Abstract
Sclerotinia stem rot caused by Sclerotinia sclerotiorum is a devastating disease of Brassica crops, but not in rice. The leaves of a rice line, a partial resistant (R) and a susceptible (S) Brassica oleracea pool that bulked from a resistance-segregating F2 population were employed for transcriptome sequencing before and after inoculation by S. sclerotiorum for 6 and 12 h. Distinct transcriptome profiles were revealed between B. oleracea and rice in response to S. sclerotiorum. Enrichment analyses of GO and KEGG indicated an enhancement of antioxidant activity in the R B. oleracea and rice, and histochemical staining exhibited obvious lighter reactive oxygen species (ROS) accumulation and cell death in rice and the R B. oleracea as compared to that in the S B. oleracea. Significant enhancement of Ca2+ signalling, a positive regulator of ROS and cell death, were detected in S B. oleracea after inoculation, while it was significantly repressed in the R B. oleracea group. Obvious difference was detected between two B. oleracea groups for WRKY transcription factors, particularly for those regulating cell death. These findings suggest diverse modulations on cell death in host in response to S. sclerotiorum. Our study provides useful insight into the resistant mechanism to S. sclerotiorum.
Sclerotinia sclerotiorum is a necrotizing fungal pathogen that infects hundreds of plant species, including important crops such as soybean and rapeseed (Brassica napus L.), but it does not infect rice (Oryza sativa)1,2. Many studies have been performed to investigate the defense response and molecular mechanisms in Brassica hosts against S. sclerotiorum. Several quantitative trait loci (QTL) for S. sclerotiorum resistance have been mapped in rapeseed and its relatives3,4,5,6,7. A number of enzymes were revealed to be associated with resistance to S. sclerotiorum, such as polygalacturonase-inhibiting protein, germin-like proteins, several mitogen-activated protein kinases (MAPKs), and WRKY transcription factors (TFs)8,9,10,11,12,13. Transcriptional studies in rapeseed revealed the biological alterations in response to S. sclerotiorum, including oxidative burst, cell wall enforcement or modification, secondary metabolism, calcium binding, antioxindation, glutathiones (GSHs) metabolism, glucosinolates metabolism, biosynthesis of lignins, TFs, carbohydrate metabolism, and energy metabolism4,11,12,14,15,16,17,18,19. Due to the lack of high resistance source, however, the molecular mechanism of resistance against S. sclerotiorum was still poorly understood.
Recently, a wild B. oleracea genotype, ‘C01’, has been identified to be with partial resistance against S. sclerotiorum, exhibiting significant higher resistance level than that of a partial resistant B. napus variety ‘Zhongshuang 9’20. The resistance QTL have been identified from an F2 population derived from ‘C01’3 and it has been proved that ‘C01’ was effective in improving Sclerotinia resistance of Brassica species21,22. In order to investigate the response to S. sclerotiorum in hosts with different resistance levels, rice with nonhost resistance to S. sclerotiorum, and a partial resistant (R) and a susceptible (S) pool that bulked from the extreme resistant and susceptible genotypes in the F2 population derived from ‘C01’ were compared for gene expression alterations in leaf before and after inoculation. Significant differences were found for antioxidation, Ca2+ signalling, WRKY TFs and ribosome which are in association with modulating cell death of host in defense to pathogens. Our study provides new insights into the resistant mechanism to S. sclerotiorum.
Results
Resistance performance and transcriptome sequencing
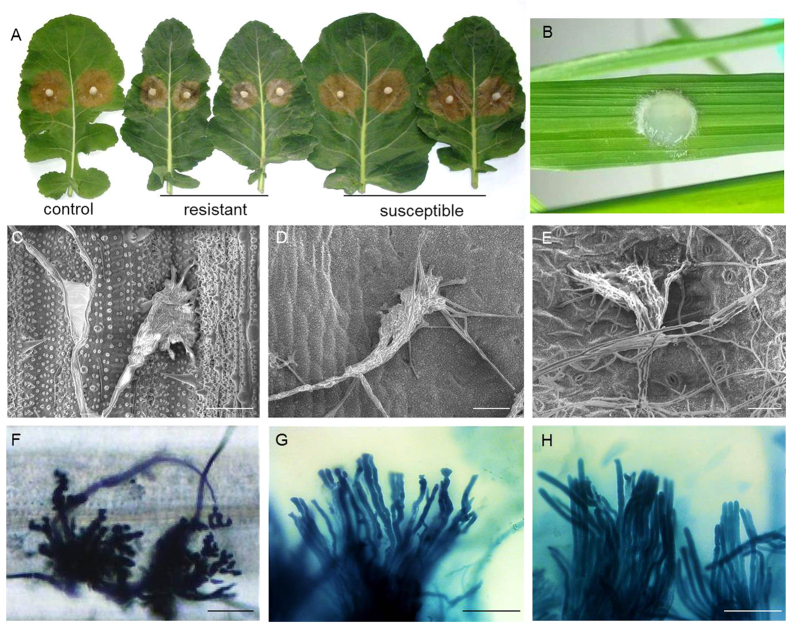
Different phenotypic responses were observed among rice, the R and the S B. oleracea group in response to S. sclerotiorum (Fig. 1). No symptom was visible on the inoculated rice leaf at 3 days after inoculation (dai), while the lesion size of the R and the S B. oleracea group averaged on 5.4 cm2 (4.5~6.2 cm2) and 9.8 cm2 (8.5~12.3 cm2) at 3 dai, respectively. Though infection cushions could be found from the inoculated leaves of rice, the partial resistant and the susceptible B. oleracea at 12 hours post-inoculation (hpi), it seems that the infection cushions on susceptible B. oleracea growed faster (already formed many hyphae) and stronger (in a bigger shape and with more ‘fingers’ which will turn into invasive hyphae soon) than that on rice and the resistant line. At 36 hpi, infection cushions were still the leading structures on the leaf surface of rice, while benched and top-aberrant hyphae were dominant at the lesion edge of the partial resistant B. oleracea, differing from the thick and fluent hyphae in the susceptible B. oleracea. These observations indicate that the propagation of S. sclerotiorum was hindered entirely in rice and partially in the R B. oleracea.
