Summary
Mycobacterium tuberculosis proteins that are exported out of the bacterial cytoplasm are ideally positioned to be virulence factors; however, the functions of individual exported proteins remain largely unknown. Previous studies identified Rv0199 as an exported membrane protein of unknown function. Here, we characterized the role of Rv0199 in M. tuberculosis virulence using an aerosol model of murine infection. Rv0199 appears to be a member of a Mce-associated membrane (Mam) protein family leading us to rename it OmamA, for orphaned Mce-associated membrane protein A. Consistent with a role in Mce transport, we showed OmamA is required for cholesterol import, which is a Mce4-dependent process. We further demonstrated a function for OmamA in stabilizing protein components of the Mce1 transporter complex. These results indicate a function of OmamA in multiple Mce transporters and one that may be analogous to the role of VirB8 in stabilizing Type IV secretion systems, as structural similarities between Mam proteins and VirB8 proteins are predicted by the Phyre 2 program. In this study, we provide functional information about OmamA and shed light on the function of Mam family proteins in Mce transporters.
Keywords: Mycobacterium tuberculosis, Mce, virulence, cholesterol, transporter, Mam
Introduction
Mycobacterium tuberculosis is a human pathogen with a significant impact on world health. Current estimates suggest that 2 billion people worldwide have been infected with M. tuberculosis and 1.5 million people die per year from tuberculosis (World Health Organization, 2014). Tuberculosis is spread by inhaled droplets and, upon entering the lungs, M. tuberculosis is phagocytosed by macrophages where it then replicates intracellularly. When inside macrophages, M. tuberculosis inhibits phagosome maturation and apoptosis, resists reactive radicals, and acquires nutrients through specialized systems (Deretic, 2008; Sturgill-Koszycki et al., 1994; Briken, 2013; Hinchey et al., 2007; Darwin et al., 2003; Niederweis, 2008). In addition, M. tuberculosis produces penetrations in the phagosome membrane providing the pathogen with cytosolic access (van der Wel et al., 2007; Simeone et al., 2015; Manzanillo et al., 2012). While these activities help explain M. tuberculosis survival in macrophages, the molecular basis of these events is not clear. Many intracellular pathogens, including M. tuberculosis, survive in macrophages with the help of proteins that are exported from the bacterial cytoplasm to the bacterial cytoplasmic membrane, cell wall, or into the host environment.(Ligon et al., 2012; Hicks & Galan, 2013; Isaac & Isberg, 2014). Because of their extracytoplasmic location, exported proteins of pathogens are ideally positioned for host interactions and for specific roles in controlling the immune response and surviving in macrophages (Forrellad et al., 2013; McCann, 2009). While exported proteins are known to play a critical role in M. tuberculosis virulence, up to 69% of M. tuberculosis exported proteins have no assigned function (Perkowski, E. and Braunstein, M. manuscript in preparation). To better understand how M. tuberculosis interacts with the host and causes disease, it is critical to identify the function of exported proteins.
In a previous study using a transposon carrying a β-lactamase reporter of export, the M. tuberculosis Rv0199 protein was identified as an exported protein (McCann et al., 2011). Rv0199 is a small 24kDa (219 amino acid) protein with a single TMHMM predicted transmembrane (TM) domain (amino acids 42-64) (Krogh et al., 2001) near its N-terminus (Figure 1A). When fused to a membrane protein, the β-lactamase reporter identifies exported domains that are positioned on the extracytoplasmic side of the membrane by producing β-lactam resistance. The site of the β-lactam resistant transposon insertion in omamA produces a hybrid protein with β-lactamase fused at amino acid 74 of OmamA, which indicates the larger C-terminal portion of the protein, following the TM domain, is on the periplasmic/cell wall side of the membrane (Figure 1A) (McCann et al., 2011).
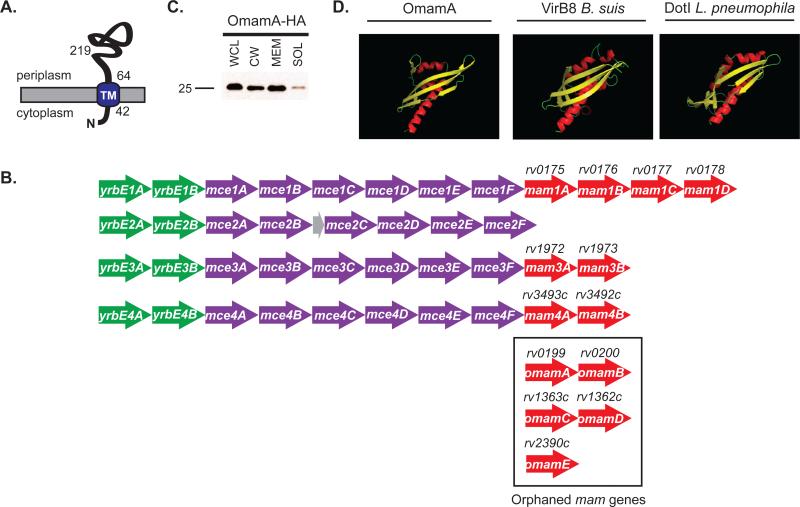
Figure 1.
Rv0199 (OmamA) is a transmembrane protein predicted to be a Mce-associated protein. A. OmamA is predicted to have a single N-terminal transmembrane domain (TM) at amino acid 42-64 (Krogh et al., 2001) and C-terminal domain exposed to the cell wall side of the membrane (McCann et al., 2011). B. OmamA is predicted to be a Mce-associated membrane (Mam) protein, however, rv0199 is not located in a mce operon. Mce operons are typically organized by two yrbE genes upstream (green), six mce genes (purple) and most have pairs of mam genes (red) downstream. Genes encoding putative orphaned mam genes are boxed. Genes encoding Omam proteins are distinguished by being distally located from mce operons (Casali & Riley, 2007). The mce2 operon additionally contains a small predicted pseudogene (grey). C. The omamAmtb gene was engineered in frame with an HA tag and expressed in M. smegmatis. Cells were lysed to generate whole cell lysates (WCL) and fractionated by differential ultracentrifugation into cell wall (CW), cell membrane (MEM), and cytoplasmic containing soluble (SOL) fractions. Results are representative of at least three independent replicates. D. Phyre 2, an online structural prediction program, predicted with high confidence (96%) that OmamA forms a NTF2-like fold. Ribbon diagrams shown represent the Phyre 2 predicted structures of OmamA colored by secondary structure in Pymol. Ribbon diagrams representing the solved crystal structures of VirB8 from Brucella suis and DotI from Legionella pneumophila are shown for comparison. Alpha helices are colored in red, Beta-strands in yellow, and turns in green.
Studies of pooled mutant libraries in macrophages or mice, predict Rv0199 to play a role in virulence (Sassetti & Rubin, 2003; Zhang et al., 2013; Stewart et al., 2005), and experiments directly testing a rv0199 transposon mutant and complemented strain in cultured macrophages confirm a role for Rv0199 in intracellular growth (McCann et al., 2011). The rv0199 gene is a core mycobacterial gene (Marmiesse et al., 2004), which means that it is highly conserved throughout pathogenic and non-pathogenic mycobacterial species but not conserved outside of actinomycetes. Yet, the function of Rv0199 is not clear, and Rv0199 is annotated as a membrane protein of unknown function (Lew et al., 2011). There is, however, limited sequence homology between Rv0199 and a family of proteins encoded by genes linked to mce operons, which express components of Mce transporters. When first noted, these linked genes were referred to as Mce-associated (mas) genes (Casali & Riley, 2007). However, to avoid confusion with the mas gene encoding mycocerosic acid synthase of M. tuberculosis, we refer to them as Mce-associated membrane (mam) genes. Like Rv0199, the encoded Mam proteins have predicted transmembrane domains near their N-terminus (Perkowski, E. and Braunstein, M. manuscript in preparation). In M. tuberculosis, there are eight mam genes linked to mce operons and an additional five genes encoding proteins with low levels of homology to Mam proteins that are scattered elsewhere in the genome (Figure 1B) (Casali & Riley, 2007). In this report, we will refer to these orphaned mam genes as omam genes to indicate their unlinked nature. Rv0199 is encoded by one of these orphaned mam genes and is referred to as OmamA in this study.
Mce transporters are multi-protein complexes considered to be functionally analogous to ABC transporters (Casali & Riley, 2007). The four Mce transporter systems in M. tuberculosis all play roles in virulence (Gioffre et al., 2005; Shimono et al., 2003; Marjanovic et al., 2010; Lima et al., 2007; McCann et al., 2011; Senaratne et al., 2008; Pandey & Sassetti, 2008) and are thought to function in lipid uptake. The best characterized Mce transporter is Mce4. Mce4 is required for cholesterol uptake (Pandey & Sassetti, 2008; Mohn et al., 2008), cholesterol being an important nutrient during M. tuberculosis infection (Pandey & Sassetti, 2008). Emerging evidence suggests that Mce1 is responsible for import of mycolic acids, long chain fatty acids characteristic of mycobacteria (Forrellad et al., 2014; Cantrell et al., 2013). Each mce operon encodes two YrbE proteins with similarity to ABC transporter permeases and six Mce proteins that are considered functionally similar to substrate binding proteins of ABC transporters (Casali & Riley, 2007). Additionally, Mce transporters are thought to share a common ATPase, MceG. Interestingly, the mceG gene is not located near any mce operon (Casali & Riley, 2007; Joshi et al., 2006). Nearly all mce operons also contain genes encoding Mam proteins (Figure 1B). Unlike the YrbE and Mce components, Mam proteins share no analogous features with ABC transporter components. Mam proteins are speculated to have a role in Mce transporter systems, but this idea is solely based on the genomic location of mam genes. To date, there have been no functional studies of any Mam protein. Consequently, the function of potential orphaned Mam proteins such as Rv0199, whose genes are distal to mce operons, is even less clear.
Here, we further characterized the role of Rv0199 in M. tuberculosis virulence using a low dose aerosol model of murine infection. We additionally showed that Rv0199 has a role in Mce lipid transport, leading us to rename Rv0199 as OmamA (orphaned Mce-associated membrane protein A), and we demonstrated a role for OmamA in stabilizing Mce1 transporter complexes. The stabilization function of OmamA may be analogous to the role of VirB8 in stabilizing Type IV secretion systems, as structural similarities between Mam proteins and VirB8 proteins are predicted by the structural prediction program Phyre 2 (Kelley & Sternberg, 2009). Our results provide important functional information about an exported protein with a role in virulence and provide the first evidence for any Mam protein functioning with Mce transporters. Finally, our results suggest that OmamA, and possibly other Mam proteins as well, have a structural role important for the stability of Mce transporters.
