026 | chr17 | 57170571 | 57170930 | − | 0.5 | 4.6 | 8.2E–06 | 9.5E–03 |
| ENSMUSG00000034064 | Poglut1 | 008 | chr16 | 38531927 | 38532578 | − | 0.5 | 4.6 | 8.2E–06 | 9.6E–03 |
| ENSMUSG00000022514 | Il1rap | 020 | chr16 | 26723018 | 26723306 | + | 0.5 | 4.5 | 8.6E–06 | 9.9E–03 |
| ENSMUSG00000047617 | BC029214 | 023 | chr2 | 25316015 | 25316141 | − | −0.5 | −4.5 | 8.7E–06 | 1.0E–02 |
| ENSMUSG00000026209 | Dnpep | 004 | chr1 | 75305285 | 75305873 | − | 0.4 | 4.6 | 8.7E–06 | 1.0E–02 |
| ENSMUSG00000029047 | Pex10 | 012 | chr4 | 154443226 | 154444495 | + | 0.6 | 4.6 | 9.0E–06 | 1.0E–02 |
| ENSMUSG00000026103 | Gls | 052 | chr1 | 52289656 | 52290067 | − | 0.4 | 4.5 | 9.1E–06 | 1.0E–02 |
| ENSMUSG00000006958 | Chrd | 016 | chr16 | 20736516 | 20736981 | + | −0.4 | −4.5 | 9.1E–06 | 1.0E–02 |
| ENSMUSG00000023952 | Gtpbp2 | 031 | chr17 | 46302947 | 46303259 | + | 0.4 | 4.5 | 9.2E–06 | 1.0E–02 |
| ENSMUSG00000020376 | Rnf130 | 001 | chr11 | 49838848 | 49838849 | + | 2.8 | 4.6 | 9.4E–06 | 1.1E–02 |
| ENSMUSG00000022433 | Csnk1e | 035 | chr15 | 79272430 | 79272432 | − | 0.8 | 4.5 | 9.4E–06 | 1.1E–02 |
| ENSMUSG00000018501 | Ncor1 | 099 | chr11 | 62270650 | 62270825 | − | 0.8 | 4.5 | 9.5E–06 | 1.1E–02 |
| ENSMUSG00000079737 + ENSMUSG00000022684 | 3110001I22Rik/ Bfar |
006 | chr16 | 13676927 | 13678498 | + | 0.5 | 4.6 | 9.6E–06 | 1.1E–02 |
| ENSMUSG00000030852 | Tacc2 | 003 | chr7 | 137765235 | 137770355 | + | 1.3 | 4.5 | 9.6E–06 | 1.1E–02 |
| ENSMUSG00000020684 | Rasl10b | 003 | chr11 | 83223190 | 83223383 | + | 0.6 | 4.7 | 9.7E–06 | 1.1E–02 |
| ENSMUSG00000030058 | Copg | 028 | chr6 | 87852761 | 87853478 | + | 0.4 | 4.5 | 9.7E–06 | 1.1E–02 |
| ENSMUSG00000023984 + ENSMUSG00000090115 + ENSMUSG00000092558 |
Gm20517/ Usp49/Med20 |
013 | chr17 | 47756513 | 47759833 | + | 0.4 | 4.5 | 9.9E–06 | 1.1E–02 |
| ENSMUSG00000090213 + ENSMUSG00000089739 + ENSMUSG00000078923 |
Tmem189/ Gm20431/ Ube2v1 |
032 | chr2 | 167487055 | 167487056 | − | 1.9 | 4.5 | 9.9E–06 | 1.1E–02 |
| ENSMUSG00000071984 | Fndc1 | 001 | chr17 | 7931434 | 7932195 | − | 0.3 | 4.5 | 1.0E–05 | 1.1E–02 |
| ENSMUSG00000027674 | Pex5l | 031 | chr3 | 33041999 | 33042004 | − | 0.7 | 4.5 | 1.1E–05 | 1.1E–02 |
| ENSMUSG00000029550 | Sppl3 | 002 | chr5 | 115461155 | 115461624 | + | 0.4 | 4.5 | 1.1E–05 | 1.1E–02 |
| ENSMUSG00000036940 | Kdm1a | 039 | chr4 | 136158224 | 136158602 | − | 0.5 | 4.5 | 1.1E–05 | 1.1E–02 |
| ENSMUSG00000030603 | Psmc4 | 021 | chr7 | 28834222 | 28834719 | − | 0.4 | 4.5 | 1.1E–05 | 1.2E–02 |
| ENSMUSG00000050357 | Rltpr | 040 | chr8 | 108219675 | 108220760 | + | 0.3 | 4.5 | 1.1E–05 | 1.2E–02 |
| ENSMUSG00000028041 | Adam15 | 014 | chr3 | 89144102 | 89144274 | − | 0.4 | 4.4 | 1.1E–05 | 1.2E–02 |
| ENSMUSG00000091509 + ENSMUSG00000022119 | Gm17066/ Rbm26 |
013 | chr14 | 105515352 | 105516202 | − | −0.7 | −4.5 | 1.1E–05 | 1.2E–02 |
| ENSMUSG00000022456 | Sept3 | 002 | chr15 | 82105677 | 82105916 | + | 0.3 | 4.6 | 1.1E–05 | 1.2E–02 |
| ENSMUSG00000030204 | Ddx47 | 025 | chr6 | 134969130 | 134970633 | + | −0.4 | −4.5 | 1.2E–05 | 1.2E–02 |
| ENSMUSG00000023952 | Gtpbp2 | 045 | chr17 | 46304971 | 46305094 | + | 0.3 | 4.4 | 1.2E–05 | 1.2E–02 |
| ENSMUSG00000021018 | Polr2h | 002 | chr16 | 20718245 | 20718742 | + | 0.6 | 4.7 | 1.2E–05 | 1.2E–02 |
| ENSMUSG00000015467 + ENSMUSG00000015474 + ENSMUSG00000092176 |
Egfl8/Ppt2/ Gm20460 |
020 | chr17 | 34752267 | 34752276 | − | 2.4 | 4.4 | 1.2E–05 | 1.2E–02 |
| ENSMUSG00000039219 | Arid4b | 044 | chr13 | 14284369 | 14285632 | + | −0.5 | −4.4 | 1.3E–05 | 1.4E–02 |
| ENSMUSG00000057672 | Pkn1 | 043 | chr8 | 86223035 | 86223066 | − | 1.4 | 4.4 | 1.3E–05 | 1.4E–02 |
| ENSMUSG00000034863 | Ano8 | 001 | chr8 | 73999922 | 74000820 | − | 0.3 | 4.5 | 1.3E–05 | 1.4E–02 |
| ENSMUSG00000003778 | Brd8 | 025 | chr18 | 34771145 | 34772690 | − | −1.1 | −4.4 | 1.3E–05 | 1.4E–02 |
| ENSMUSG00000004947 | Dtx2 | 015 | chr5 | 136495428 | 136497624 | + | 0.8 | 4.4 | 1.3E–05 | 1.4E–02 |
| ENSMUSG00000068917 | Clk2 | 019 | chr3 | 88976087 | 88977298 | + | 0.8 | 4.4 | 1.4E–05 | 1.4E–02 |
| ENSMUSG00000064267 | Hvcn1 | 013 | chr5 | 122688520 | 122688646 | + | 0.5 | 4.5 | 1.4E–05 | 1.4E–02 |
| ENSMUSG00000057788 | Ddx49 | 011 | chr8 | 72819719 | 72820281 | − | 0.4 | 4.5 | 1.4E–05 | 1.4E–02 |
| ENSMUSG00000057672 | Pkn1 | 042 | chr8 | 86222997 | 86223034 | − | 1.2 | 4.4 | 1.4E–05 | 1.4E–02 |