Figure 1. Performances of Sclerotinia sclerotiorum-infected leaves of Brassica oleracea and rice.
(A) Symptoms of resistant and susceptible plants of Brassica oleracea at 3 days after inoculation by S. sclerotiorum. The left side exhibits the performance of control ‘Zhongshuang 9’ which is a partial resistant rapeseed variety against S. sclerotiorum. (B) Performance of rice at 3 days after inoculation by S. sclerotiorum. (C–E) Infection cushions on leaves of rice, the partial resistant and the susceptible B. oleracea at 12 hpi, respectively (observed using electron microscope). (F–H) Infection cushion on rice leaf and invasive hyphae in leaves of the partial resistant and the susceptible B. oleracea at 36 hpi, respectively. Scale bar = 50 μm.
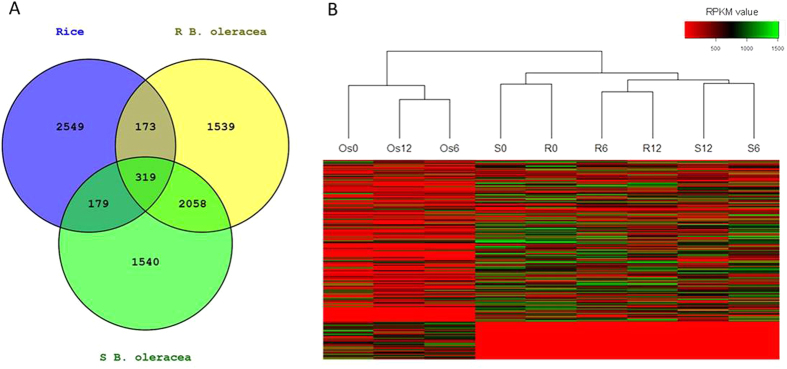
Each of three libraries of rice (constructed at 0, 6 and 12 hpi, named as Os0, Os6 and Os12, respectively), the R (R0, R6 and R12) and the S (S0, S6 and S12) B. oleracea group were sequenced on an Illumina Hiseq 2000TM platform, yielding more than 25 million clean reads for each B. oleracea sample and over 15 million for rice. Of these, 72.3% and 76.8% on average were aligned to the reference genome of B. oleracea and rice, respectively (Supplementary Table 1). A total of 34,550 and 27,405 unigenes were detected from B. oleracea and rice, of which 7,230 B. oleracea genes (4,961 from R group, 4,890 from S group, with 2,621 overlap genes) and 3,470 rice genes exhibited more than two-fold transcriptional differences between neighboring time points. The real-time qRT-PCR (qRT-PCR) analysis for 46 differentially expressed genes (DEGs) from B. oleracea and five DEGs of interest showed general agreement with the RNA-seq data (Supplementary Figure 1).
Comparison of gene expression alterations after inoculation between B. oleracea and rice
After aligning all DEGs to Arabidopsis homologous genes, 671 overlapped genes were found in rice and B. oleracea (Fig. 2A, Supplementary Table 2). These genes significantly enriched in Gene Ontology (GO) terms related to response to abiotic stimuli, such as response to light stimulus, chemical stimulus and stress (Supplementary Figure 2). However, an obvious difference for transcriptome profile was revealed between B. oleracea and rice by the heatmap upon all DEGs (Fig. 2B). In order to get insight into the resistance mechanism that overlapped in rice and resistant B. oleracea, GO analysis was conducted separately on the up- and down-regulated DEGs, and in two B. oleracea groups on the up- and down-regulated DGEs that specifically found in each group (Fig. 3, Supplementary Table 3).
Figure 2. DEGs in rice and Brassica oleracea in response to Sclerotinia sclerotiorum.
(A) Venn diagram of DEGs in rice and B. oleracea during whole infection. (B) Heatmap of inoculated and non-inoculated B. oleracea and rice according to the RPKM values of DEGs. Os, rice; R, Resistant B. oleracea; S, susceptible B. oleracea; Os0, R0 and S0, non-inoculated controls; Os6, R6 and S6, samples collected 6 hpi; Os12, R12 and S12, samples collected 12 hpi.
Figure 3.
Significantly enriched GO (in black) and KEGG (in red) terms among up-regulated (A) and down-regulated DEGs (B) in rice, resistance (R) and susceptible (S) Brassica oleracea.
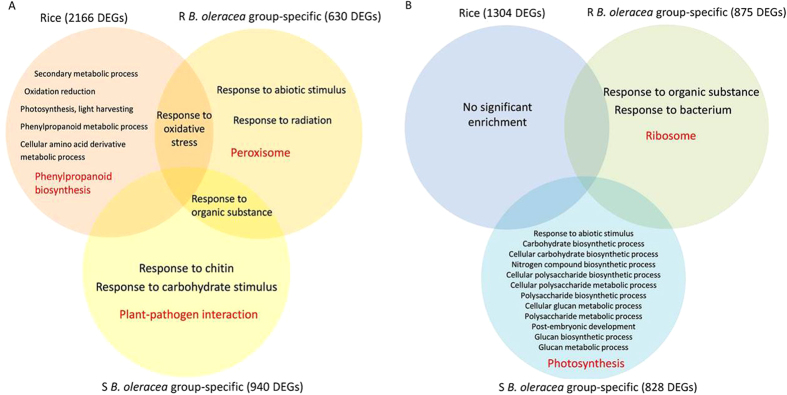
No significant biology process was enriched in rice by analyzing 1,304 down-regulated DEGs, while 2,166 up-regulated DEGs significantly enriched in ‘response to oxidative stress’, ‘secondary metabolic process’, ‘oxidation reduction’, ‘photosynthesis, light harvesting’, ‘phenylpropanoid metabolic process’ and ‘cellular amino acid derivative metabolic process’. Among these, the term ‘response to oxidative stress’ was also significantly enriched in the R B. oleracea group by analyzing 630 up-regulated DEGs that were R-group specific. No common biology process was detected between rice and the S B. oleracea group (Fig. 3).