Results
OmamA is important for murine infection
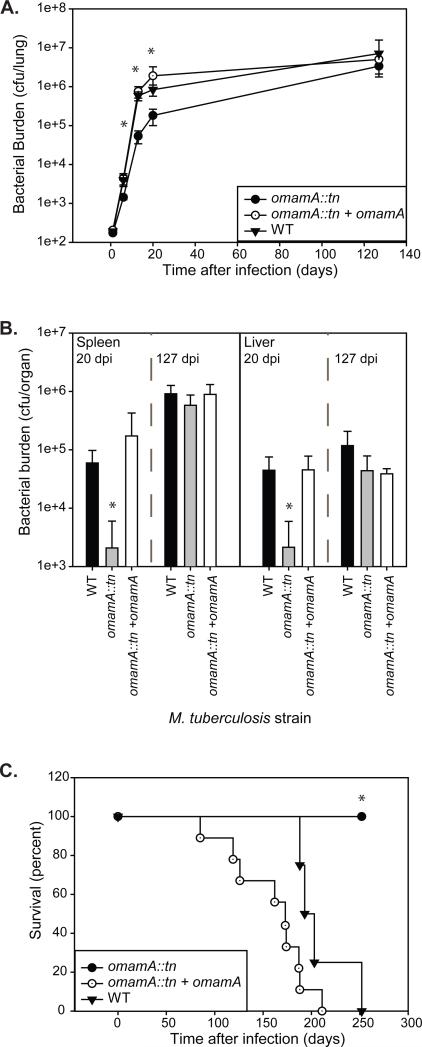
Previous studies revealed a transposon insertion in the M. tuberculosis rv0199 gene, hereafter referred to as omamA, results in a growth defect in resting murine bone-marrow derived macrophages (McCann et al., 2011) and TraSH analysis of a pooled M. tuberculosis mutant library in mice predicts a role for OmamA during host infection (Sassetti & Rubin, 2003; Zhanget al., 2013). To further explore the role of the OmamA protein in M. tuberculosis infection, we evaluated the course of murine infection with the omamA transposon mutant (omamA::tn) and compared it to infection with an omamAWT strain, hereon referred to as wild type (WT) (McCannet al., 2011). Groups of C57BL/6 mice were infected by low dose aerosol with WT, omamA::tn, or a complemented omamA::tn +omamA strain. Mice infected with the omamA mutant had lower bacterial burden in the lungs at 6, 13, and 20 days post-infection compared to WT (Figure 2A). However, by 127 days post-infection there was no longer any difference in bacterial burden in the lungs of mice infected with the omamA mutant strain as compared to WT infected mice (Figure 2A). Because aerosol delivered M. tuberculosis is not detected in the spleen or liver at the early time points, we quantitated the bacterial burden in these organs at 20 and 127 days post-infection only. The omamA infected mice had reduced bacterial burden compared to WT in spleen and liver at 20 days post-infection and, like the burden in the lungs, the number of omamA mutant bacteria reached equivalent levels to WT by 127 days post-infection (Figure 2B). Importantly, all defects in bacterial burden were fully restored in mice infected with the complemented strain. These data indicate that OmamA is important for early exponential phase growth in the mouse model of infection.
Figure 2.
OmamA is required for early growth and virulence during murine infection. C57BL/6 mice were infected with a low dose aerosol of WT, omamA::tn, or omamA::tn +omamA complemented strains. Groups of four mice per strain were sacrificed at various days post infection (dpi) and bacterial burden (colony forming units, cfu) was assessed by plating from A. lung homogenates or B. spleen and liver homogenates. No significant differences were observed in the initial lung burden determined one day post infection (WT 193 +/− 10, omamA::tn 174 +/− 9 and omamA::tn +omamA 206 +/− 18). C. Groups of mice were monitored for survival. * indicates p<0.05 as compared to WT. tn indicates transposon insertion. Error bars represent standard deviation. Results are representative of two independent experiments comparing WT (MBTB178), omamA::tn (MBTB319), and omamA::tn + omamA (MBTB320).
We also assessed long-term survival of mice infected with the strains described above. The omamA mutant infected animals survived significantly longer (>250 days) compared to WT (193 days average) and the complemented strain (173 days average) (Figure 2C). The complemented strain not only alleviated the attenuated phenotype of the omamA mutant, but also appeared to potentially accelerate time to death in comparison to WT M. tuberculosis (p=0.05). The behavior of the complemented strain may be due to non-physiological levels of OmamA, as the gene is expressed off the constitutive hsp60 promoter on a multi-copy plasmid.
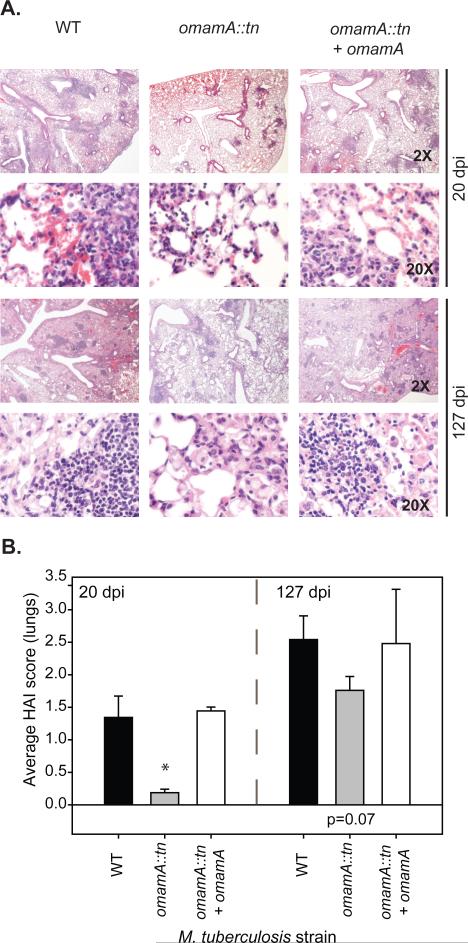
H&E stained lung sections demonstrated that mice infected with the omamA mutant displayed reduced inflammatory infiltration and increased open alveolar spaces in comparison to WT infected mice. The omamA mutant showed this reduced histopathology in both early (Day 20) and late (Day 127) timepoints, and the phenotypes were fully restored in the complemented strain (Figure 3A). Blinded scoring of these sections demonstrated that the omamA mutant infected mice had lower histopathology scores (histological activity index, HAI) early during infection compared to WT infected mice (Figure 3B). Even after the bacterial burden in omamA infected mice caught up to WT levels (Day 127) the HAI scores trended lower in omamA mutant infected mice compared to WT infected mice (p=0.07). The lower histopathology of the omamA mutant infected mice may help account for their longer survival time in comparison to WT infected mice.
Figure 3.
Mice infected with the omamA mutant have reduced histopathology compared to WT infected mice. A. A single lung lobe from was fixed and H&E stained for histology. Shown are representative images captured under 2X and 20X magnification from 20 and 127 days post infection comparing WT (MBTB178), omamA::tn (MBTB319), and omamA::tn + omamA (MBTB320). B. Average histological activity index (HAI) scores were determined by an experienced blinded reviewer.
OmamA is an exported protein with predicted structural similarity to VirB8 and Mce-associated membrane proteins
A β-lactamase reporter was previously used to identify OmamA as an exported protein of M. tuberculosis (McCann et al., 2011). To confirm the exported nature of OmamA, subcellular fractions of a Mycobacterium smegmatis strain engineered to express a C-terminal HA tagged OmamA were prepared for Western blot analysis. OmamA-HA primarily localized to the membrane and cell wall fractions of M. smegmatis, with a smaller fraction of OmamA-HA being detected in the soluble fraction, which includes cytoplasmic material (Figure 1C). This result supports the identification of OmamA as an exported protein.
Consistent with a prior bioinformatics analysis (Casali & Riley, 2007), ClustalW2 (Goujon et al., 2010; Thompson et al., 2002) revealed the C-terminal region of OmamA to have a low level of sequence identity (~10-25%) with Mce-associated membrane (Mam) proteins of actinomycetes. Mam proteins are uncharacterized proteins found downstream of mce operons (Casali & Riley, 2007). However, the omamA gene is not linked to a mce operon, leading us to call it an orphaned mam gene (omam). ClustalW2 reveals 10-25% identity between any two Mam proteins, which is similar to the low homology shared between OmamA and Mam proteins (Supplemental Figure 1). To gain more insight into potential functional domains of the OmamA protein, we used an online 3D structural prediction program Phyre 2 (protein homology/analogy recognition engine version 2.0) (Kelley & Sternberg, 2009). Phyre 2 predicted that the C-terminal domain of OmamA (aa 69-212) folds similarly to NTF2 family proteins (Supplemental Figure 2) (Chaillan-Huntington et al., 2001). Phyre 2 also predicted structural similarity between the C-terminal domain of OmamA and NTF2-like domains in some bacterial proteins. Notable matches were to the structures of VirB8 proteins from Brucella suis and Agrobacterium tumefaciens and DotI of Legionella pneumophila (Figure 1D, Supplemental Figure 2) (Smith et al., 2012; Bailey et al., 2006; Terradot et al., 2005; Kuroda et al., 2015). Like OmamA, VirB8 is a small protein, 26 kDa, with an N-terminal transmembrane domain, and the majority of the protein localized to the periplasm. Additionally, like OmamA, in B. abortus VirB8 plays an important role during infection of both mice and macrophages (den Hartigh et al., 2008). VirB8 is a component of the type IV secretion system, a large multi-protein transporter, and it is important to both the stability and function of the transporter complex (Kumar et al., 2000; Sivanesan & Baron, 2011; den Hartigh et al., 2008). DotI is the presumed VirB8 counterpart of the L. pneumophila type IV secretion system, Dot/Icm (Kuroda et al., 2015). To determine whether these structural predictions for OmamA are shared with Mam family proteins, we used Phyre 2 to predict the structure of all M. tuberculosis Mam proteins. Strikingly, like OmamA, all M. tuberculosis Mam family proteins had high confidence structural predictions to NTF2 domain containing proteins, including VirB8 and DotI (Supplemental Figure 2). Given the similarity between the structural predictions of OmamA, Mam and VirB8 proteins, we hypothesized a function of OmamA in Mce transporters, possibly a function analogous to that of VirB8 stabilizing multi-protein transporter complexes.