| ENSMUSG00000027710 | Acad9 | 015 | chr3 | 35979219 | 35979628 | + | 0.8 | 4.4 | 1.5E–05 | 1.4E–02 |
| ENSMUSG00000031447 | Lamp1 | 001 | chr8 | 13159135 | 13159434 | + | 0.6 | 4.6 | 1.5E–05 | 1.4E–02 |
| ENSMUSG00000039470 | Zdhhc2 | 005 | chr8 | 41509213 | 41509353 | + | 0.9 | 4.4 | 1.5E–05 | 1.4E–02 |
| ENSMUSG00000021493 | Pdlim7 | 021 | chr13 | 55609242 | 55609568 | − | 0.3 | 4.4 | 1.5E–05 | 1.4E–02 |
| ENSMUSG00000071793 | 2610005L07Rik | 002 | chr8 | 19981360 | 19983980 | − | 0.3 | 4.5 | 1.5E–05 | 1.4E–02 |
| ENSMUSG00000044060 + ENSMUSG00000089798 | A830010M20Rik/ 1700028K03Rik |
003 | chr5 | 107926397 | 107926739 | + | 1.6 | 4.4 | 1.5E–05 | 1.4E–02 |
| ENSMUSG00000058301 | Upf1 | 024 | chr8 | 72876677 | 72877172 | − | 0.4 | 4.4 | 1.5E–05 | 1.5E–02 |
| ENSMUSG00000008976 | Gabpa | 011 | chr16 | 84856285 | 84856706 | + | 1.1 | 4.4 | 1.5E–05 | 1.5E–02 |
| ENSMUSG00000007617 | Homer1 | 016 | chr13 | 94136198 | 94136232 | + | 1.6 | 4.4 | 1.5E–05 | 1.5E–02 |
| ENSMUSG00000074247 | Dda1 | 013 | chr8 | 73996515 | 73996681 | + | 0.6 | 4.5 | 1.5E–05 | 1.5E–02 |
| ENSMUSG00000029478 | Ncor2 | 011 | chr5 | 125503295 | 125503590 | − | 1.3 | 4.3 | 1.6E–05 | 1.5E–02 |
| ENSMUSG00000055805 | Fmnl1 | 035 | chr11 | 103059093 | 103059190 | + | 0.4 | 4.4 | 1.6E–05 | 1.5E–02 |
| ENSMUSG00000036459 | Wtip | 014 | chr7 | 34917838 | 34918287 | − | 1.5 | 4.5 | 1.6E–05 | 1.5E–02 |
| ENSMUSG00000028041 | Adam15 | 027 | chr3 | 89146654 | 89147034 | − | 0.5 | 4.4 | 1.6E–05 | 1.6E–02 |
| ENSMUSG00000035569 | Ankrd11 | 023 | chr8 | 125565849 | 125565897 | − | 0.7 | 4.4 | 1.6E–05 | 1.6E–02 |
| ENSMUSG00000024068 | Spast | 001 | chr17 | 74738327 | 74738785 | + | 0.4 | 4.4 | 1.7E–05 | 1.6E–02 |
| ENSMUSG00000038502 | Ptov1 | 035 | chr7 | 52124721 | 52125158 | − | 0.3 | 4.4 | 1.7E–05 | 1.6E–02 |
| ENSMUSG00000002010 | Idh3g | 029 | chrX | 71027708 | 71028013 | − | 0.4 | 4.4 | 1.7E–05 | 1.6E–02 |
| ENSMUSG00000028967 | Errfi1 | 003 | chr4 | 150229577 | 150229923 | + | −1.6 | −4.6 | 1.7E–05 | 1.6E–02 |
| ENSMUSG00000089704 + ENSMUSG00000031826 + ENSMUSG00000092329 |
Galnt2/Usp10/ Gm20388 |
031 | chr8 | 122478834 | 122479217 | + | 0.5 | 4.3 | 1.8E–05 | 1.7E–02 |
| ENSMUSG00000022514 | Il1rap | 027 | chr16 | 26728019 | 26728314 | + | −0.5 | −4.4 | 1.9E–05 | 1.7E–02 |
| ENSMUSG00000089012 + ENSMUSG00000017421 | /Zfp207 | 026 | chr11 | 80211918 | 80211972 | + | −0.4 | −4.4 | 1.9E–05 | 1.7E–02 |
| ENSMUSG00000031392 + ENSMUSG00000076127 + ENSMUSG00000092907 |
Irak1/Mir718/ Mir5132 |
058 | chrX | 71269171 | 71269187 | − | 1.9 | 4.3 | 1.9E–05 | 1.7E–02 |
| ENSMUSG00000045009 | Prrt3 | 002 | chr6 | 113446294 | 113446925 | − | 0.5 | 4.6 | 1.9E–05 | 1.7E–02 |
| ENSMUSG00000022710 | Usp7 | 020 | chr16 | 8697013 | 8697568 | − | 0.5 | 4.3 | 1.9E–05 | 1.8E–02 |
| ENSMUSG00000037300 | Ttc13 | 021 | chr8 | 127206060 | 127206430 | − | −0.5 | −4.3 | 1.9E–05 | 1.8E–02 |
| ENSMUSG00000089661 + ENSMUSG00000053291 + ENSMUSG00000093348 |
Mia1/Rab4b/ Mir3101 |
022 | chr7 | 27961113 | 27961176 | − | −0.6 | −4.3 | 1.9E–05 | 1.8E–02 |
| ENSMUSG00000050640 | Tmem150c | 004 | chr5 | 100512966 | 100515024 | − | −0.3 | −4.4 | 2.0E–05 | 1.8E–02 |
| ENSMUSG00000027692 | Tnik | 046 | chr3 | 28565151 | 28565371 | + | −0.5 | −4.3 | 2.0E–05 | 1.8E–02 |
| ENSMUSG00000092367 + ENSMUSG00000011751 + ENSMUSG00000089832 |
Gm20479/ Spnb4/Shkbp1 |
015 | chr7 | 28132192 | 28132473 | − | 0.7 | 4.3 | 2.0E–05 | 1.8E–02 |
| ENSMUSG00000029064 | Gnb1 | 001 | chr4 | 154865470 | 154865478 | + | 1.9 | 4.4 | 2.1E–05 | 1.8E–02 |
| ENSMUSG00000032737 | Inppl1 | 010 | chr7 | 108976713 | 108976802 | − | −0.4 | −4.3 | 2.1E–05 | 1.8E–02 |
| ENSMUSG00000044857 | Lemd2 | 009 | chr17 | 27340604 | 27341383 | − | 0.6 | 4.5 | 2.1E–05 | 1.8E–02 |
| ENSMUSG00000055313 | Pgbd1 | 008 | chr13 | 21526220 | 21526540 | − | −1.2 | −4.4 | 2.1E–05 | 1.8E–02 |
| ENSMUSG00000021910 | Nisch | 032 | chr14 | 31994136 | 31994656 | − | 0.3 | 4.3 | 2.1E–05 | 1.8E–02 |
| ENSMUSG00000025060 | Slk | 001 | chr19 | 47654168 | 47654508 | + | 1.2 | 4.3 | 2.1E–05 | 1.9E–02 |
| ENSMUSG00000052423 | B4galt3 | 011 | chr1 | 173201123 | 173201236 | + | −0.6 | −4.3 | 2.2E–05 | 1.9E–02 |
| ENSMUSG00000071644 | Eef1g | 008 | chr19 | 9044478 | 9044691 | + | −0.8 | −4.4 | 2.2E–05 | 1.9E–02 |