In details, a total of 26 DEGs were involved in the term of ‘response to oxidative stress’ in rice (FDR = 3.91e−4), including 25 peroxidases (Supplementary Table 4). These antioxidant genes were sharply induced at 6 hpi, and 16 of these were maintained at high expression levels at 12 hpi. For example, a precursor of cationic peroxidase SPC4 (LOC_Os01g73200) was up regulated for 390 folds at 6 hpi and even increased for 420 folds at 12 hpi. Being similarly, the GO term ‘response to oxidative stress’ that significantly enriched in the R B. oleracea group (FDR = 1.01e−02) involves 19 antioxidant genes, including 9 peroxidases, 7 catalases (CATs) and 3 superoxide dismutases (SODs). These genes were increased for abundance by up to 45 folds in the R group at 6 hpi. These imply a notable inducement of antioxidant activity in rice and the resistant B. oleracea after inoculation.
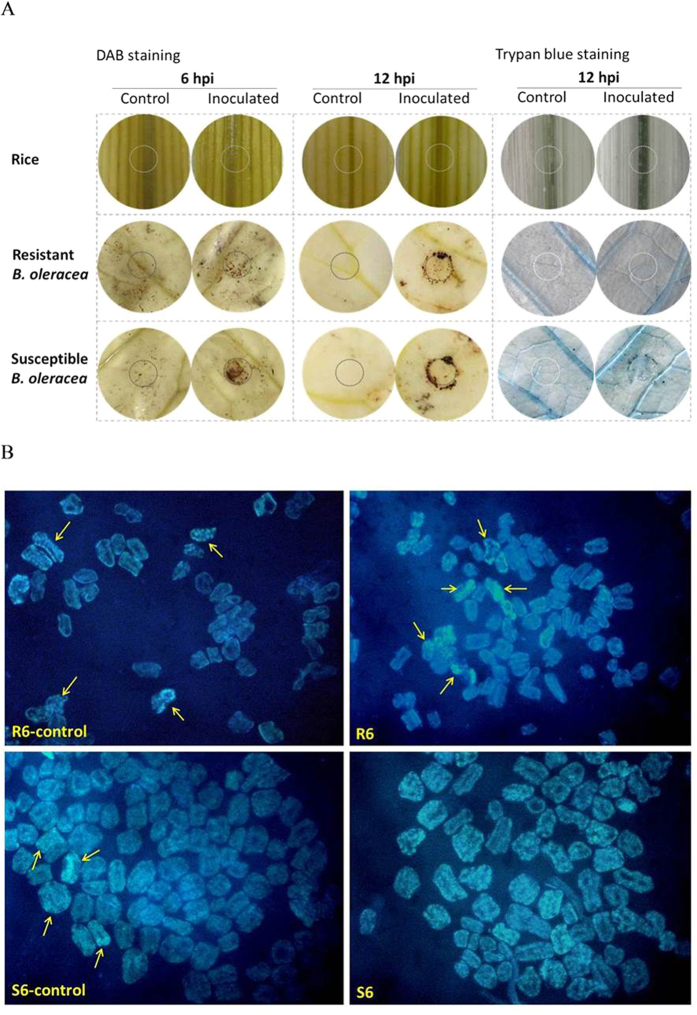
The high antioxidant activity was also observed in rice and the resistant B. oleracea by histochemical staining study and enzyme activity study. H2O2 is one of the prominent reactive oxygen species (ROS) which cause cell death in host23. By staining leaves with 3,0-diaminobenzidine (DAB) which visualizes the accumulation of H2O2, we found obvious stained dots at both 6 and 12 hpi in the susceptible B. oleracea, but no stained dot in rice during the whole infection and only a few stained dots on the edge of inoculation point in the resistant B. oleracea at 12 hpi (Fig. 4). This result was supported by the total antioxidant capacity assay, which revealed singificant enhanced antioxidant enzyme activity in rice (6 hpi) and R B. oleracea (both 6 and 12 hpi) after inoculation (P < 0.01) (Fig. 5). Correspondingly, the trypan blue staining revealed heavier necrosis in the susceptible B. oleracea than that in the resistant B. oleracea and rice at 12 hpi, and the DAPI staining presented obvious higher ratio of apoptosis in the susceptible B. oleracea than that in the resistant B. oleracea at 6 hpi (Fig. 4).
Figure 4. H2O2 accumulation and cell death in rice and Brassica oleracea after inoculation with Sclerotinia sclerotiorum.
Controls (inoculated with PDA agar discs) and infected leaves were stained with 3,0-diaminobenzidine (DAB) to visualize the H2O2 accumulation (A), with trypan blue for necrosis (A), and with DAPI for apoptosis (B), respectively. Open cycle on each leaf in A indicates the inoculation site. Arrows in B indicates typical apoptosis cells (all cells of S6 are apoptosis, thus are not indicated).