Deletion of omamA in Mycobacterium smegmatis leads to a mce mutant morphology phenotype
With the goal of assigning a function to OmamA, we first explored the potential for OmamA to contribute to Mce transport in the mycobacterial model organism M. smegmatis. M. smegmatis has six mce operons with nine mce-associated mam genes and ten orphaned omam genes (Casali & Riley, 2007) (Supplemental Figure 3). In M. smegmatis, msmeg0235 is the ortholog of omamA, and will be referred to as omamAms. Like OmamAmtb, the OmamAms protein has a predicted transmembrane domain near the N-terminus, and OmamAms has 55% identity and 76% similarity to OmamAmtb in the C-terminal domain according to BLAST (Altschul et al., 1990). We constructed a deletion mutant of omamAms and compared phenotypes of the omamAms mutant to those of M. smegmatis mutants lacking mce4 or all six M. smegmatis mce operons (mce6X) (Klepp et al., 2012).
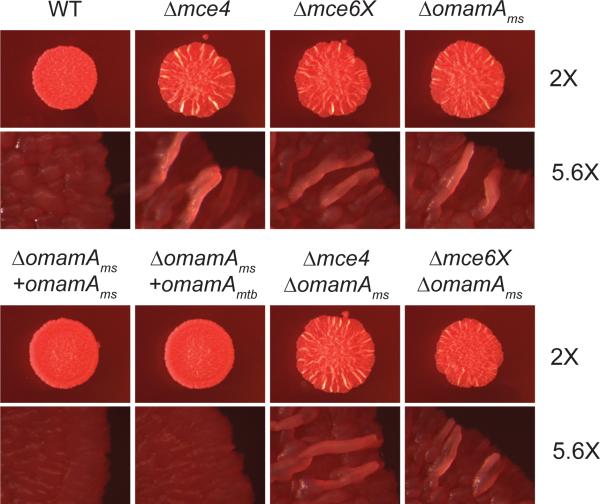
Previous studies revealed a rugose morphology for the mce6X M. smegmatis mutant growing on Mueller Hinton agar plates containing Congo red (Klepp et al., 2012). Consequently, we tested whether the omamAms mutant displays a similar morphology. Plates were incubated at 37°C for two days and morphology was assessed by low-magnification microscopy. WT M. smegmatis displayed flat, shiny colonies, but mce4, mce6X, and the omamAms mutants displayed rugose morphology (Figure 4). The rugose phenotype of the omamAms mutant could be complemented by either expression of omamAmtb or omamAms from a plasmid (Figure 4). While the basis of the mce mutant rugose phenotype is not currently understood, the appearance of a similar phenotype for the omamAms M. smegmatis mutant is consistent with a role for OmamA in Mce transporters.
Figure 4.
The omamAms mutant shares a morphology phenotype with mce operon mutants. Two μL spots of culture were plated on Mueller Hinton plates containing glucose and Congo red. The resulting colonies were visualized after 2 days at 2X and 5.6X magnification (Leica M420 macroscope). Results are representative of at least three independent experiments comparing WT +pMV261 (EP1182), Δmce4 +pMV261 (EP1204), Δmce6X +pMV261 (EP1208), ΔomamA +pMV261 (EP1193), ΔomamA +omamAms (EP1194), ΔomamA +omamAmtb (EP1203), ΔomamAΔmce4 +pMV261 (EP1206), and ΔomamAΔmce6X +pMV261 (EP1210). pMV261 is an empty vector, omamA expression constructs are cloned in pMV261.
Double mutants omamAmsmce4 and omamAmsmce6X mutants were also constructed and tested for possible epistatic interactions. Double mutants were spotted and compared to single mce4 or mce6X mutants (Figure 4). If the rugose phenotype of the omamAms mutant is due to the effective loss of Mce transport, the double omamAmsmce4 and omamAmsmce6X should look like single mce4 or mce6X mutants. If rugosity of the omamAms mutant is independent of Mce transporter function, an additive effect on rugose morphology from losing both mce operons and omamAms may occur. The double mutant phenotype was indistinguishable from that of the single mutants, suggesting that OmamA functions in the Mce transporter pathway.
OmamA is required for cholesterol utilization
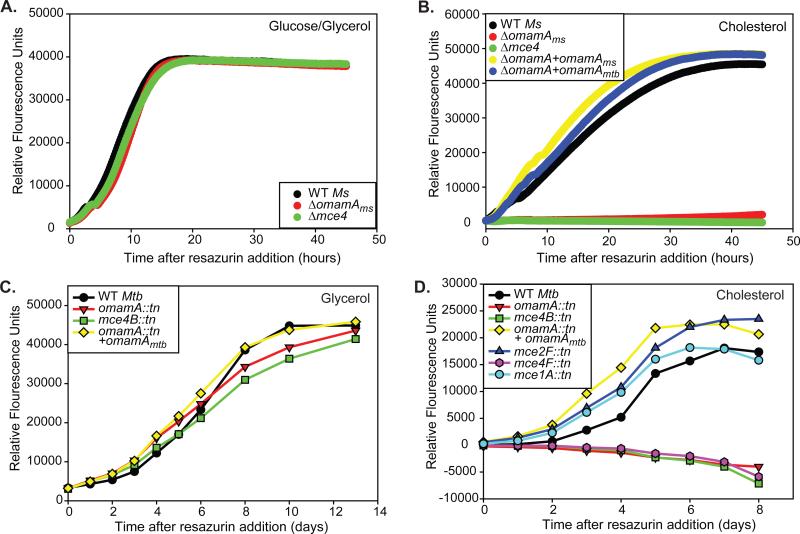
The Mce4 transporter is the best characterized Mce system with a demonstrated function in cholesterol import. Mycobacterial mutants lacking the mce4 operon are defective in cholesterol uptake and growth on cholesterol as a sole carbon source (Klepp et al., 2012; Pandey & Sassetti, 2008). To test whether OmamA contributes to Mce4 function, we assayed the omamAms mutant for its ability to utilize cholesterol as a sole carbon source. For these experiments, we followed the metabolic activity of mycobacteria using resazurin as previously described (Hayden et al., 2013). Resazurin is a blue dye that converts to a pink fluorescent compound when reduced by metabolically active cells. M. smegmatis strains were grown in minimal media supplemented with standard glucose and glycerol carbon sources or cholesterol as the sole carbon source. In glucose and glycerol containing media, both WT and the omamAms mutant reduced resazurin over time (Figure 5A). However, in media with cholesterol as a sole carbon source, resazurin reduction was observed with WT M. smegmatis but the omamAms mutant showed very little to no resazurin reduction. Strikingly, the behavior of the omamAms mutant in cholesterol media was equivalent to that of the mce4 M. smegmatis mutant (Figure 5B). The cholesterol phenotype of the omamAms mutant could be fully complemented by expression of either omamAms or omamAmtb from a plasmid (Figure 5B).
Figure 5.
OmamA is required for M. smegmatis and M. tuberculosis to utilize cholesterol. A. 104 colony forming units (cfu) of M. smegmatis strains were added to M9 glucose/glycerol and metabolic activity was monitored by resazurin conversion over time. B. 104 cfu of M. smegmatis were added to minimal M9 media plus cholesterol, and metabolic activity was monitored by resazurin conversion over time. Relative fluorescence unit measurements in cholesterol media are reported after subtraction of the minimal signal from no carbon source. 104 cfu of M. tuberculosis were added to minimal Sauton's media supplemented with C. glycerol or D. cholesterol, and metabolic activity was monitored by resazurin conversion over time. Relative fluorescence unit measurements in cholesterol media are reported after subtraction of the minimal signal from no carbon source. Results are representative of at least three independent experiments. M. smegmatis (Ms) strains: WT +pMV261 (EP1182), Δmce4 +pMV261 (EP1204), ΔomamA +pMV261 (EP1193), ΔomamA +omamAms (EP1194), and ΔomamA +omamAmtb (EP1203). M. tuberculosis (Mtb) strains: WT (MBTB178), omamA::tn (MBTB319), omamA::tn + omamA (MBTB320), mce2F::tn (MBTB156), mce1A::tn (MBTB204), mce4B::tn (MBTB329), mce4F::tn (MBTB288).
To determine whether OmamA also contributes to Mce4 function in M. tuberculosis, we similarly tested the M. tuberculosis omamA mutant for a defect in utilization of cholesterol as a sole carbon source. M. tuberculosis transposon mutants in several mce operons, including the mce4 operon (insertion mutants in the mce4B and mce4F genes), mce1 (mce1B), and mce2 (mce2F) were tested in parallel with the omamA mutant and complemented strains. In glycerol media, the omamA mutant behaved like WT in reducing resazurin over time (Figure 5C). However, in cholesterol media, the omamA mutant and mce4 mutants with transposon insertions in mce4B or mce4F were unable to utilize cholesterol as a sole carbon source (Figure 5D). As with the M. smegmatis cholesterol experiments, the omamA and the mce4 mutants of M. tuberculosis exhibited the same level of defect in cholesterol media. The omamA mutant was fully complemented by expression of omamAmtb in the complemented strain. Transposon mutants interrupting mce1 and mce2 operons displayed no defect for utilization of cholesterol (Figure 5D), consistent with previous reports (Pandey & Sassetti, 2008; Griffin et al., 2011). These data demonstrate that OmamA is required for cholesterol utilization in both M. smegmatis and M. tuberculosis.
