Abstract
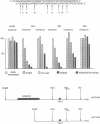
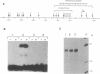
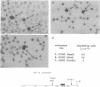
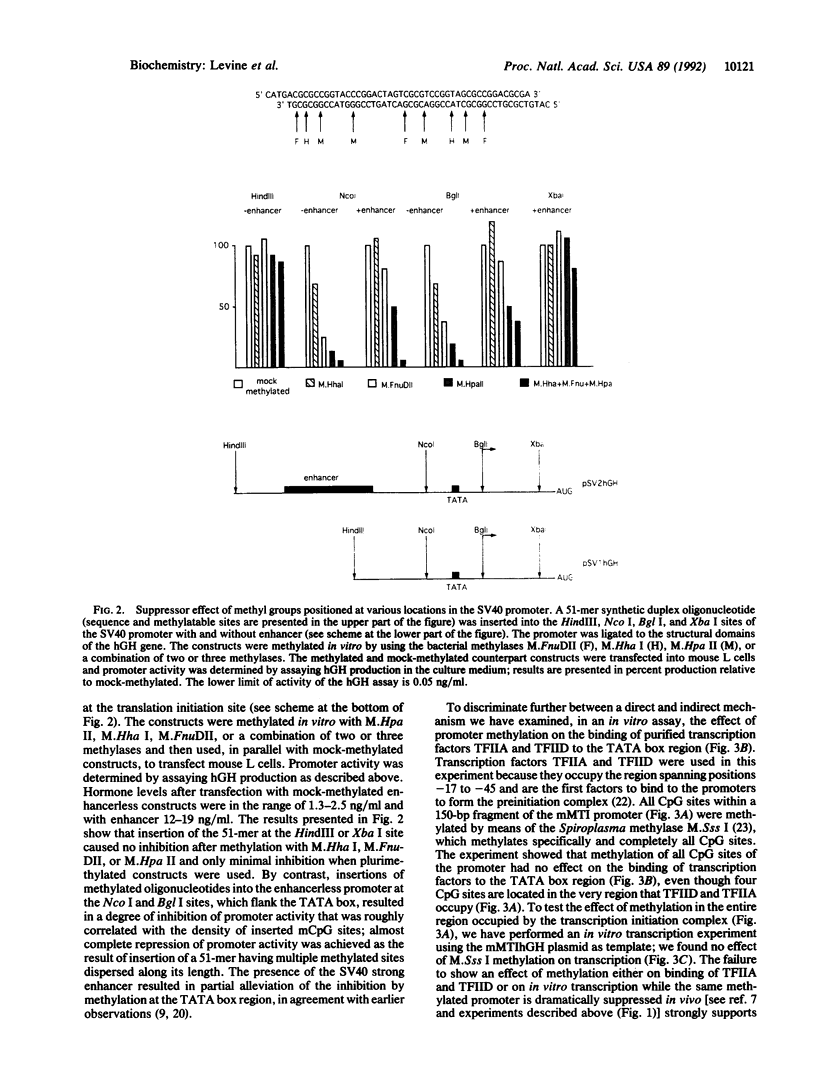
Although the first observations of the inhibitory effect of methylation on gene activity were made almost a decade ago, the mechanism by which methyl groups affect transcription is still obscure. Here we use engineered promoters methylated in vitro in transient transfections to study the mechanism by which methylation mediates promoter repression. The results clearly show that the location of the methyl groups within the promoter region determines the extent of promoter repression. The most effective suppression was observed when methylation was in the preinitiation domain. The results also support a previous suggestion that a mediator protein is involved in the mechanism of promoter inhibition. The suppressor effect of methylation at sequences flanking the TATA box can be partially overcome in the presence of the simian virus 40 enhancer. In addition, results obtained by transient thymidine labeling of Ltk- cells that were transfected with a methylated thymidine kinase gene from herpes simplex virus, at the level of approximately one template per cell, further support the conclusion that methylation affects primarily transcription preinitiation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992 Jan;11(1):327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhausen G., Wittig B., Graessmann M., Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. The major late transcription factor binds to and activates the mouse metallothionein I promoter. Genes Dev. 1987 Nov;1(9):973–980. doi: 10.1101/gad.1.9.973. [DOI] [PubMed] [Google Scholar]
- Fradin A., Manley J. L., Prives C. L. Methylation of simian virus 40 Hpa II site affects late, but not early, viral gene expression. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5142–5146. doi: 10.1073/pnas.79.17.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höller M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988 Sep;2(9):1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989 May;3(5):612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Levine A., Cantoni G. L., Razin A. Inhibition of promoter activity by methylation: possible involvement of protein mediators. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J., Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991 Jul 22;285(2):155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- Maldonado E., Ha I., Cortes P., Weis L., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol Cell Biol. 1990 Dec;10(12):6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Samuels M., Sharp P. A. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Salser S. J., Wold B. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev. 1988 Apr;2(4):412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- Murray E. J., Grosveld F. Site specific demethylation in the promoter of human gamma-globin gene does not alleviate methylation mediated suppression. EMBO J. 1987 Aug;6(8):2329–2335. doi: 10.1002/j.1460-2075.1987.tb02508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Selden R. F., Howie K. B., Rowe M. E., Goodman H. M., Moore D. D. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol Cell Biol. 1986 Sep;6(9):3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G., Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988 Sep 9;241(4871):1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- Watt F., Molloy P. L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988 Sep;2(9):1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Weih F., Nitsch D., Reik A., Schütz G., Becker P. B. Analysis of CpG methylation and genomic footprinting at the tyrosine aminotransferase gene: DNA methylation alone is not sufficient to prevent protein binding in vivo. EMBO J. 1991 Sep;10(9):2559–2567. doi: 10.1002/j.1460-2075.1991.tb07796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B., Langner K. D., Jüttermann R., Müller U., Zock C., Klimkait T., Doerfler W. Reactivation of the methylation-inactivated late E2A promoter of adenovirus type 2 by E1A (13 S) functions. J Mol Biol. 1988 Jul 20;202(2):255–270. doi: 10.1016/0022-2836(88)90456-1. [DOI] [PubMed] [Google Scholar]