Figure 5. The total antioxidant capacity of rice and resistant (R) and susceptible (S) Brassica oleracea after inoculation with Sclerotinia sclerotiorum.
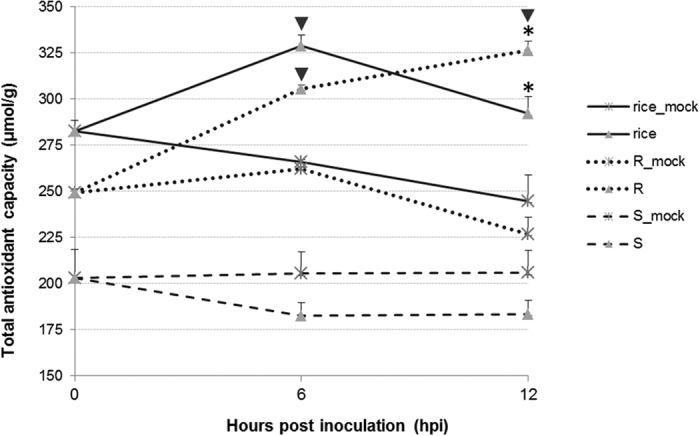
Mocks were inoculated with agar discs. Asterisks indicate significant difference of inoculated samples with corresponding mocks at a same time point, and inverted triangles represent significant difference of samples at a certain time point comparing to a former time point (P < 0.01).
Transcriptome alterations during inoculation within B. oleracea
The heatmap of DEGs revealed similar transcriptome pattern between the R and S B. oleracea groups prior to inoculation (Fig. 2), suggesting a high similarity in their genetic background. However, distinct expression profiles were exhibited between the two groups after inoculation (Fig. 2), reflecting different responses to S. sclerotiorum between resistant and susceptible B. oleracea.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the term ‘ribosome’ was significantly enriched among 875 down-regulated DEGs specific to the R group (PBonferroni = 6.69e−21), while the term ‘Photosynthesis’ was significantly enriched among 828 down-regulated DEGs specific to the S group (PBonferroni = 1.13e−03) (Fig. 3, Supplementary Table 3). It suggests a fast repression on translation in the resistant B. oleracea, and a significant photosynthetic damage in the susceptible B. oleracea during infection.
By analyzing the R-group-specific up-regulated DEGs, significant enrichment was detected on ‘Peroxisome’ (PBonferroni = 2.99e−04), which mainly performs antioxidant function in response to stress24. These DEGs involved in ‘Peroxisome’ were the same as those in the GO term ‘response to oxidative stress’ in the R group which has been mentioned above. The term of ‘Plant-pathogen interaction’ was significantly enriched by the S-group-specific up-regulated DEGs (PBonferroni = 1.91e−05) (Fig. 3, Supplementary Table 3). By deep comparison of DEGs in ‘Plant-pathogen interaction’ between the two groups, we found obvious different alterations on two sub-terms of ‘Plant-pathogen interaction’, i.e., the Ca2+ signalling and the WRKY TFs. The details were shown below.
Ca2+ signalling
Among 196 genes that involved in Ca2+ signalling in the reference genome of B. oleracea (http://www.genome.jp/kegg-bin/show_pathway?ko04626+K13448), 37 exhibited expression changes after infection in the R and S groups (Supplementary Table 4). As compared with 0 hpi, one CNGC12 (Bol030676) which transports Ca2+ from the apoplast into the cell25,26 was up-regulated in the S group for 14 folds at 6 hpi and additional five folds at 12 hpi, while it was slightly repressed in the R group at 6 hpi and up-regulated only for three folds at 12 hpi. The other 36 DEGs were found to encode Ca2+ -sensor proteins, such as calmodulin (CaM), CaM-like proteins (CMLs) and calcium-dependent protein kinases (CDPKs)27. Of these, 22 genes were up regulated among 25 DEGs that detected in the S group, while 15 genes were down regulated among 21 DEGs that detected in the R group. Two of these DEGs, CAM5 (Bol020573) and a CML37 (Bol035298), were selected and certified for gene expression profile by qRT-PCR (Supplementary Figure 1). These findings indicate that the Ca2+ signalling was possibly promoted in the susceptible group but suppressed in the resistant B. oleracea after inoculation with S. sclerotiorum.
WRKY TFs
A total of 50 DEGs encoding WRKY TFs were found in B. oleracea, including 34 from the S group and 32 from the R group (Supplementary Table 4). More WRKYs were up-regulated in the S group (25 in 28 DEGs at 6 hpi and six in eight DEGs at 12 hpi) than in the R group (11 in 24 DEGs at 6 hpi and seven in ten DEGs at 12 hpi). Of these, only eight up-regulated DEGs overlapped in the two groups, including WRKY6, 28, 30 (2 copies), 33, 45, 53 and 75 which have been verified to positively regulate the resistance to S. sclerotiorum11,12,13,15,16,19. It suggested diverse activation on WRKY TFs in the resistant and susceptible B. oleracea.
Interestingly, converse regulation patterns were observed between the two groups at 6 hpi for WRKY50 (Bol022304 and Bol012741), WRKY51 (Bol015902 and Bol019066) and WRKY70 (Bol044275 and Bol002548) which are positively modulators of the downstream salicylic acid (SA)-mediated signaling28,29,30. Among these, five were down-regulated in the R group, whereas four were significantly up-regulated in the S group (Supplementary Table 4). The expression of two WRKY50 (Bol022304 and Bol012741), a WRKY51 (Bol015902), two WRKY70 (Bol044275 and Bol002548) and other four WRKYs (Bol000927, Bol036034, Bol040536 and Bol042130) were certified by qRT-PCR (Supplementary Figure 1). Accordantly, two homologs of PR-1 (Bol019271 and Bol006477), the defense marker in SA signalling, were repressed specifically in the R group at 6 hpi for 2.3 and 25.2 folds, respectively. These findings seem to indicate a potential repression on SA signalling in the resistant B. oleracea.