OmamA is required for cholesterol uptake
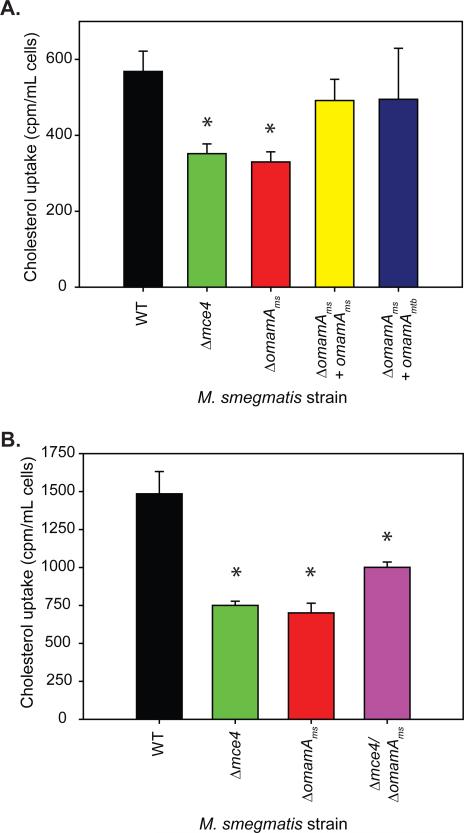
The cholesterol growth defects of omamA mutants of M. smegmatis or M. tuberculosis were indistinguishable from those of mce4 mutants, suggesting a role of OmamA in Mce4 cholesterol import. To directly test whether OmamA is required for cholesterol import, as opposed to playing a role in downstream cholesterol metabolism, we tested the ability of WT M. smegmatis, the omamAms mutant, and complemented strains to import radioactively labeled cholesterol. M. smegmatis strains were grown overnight in media with glucose and glycerol and then incubated for two hours in minimal media with C14 labeled cholesterol as the sole carbon source. After incubation, cells were washed extensively, and the level of accumulated cholesterol in the cells was quantified. In these experiments, the mce4 mutant exhibited a two-fold reduction in cholesterol uptake in comparison to WT, consistent with previous reports (Pandey & Sassetti, 2008; Klepp et al., 2012). The omamAms mutant also revealed a defect in cholesterol uptake in comparison to WT, and this defect was equivalent to that observed with the M. smegmatis mce4 mutant (Figure 6A). The cholesterol uptake defect of the omamA mutant could be complemented by either omamAmtb or omamAms (Figure 6A). While both the mce4 and omamAms mutants exhibited a significant reduction in cholesterol uptake, there remained detectable levels of cell-associated C14 cholesterol with both mutants. Previous uptake studies also report residual levels of cholesterol associated with mce4 mutants, leading to the suggestion that additional cholesterol importers may exist in mycobacteria (Pandey & Sassetti, 2008; Klepp et al., 2012). When we examined the double omamAmsmce4 mutant it was no more defective than single mce4 or omamAms mutants. In fact, the double mutant showed slightly improved cholesterol uptake in comparison to the single mce4 and omamAms mutations alone (Figure 6B). The lack of an additive effect of the mce4 and omamAms mutations on the cholesterol uptake phenotype is consistent with OmamA functioning in concert with Mce4 to import cholesterol, as opposed to being part of an independent cholesterol uptake pathway.
Figure 6.
OmamA is required for cholesterol uptake. A. and B. M. smegmatis strains were grown overnight in M9 glucose/glycerol, and washed extensively in M9 no carbon source. Cells were incubated with 4-C14-cholesterol for two hours, washed extensively, and cell associated radioactivity levels were measured by scintillation counter. * indicates p<0.05 compared to WT. Error bars represent standard deviation. Results are representative of at least three independent experiments. M. smegmatis strains: WT +pMV261 (EP1182), Δmce4 +pMV261 (EP1204), ΔomamA +pMV261 (EP1193), ΔomamA +omamAms (EP1194), ΔomamA +omamAmtb (EP1203), and ΔomamAΔmce4+pMV261 (EP1206).
OmamA impacts the levels of Mce1 proteins
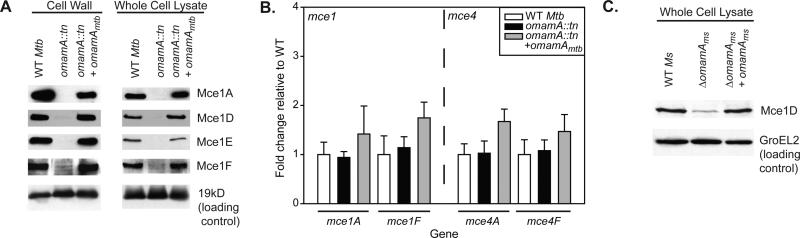
The above studies demonstrated that OmamA contributes to Mce4 cholesterol import and utilization. However, a function of OmamA beyond Mce4 seems likely. This is because the role of OmamA in promoting M. tuberculosis growth in resting murine macrophages (McCann et al., 2011) cannot be explained by an effect on Mce4, as there is no obvious role for Mce4 in promoting growth in resting macrophages (Pandey & Sassetti, 2008; Stewart et al., 2005; Rengarajan et al., 2005; McCann et al., 2011). However, because M. tuberculosis mce1 mutantsare reported in several studies to be defective for growth in macrophages (Rengarajan et al., 2005; Stewart et al., 2005; McCann et al., 2011), we hypothesized that OmamA contributes to Mce1 transporter function in addition to Mce4 function. Due to the predicted structural similarities between OmamA and VirB8, and the role of VirB8 in stabilizing the multi-protein type IV secretion complex (den Hartigh et al., 2008; Sivanesan & Baron, 2011), we further hypothesized that OmamA stabilizes proteins within Mce transporter complexes. Thus, to investigate the potential contribution of OmamA to the Mce1 transporter system and stability of Mce complexes we performed Western blot analysis of four M. tuberculosis Mce1 proteins (Mce1A, Mce1D, Mce1E, and Mce1F) in M. tuberculosis WT, the omamA mutant, and the complemented strain. Mce1A, Mce1D, Mce1E, and Mce1F were localized to the cell wall in M. tuberculosis WT and the complemented strain, consistent with previous subcellular localization experiments performed in M. smegmatis (Forrellad et al., 2014). However, Mce protein levels were dramatically diminished in the cell wall of the omamAmtb mutant (Figure 7A). Further, Mce1A, Mce1D, Mce1E, and Mce1F levels were significantly decreased in the whole cell lysate of the omamAmtb mutant, demonstrating that they were not merely mislocalized in the omamAmtb mutant. Mce1 protein levels were fully restored in the complemented strain. The effect of the omamA mutation on Mce proteins was not due to a broad defect on cell wall proteins, as shown by equivalent levels of the exported 19kD lipoprotein in cell wall fractions of all three strains (Figure 7A).
Figure 7.
Mce1 protein levels are reduced in the absence of OmamA. A. M. tuberculosis (Mtb)cells were irradiated and lysed by French press to generate whole cell lysates (WCL) and fractionated by differential ultracentrifugation into cell wall fractions. Western blots were performed for Mce1A, Mce1D, Mce1E, Mce1F, and the 19kD lipoprotein with equal protein amount loaded across strains. Results are representative of at least three independent replicates. B. RNA was collected from M. tuberculosis WT, omamA::tn, and omamA::tn +omamA complemented strains and transcript levels of mce1A, mce1F, mce4A, and mce4F were determined by Quantitative Real-Time PCR and normalized to expression of the housekeeping protein sigA (Manganelli et al., 1999). Reported are fold change values for each gene relative to expression in WT M. tuberculosis. Error bars represent standard deviation. Results are representative of at least three independent experiments with WT (MBTB178), omamA::tn (MBTB319), and omamA::tn + omamA (MBTB320). C. M. smegmatis (Ms) cells were lysed by glass beads to generate whole cell lysates (WCL). Western blots were performed for Mce1D and GroEL2 with equal protein amount loaded across strains. Results are representative of at least three independent experiments with M. smegmatis WT +pMV261 (EP1182), ΔomamA +pMV261(EP1193) and ΔomamA +omamAms (EP1194) strains.
The Western blot results are consistent with Mce1 proteins being unstable in the absence of OmamA, however; an alternate explanation is that OmamA is required for the expression of genes in the mce1 operon. To rule out the possibility that the dramatic reduction of Mce1 proteins in the omamA mutant is due to a transcriptional effect, we measured the level of mce1 transcripts in WT, omamAmtb mutant, and complemented strains using Quantitative Real-Time PCR. All three strains harbored equivalent amounts of mce1A and mce1F transcripts. Thus, the dramatic decrease in of Mce1 protein levels in the omamA mutant is not a consequence of lower transcript levels. We similarly quantified mce4 transcript levels in the omamAmtb mutant and again observed equivalent levels of mce4A and mce4F transcripts in the omamAmtb mutant compared to WT and complemented strains (Figure 7B).
Although the anti-Mce1D antibody was raised against a peptide from the M. tuberculosis protein, this antibody was also able to recognize the M. smegmatis Mce1D, which allowed us to similarly evaluate the level of Mce1D in the omamAms mutant of M. smegmatis. The omamAms mutant had less Mce1D protein in the whole cell lysate in comparison to WT or complemented strains while the level of GroEL2 protein, as a control, was equivalent across the three strains (Figure 7C). Thus, the effect of OmamA on Mce1 protein levels is observed in both M. smegmatis and M. tuberculosis.
OmamA stabilizes members of the Mce1 transport complex
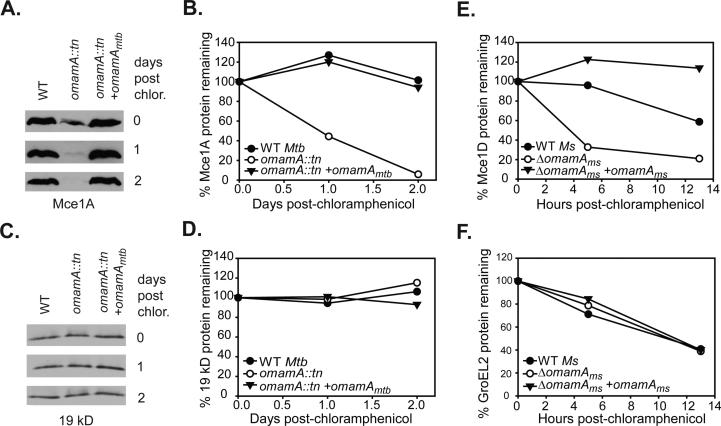
The reduced levels of multiple Mce1 proteins in the omamA mutant is consistent with OmamA having a function, similar to that of VirB8, in stabilizing multi-component transporters. To more directly test for a role of OmamA in stabilizing Mce proteins, we added chloramphenicol to cultures of M. tuberculosis or M. smegmatis omamA mutant, WT and complemented strains to prevent new protein synthesis and then followed the decay of Mce proteins in whole cell lysates by Western blot. As expected from the previous experiments (Figure 7), in the omamA mutant of M. tuberculosis the level of Mce1A was reduced even prior to chloramphenicol treatment; however, by loading more protein, we were able to detect the protein and monitor its degradation. After 2 days of chloramphenicol treatment, Mce1A protein levels were unchanged in WT and complemented strains of M. tuberculosis. In contrast, Mce1A protein in the omamA mutant was highly unstable and over a two day time course the level of Mce1A was dramatically reduced (Figure 8A-B). The instability of Mce1A in the omamA mutant did not reflect general protein instability in our experimental conditions, as the exported 19 kD lipoprotein was equally stable in all three strains (Figure 8C-D). Similar experiments were performed in M. smegmatis, monitoring the stability of Mce1D. Following chloramphenicol addition, the level of Mce1D protein decreased at a faster rate in the omamAms mutant than in the WT or complemented strains (Figure 8E), while degradation of a control protein, GroEL2, occurred at a similar rate in all strains (Figure 8F). Together, these data provide evidence for OmamA of M. tuberculosis and M. smegmatis playing a role in stabilizing components of the Mce1 transporter.