| ENSMUSG00000001763 | Tspan33 | 001 | chr6 | 29644222 | 29644233 | + | 1.7 | 4.4 | 2.2E–05 | 1.9E–02 |
| ENSMUSG00000022656 | Pvrl3 | 027 | chr16 | 46496966 | 46497080 | − | 2.0 | 4.3 | 2.2E–05 | 1.9E–02 |
| ENSMUSG00000035901 | Dennd5a | 034 | chr7 | 117103791 | 117103837 | − | 1.1 | 4.3 | 2.2E–05 | 1.9E–02 |
| ENSMUSG00000019854 | Reps1 | 002 | chr10 | 17775712 | 17775745 | + | 0.8 | 4.3 | 2.2E–05 | 1.9E–02 |
| ENSMUSG00000000568 | Hnrnpd | 009 | chr5 | 100391153 | 100391223 | − | 0.4 | 4.3 | 2.2E–05 | 1.9E–02 |
| ENSMUSG00000089715 + ENSMUSG00000089837 + ENSMUSG00000022421 |
Cbx6/Npcd/ Nptxr |
012 | chr15 | 79634889 | 79635058 | − | 0.9 | 4.3 | 2.3E–05 | 1.9E–02 |
| ENSMUSG00000026839 | Upp2 | 002 | chr2 | 58419729 | 58419797 | + | 0.9 | 4.4 | 2.3E–05 | 2.0E–02 |
| ENSMUSG00000026426 | Arl8a | 001 | chr1 | 137043401 | 137043677 | + | 0.2 | 4.4 | 2.3E–05 | 2.0E–02 |
| ENSMUSG00000028381 | Ugcg | 001 | chr4 | 59202129 | 59202429 | + | 1.7 | 4.4 | 2.4E–05 | 2.0E–02 |
| ENSMUSG00000041506 | Rrp9 | 023 | chr9 | 106386851 | 106387407 | + | 0.4 | 4.3 | 2.5E–05 | 2.1E–02 |
| ENSMUSG00000054934 | Kcnmb4 | 001 | chr10 | 115854917 | 115855462 | − | −0.6 | −4.5 | 2.5E–05 | 2.1E–02 |
| ENSMUSG00000063160 + ENSMUSG00000003762 | Numbl/Adck4 | 037 | chr7 | 28047272 | 28049894 | + | 0.3 | 4.3 | 2.5E–05 | 2.1E–02 |
| ENSMUSG00000072770 + ENSMUSG00000030330 | Acrbp/Ing4 | 037 | chr6 | 125001261 | 125001378 | + | 0.5 | 4.3 | 2.6E–05 | 2.2E–02 |
| ENSMUSG00000024182 | Axin1 | 004 | chr17 | 26275848 | 26275894 | + | 1.3 | 4.3 | 2.6E–05 | 2.2E–02 |
| ENSMUSG00000024835 | Coro1b | 021 | chr19 | 4152580 | 4152808 | + | −0.3 | −4.3 | 2.6E–05 | 2.2E–02 |
| ENSMUSG00000033059 | Pygb | 019 | chr2 | 150649343 | 150649711 | + | 0.8 | 4.3 | 2.6E–05 | 2.2E–02 |
| ENSMUSG00000025571 | Tnrc6c | 016 | chr11 | 117603815 | 117604240 | + | −0.6 | −4.3 | 2.6E–05 | 2.2E–02 |
| ENSMUSG00000035953 | Tmem55b | 005 | chr14 | 51547713 | 51548495 | − | 0.3 | 4.3 | 2.6E–05 | 2.2E–02 |
| ENSMUSG00000059248 | Sept9 | 021 | chr11 | 117218024 | 117218313 | + | 0.9 | 4.3 | 2.7E–05 | 2.2E–02 |
| ENSMUSG00000004846 + ENSMUSG00000076240 | Plod3/Mir702 | 033 | chr5 | 137470929 | 137471047 | + | 0.7 | 4.3 | 2.7E–05 | 2.2E–02 |
| ENSMUSG00000039069 | Gtpbp5 | 017 | chr2 | 179818166 | 179818768 | + | 0.4 | 4.3 | 2.8E–05 | 2.2E–02 |
| ENSMUSG00000031979 | Cog2 | 014 | chr8 | 127066851 | 127067909 | + | 0.4 | 4.3 | 2.8E–05 | 2.3E–02 |
| ENSMUSG00000031392 + ENSMUSG00000076127 + ENSMUSG00000092907 |
Irak1/Mir718/ Mir5132 |
055 | chrX | 71269085 | 71269147 | − | 0.9 | 4.2 | 2.8E–05 | 2.3E–02 |
| ENSMUSG00000009207 | Lnp | 022 | chr2 | 74389952 | 74393044 | − | −0.7 | −4.2 | 2.8E–05 | 2.3E–02 |
| ENSMUSG00000030374 | Strn4 | 021 | chr7 | 17417980 | 17418274 | + | 0.3 | 4.2 | 2.8E–05 | 2.3E–02 |
| ENSMUSG00000019927 | Ube2d1 | 008 | chr10 | 70722568 | 70724831 | − | 0.3 | 4.4 | 2.8E–05 | 2.3E–02 |
| ENSMUSG00000052949 | Rnf157 | 037 | chr11 | 116274319 | 116274346 | − | 1.7 | 4.2 | 2.9E–05 | 2.4E–02 |
| ENSMUSG00000036052 | Dnajb5 | 003 | chr4 | 42962786 | 42962876 | + | −0.4 | −4.3 | 3.0E–05 | 2.4E–02 |
| ENSMUSG00000040048 | Ndufb10 | 002 | chr17 | 24859166 | 24859325 | − | 0.5 | 4.3 | 3.0E–05 | 2.4E–02 |
| ENSMUSG00000038848 | Ythdf1 | 009 | chr2 | 180655389 | 180655654 | − | 0.8 | 4.4 | 3.0E–05 | 2.4E–02 |
| ENSMUSG00000020610 | Amz2 | 020 | chr11 | 109298566 | 109299462 | + | 0.2 | 4.3 | 3.0E–05 | 2.4E–02 |
| ENSMUSG00000075876 + ENSMUSG00000064791 + ENSMUSG00000075924 + ENSMUSG00000015656 |
Snord14c/ Snord14e/ Snord14d/Hspa8 |
024 | chr9 | 40611297 | 40611462 | + | −0.6 | −4.2 | 3.1E–05 | 2.4E–02 |
| ENSMUSG00000063659 | Zfp238 | 002 | chr1 | 179375952 | 179377219 | + | 0.4 | 4.7 | 3.1E–05 | 2.4E–02 |
| ENSMUSG00000004865 | Srpk1 | 026 | chr17 | 28757088 | 28757398 | − | 2.6 | 4.2 | 3.1E–05 | 2.4E–02 |
| ENSMUSG00000002325 + ENSMUSG00000047098 | Irf9/Rnf31 | 062 | chr14 | 56227758 | 56228864 | + | 0.3 | 4.2 | 3.1E–05 | 2.4E–02 |
| ENSMUSG00000038244 | Mical2 | 012 | chr7 | 119459543 | 119461899 | + | −0.5 | −4.2 | 3.1E–05 | 2.5E–02 |
| ENSMUSG00000041740 | Rnf10 | 012 | chr5 | 115693701 | 115694029 | − | 0.6 | 4.2 | 3.2E–05 | 2.5E–02 |
| ENSMUSG00000045205 | Dpy19l4 | 001 | chr4 | 11188462 | 11192225 | − | 0.3 | 4.2 | 3.2E–05 | 2.5E–02 |
| ENSMUSG00000026860 | Sh3glb2 | 023 | chr2 | 30205260 | 30205807 | − | 0.4 | 4.2 | 3.2E–05 | 2.5E–02 |