Discussion
Many attempts have been made to understand the resistance mechanism of Brassica crops against S. sclerotiorum and revealed various aspects of defense response to this necrotrophic pathogen4,11,12,14,15,16,17,18,19. We also found great alterations in B. oleracea in response to S. sclerotiorum, such as plant hormone signal transduction, plant–pathogen interaction, and biosynthesis of glucosinolate, phenylpropanoid, flavonoid and other secondary metabolites (Supplementary Table 5). However, when comparing the hosts with various level of resistance, we found obvious difference for antioxidant activity between the resistant (rice and the R B. oleracea) and the susceptible host (the S B. oleracea) at the early infection phase by S. sclerotiorum.
ROS play a crucial role in defense reaction against pathogens but also cause damages and cell death in host23. In previous studies, several antioxidation-related proteins or genes have been revealed to be associated to the defence to S. sclerotiorum in B. napus, such as GSTU11, GSTU12, SOD, and peroxidase4,15,17,18,31. An enzyme assay indicated a slight higher activity of antioxidant enzymes in resistant B. napus genotype than that in the susceptible genotype18. In our study, dozens of genes encoding ROS scavenging enzymes such as peroxidase, SOD and CAT were strongly increased in rice and the R B. oleracea. In addition, eight glutathione transferases (GSTs) which positively regulate GSH content and reduce ROS32 were up-regulated specifically in the R group (Supplementary Table 4). The histochemical staining and the antioxidant capacity assay further proved stronger antioxidant activity and lighter cell death in rice and the R B. oleracea than that in the susceptible B. oleracea. Therefore, we reason that the effective ROS scavenging systems in the resistant hosts possibly help to alleviate and delay the cell death, and thus defend the invasion of the necrotrophic fungus which obtains nutrients from necrotic host tissues33,34.
By further comparison within B. oleracea, we detected an enhanced Ca2+ signalling in the S B. oleracea group according to the great up-regulation of CNGC12, CaMs and CDPKs. In a previous report15, several genes involved in calcium signal transduction were found to be up-regulated in a susceptible B. napus variety, including one gene (AT3G50770) which is homologous to a calmodulin-related gene (Bol002305) induced specifically in the S group in our study. Conversely, though the CNGC12 (Bol030676) was slightly enhanced at 12 hpi in the R group, the CaMs and CDPKs were repressed in the R group. Transient elevation of cytosolic Ca2+ always occurs after pathogen perception to activate downstream signalling cascade which includes activation of CaM and protein kinases, and generation of nitric oxide (NO) and ROS35,36,37,38,39. The activation of CDPKs, generation of NO and ROS, and increase in Ca2+ binding have been demonstrated to positively regulate programmed cell death (PCD)40. In multicellular organisms, apoptosis and autophagy are recognized as two principal means by which these genetically determined cell deaths occur. It was found that S. sclerotiorum hijacks host pathways via oxalic acid (OA) and induces cell death in host plant tissue resulting in hallmark apoptotic features in a time and dose dependent manner, while the autophagy is a defense response of plant in S. slerotiorum-plant interaction41. In the present study, heavier apoptosis was caused by S. sclerotiorum in susceptible B. oleracea as compared with that in resistant B. oleracea. Therefore, the enhanced Ca2+ signalling in the S group possibly promotes apoptosis and thus helps the propagation of S. sclerotiorum, while the suppressed Ca2+ signalling in the R group may contribute to the inhibition the expansion of nectrophic hyphae.
In this study, diverse WRKY TFs were activated in the resistant and susceptible B. oleracea. Except for a few WRKYs that activated in both groups, we further found specific changes for some WRKYs between the two groups. For example, we found up-regulation of WRKY4 (Bol010361) specifically in the R group (Supplement Table 4). It is reported that WRKY4 in Tamarix hispida activated SODs and peroxidase under stress, thus protected cells from death42. Whereas, two copies of WRKY11 (Bol017448, Bol033735) and one WRKY17 (Bol041910) were up-regulated only in the S group (Supplement Table 4). In previous studies, WRKY17 from cotton (Gossypium hirsutum) increased ROS level and enhanced oxidative damage in transgenic tobacco (Nicotiana benthamiana) in response to stresses43, and WRKY11 also positively regulated ROS and caused severe cell death in tobacco leaves44. It provides another proof on that, the resistant B. oleracea possibly activated antioxidant enzymes to scavenge ROS, while the susceptible B. oleracea promoted the accumulation of ROS. In addition, WRKY50, WRKY51 and WRKY70 were revealed to positively modulate the downstream SA-signalling which regulates the HR based PCD and negatively regulate the resistance against necrotrophic pathogens28,29,30. The unique repression of these WRKYs (Bol012741, Bol022304, Bol015902, Bol019066 and Bol044275) and the PR-1 homologs (Bol019271 and Bol006477) in the R group indicated a repression of SA signalling in resistant B. oleracea in response to S. sclerotiorum. These data suggest a more effective WRKY network in resistant B. oleracea in protecting the host cells from death.
Ribosome is the place to biosynthesize proteins in eukaryotes, and the inactivation of ribosome by ribosome-inactivating proteins (RIPs) in plants is association with broad-spectrum resistance to pathogens45,46,47. In this study, a fast and severe repression on genes encoding ribosome proteins was detected in the resistant B. oleracea after inoculation. It seems that the RIP-dependent resistance may be also associated with modulation of cell death in host46,47. Further investigations are needed in future research.