Figure 8.
Mce1 proteins are stabilized by the presence of OmamA. Cultures of M. tuberculosis (Mtb) and M. smegmatis (Ms) were treated with chloramphenicol to prevent further protein synthesis and protein stability was monitored over time by Western blot analysis of whole cell lysates (WCL) with equal protein amount loaded across strains and timepoints. Samples were removed at specific time points, formalin fixed (for M. tuberculosis) and lysed by glass beads to generate WCL. A. The stability of Mce1A in M. tuberculosis strains was followed over a two day time course by Western blot analysis. B. Mce1A protein decay was quantified by measuring band intensity on Western blots using ImageJ. C. The stability of the exported 19 kD lipoprotein in M. tuberculosis over time as monitored by Western blot. D. 19 kD protein abundance was quantitated by ImageJ. Results are representative of at least two independent experiments with M. tuberculosis WT (MBTB178), omamA::tn (MBTB319), and omamA::tn + omamA (MBTB320) strains. E. Decay of Mce1D was quantified in M. smegmatis cultures over 13 hours by measuring band intensity on Western blots using ImageJ. F. Decay of GroEL2 was quantified in M. smegmatis cultures over 13 hours by measuring band intensity on Western blots using ImageJ. Results are representative of at least three independent experiments with M. smegmatis WT +pMV261 (EP1182), ΔomamA +pMV261 (EP1193) and ΔomamA +omamAms (EP1194) strains.
Discussion
The goal of this work was to extend our previous identification of OmamA as an exported protein of unknown function with a role in promoting growth in macrophages (McCann et al., 2011). Here, we investigated the potential significance of similarities between OmamA and Mam proteins and we demonstrated a role for OmamA in cholesterol uptake, which is a Mce4 transporter-dependent process. We further showed that the contribution of OmamA to Mce transporters extends beyond Mce4 to Mce1, as revealed by the reduced levels and instability of Mce1 proteins in both M. smegmatis and M. tuberculosis. While our relatively limited understanding of the Mce1 transporter prevented us from more direct testing of an effect of OmamA on Mce1 function, the dramatic reduction in at least four Mce1 proteins in the omamA mutant of M. tuberculosis argues strongly for a role of OmamA in Mce1 transporter function, as is the case for Mce4. Our demonstration of a role of OmamA in Mce transporters is particularly striking given that the omamA gene is not linked to any mce operon and no Mam proteins have been functionally characterized previously. The lack of assayable in vitro phenotypes similarly prevented us from testing a role for OmamA in the Mce2 and Mce3 systems. It remains a possibility that OmamA is also involved in these additional Mce transporter systems.
Both M. smegmatis and M. tuberculosis were used in this study. In particular, for logistical reasons, the cholesterol uptake experiment was performed in M. smegmatis as a model for M. tuberculosis. Although there is a risk that results may not translate between these species this seems unlikely to be the case here because the phenotypes of the M. tuberculosis omamA mutant are fully consistent with a role of OmamA in cholesterol uptake. In addition, our demonstration of M. tuberculosis omamA being able to complement the M. smegmatis omamA mutant phenotypes indicates conservation of function of the M. smegmatis and M. tuberculosis gene products.
Prior TraSH analysis predicted omamA to play a role during murine infection (Sassetti & Rubin, 2003; Zhang et al., 2013). Our evaluation of mice infected with a single omamA mutant or a complemented strain in a low-dose aerosol model provides important validation of the TraSH prediction while also providing a more detailed picture of the contribution of omamA to M. tuberculosis infection. Our animal studies revealed the omamA mutant to have a reduced bacterial burden during the growth-in-vivo phase of infection (first 3 weeks), which is consistent with the role of OmamA in promoting M. tuberculosis growth in macrophages (McCann et al., 2011). However, later in infection the organ burden of the mutant was no different than WT or complemented strains (as seen in independent experiments). Interestingly, despite the equalized bacterial burden later in infection the omamA mutant infected animals exhibited reduced pathology at late time points and prolonged survival compared to WT and complemented strains. It is worth noting that there are prior examples of M. tuberculosis mutants (i.e. whiB3 and sigH) that elicit reduced immunopathology and prolonged survival of mice despite having a normal bacterial burden (Steyn et al., 2002; Kaushal et al., 2002). In these cases, the mutants are thought to be defective in the inducing harmful immunopathology, which could possibly be the case with the omamA mutant, as well. Alternatively, there may be long term consequences of the delayed growth phenotype of the omamA mutant that persist even after the bacterial burden catches up.
Because of the connection we made between OmamA and Mce systems, we compared the mouse phenotypes of the omamA mutant to infection phenotypes reported for mce mutants. Unfortunately, such comparisons are complicated by discrepant results between studies of mce mutants and the wide variety of models employed (infection route, single or pooled mutants, nature of the mutation, mouse strain, etc.) (Forrellad et al., 2013; Casali & Riley, 2007). For mce1 mutants, in particular, there are reports of attenuated as well as hypervirulent phenotypes (Gioffre et al., 2005; Shimono et al., 2003; Joshi et al., 2006; Marjanovic et al., 2010; Lima et al., 2007; Sassetti & Rubin, 2003). Nonetheless, there are published reports of attenuated phenotypes of mce1, mce2 and mce3 mutants in mice that resemble those of the omamA mutant: reduced bacterial burden early in infection, increased survival time, and/or reduced lung pathology (Marjanovic et al., 2010; Senaratne et al., 2008; Sassetti & Rubin, 2003; Gioffre et al., 2005; Joshi et al., 2006). In contrast, mce4 mutants are not reported to have defects early in infection but to have defects later during the persistence phase, in particular when tested in an intravenous infection with a 1:1 mixture of WT:mce4 strains (Joshi et al., 2006; Pandey & Sassetti, 2008; Sassetti & Rubin, 2003). When tested in a single strain aerosol infection, similar to the one used in our study, a mce4 mutant only exhibits a subtle defect in bacterial burden during the persistence phase of infection (Senaratne et al., 2008), which may explain why we did not observe any defect in persistence. However, in the aerosol model reduced pathology late in infection and prolonged survival of mice infected with a mce4 mutant is observed (Senaratne et al., 2008). Thus, the in vivo growth defect of the omamA mutant is unlikely to result from an effect on Mce4, rather, a role of OmamA in additional Mce systems may account for this specific in vivo phenotype. However, future studies will be required to more clearly understand the omamA mutant phenotypes observed in animals.
The omamA mutant phenotypes we observed in cholesterol-containing media were indistinguishable from mce4 mutants. These results not only support a role for OmamA in Mce4 transport, but they additionally reveal OmamA to be a new protein required for cholesterol utilization in vitro. In a Tn-seq mutagenesis study to identify M. tuberculosis genes required for in vitro growth on cholesterol, all genes in the mce4 operon were identified, including mam4A and mam4B, but omamA was not identified (Griffin et al., 2011). Interestingly, omamA barely missed the cutoff for statistical significance in this study (p=0.06), consistent with a role in cholesterol utilization.
It is also interesting to compare our results indicating a role for OmamA in Mce transport pathways to the results of a transposon mutagenesis screen conducted in mce1 or mce4 mutant backgrounds (Joshi et al., 2006). In this genetic interaction screen, genes that are members of the same Mce transport pathway or genes in redundant parallel pathways were uncovered. Once again, although omamA was not predicted as having genetic interactions with mce1 or mce4 in this earlier study, inspection of the supplemental data revealed the behavior of omamA mutations in mce1 and mce4 backgrounds to be consistent with omamA being part of these Mce pathways (Joshi et al., 2006).
Given the many mce-linked mam genes (eight) and unlinked omam (five) genes in M. tuberculosis, our finding that deletion of omamA yielded phenotypes as dramatic as complete deletion of the mce4 operon was surprising, as was the discovery that OmamA impacted more than one Mce system. The dramatic phenotypes of the omamA mutant raise questions about whether other Mam proteins of M. tuberculosis will also have such broad effects. Data from transposon mutagenesis screens predicts similar phenotypes for mutations in mam genes and the adjoining mce operons (Rengarajan et al., 2005; Sassetti & Rubin, 2003; Griffin et al., 2011), which supports the idea of mam genes functioning with their linked mce system. It is possible that the function of mce operon associated mam genes may not extend to unlinked mce loci. For example, the mce1-associated mam genes (mam1A-D) are not predicted to be required for growth on cholesterol like mce4 mutants (Griffin et al., 2011). Furthermore, it remains unclear whether all orphaned Mam proteins are required for multiple Mce transporters or whether they are even involved in Mce transport at all. Individual mam and omam mutants will need to be constructed and characterized in order to determine if the dramatic role of OmamA in Mce function is unique or representative of the overall importance of all Mam family members.