| ENSMUSG00000022974 | Gcfc1 | 033 | chr16 | 91044251 | 91044409 | − | 0.7 | 4.2 | 3.2E–05 | 2.5E–02 |
| ENSMUSG00000028863 | Meaf6 | 014 | chr4 | 124780674 | 124780747 | + | 0.6 | 4.2 | 3.2E–05 | 2.5E–02 |
| ENSMUSG00000052997 | Uba2 | 007 | chr7 | 34927894 | 34928288 | − | 0.5 | 4.2 | 3.3E–05 | 2.5E–02 |
| ENSMUSG00000001911 | Nfix | 031 | chr8 | 87324040 | 87324243 | − | 2.5 | 4.2 | 3.3E–05 | 2.5E–02 |
| ENSMUSG00000034656 | Cacna1a | 067 | chr8 | 87163145 | 87163333 | + | 0.4 | 4.2 | 3.3E–05 | 2.5E–02 |
| ENSMUSG00000019843 | Fyn | 004 | chr10 | 39089609 | 39089996 | + | 0.3 | 4.2 | 3.3E–05 | 2.5E–02 |
| ENSMUSG00000031167 | Rbm3 | 001 | chrX | 7716101 | 7716103 | − | 2.0 | 4.2 | 3.6E–05 | 2.7E–02 |
| ENSMUSG00000022514 | Il1rap | 026 | chr16 | 26727733 | 26728018 | + | −0.5 | −4.2 | 3.6E–05 | 2.8E–02 |
| ENSMUSG00000004233 | Wars2 | 014 | chr3 | 99039402 | 99040231 | + | 2.3 | 4.2 | 3.7E–05 | 2.8E–02 |
| ENSMUSG00000024571 + ENSMUSG00000057130 | Gm16286/Txnl4a | 017 | chr18 | 80404083 | 80404423 | + | −0.7 | −4.2 | 3.7E–05 | 2.8E–02 |
| ENSMUSG00000020978 | Klhdc2 | 002 | chr12 | 70397709 | 70397741 | + | 0.7 | 4.2 | 3.7E–05 | 2.8E–02 |
| ENSMUSG00000024335 | Brd2 | 037 | chr17 | 34258693 | 34258933 | − | 2.3 | 4.2 | 3.8E–05 | 2.9E–02 |
| ENSMUSG00000026155 | Smap1 | 025 | chr1 | 23928877 | 23929156 | − | 0.3 | 4.2 | 3.8E–05 | 2.9E–02 |
| ENSMUSG00000004561 + ENSMUSG00000072572 | Mettl17/Slc39a2 | 055 | chr14 | 52514182 | 52514573 | + | 0.3 | 4.2 | 3.8E–05 | 2.9E–02 |
| ENSMUSG00000019254 | Ppp1r12c | 053 | chr7 | 4453267 | 4453282 | − | 1.1 | 4.2 | 3.9E–05 | 2.9E–02 |
| ENSMUSG00000040331 | Nsmce4a | 020 | chr7 | 137690506 | 137690895 | − | 0.3 | 4.2 | 3.9E–05 | 2.9E–02 |
| ENSMUSG00000005882 | Uqcc | 030 | chr2 | 155736341 | 155737476 | − | −0.7 | −4.2 | 4.0E–05 | 2.9E–02 |
| ENSMUSG00000021234 | Fam161b | 004 | chr12 | 85687442 | 85687736 | − | 0.6 | 4.2 | 4.0E–05 | 3.0E–02 |
| ENSMUSG00000008153 | Clstn3 | 005 | chr6 | 124386790 | 124386835 | − | 1.0 | 4.2 | 4.1E–05 | 3.0E–02 |
| ENSMUSG00000021610 | Clptm1l | 001 | chr13 | 73741554 | 73741847 | + | 0.2 | 4.2 | 4.3E–05 | 3.1E–02 |
| ENSMUSG00000027303 | Ptpra | 002 | chr2 | 130276280 | 130276294 | + | 0.5 | 4.2 | 4.3E–05 | 3.2E–02 |
| ENSMUSG00000038546 | Ranbp9 | 017 | chr13 | 43576247 | 43576297 | − | 1.7 | 4.2 | 4.4E–05 | 3.2E–02 |
| ENSMUSG00000030374 | Strn4 | 020 | chr7 | 17416930 | 17416988 | + | 0.5 | 4.1 | 4.4E–05 | 3.2E–02 |
| ENSMUSG00000025198 | Erlin1 | 024 | chr19 | 44144181 | 44144186 | − | −1.0 | −4.2 | 4.4E–05 | 3.2E–02 |
| ENSMUSG00000031292 | Cdkl5 | 021 | chrX | 157432523 | 157432634 | − | −1.8 | −4.2 | 4.5E–05 | 3.3E–02 |
| ENSMUSG00000086285 | D630044L22Rik | 006 | chr17 | 26099007 | 26099155 | + | −1.4 | −4.3 | 4.6E–05 | 3.3E–02 |
| ENSMUSG00000019189 | Rnf145 | 012 | chr11 | 44365322 | 44368479 | + | −0.5 | −4.2 | 4.6E–05 | 3.3E–02 |
| ENSMUSG00000028849 | Mtap7d1 | 017 | chr4 | 125914140 | 125914303 | − | 0.2 | 4.1 | 4.6E–05 | 3.3E–02 |
| ENSMUSG00000027642 | Rpn2 | 034 | chr2 | 157147897 | 157148207 | + | 0.4 | 4.1 | 4.6E–05 | 3.3E–02 |
| ENSMUSG00000053907 | Mat2a | 002 | chr6 | 72384390 | 72384538 | − | 0.3 | 4.2 | 4.6E–05 | 3.3E–02 |
| ENSMUSG00000001416 | Cct3 | 008 | chr3 | 88103326 | 88104721 | + | 0.5 | 4.1 | 4.6E–05 | 3.3E–02 |
| ENSMUSG00000022265 | Ank | 001 | chr15 | 27396432 | 27396854 | + | 0.3 | 4.2 | 4.6E–05 | 3.3E–02 |
| ENSMUSG00000025103 | Btbd1 | 008 | chr7 | 88973859 | 88974317 | − | 0.3 | 4.3 | 4.7E–05 | 3.3E–02 |
| ENSMUSG00000085609 | 1700016P03Rik | 002 | chr11 | 74987480 | 74987663 | + | 1.4 | 4.6 | 4.7E–05 | 3.3E–02 |
| ENSMUSG00000044783 | Hjurp | 023 | chr1 | 90171673 | 90173793 | − | 0.5 | 4.1 | 4.7E–05 | 3.3E–02 |
| ENSMUSG00000031245 | Hmgn5 | 001 | chrX | 106199873 | 106201410 | − | 0.4 | 4.4 | 4.7E–05 | 3.3E–02 |
| ENSMUSG00000034135 | Sik3 | 002 | chr9 | 45820947 | 45821082 | + | 0.8 | 4.1 | 4.8E–05 | 3.3E–02 |
| ENSMUSG00000029364 | Wsb2 | 001 | chr5 | 117807313 | 117807313 | + | 2.2 | 4.2 | 4.8E–05 | 3.4E–02 |
| ENSMUSG00000000568 | Hnrnpd | 008 | chr5 | 100391117 | 100391152 | − | 0.4 | 4.1 | 4.9E–05 | 3.4E–02 |
| ENSMUSG00000028347 | Tmeff1 | 002 | chr4 | 48598065 | 48598236 | + | 1.1 | 4.2 | 4.9E–05 | 3.4E–02 |
| ENSMUSG00000025198 | Erlin1 | 025 | chr19 | 44144187 | 44144234 | − | −1.0 | −4.1 | 4.9E–05 | 3.4E–02 |