Conclusions
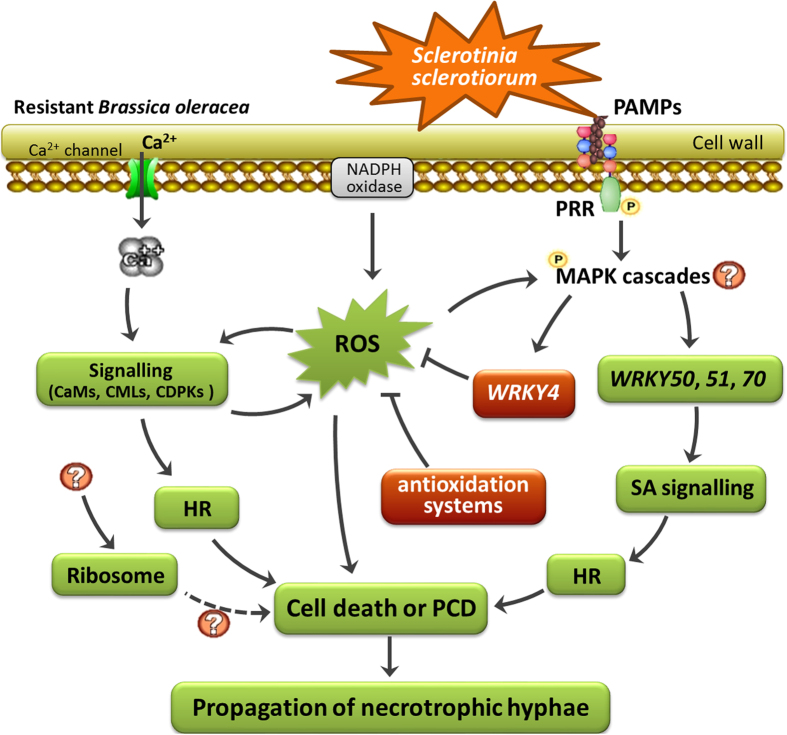
High antioxidant activity was induced in rice and the resistant B. oleracea during the inoculation of S. sclerotiorum. The resistant B. oleracea possesses an efficient network to impair cell death, including the activation on antioxidant genes and WRKYs, and the suppression on Ca2+ signalling and perhaps ribosome (Fig. 6). This study reveals diverse modulations on cell death among hosts, and suggests an important role of impairing cell death via multiple ways in the resistant B. oleracea to defend against S. sclerotiorum.
Figure 6. Putative network in resistant Brassica oleracea to impair propagation of Sclerotinia sclerotiorum during early infection phase (0 to 12 hpi) via modulating cell death and programmed cell death (PCD).
The enhanced and repressed nodes were indicated in red and green backgrounds, respectively.
Methods
Plant materials
The resistance value against S. sclerotiorum was previously recorded over two years in an F2 population that composed of 149 vegetatively propagated lines derived from a cross between ‘C01’ and a susceptible cultivated genotype ‘C41’3,20. Based on the resistance performance, three extrem resistant F2 lines were selected and pooled with ‘C01’ as the R group, and six highly susceptible F2 lines were bulked with ‘C41’ as the S group. The two groups exhibited 94.7% genetic difference within the QTL region (18 of 19 QTL-linked markers exhibited different band patterns in the two groups), and 94.9% genetic similarity in the non-QTL region (410 of 432 markers showed the same band patterns in the two groups). The plants were grown in the field, with 30 cm inter-row and 25 cm intra-row spacing. A rice line (O. sativa ssp. indica), ‘93–11’ was grown in the field under normal management. All materials were moved into growth chamber one week before inoculation.
Inoculation and tissue harvest
A S. sclerotiorum isolate from a previous study3 was used. Mycelium-agar plugs (6-mm-diameter) punched from the growing margin of 3-day-old cultures of S. sclerotiorum on potato dextrose agar medium were used as inoculum. The third true leaf at nine-leaf stage of B. oleracea and the second leaf of rice seedling were inoculated by attachment of the prepared fungal plugs. The inoculated leaves were sealed with cling film and incubated at 22 °C and 85% relative humidity. As a criterion of resistance reaction, lesion size was measured at 3 dai. The resistance evaluation was performed across two years with three replications. Leaf tissues extending 10 mm beyond the inoculation site at 0, 6, and 12 hpi were excised for RNA extraction.
Transcriptome sequencing and qRT-PCR analysis
Total RNA was extracted using TRNzol-A + Reagent (TianGen, Beijing, China) and the B. oleracea RNA were pooled according to resistance level at each time point, resulting in three R and three S samples, i.e. R0, R6, R12, S0, S6 and S12. Accordingly, the RNA samples of rice were encoded as Os0, Os6 and Os12. Nine library preparations were generated and sequenced on an Illumina Hiseq 2000TM platform. The sequences were base-called and quality-checked by the Illumina data processing pipeline. The raw reads were filtered to obtain high-quality clean reads by removing adaptor sequences, duplicated sequences, reads containing more than 5% “N” (i.e., ambiguous bases in reads), and reads in which more than 50% of the bases showed a Q-value (i.e., Bonferroni-adjusted P value) ≤ 5. Clean reads were aligned to B. oleracea v1.0 genome sequence (http://brassicadb.org/brad/index.php) and the rice genome annotation project data (http://rice.plantbiology.msu.edu/downloads_gad.shtml) using SOAP2, respectively. The uniquely mapped reads were used for gene expression analysis. The abundances of all unigenes were estimated using RPKM (reads per kilobyte per million mapped reads)48. DEGs were identified according to Audic and Claverie49 and Dang et al.50. The threshold determining the significance of DEGs among multiple tests was set at a false discovery rate (FDR) ≤ 0.001 and |log2(folds change)| ≥ 1.