The Mycobacterium leprae genome is highly reduced in comparison to other mycobacterial species and is thought to have only maintained a minimal set of genes required for its intracellular lifestyle (Moran, 2002; Singh & Cole, 2011). Interestingly, omamA and the downstream omamB are only two omam genes conserved in the M. leprae genome, which contains a single mce operon, mce1 (Supplemental Figure 3). Conservation of omamA in M. leprae supports the importance of omamA in intracellular growth and virulence. Additionally, the conserved presence and arrangement of omamA and omamB suggests that the corresponding proteins may function together. Like omamA and omamB, Mam family proteins are usually encoded in pairs (Figure 1B, Supplemental Figure 3) (Casali & Riley, 2007), although the significance of this arrangement is unknown. Future study of OmamB, the protein encoded by rv0200, could help to determine whether OmamB also has a broad role in Mce transport like OmamA.
While crystal structures for Mam proteins will be necessary to prove structural similarity, the unexpected Phyre2 predictions were helpful for identifying a function for OmamA. VirB8 is an essential component of bacterial type IV secretion systems, with roles assembling the core complex of the transport machinery, providing stability to multiple proteins within the complex and potentially anchoring the transporter to the cytoplasmic membrane (Paschos et al., 2006; Fronzes et al., 2009; Kumar et al., 2000; Baron, 2006). In the absence of VirB8, many proteins within the type IV secretion apparatus become destabilized and degraded (den Hartigh et al., 2008; Sivanesan & Baron, 2011). Similarly, in the absence of OmamA all four of the M. tuberculosis Mce1 proteins and one M. smegmatis Mce1 protein we monitored were present at dramatically reduced levels. These reduced protein levels are due to protein instability, as shown by monitoring the stability of Mce1A in M. tuberculosis and Mce1D in M. smegmatis, as representative proteins. These results suggest that OmamA, and Mam proteins in general, may play analogous roles to VirB8 in the formation and stabilization of the core Mce1 transport complex, resulting in destabilization of the complex and individual Mce1 proteins in their absence. Due to a lack of anti-Mce4 antibodies, we were unable to test the effect of OmamA on the stability of proteins that comprise the Mce4 transporter. However, we propose that a similar function of OmamA in stabilization of the Mce4 transporter accounts for the defects in cholesterol uptake and metabolism observed in omamA mutants.
Interestingly, VirB8 also plays a role in substrate transport during type IV secretion (Cascales & Christie, 2004), which raises the possibility that Mam proteins may also have an additional role in Mce substrate movement. Due to structural and functional analogies between OmamA and VirB8, we propose a model wherein OmamA interacts with Mce proteins, potentially driving Mce complex formation and ultimately providing stability to Mce proteins within the complex (Figure 9). However, it is important to emphasize the speculative nature of the interactions depicted in this model, as there have yet to be any direct studies of protein-protein interactions in the presumed Mce macromolecular complex. Additionally, the stoichiometry of the proposed macromolecular complex remains unknown. Because there are thirteen Mam family proteins in M. tuberculosis and only four Mce transporters, we predict that each transporter may be stabilized by multiple Mam family members.
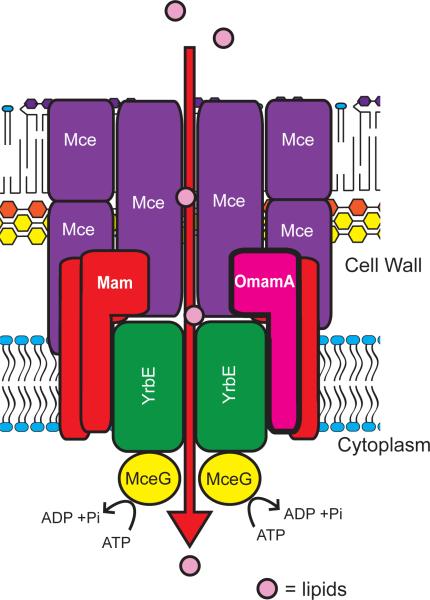
Figure 9.
OmamA is an integral membrane protein that is important to Mce transporter stability and function. OmamA (pink) and other Mam proteins (red) are embedded in the inner membrane by an N-terminal transmembrane domain with the majority of the protein being localized on the cell wall side of the membrane. Mce permease proteins (YrbE), shown in green, are multi-membrane spanning proteins localized to the inner membrane. Some Mce proteins contain predicted TM domains; however, localization from this and other studies (Klepp et al., 2012) suggests that Mce proteins are located within the cell wall (shown in purple). MceG, shown in yellow, is the cytoplasmic Mkl family ATPase predicted to be responsible for ATP-hydrolysis that powers the transport of substrates, shown in light pink, through the complex.
Although Mce transporters are of clear importance to M. tuberculosis virulence and a core component of the M. tuberculosis genome (Gioffre et al., 2005; Shimono et al., 2003; Marjanovic et al., 2010; Lima et al., 2007; Sassetti & Rubin, 2003; Rengarajan et al., 2005; Stewart et al., 2005; McCann et al., 2011; Senaratne et al., 2008; Pandey & Sassetti, 2008), there has yet to be a systematic genetic or biochemical analysis of the individual Mce transporter proteins in terms of their contribution to virulence or their function in the transport mechanism. Mce transporter components are assigned potential functions by analogy to classic ABC transporters (ex. ATPase, permease, or solute binding proteins) (Casali & Riley, 2007). However, Mce transporters are distinguished from ABC transporters in the multitude of individual proteins predicted to be involved: two YrbE permeases, six predicted Mce solute binding proteins, and a shared ATPase MceG. The function of all of these individual transporter components requires validation. Because Mam proteins share no obvious ABC transporter counterpart their function was an even bigger mystery. The results of this study provide an essential framework for studying the role of Mam family proteins in the stabilization of Mce transporter systems.
Experimental Procedures
Bacterial strains and plasmids
In this study, we used the bacterial strains listed in Supplemental Table 1 and plasmids as listed in Supplemental Table 2. The M. tuberculosis omamA (rv0199) mutant was generated in a previous transposon mutagenesis study performed in a M. tuberculosis β-lactamase (ΔblaC) background (McCann et al., 2011; Flores et al., 2005). The omamA::tn mutant has a hygromycin resistant Tn’blaTEM-1 transposon inserted in the omamA coding sequence at amino acid position 74 and it expresses an exported OmamA-‘BlaTEM-1 fusion protein. The omamA::tn mutant (omamA::tn, ΔblaC) used in this study (MBTB319) additionally carries the empty pMV261.kan plasmid. For mutant characterization, omamA::tn was compared to strain MBTB178 (omamAWT, ΔblaC, pJES137, pMV261.kan). Plasmid pJES137 is an integrating hygromycin resistant plasmid that expresses ‘blaTEM-1. MBTB178 is referred to as WT in the text. The M. tuberculosis complemented strain (omamA::tn, ΔblaC, pJES178) expresses omamA from the hsp60 promoter of the kanamycin resistant plasmid pJES178 (McCann et al., 2011). This series of omamA::tn (MBTB319), omamAWT (MBTB178) and complemented (MBTB320) strains are all hygromycin and kanamycin resistant to enable growth in identical media conditions.
Bacterial growth
M. tuberculosis strains were grown in Middlebrook 7H9 broth (Difco) supplemented with 1X albumin dextrose saline (ADS), 0.5% glycerol and either 0.025% Tween 80 (Tw) or 0.025% tyloxapol (Ty). M. smegmatis strains were grown in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glucose, 0.5% glycerol, and either 0.05% Tween 80 (Tw) or 0.05% Tyloxapol (Ty). Medium was supplemented with 20μg mL−1 kanamycin or 50μg mL−1 hygromycin as needed for mycobacterial cultures. E. coli strains were grown in Luria-Bertani medium (Fisher) supplemented as necessary with 40μg mL−1 kanamycin.
Mouse experiments
Female C57BL/6 mice aged 7-10 weeks were infected with ~200 cfu of M. tuberculosis by aerosol using a Madison chamber (Mechanical Engineering Workshop, Madison, WI), and bacterial burden was determined, as previously described (Kurtz et al., 2006). Groups of four mice per strain were sacrificed, organs homogenized, and diluted and plated to determine bacterial burden at various times after infection. The lower right lobe of the lungs was inflated and fixed in 10% formalin for histology.
Histopathology
Inflammation was determined in 5 μm sections following hematoxylin and eosin (H&E) staining. Paraffin embedded sections were set and cut to reveal the maximum longitudinal visualization of the intrapulmonary main axial airway. Histopathology was evaluated and scored by an experienced blinded reviewer (I.C.A.) on a scale of 0 (absent) to 3 (severe), as previously described (McElvania Tekippe et al., 2010; Allen et al., 2013; Allen et al., 2009). The parameters assessed included overall leukocyte infiltration, perivascular and peribroncheolar cuffing, extravasation, and the estimated percent of lung area involved with inflammation. Each individual parameter was scored and averaged to generate the histology score.
Mutant construction
M. smegmatis mutants were constructed by recombineering, as previously described (van Kessel & Hatfull, 2008; van Kessel & Hatfull, 2007). Briefly, upstream and downstream flanks were PCR amplified and cloned into pMP614 (kind gift from Martin Pavelka), which was then linearized to produce the final recombineering fragment, carrying a hygromycin resistance marker flanked by DNA sequences upstream and downstream of msmeg0235. Parental strains carrying a kanamycin marked plasmid expressing a recombinase, pJV53, (van Kessel & Hatfull, 2007; van Kessel & Hatfull, 2008) were used for recombineering. Following three hour induction of the recombinase with acetamide, electroporation was used to introduce the linear recombineering fragment. Allelic exchange recombinants were selected for double resistance to hygromycin and kanamycin. Strains were cured of pJV53 by passaging 3-4 times in the absence of kanamycin. Plasmid cured strains were then transformed with the resolvase expressing pMP854 plasmid (kind gift from Martin Pavelka), to remove the hygromycin marker in the deletion cassette. Hygs strains were cured for pMP854 as described above to generate the final unmarked deletion strains. Mutant construction was confirmed by Southern blot (data not shown).
OmamAms complementation and OmamAmtb-HA vector construction
The msmeg0235 gene (omamAms) was PCR amplified by msmeg0235_F1 x msmeg0235_R1, the rv0199 gene (omamAmtb) was PCR amplified by rv0199HA_F_MscI x rv0199HA_R_HindIII, and PCR fragments were cloned into pCR2.1 (Invitrogen). The resulting plasmids were sequenced to confirm they were error-free. The omamAms fragment was digested from pCR2.1 with EcoRI, gel purified, and ligated into EcoRI digested pMV261.kan (Stover et al., 1991). The omamAmtb fragment was digested from pCR2.1 with MscI and HindIII, gel purified, and ligated into MscI/HindIII digested JSC77 (Glickman et al., 2000), containing an in-frame C-terminal HA tag. Primer sequences are provided (Supplemental Table 3).