| ENSMUSG00000033904 | 6330503K22Rik | 022 | chr7 | 125876487 | 125878781 | + | 0.3 | 4.1 | 4.9E–05 | 3.4E–02 |
| ENSMUSG00000023087 | Ccrn4l | 003 | chr3 | 51044128 | 51051726 | + | 0.4 | 4.3 | 4.9E–05 | 3.4E–02 |
| ENSMUSG00000067713 | Prkag1 | 001 | chr15 | 98643229 | 98643911 | − | −0.3 | −4.2 | 4.9E–05 | 3.4E–02 |
| ENSMUSG00000090266 + ENSMUSG00000020818 | 1110005A03Rik/ Mfsd11 |
033 | chr11 | 116719896 | 116720427 | + | −0.8 | −4.1 | 5.0E–05 | 3.4E–02 |
| ENSMUSG00000028412 | Slc44a1 | 002 | chr4 | 53453543 | 53453555 | + | 0.7 | 4.1 | 5.0E–05 | 3.4E–02 |
| ENSMUSG00000027351 | Spred1 | 001 | chr2 | 116947110 | 116947373 | + | 0.8 | 4.3 | 5.0E–05 | 3.4E–02 |
| ENSMUSG00000028047 | Thbs3 | 020 | chr3 | 89025074 | 89025135 | + | 1.1 | 4.1 | 5.1E–05 | 3.5E–02 |
| ENSMUSG00000003279 | Dlgap1 | 005 | chr17 | 70318742 | 70318760 | + | 0.6 | 4.1 | 5.3E–05 | 3.6E–02 |
| ENSMUSG00000030584 | Dpf1 | 006 | chr7 | 30093118 | 30093973 | + | 0.5 | 4.1 | 5.3E–05 | 3.6E–02 |
| ENSMUSG00000045994 | B3gat1 | 002 | chr9 | 26541388 | 26541408 | + | 1.9 | 4.1 | 5.3E–05 | 3.6E–02 |
| ENSMUSG00000003360 | Ddx23 | 020 | chr15 | 98482970 | 98483318 | − | 0.5 | 4.1 | 5.4E–05 | 3.6E–02 |
| ENSMUSG00000004994 | Ccdc130 | 011 | chr8 | 86785700 | 86785805 | − | 0.9 | 4.1 | 5.4E–05 | 3.6E–02 |
| ENSMUSG00000063873 | Slc24a3 | 002 | chr2 | 144993655 | 144993933 | + | 0.4 | 4.1 | 5.6E–05 | 3.7E–02 |
| ENSMUSG00000078919 + ENSMUSG00000051149 | Dpm1/Adnp | 009 | chr2 | 168032210 | 168032422 | − | 0.9 | 4.1 | 5.6E–05 | 3.7E–02 |
| ENSMUSG00000052423 | B4galt3 | 030 | chr1 | 173204520 | 173206012 | + | 0.3 | 4.1 | 5.6E–05 | 3.7E–02 |
| ENSMUSG00000058239 | Usf2 | 007 | chr7 | 31731875 | 31731898 | − | −0.3 | −4.1 | 5.7E–05 | 3.7E–02 |
| ENSMUSG00000027639 | Samhd1 | 004 | chr2 | 156925240 | 156927056 | − | 0.4 | 4.1 | 5.7E–05 | 3.7E–02 |
| ENSMUSG00000032557 | Uba5 | 016 | chr9 | 103962489 | 103962547 | − | 0.8 | 4.1 | 5.7E–05 | 3.8E–02 |
| ENSMUSG00000049092 | Gpr137c | 001 | chr14 | 45839394 | 45839667 | + | 1.2 | 4.2 | 5.8E–05 | 3.8E–02 |
| ENSMUSG00000019970 | Sgk1 | 002 | chr10 | 21602175 | 21602513 | + | −0.5 | −4.1 | 5.8E–05 | 3.8E–02 |
| ENSMUSG00000072770 + ENSMUSG00000030330 | Acrbp/Ing4 | 041 | chr6 | 125001872 | 125001908 | + | 2.4 | 4.1 | 5.8E–05 | 3.8E–02 |
| ENSMUSG00000024843 | Chka | 032 | chr19 | 3889989 | 3891730 | + | 0.4 | 4.1 | 5.9E–05 | 3.8E–02 |
| ENSMUSG00000063873 | Slc24a3 | 001 | chr2 | 144993490 | 144993654 | + | 0.8 | 4.1 | 5.9E–05 | 3.9E–02 |
| ENSMUSG00000020719 + ENSMUSG00000093273 + ENSMUSG00000093133 |
Ddx5/Mir3064/ | 020 | chr11 | 106645047 | 106645272 | − | 0.2 | 4.1 | 6.0E–05 | 3.9E–02 |
| ENSMUSG00000020893 | Per1 | 026 | chr11 | 68919799 | 68919954 | + | 0.8 | 4.1 | 6.0E–05 | 3.9E–02 |
| ENSMUSG00000058239 | Usf2 | 021 | chr7 | 31741401 | 31741447 | − | 0.4 | 4.1 | 6.2E–05 | 4.0E–02 |
| ENSMUSG00000003438 | Timm50 | 013 | chr7 | 29094525 | 29095097 | − | 0.7 | 4.1 | 6.2E–05 | 4.0E–02 |
| ENSMUSG00000019254 | Ppp1r12c | 051 | chr7 | 4452964 | 4453131 | − | 0.4 | 4.0 | 6.3E–05 | 4.0E–02 |
| ENSMUSG00000032382 | Snx1 | 006 | chr9 | 65938538 | 65939154 | − | 0.3 | 4.1 | 6.3E–05 | 4.0E–02 |
| ENSMUSG00000000902 | Smarcb1 | 009 | chr10 | 75367320 | 75368774 | − | −0.4 | −4.1 | 6.4E–05 | 4.1E–02 |
| ENSMUSG00000015467 + ENSMUSG00000015474 + ENSMUSG00000092176 |
Egfl8/Ppt2/ Gm20460 |
019 | chr17 | 34752266 | 34752266 | − | 2.2 | 4.0 | 6.6E–05 | 4.2E–02 |
| ENSMUSG00000056692 | D17Wsu92e | 009 | chr17 | 27957437 | 27957487 | − | 1.4 | 4.2 | 6.6E–05 | 4.2E–02 |
| ENSMUSG00000020719 + ENSMUSG00000093273 + ENSMUSG00000093133 |
Ddx5/Mir3064/ | 015 | chr11 | 106644586 | 106644625 | − | 0.2 | 4.0 | 6.6E–05 | 4.2E–02 |
| ENSMUSG00000060216 | Arrb2 | 011 | chr11 | 70249075 | 70249498 | + | 0.6 | 4.0 | 6.7E–05 | 4.3E–02 |
| ENSMUSG00000047030 | Spata2 | 002 | chr2 | 167306933 | 167307058 | − | −0.6 | −4.1 | 6.7E–05 | 4.3E–02 |
| ENSMUSG00000020128 | Vps54 | 001 | chr11 | 21138892 | 21139283 | + | 0.7 | 4.0 | 6.8E–05 | 4.3E–02 |
| ENSMUSG00000047368 | Fam108b | 001 | chr19 | 21727799 | 21728083 | + | 0.7 | 4.4 | 7.0E–05 | 4.4E–02 |
| ENSMUSG00000038084 | Opa1 | 047 | chr16 | 29652577 | 29654970 | + | 0.2 | 4.0 | 7.0E–05 | 4.4E–02 |
| ENSMUSG00000020333 | Acsl6 | 029 | chr11 | 54150651 | 54151230 | + | 0.4 | 4.0 | 7.1E–05 | 4.5E–02 |