Heatmap was constructed among all samples by the ‘gplots’ package of R using the function ‘heatmap.2.’ . All DEGs of rice and B. oleracea were firstly aligned to Arabidopsis genes according to information from http://plants.ensembl.org/Oryza_sativa/Info/Index and http://www.ocri-genomics.org/bolbase/at_synviewer.htm, and the expressions of the corresponding Arabidopsis homologs were then used to build the heatmap.
Functional enrichment analyses, consisting of GO and KEGG analyses, were performed using the ultra-geometric test51. A FDR-value ≤ 0.05 was set as the threshold to determine significant enrichments of GO terms and KEGG pathways in B. oleracea and rice.
Analysis of qRT-PCR was performed with three replications using a CFX96™ Real-Time PCR Detection System to validate the transcription patterns. The PCR cycling conditions comprised 1 cycle of 95 °C for 30 s, then 39 cycles of 95 °C for 5 s and 55–70 °C for 1 min, followed by a melting curve ramping from 65 °C to 95 °C with temperature increasing by 0.5 °C every 5 s (1 cycle). Actin3 and Actin7 were used as the internal control.
Histochemical staining
To detect hydrogen peroxide in situ, inoculated leaves and mocks (inoculated with PDA agar discs) were infiltrated with DAB (Sigma-Aldrich; 1 mg/ml DAB-HCl, pH 3.8) solution at 6 and 12 hpi according to Fryer et al.52. Trypan blue staining was applied as described by Frye and Innes53 to visualize the mycelium of pathogen in host leaves at 36 hpi, and to reveal the dead cells in hosts at 12 hpi. Apoptosis was visualized in B. oleracea at 6 hpi with DAPI (Beyotime, China). Leaf discs under inoculums were fixed with methanol-acetone (3:1) for 24 h at room temperature, and then treated with HCl (1 mol/L) at 60 °C for 8 min. After 5-min incubation with DAPI at room temperature, leaf discs were washed with PBS buffer (0.05 mol/L, pH 7.8) for three times. Tissues from the edge of leaf discs were viewed using a fluorescence microscope (Olympus, Japan).
Antioxidant enzyme activity assay
Total antioxidant capacity was tested in the pathogen- and agar disc-inoculated leaf samples in rice, R and S B. oleracea at 0, 6 and 12 hpi by using a 2,2-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid (ABTS) method, with three biological replicates. All the samples were immediately ground to fine powders in liquid nitrogen. Approximately 0.2 g powder of each sample was added to 10 ml cold phosphate buffer (0.05 mol/L, pH 7.8), and centrifuged (6000 r/min) for 15 min. The liquid was then tested using a Total Antioxidant Capacity Assay Kit (Beyotime, China) following the manufacturer’s instruction.
Additional Information
How to cite this article: Mei, J. et al. Transcriptomic comparison between Brassica oleracea and rice (Oryza sativa) reveals diverse modulations on cell death in response to Sclerotinia sclerotiorum. Sci. Rep. 6, 33706; doi: 10.1038/srep33706 (2016).
Supplementary Material
Acknowledgments
We thank Zhonglin Mou (University of Florida) and Joseph Onwusemu Disi (Auburn University) for kindly comments on the manuscript. This study was financially supported by the 973 Program (2015CB150201), National Nature Science Foundation of China (31401861, and 31401411), Key Projects in National Science and Technology (2014BAD01B07) and Fundamental Research Funds for the Central Universities (XDJK2014B036 and 2362015XK05).
Footnotes
Author Contributions J.M., Y.D. and Y.L. conducted the experiment; J.M., Y.D. and W.Q. wrote the manuscript; C.T., H.D. and Q.X. analyzed the sequencing data; Y.Y., H.W., J.Y., S.L. and J.L. performed data analysis; W.Q. directed the project.
References
- Boland G. & Hall R. Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol 16, 93–108 (1994). [Google Scholar]
- Purdy L. Sclerotinia sclerotiorum: history diseases and symptomatology, host range, geographic distribution and impact. Phytopathology 69, 875–890 (1979). [Google Scholar]
- Mei J. Q. et al. Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor Appl Genet 126, 549–556 (2013). [DOI] [PubMed] [Google Scholar]
- Wei L. et al. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. Identification of QTLs for resistance to sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. Plos One 8, e67740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. W. et al. Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor Appl Genet 112, 509–516 (2006). [DOI] [PubMed] [Google Scholar]
- Wei D. Y. et al. Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol Breeding 34, 1797–1804 (2014). [Google Scholar]
- Chen X. et al. Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep 32, 1589–1599 (2013). [DOI] [PubMed] [Google Scholar]
- Rietz S., Bernsdorff F. E. M. & Cai D. G. Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J Exp Bot 63, 5507–5519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. Overexpression of Brassica napus MPK4 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Mol Plant Microbe Interact 22, 235–244 (2009). [DOI] [PubMed] [Google Scholar]
- Yang B., Jiang Y. Q., Rahman M. H., Deyholos M. K. & Kav N. N. V. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. Bmc Plant Biol 9, 68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. W. et al. Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Molecular Plant Pathology 10, 635–649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Mol Plant Pathol 15, 677–689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Sci 184, 75–82 (2012). [DOI] [PubMed] [Google Scholar]
- Yang B., Sivastava S., Deyholos M. K. & Kav N. N. V. Transcriptional profiling of canola (Brassica napus L.) responses to the fungal pathogen Sclerotinia sclerotiorum. Plant Sci 173, 156–171 (2007). [Google Scholar]
- Zhao J. W. et al. Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 227, 13–24 (2007). [DOI] [PubMed] [Google Scholar]
- Liang Y., Srivastava S., Rahman M. H., Strelkov S. E. & Kav N. N. Proteome changes in leaves of Brassica napus L. as a result of Sclerotinia sclerotiorum challenge. J Agric Food Chem 56, 1963–1976 (2008). [DOI] [PubMed] [Google Scholar]