Transformation
M. smegmatis strains were transformed by electroporation, as previously described (Snapper et al., 1990).
Morphology
Congo red assays were performed, as previously described (Klepp et al., 2012). Mueller Hinton agar plates were supplemented with 0.2% glucose and 100μg mL−1 Congo red (Sigma). Colony morphology was analyzed by plating 2 μL spots of OD600 1.0 M. smegmatis strains. Plates were incubated at 37°C for two days and visualized using a low-magnification Leica M420 macroscope with 2X and 5.6X magnification.
Cholesterol Metabolism Assays for M. tuberculosis
A cholesterol stock solution was prepared by solubilizing cholesterol in ethanol and tyloxapol, as follows. A 1:1 solution of 200 proof ethanol:Tyloxapol (Sigma) was prepared, filtered, and heated to 50°C. 200mg mL−1 cholesterol was dissolved in 3:1 chloroform:methanol, and added dropwise to the 50°C tyloxapol solution until reaching 20% final volume. Sauton's media was prepared and pH adjusted to 7.4: 1L dH2O, 4g DL asparagine, 2g sodium citrate, 0.5g K2HPO4, 0.5g MgSO4-7H2O, 0.05g ferric ammonium citrate, 0.025% Tyloxapol, and supplemented with either 6% glycerol or 0.5 mM cholesterol from stock solution. M. tuberculosis strains were diluted to 105 cfu mL−1 in Sauton's +Ty and 104 cfu were aliquoted into 96 well plates with Sauton's supplemented with glycerol or cholesterol, incubated shaking at 37°C for seven days, then resazurin (Sigma) was added to a final concentration 0.0125 mg mL−1. Resazurin conversion was followed using fluorescence and was monitored daily by a Tecan Infinite 200 Pro at hv=544 nm excitation and hv=590 nm emission.
Cholesterol Metabolism Assays for M. smegmatis
A cholesterol stock solution was prepared by solubilizing cholesterol in cyclodextrin, as previously described (Klein et al., 1995). Briefly, 1g methyl-ß-cyclodextrin (C4555 Sigma) was dissolved in 11mL PBS (0.09g mL−1) and heated to 80°C with continuous stirring. 30 mg cholesterol (Sigma) was dissolved in 400μL 2:1 isopropanol/chloroform. The cholesterol solution was added to the cyclodextrin in 50μL aliquots, stirring continuously. The solution was cooled slowly, filtered for sterility, and kept at room temperature. M9 minimal media was prepared as follows: 1L dH2O, 12.8g Na2HPO4, 3g KH2PO4, 0.5g NaCl, 1g NH4Cl, 25 μL 1M CaCl2, 500μL 1M MgSO4, and 2.5 mL 10% Tyloxapol (Ty, Sigma), and supplemented with 0.2% glucose and 0.5% glycerol or 0.5mM cholesterol from stock solution. M. smegmatis strains were grown to OD600 1.0 in M9 supplemented with 0.2% glucose and 0.5% glycerol + 0.05% Ty. Strains were washed in M9 +Ty three times by pelleting cells at 1,900 × g for 10 minutes at 4°C, and diluted to 105 cfu mL−1 in M9 +Ty, and 104 cfu were plated into 96 well plates with M9 containing glycerol or cholesterol. Plates were incubated shaking at 37°C overnight, after which resazurin (Sigma) was added to a final concentration 0.0125 mg mL−1. Florescence was monitored every 10 minutes by a Spectramax M2 using hv=544 nm excitation and hv=590 nm emission.
Cholesterol uptake
Cholesterol uptake experiments were performed, similar to previously reported (Klepp et al., 2012). M. smegmatis strains were grown to OD600 1.0 in M9 supplemented with 0.2% glucose and 0.5% glycerol + 0.05% Ty. Strains were washed in M9 + Ty three times by pelleting cells at 1,900 × g for 10 minutes at 4°C, and then equalized to OD600 0.5 in M9 + Ty, and incubated with 0.04μCi 4-C14 cholesterol (Perkin Elmer NEC018050UC) for 2 hours at 37°C. After incubation, cells were pelleted and washed three times with M9 + Ty, and cell associated radioactivity was measured by scintillation counter.
Subcellular fractionation
M. tuberculosis cells were pelleted by centrifugation (1,900 × g) and sterilized by irradiation (JL Shephard Mark I 137Cs irradiator, Department of Radiobiology, University of North Carolina at Chapel Hill). After sterilization, M. tuberculosis cells were removed from BSL-3 containment. M. smegmatis cells were simply pelleted by centrifugation for 10 minutes at 1,900 × g. Subcellular fractionation was then performed, as previously described (Gibbons et al., 2007). Briefly, cells were resuspended in PBS containing protease inhibitors, lysed in a French pressure cell, and unlysed cells were removed by centrifugation (1,900 × g). The clarified whole cell lysates (WCL) were subjected to differential ultracentrifugation, 27,000 × g for 30 minutes to pellet the cell wall (CW), 100,000 × g for 2 hours to pellet the membrane (MEM), and remaining soluble (SOL) fraction containing the cytoplasm.
Western blotting
Equal protein amounts, as determined by Bicinchonic acid assay (Pierce), for all fractions and strains were separated by SDS-PAGE and transferred to nitrocellulose membranes. Proteins were detected using the following antibodies: Mce1 antibodies (a gift from Christopher Sassetti, University of Massachusetts Medical School (Feltcher et al., 2015)): anti-Mce1A (1:10,000), anti-Mce1D (1:5,000), anti-Mce1E/Lprk (1:5,000), anti-Mce1F (1:10,000), anti-19kD (1:20,000) (a gift from Douglas Young, Imperial College, United Kingdom), and anti-HA (1:25,000) (Covance). GroEL2 was detected using an anti-HIS (1:10,000) (Abgent) antibody, as previously described (Feltcher et al., 2013), which recognizes a string of histidines in GroEL2 (Rengarajan et al., 2008). Anti-mouse and Anti-rabbit IgG conjugated HRP (Biorad) were used as secondary antibodies, as appropriate. HRP signal was detected using Western Lighting Chemiluminescent detection reagent (Perkin-Elmer). Quantitation of Western blots was calculated by densitometry using ImageJ (Schneider et al., 2012).
Quantitative Real-Time PCR
Triplicate M. tuberculosis cultures were grown to OD600 of 1.0 and pelleted by centrifugation for 10 minutes at 1,900 × g, and qRT-PCR was performed. Bacteria were lysed by 3:1 chloroform methanol, mixed with Trizol (Invitrogen), and the upper phase was separated and RNA precipitated overnight in isopropanol. RNA samples were pelleted and washed in 70% ethanol, and resuspended in RNase-free H2O. RNA samples were treated with DNase (Promega), purified (Zymo RNA Clean and Concentrator Kit), and converted to cDNA using iScript cDNA Synthesis Kit (BioRad). Triplicate biological and triplicate technical replicates of cDNA from 40 ng RNA each were used for qRT-PCR using the Sensimix SYBR and Flourescein kit (Bioline). Transcript copy number for each gene was calculated as compared to known concentrations of genomic DNA, and each sample was normalized to housekeeping gene sigA transcript levels. Primer sequences are provided (Supplemental Table 3).
Protein stability experiments
Cultures of M. tuberculosis or M. smegmatis were first grown to approximately OD600 1.0 and diluted to OD600 0.5 in media containing 20 ug mL−1 or 35 ug mL−1 of chloramphenicol (Sigma) for M. tuberculosis and M. smegmatis, respectively. At specific time points, samples were taken to measure the amount of Mce1 protein present in WCL. For M. tuberculosis samples, prior to cell lysis the cells were washed twice with PBS + 0.02% Tween 80, sterilized by fixation in an equal volume of 10% formalin (Fisher) for 1 hour and washed post-fixation. Whole cell lysates were prepared as previously described (Braunstein et al., 2001). Briefly, cells were resuspended in extraction buffer, lysed by MagNA Lyser (Roche) with glass beads, and denatured by boiling. Equal protein amounts as determined by Bicinchonic acid assay (Pierce) were loaded for Western blot analysis, as described above. Quantitation of Western blots was calculated by densitometry using ImageJ (Schneider et al., 2012).
Statistics
Statistics were performed in SigmaPlot. Normality testing (Shapiro-Wilk) and equal variance testing was done to determine correct statistical methods. Comparisons passing normality and equal variance with two groups were performed by two-tailed Student's t-test. Comparisons not passing normality with two groups were performed by Mann-Whitney rank sum test. Comparisons passing normality and equal variance with more than two groups used one way analysis of variance (ANOVA), followed by multiple comparisons with the Holm-Sidak method as appropriate. Comparisons not passing normality with more than two groups used Kruskal-Wallis one way analysis of variance on ranks, followed by multiple comparisons with Student-Newman-Keuls. Survival was analyzed by Log-rank test followed by multiple comparisons with the Holm-Sidak method.
Supplementary Material
Acknowledgements
This work was supported by a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease Award, by NIH R21 AI076685 and NIH RO1 AI054540 Awards (to M.B.). E.F.P. was supported by a University of North Carolina Dissertation Completion Fellowship. J.R.M. was supported by training grant NIH 5-T32-GM008581 and a Society of Fellows graduate dissertation fellowship. J.T.S. was supported by a University of North Carolina Morehead Fellowship.
We would like to thank Sara Johnson for assistance with this work. We would like to thank Fabiana Bigi for providing the mce4 and mce6X M. smegmatis strains, Chris Sassetti and Jennifer Griffin for providing the Mce1 antibodies, and Douglas Young for providing the 19kD antibody. We would like to thank Martin Pavelka for providing the plasmids pMP614 and pMP854. We also thank Peggy Cotter, Mary Hondalus and all members of the Braunstein lab for critical reading of the manuscript.