| ENSMUSG00000032322 | Pstpip1 | 008 | chr9 | 55969742 | 55969829 | + | 2.1 | 4.0 | 7.1E–05 | 4.5E–02 |
| ENSMUSG00000061207 | Stk19 | 010 | chr17 | 34961794 | 34962547 | − | 0.3 | 4.1 | 7.2E–05 | 4.5E–02 |
| ENSMUSG00000022974 | Gcfc1 | 034 | chr16 | 91044410 | 91044599 | − | 0.7 | 4.0 | 7.2E–05 | 4.5E–02 |
| ENSMUSG00000016933 | Plcg1 | 002 | chr2 | 160557046 | 160557080 | + | 0.8 | 4.0 | 7.4E–05 | 4.6E–02 |
| ENSMUSG00000020333 | Acsl6 | 030 | chr11 | 54151231 | 54151913 | + | 0.4 | 4.0 | 7.5E–05 | 4.6E–02 |
| ENSMUSG00000059316 | Slc27a4 | 004 | chr2 | 29662518 | 29662875 | + | 0.4 | 4.1 | 7.5E–05 | 4.6E–02 |
| ENSMUSG00000003848 | Nob1 | 013 | chr8 | 109942343 | 109942566 | − | 0.8 | 4.0 | 7.5E–05 | 4.7E–02 |
| ENSMUSG00000078789 + ENSMUSG00000038268 | Dph1/Ovca2 | 016 | chr11 | 74994164 | 74994191 | − | −0.8 | −4.0 | 7.6E–05 | 4.7E–02 |
| ENSMUSG00000004929 | Thop1 | 006 | chr10 | 80536111 | 80537622 | + | 0.6 | 4.0 | 7.6E–05 | 4.7E–02 |
| ENSMUSG00000038644 | Pold1 | 017 | chr7 | 51789557 | 51789559 | − | 1.7 | 4.0 | 7.7E–05 | 4.7E–02 |
| ENSMUSG00000035901 | Dennd5a | 035 | chr7 | 117103838 | 117103914 | − | 1.0 | 4.0 | 7.9E–05 | 4.8E–02 |
| ENSMUSG00000089715 + ENSMUSG00000089837 + ENSMUSG00000022421 |
Cbx6/Npcd/ Nptxr |
014 | chr15 | 79635063 | 79635194 | − | 1.0 | 4.0 | 7.9E–05 | 4.8E–02 |
| ENSMUSG00000075876 + ENSMUSG00000064791 + ENSMUSG00000075924 + ENSMUSG00000015656 |
Snord14c/ Snord14e/ Snord14d/Hspa8 |
031 | chr9 | 40612078 | 40612163 | + | 1.5 | 4.0 | 7.9E–05 | 4.8E–02 |
| ENSMUSG00000037266 | D4Wsu53e | 018 | chr4 | 134481348 | 134481360 | + | −0.3 | −4.0 | 7.9E–05 | 4.8E–02 |
| ENSMUSG00000044700 | Tmem201 | 003 | chr4 | 149091187 | 149092128 | − | 0.4 | 4.0 | 8.0E–05 | 4.9E–02 |
| ENSMUSG00000027692 | Tnik | 051 | chr3 | 28569508 | 28574780 | + | −0.2 | −4.0 | 8.1E–05 | 4.9E–02 |
| ENSMUSG00000093290 + ENSMUSG00000035632 | Mir3572/Cnot3 | 029 | chr7 | 3611109 | 3612278 | + | 0.3 | 4.0 | 8.1E–05 | 4.9E–02 |
| ENSMUSG00000009394 | Syn2 | 001 | chr6 | 115084920 | 115085478 | + | 0.2 | 4.1 | 8.1E–05 | 4.9E–02 |
| ENSMUSG00000053754 | Chd8 | 006 | chr14 | 52821423 | 52821818 | − | −0.5 | −4.0 | 8.2E–05 | 4.9E–02 |
| ENSMUSG00000037331 | Larp1 | 001 | chr11 | 57822566 | 57823049 | + | 0.5 | 4.0 | 8.2E–05 | 4.9E–02 |
| ENSMUSG00000011658 | Fuz | 044 | chr7 | 52157792 | 52158001 | + | 0.4 | 4.0 | 8.3E–05 | 4.9E–02 |
| ENSMUSG00000074657 | Kif5a | 002 | chr10 | 126666106 | 126666800 | − | 0.5 | 4.0 | 8.3E–05 | 4.9E–02 |
| ENSMUSG00000022394 | L3mbtl2 | 004 | chr15 | 81494386 | 81494430 | + | −0.5 | −4.0 | 8.3E–05 | 4.9E–02 |
| ENSMUSG00000038453 | Srcin1 | 034 | chr11 | 97436441 | 97436529 | − | 1.8 | 4.0 | 8.3E–05 | 4.9E–02 |
| ENSMUSG00000065438 + ENSMUSG00000065497 + ENSMUSG00000065561 + ENSMUSG00000091158 + ENSMUSG00000065570 |
Mir377/Mir410/ Mir369/Mirg/ Mir412 |
018 | chr12 | 110987305 | 110987665 | + | −0.3 | −4.0 | 8.4E–05 | 5.0E–02 |
Upregulated bins during memory consolidation included Snord14e, which reside in the introns of the Hspa8 gene. We have recently validated Snord14e upregulation after detecting differences by microarray (Peixoto, Wimmer et al., 2015). We also discovered that a poorly studied short isoform of Homer1 known as Ania-3 (Ensembl Homer1-005) (Bottai et al., 2002) is upregulated after contextual fear conditioning. Homer1a has previously been shown to be upregulated by fear conditioning (Mahan et al., 2012), but Ania-3 has not been studied. To validate our results, we performed qPCR in a separate cohort of mice, comparing the bins observed to change to a bin of the same gene that was unchanged. Ania-3 was found to be upregulated independently of the entire Homer1 gene (Fig. 1a). Ribosome biogenesis protein Las1l exhibited bin-specific downregulation in response to contextual fear conditioning and this was also confirmed by qPCR (Fig. 1b). RNA-binding protein Rbm3, which our lab has shown to change in the hippocampus after sleep deprivation (Vecsey et al., 2012), displays complex regulation with both upregulated and downregulated bins after learning. Both the upregulated and downregulated bins were confirmed by qPCR in a separate cohort of animals (Fig. 1c). In all cases, the bin predicted to change was significantly regulated while a control bin in the same gene was unchanged.
Fig. 1.