- Wen L. et al. Using proteomic analysis to find the proteins involved in resistance against Sclerotinia sclerotiorum in adult Brassica napus. Eur J Plant Pathol 137, 505–523 (2013). [Google Scholar]
- Wu J. et al. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci Rep 6, 19007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J. et al. Identification of resistant sources against Sclerotinia sclerotiorum in Brassica crops with emphasis on B. oleracea. Euphytica 177, 393–400 (2011). [Google Scholar]
- Mei J. et al. Transfer of sclerotinia resistance from wild relative of Brassica oleracea into Brassica napus using a hexaploidy step. Theor Appl Genet 128, 639–644 (2015). [DOI] [PubMed] [Google Scholar]
- Ding Y. et al. Transfer of sclerotinia stem rot resistance from wild Brassica oleracea into B. rapa. Mol Breeding 35, 225 (2015). [Google Scholar]
- Beckman K. B. & Ames B. N. The free radical theory of aging matures. Physiol Rev 78, 547–581 (1998). [DOI] [PubMed] [Google Scholar]
- Tiew T. W., Sheahan M. B. & Rose R. J. Peroxisomes contribute to reactive oxygen species homeostasis and cell division induction in Arabidopsis protoplasts. Front Plant Sci 6, 658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart W., Chin K., Ung H., Moeder W. & Yoshioka K. The cyclic nucleotide-gated channels AtCNGC11 and 12 are involved in multiple Ca2+ -dependent physiological responses and act in a synergistic manner. J Exp Bot 62, 3671–3682 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart W. et al. The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Molecular Biology 65, 747–761 (2007). [DOI] [PubMed] [Google Scholar]
- Poovaiah B. W., Du L., Wang H. & Yang T. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol 163, 531–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G. & Palva E. T. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Kariola T. & Palva E. T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46, 477–491 (2006). [DOI] [PubMed] [Google Scholar]
- Gao Q. M., Venugopal S., Navarre D. & Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 155, 464–476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg H., Li H., Sivasithamparam K. & Barbetti M. J. Differentially expressed proteins and associated histological and disease progression changes in cotyledon tissue of a resistant and susceptible genotype of Brassica napus infected with Sclerotinia sclerotiorum. Plos One 8, e65205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson D. J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14, 1511–1527 (1998). [DOI] [PubMed] [Google Scholar]
- Dickman M. B. et al. Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc Natl Acad Sci USA 98, 6957–6962 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin E. M. & Levine A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10, 751–757 (2000). [DOI] [PubMed] [Google Scholar]
- Bindschedler L. V. et al. Early signalling events in the apoplastic oxidative burst in suspension cultured french bean cells involve cAMP and Ca+. New Phytol 151, 185–194 (2001). [DOI] [PubMed] [Google Scholar]
- Davies D. R., Bindschedler L. V., Strickland T. S. & Bolwell G. P. Production of reactive oxygen species in Arabidopsis thaliana cell suspension cultures in response to an elicitor from Fusarium oxysporum: implications for basal resistance. J Exp Bot 57, 1817–1827 (2006). [DOI] [PubMed] [Google Scholar]
- Harding S. A., Oh S. H. & Roberts D. M. Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. Embo J 16, 1137–1144 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T. et al. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca+ binding motifs. Plant Cell 10, 255–266 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Mizoguchi T., Yoshida R., Ichimura K. & Shinozaki K. Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Molecular Cell 41, 649–660 (2011). [DOI] [PubMed] [Google Scholar]
- Clarke A., Desikan R., Hurst R. D., Hancock J. T. & Neill S. J. NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24, 667–677 (2000). [DOI] [PubMed] [Google Scholar]
- Kabbage M., Williams B. & Dickman M. B. Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog 9, e1003287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. et al. A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol Biol 82, 303–320 (2013). [DOI] [PubMed] [Google Scholar]
- Yan H. et al. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol 55, 2060–2076 (2014). [DOI] [PubMed] [Google Scholar]
- Adachi H. et al. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27, 2645–2663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra R. & Saini R. Transformation of blackgram (Vigna mungo (L.) Hepper) by barley chitinase and ribosome-inactivating protein genes towards improving resistance to Corynespora leaf spot fungal disease. Appl Biochem Biotechnol 174, 2791–2800 (2014). [DOI] [PubMed] [Google Scholar]
- Corrado G. et al. Inducible expression of a Phytolacca heterotepala ribosome-inactivating protein leads to enhanced resistance against major fungal pathogens in tobacco. Phytopathology 95, 206–215 (2005). [DOI] [PubMed] [Google Scholar]
- Qian Q., Huang L., Yi R., Wang S. & Ding Y. Enhanced resistance to blast fungus in rice (Oryza sativa L.) by expressing the ribosome-inactivating protein alpha-momorcharin. Plant Sci 217–218, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., Mccue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Audic S. & Claverie J. M. The significance of digital gene expression profiles. Genome Res 7, 986–995 (1997). [DOI] [PubMed] [Google Scholar]
- Dang Z. H. et al. Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. Bmc Genomics 14, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z. & Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38, 64–70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer M. J., Oxborough K., Mullineaux P. M. & Baker N. R. Imaging of photo-oxidative stress responses in leaves. J Exp Bot 53, 1249–1254 (2002). [PubMed] [Google Scholar]
- Frye C. A. & Innes R. W. An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.