References
- Allen IC, McElvania-TeKippe E, Wilson JE, Lich JD, Arthur JC, Sullivan JT, Braunstein M, Ting JP. Characterization of NLRP12 during the in vivo host immune response to Klebsiella pneumoniae and Mycobacterium tuberculosis. PLoS One. 2013;8:e60842. doi: 10.1371/journal.pone.0060842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey S, Ward D, Middleton R, Grossmann JG, Zambryski PC. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc Natl Acad Sci U S A. 2006;103:2582–2587. doi: 10.1073/pnas.0511216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2006;84:890–899. doi: 10.1139/o06-148. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Brown AM, Kurtz S, Jacobs WR., Jr. Two nonredundant SecA homologues function in mycobacteria. J Bacteriol. 2001;183:6979–6990. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briken V. Mycobacterium tuberculosis genes involved in regulation of host cell death. Adv Exp Med Biol. 2013;783:93–102. doi: 10.1007/978-1-4614-6111-1_5. [DOI] [PubMed] [Google Scholar]
- Cantrell SA, Leavell MD, Marjanovic O, Iavarone AT, Leary JA, Riley LW. Free mycolic acid accumulation in the cell wall of the mce1 operon mutant strain of Mycobacterium tuberculosis. Journal of microbiology. 2013;51:619–626. doi: 10.1007/s12275-013-3092-y. [DOI] [PubMed] [Google Scholar]
- Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics. 2007;8:60. doi: 10.1186/1471-2164-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillan-Huntington C, Butler PJ, Huntington JA, Akin D, Feldherr C, Stewart M. NTF2 monomer-dimer equilibrium. J Mol Biol. 2001;314:465–477. doi: 10.1006/jmbi.2001.5136. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- den Hartigh AB, Rolan HG, de Jong MF, Tsolis RM. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J Bacteriol. 2008;190:4427–4436. doi: 10.1128/JB.00406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy, an immunologic magic bullet: Mycobacterium tuberculosis phagosome maturation block and how to bypass it. Future Microbiol. 2008;3:517–524. doi: 10.2217/17460913.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltcher ME, Gibbons HS, Ligon LS, Braunstein M. Protein export by the mycobacterial SecA2 system is determined by the preprotein mature domain. J Bacteriol. 2013;195:672–681. doi: 10.1128/JB.02032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltcher ME, Gunawardena HP, Zulauf KE, Malik S, Griffin JE, Sassetti CM, Chen X, Braunstein M. Label-free quantitative proteomics reveals a role for the Mycobacterium tuberculosis SecA2 pathway in exporting solute binding proteins and Mce transporters to the cell wall. Molecular & cellular proteomics : MCP. 2015 doi: 10.1074/mcp.M114.044685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores AR, Parsons LM, Pavelka MS., Jr. Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology. 2005;151:521–532. doi: 10.1099/mic.0.27629-0. [DOI] [PubMed] [Google Scholar]
- Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, de la Paz Santangelo M, Cataldi AA, Bigi F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrellad MA, McNeil M, Santangelo Mde L, Blanco FC, Garcia E, Klepp LI, Huff J, Niederweis M, Jackson M, Bigi F. Role of the Mce1 transporter in the lipid homeostasis of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2014;94:170–177. doi: 10.1016/j.tube.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons HS, Wolschendorf F, Abshire M, Niederweis M, Braunstein M. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J Bacteriol. 2007;189:5090–5100. doi: 10.1128/JB.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioffre A, Infante E, Aguilar D, Santangelo MP, Klepp L, Amadio A, Meikle V, Etchechoury I, Romano MI, Cataldi A, Hernandez RP, Bigi F. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 2005;7:325–334. doi: 10.1016/j.micinf.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Glickman MS, Cox JS, Jacobs WR., Jr. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden JD, Brown LR, Gunawardena HP, Perkowski EF, Chen X, Braunstein M. Reversible acetylation regulates acetate and propionate metabolism in Mycobacterium smegmatis. Microbiology. 2013;159:1986–1999. doi: 10.1099/mic.0.068585-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SW, Galan JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol. 2013;11:316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchey J, Lee S, Jeon BY, Basaraba RJ, Venkataswamy MM, Chen B, Chan J, Braunstein M, Orme IM, Derrick SC, Morris SL, Jacobs WR, Jr., Porcelli SA. Enhanced priming of adaptive immunity by a proapoptotic mutant of Mycobacterium tuberculosis. The Journal of clinical investigation. 2007;117:2279–2288. doi: 10.1172/JCI31947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac DT, Isberg R. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol. 2014;9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci U S A. 2006;103:11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe YC, Fleischmann RD, Bishai WR. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci U S A. 2002;99:8330–8335. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Klein U, Gimpl G, Fahrenholz F. Alteration of the myometrial plasma membrane cholesterol content with beta-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- Klepp LI, Forrellad MA, Osella AV, Blanco FC, Stella EJ, Bianco MV, Santangelo Mde L, Sassetti C, Jackson M, Cataldi AA, Bigi F, Morbidoni HR. Impact of the deletion of the six mce operons in Mycobacterium smegmatis. Microbes Infect. 2012;14:590–599. doi: 10.1016/j.micinf.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kumar RB, Xie YH, Das A. Subcellular localization of the Agrobacterium tumefaciens TDNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol Microbiol. 2000;36:608–617. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Kubori T, Thanh Bui X, Hyakutake A, Uchida Y, Imada K, Nagai H. Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion. Scientific reports. 2015;5:10912. doi: 10.1038/srep10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, McKinnon KP, Runge MS, Ting JP, Braunstein M. The SecA2 secretion factor of Mycobacterium tuberculosis promotes growth in macrophages and inhibits the host immune response. Infect Immun. 2006 doi: 10.1128/IAI.01022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList--10 years after. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Ligon LS, Hayden JD, Braunstein M. The ins and outs of Mycobacterium tuberculosis protein export. Tuberculosis (Edinb) 2012;92:121–132. doi: 10.1016/j.tube.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima P, Sidders B, Morici L, Reader R, Senaratne R, Casali N, Riley LW. Enhanced mortality despite control of lung infection in mice aerogenically infected with a Mycobacterium tuberculosis mce1 operon mutant. Microbes Infect. 2007;9:1285–1290. doi: 10.1016/j.micinf.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanovic O, Miyata T, Goodridge A, Kendall LV, Riley LW. Mce2 operon mutant strain of Mycobacterium tuberculosis is attenuated in C57BL/6 mice. Tuberculosis (Edinb) 2010;90:50–56. doi: 10.1016/j.tube.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmiesse M, Brodin P, Buchrieser C, Gutierrez C, Simoes N, Vincent V, Glaser P, Cole ST, Brosch R. Macro-array and bioinformatic analyses reveal mycobacterial 'core' genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology. 2004;150:483–496. doi: 10.1099/mic.0.26662-0. [DOI] [PubMed] [Google Scholar]
- McCann JR, McDonough JA, Sullivan JT, Feltcher ME, Braunstein M. Genome-wide identification of Mycobacterium tuberculosis exported proteins with roles in intracellular growth. J Bacteriol. 2011;193:854–861. doi: 10.1128/JB.01271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann JR, Kurtz S, Braunstein M. Secreted and exported proteins important to Mycobacterium tuberculosis pathogenesis. In: Wooldridge K, editor. Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis. Caister Academic Press; Norfolk, UK: 2009. pp. 265–298. [Google Scholar]
- McElvania Tekippe E, Allen IC, Hulseberg PD, Sullivan JT, McCann JR, Sandor M, Braunstein M, Ting JP. Granuloma formation and host defense in chronic Mycobacterium tuberculosis infection requires PYCARD/ASC but not NLRP3 or caspase-1. PLoS One. 2010;5:e12320. doi: 10.1371/journal.pone.0012320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn WW, van der Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, Eltis LD. The actinobacterial mce4 locus encodes a steroid transporter. J Biol Chem. 2008;283:35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Niederweis M. Nutrient acquisition by mycobacteria. Microbiology. 2008;154:679–692. doi: 10.1099/mic.0.2007/012872-0. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos A, Patey G, Sivanesan D, Gao C, Bayliss R, Waksman G, O'Callaghan D, Baron C. Dimerization and interactions of Brucella suis VirB8 with VirB4 and VirB10 are required for its biological activity. Proc Natl Acad Sci U S A. 2006;103:7252–7257. doi: 10.1073/pnas.0600862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Murphy E, Park A, Krone CL, Hett EC, Bloom BR, Glimcher LH, Rubin EJ. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc Natl Acad Sci U S A. 2008;105:264–269. doi: 10.1073/pnas.0710601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senaratne RH, Sidders B, Sequeira P, Saunders G, Dunphy K, Marjanovic O, Reader JR, Lima P, Chan S, Kendall S, McFadden J, Riley LW. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. Journal of medical microbiology. 2008;57:164–170. doi: 10.1099/jmm.0.47454-0. [DOI] [PubMed] [Google Scholar]
- Shimono N, Morici L, Casali N, Cantrell S, Sidders B, Ehrt S, Riley LW. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc Natl Acad Sci U S A. 2003;100:15918–15923. doi: 10.1073/pnas.2433882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R, Sayes F, Song O, Groschel MI, Brodin P, Brosch R, Majlessi L. Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog. 2015;11:e1004650. doi: 10.1371/journal.ppat.1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Cole ST. Mycobacterium leprae: genes, pseudogenes and genetic diversity. Future Microbiol. 2011;6:57–71. doi: 10.2217/fmb.10.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanesan D, Baron C. The dimer interface of Agrobacterium tumefaciens VirB8 is important for type IV secretion system function, stability, and association of VirB2 with the core complex. J Bacteriol. 2011;193:2097–2106. doi: 10.1128/JB.00907-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Coincon M, Paschos A, Jolicoeur B, Lavallee P, Sygusch J, Baron C. Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem Biol. 2012;19:1041–1048. doi: 10.1016/j.chembiol.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Stewart GR, Patel J, Robertson BD, Rae A, Young DB. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1:269–278. doi: 10.1371/journal.ppat.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr., Kawakami RP, Bloom BR. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci U S A. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis ... [et al.] 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2: Unit 2 3. [DOI] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nature methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- van Kessel JC, Hatfull GF. Mycobacterial recombineering. Methods Mol Biol. 2008;435:203–215. doi: 10.1007/978-1-59745-232-8_15. [DOI] [PubMed] [Google Scholar]
- World Health Organization Global Tuberculosis Report 2014. 2014 [Google Scholar]
- Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell. 2013;155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.