Bin-specific regulation of Homer1 (Ania-3), Las1l, and Rbm3. (A) (left) diffSplice result showing the predicted significant bin changes of the Homer1 gene in red on a log2 scale. Bins 16–18 indicate the Ania-3 isoform. (right) qPCR validation of the change in Bin18 in an independent cohort of mice. Bin 21 expression was compared as a control. (B) (left) diffSplice result showing the predicted significant bin changes of the Las1l gene in red on a log2 scale. (right) qPCR validation of the change in Bin 15 in an independent cohort of mice. Expression of Bin 17 was used as a control. (C) (left) diffSplice result showing the predicted significant bin changes of the Rbm3 gene in red on a log2 scale. (right) qPCR validation of the changes in Bin 2 and Bin 22 an independent cohort of mice. Expression of Bin 7 was used as a control. HC = homecage, FC = fear conditioned. * denotes a p-value of <0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To test whether these changes are specific to the association of context and shock, a separate cohort of animals were either immediately shocked or exposed to the context with no footshock. Expression of Ania-3 shows a change in response to context only, but not after immediate shock (Fig. 2). This is not surprising given the overlap of gene expression between fear conditioning and spatial training (Keeley et al., 2006; Poplawski et al., 2014). An increase in Ania-3 may represent splicing changes in response to a novel environment. Las1l displays a non-significant trend toward a reduction in the context only, but not immediate shock (Fig. 2). Las1l bin 15 may represent a splicing change that occurs only with a context-shock association. Rbm3 bin 2 expression shows changes in both the immediate shock and context only conditions, (Fig. 2), suggesting that this alternative splicing occurs with minimal perturbation and may not reflect a learning event. Rbm3 bin 22 showed only a trend toward a decrease in both cases. Therefore, changes in Rbm3 bin 2 may represent any activity within the hippocampus, while Rbm3 bin 22 could be specific to context-shock associations. These results indicate that alternative splicing can occur in response to a variety of factors and may be a specific marker in the hippocampus of recent behavioral stimuli.
Fig. 2.
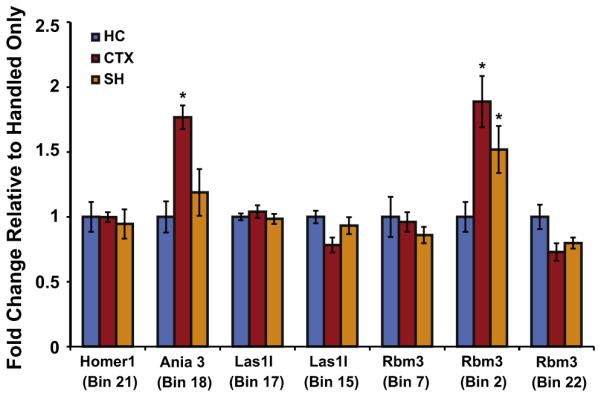
Alternative splicing changes variably in response to either component of fear conditioning alone. The same primers used in Fig. 1 were used to test gene expression after context only (n = 8) or immediate shock controls (n = 8) or homecage animals (n = 7) and analyzed by ANOVA. Ania-3 and Las1l changes in response to the context only control and Rbm3 changes with all manipulations. This may suggest Ania-3 and Las1l are markers of contextual novelty while Rbm3 responds to many stimuli. No control bins change with either context-only or immediate shock controls. HC = homecage, CTX = context only, SH = shock only. * denotes significance below an alpha of 0.05.
4. Discussion
In this study, we provide the first evidence of genome-wide regulation of alternative splicing after learning in the hippocampus. Using bin counts produced by HTSeq and the limma Bioconductor package, we compared bins representing a unique piece of a gene against expression of that entire gene to create a list of bin-level changes. We were able to detect significant gene expression changes at 171 bins occurring in response to contextual fear conditioning at 138 genes. The exact number of potential splicing sites is not known in neurons, and splicing studies have identified as low as 3110 splicing events in neurons (Zhang et al., 2014) or as high as 92–94% of all genes (Wang et al., 2008). We suspect that 138 genes showing changes is only a small fraction of the potential change. It is unclear at this time why memory retrieval shows a larger set of changes than memory consolidation. This study used whole hippocampus, so only a small fraction of all cells in the sample are being activated by learning. However, our RUV analysis removes unwanted variation including that from nonresponsive cells, so we believe that the changes observed are due to activated neurons.
Although individual examples of alternative splicing have been observed during memory consolidation (Lubin et al., 2008; Mahan et al., 2012; Rozic, Lupowitz, Piontkewitz, & Zisapel, 2011), no studies have explored this phenomenon genome-wide. We also identified candidate genes displaying alternative regulation that may be important for learning. As previously reported (Peixoto, Wimmer et al., 2015), we confirmed that Snord14e, which exists within an intron of Hspa8, is regulated by fear conditioning. It is unclear why Snord14e increases in polyadenylated RNA, but it could be due increases in intron retention during transcription or splicing and polyadenylation of a Snord14e precursor. We also implicate the selective alternative splicing of Homer1 isoform Ania-3, RNA-binding protein Rbm3 and ribosome biogenesis regulator Las1l in learning for the first time. These results emphasize the importance of using genome-wide binning techniques to identify subtle changes in splicing following fear conditioning, which would be overlooked with standard RNA-seq analysis.
It is interesting that we observed different results for Rbm3 alternative splicing in the context only and immediate shock controls. At the gene-level, gene expression changes after contextual and spatial learning are known to overlap (Poplawski et al., 2014). Previous work from our lab has highlighted similar gene expression between fear conditioning and context-only exposure in the hippocampus, but not the amygdala (Keeley et al., 2006). Thus, we anticipated similar results between our fear conditioning results and the context only control, as was the case with all splicing events tested. This confirms our previous findings that exposure to a context is sufficient to elicit similar gene expression changes that occur when context is paired with shock.
However, we found that Rbm3 had a unique response to the components of fear conditioning, with an immediate shock being able to alter alternative splicing of bin 2 of this gene. Immediate shock does not provide the subject enough time to form a contextual representation of the space, and therefore is generally thought not to cause expression changes in the hippocampus (Huff et al., 2006). Thus, the change after immediate shock in Rbm3 may suggest that splicing of Rbm3 bin 2 in the hippocampus is altered by many brain stimuli. In contrast, Rbm3 bin 22, Ania-3 and Las1l may be specific to exposure to a novel context or context-shock association. Thus, splicing changes in Rbm3 bin 2 may not be involved in forming long-term contextual fear memories, while splicing in Ania-3 and Las1l may have a role in encoding these types of memories. We hypothesize that this may be an instance of a broader phenomenon in neuronal plasticity, where certain splicing events are regulated by any neuronal activity while others only respond to specific stimuli.
In the present study, we did not determine the type of splicing that is occurring at each of these bins, which will be the subject of future analyses. Each bin observed could be the result of many types of regulation, including exon skipping, intron retention, or alternative start/stop sites. Ania-3 has previously been reported as an alternative isoform of Homer1 that responds like an immediate early gene (Bottai et al., 2002), and the change we detect here corresponds to the unique Ania-3 exon. Whether Ania-3 splicing is regulated by SRp20, as is the case for Homer1a (Wang et al., 2014), is a subject for future investigation. Las1l bin 15 appears to be a retained intron, but whether this is part of the canonical Las1l mRNA or part of a different isoform is unknown. It is also possible that this could be a small RNA that is spliced out of the final polyadenylated mRNA. Rbm3 bin 2 is one of several potential transcriptional termination sites of Rbm3 while bin 22 could either be an early termination site or a retained intron. Further study will be required to determine the exact identity and function of the isoforms that are regulated by contextual fear conditioning.
The mechanism that drives this alternative splicing is not studied within these experiments, although transcription of certain splicing proteins such as SRp20 is known to change after fear conditioning (Antunes-Martins et al., 2007). Recent studies have also highlighted the importance of Rbfox1 in splicing and mRNA regulation in neurons (Lee et al., 2016). Our data indicates regulation of a specific isoform of splicing factor Sfpq. However, it is unclear whether these transcriptional changes would be translated into protein and affect splicing by 30 min after training. The mechanism by which alternative regulation of transcripts is controlled during memory consolidation is an important question for future studies. It is possible that changes in epigenetic modifications are regulating this selective transcript usage (Zhou, Luo, Wise, & Lou, 2014), including H3K36me3 and H4K20me1 (Luco et al., 2010; Zhu, Wang, Liu, & Wang, 2013). It would be interesting to observe whether the differential bins discovered in this study show differential histone modifications as well. We hope our findings and this unique analysis method drive further study into the mechanisms of isoform-specific changes in gene expression during memory consolidation.
Acknowledgments
This research was supported by NIH R01MH087463 to T.A.
Footnotes
Conflict of interest
None.
References
- Anders S, Pyl PT, Huber W. HTSeq — A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Research. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes-Martins A, Mizuno K, Irvine EE, Lepicard EM, Giese KP. Sex-dependent up-regulation of two splicing factors, Psf and Srp20, during hippocampal memory formation. Learning & Memory. 2007;14:693–702. doi: 10.1101/lm.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P, Kirtley A, Thomas KL. Quantitatively and qualitatively different cellular processes are engaged in CA1 during the consolidation and reconsolidation of contextual fear memory. Hippocampus. 2012;22:149–171. doi: 10.1002/hipo.20879. [DOI] [PubMed] [Google Scholar]
- Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. Journal of Neuroscience. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learning & Memory. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- Cavallaro S, D’Agata V, Manickam P, Dufour F, Alkon DL. Memory-specific temporal profiles of gene expression in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD, Fung ET, O’Brien RJ, Huganir RL. Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. Journal of Neuroscience. 1998;18:720–730. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Blencowe BJ. Tissue-specific alternative splicing remodels protein–protein interaction networks. Molecular Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, Kroener BT, Manglesdorf DJ, Abel T. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. The Journal of Clinical Investigation. 2012;122:3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huff NC, Frank M, Wright-Hardesty K, Sprunger D, Matus-Amat P, Higgins E, Rudy JW. Amygdala regulation of immediate-early gene expression in the hippocampus induced by contextual fear conditioning. Journal of Neuroscience. 2006;26:1616–1623. doi: 10.1523/JNEUROSCI.4964-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Vianna MR, Medina JH, Izquierdo I. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fearmotivated learning. Journal of Neuroscience. 2002;22:6781–6789. doi: 10.1523/JNEUROSCI.22-15-06781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolski F, Coussen F, Nagarajan N, Normand E, Rosenmund C, Mulle C. Subunit composition and alternative splicing regulate membrane delivery of kainate receptors. Journal of Neuroscience. 2004;24:2506–2515. doi: 10.1523/JNEUROSCI.5116-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley MB, Wood MA, Isiegas C, Stein J, Hellman K, Hannenhalli S, Abel T. Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learning & Memory. 2006;13:135–142. doi: 10.1101/lm.86906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klur S, Muller C, Pereira de Vasconcelos A, Ballard T, Lopez J, Galani R, Certa U, Cassel JC. Hippocampal-dependent spatial memory functions might be lateralized in rats: An approach combining gene expression profiling and reversible inactivation. Hippocampus. 2009;19:800–816. doi: 10.1002/hipo.20562. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, Geschwind DH, Black DL, Martin KC. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff SE, Rosenfeld MG, Evans RM. Complex transcriptional units: Diversity in gene expression by alternative RNA processing. Annual Review of Biochemistry. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Choi S, Lee SY, Cao YA, Ahn HJ, Worley KC, Pizzi M, Liou HC, Sweatt JD. A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel. Journal of Neuroscience. 2004;24:3933–3943. doi: 10.1523/JNEUROSCI.5646-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. Journal of Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan AL, Mou L, Shah N, Hu JH, Worley PF, Ressler KJ. Epigenetic modulation of Homer1a transcription regulation in amygdala and hippocampus with pavlovian fear conditioning. Journal of Neuroscience. 2012;32:4651–4659. doi: 10.1523/JNEUROSCI.3308-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei B, Li C, Dong S, Jiang CH, Wang H, Hu Y. Distinct gene expression profiles in hippocampus and amygdala after fear conditioning. Brain Research Bulletin. 2005;67:1–12. doi: 10.1016/j.brainresbull.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes, Brain and Behavior. 2012;11:651–659. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandrikopoulou A, Doll T, Tucker RP, Garner CC, Matus A. Embryonic MAP2 lacks the cross-linking sidearm sequences and dendritic targeting signal of adult MAP2. Nature. 1989;340:650–652. doi: 10.1038/340650a0. [DOI] [PubMed] [Google Scholar]
- Peixoto L, Risso D, Poplawski SG, Wimmer ME, Speed TP, Wood MA, Abel T. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic Acids Research. 2015;43:7664–7674. doi: 10.1093/nar/gkv736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto LL, Wimmer ME, Poplawski SG, Tudor JC, Kenworthy CA, Liu S, Abel T. Memory acquisition and retrieval impact different epigenetic processes that regulate gene expression. BMC Genomics. 2015;16(Suppl. 5):S5. doi: 10.1186/1471-2164-16-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplawski SG, Schoch H, Wimmer ME, Hawk JD, Walsh JL, Giese KP, Abel T. Object-location training elicits an overlapping but temporally distinct transcriptional profile from contextual fear conditioning. Neurobiology of Learning and Memory. 2014;116:90–95. doi: 10.1016/j.nlm.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: Recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nature Biotechnology. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogelj B, Hartmann CE, Yeo CH, Hunt SP, Giese KP. Contextual fear conditioning regulates the expression of brain-specific small nucleolar RNAs in hippocampus. The European Journal of Neuroscience. 2003;18:3089–3096. doi: 10.1111/j.1460-9568.2003.03026.x. [DOI] [PubMed] [Google Scholar]
- Rozic G, Lupowitz Z, Piontkewitz Y, Zisapel N. Dynamic changes in neurexins’ alternative splicing: Role of Rho-associated protein kinases and relevance to memory formation. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0018579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. Journal of Neuroscience. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Peixoto L, Choi JH, Wimmer M, Jaganath D, Hernandez PJ, Abel T. Genomic analysis of sleep deprivation reveals translational regulation in the hippocampus. Physiological Genomics. 2012;44:981–991. doi: 10.1152/physiolgenomics.00084.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chikina MD, Pincas H, Sealfon SC. Homer1 alternative splicing is regulated by gonadotropin-releasing hormone and modulates gonadotropin gene expression. Molecular and Cellular Biology. 2014;34:1747–1756. doi: 10.1128/MCB.01401-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Luo G, Wise JA, Lou H. Regulation of alternative splicing by local histone modifications: Potential roles for RNA-guided mechanisms. Nucleic Acids Research. 2014;42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Wang G, Liu B, Wang Y. Modeling exon expression using histone modifications. PLoS ONE. 2013;8:e67448. doi: 10.1371/journal.pone.0067448. [DOI] [PMC free article] [PubMed] [Google Scholar]