Abstract
The emergence of drug resistance mutations is increasing after the implementation of highly active antiretroviral therapy. To characterize two novel mutations L228I and Y232H in the primer grip of reverse transcriptase (RT) of HIV-1 circulating recombination form 08_BC (CRF08_BC) subtype, both mutant clones were constructed to determine their impacts on viral phenotypic susceptibility and replication capacity (RC). Results showed that the novel mutation, L228I, conferred a low-level resistance to etravirine by itself. L228I in combination with Y188C displayed a high level of cross-resistance to both nevirapine (NVP) and efavirenz (EFV). The copresence of A139V and Y232H induced a moderate level of resistance to NVP and EFV. Mutations Y188C/L228I, A139V, Y232H, and A139V/Y232H reduced more than 55% of viral RC compared with that of the wild-type (WT) reference virus. Modeling study suggested that the copresence of Y188C/L228I or A139V/Y232H might induce conformational changes to RT, which might result in reduced drug susceptibility and viral RC due to abolished hydrogen bonding or complex interaction with vicinal residues. Our results demonstrated that L228I and Y232H were novel accessory nonnucleoside reverse transcriptase inhibitor resistance–related mutations and provided valuable information for clinicians to design more effective treatment to patients infected with HIV-1 subtype CRF08_BC.
Introduction
To control HIV/AIDS-related morbidity and mortality and improve the life quality of HIV-1–infected patients, highly active antiretroviral therapy (HAART) was introduced and has achieved remarkable progress.1,2 Currently, the first-line HAART treatment for HIV-infected adults includes two nucleoside reverse transcriptase inhibitors (NRTIs), such as tenofovir (TDF) with lamivudine (3TC) or emtricitabine (FTC), plus one nonnucleoside reverse transcriptase inhibitor (NNRTI), such as efavirenz (EFV). If the regimen above is contraindicated or not available, one of the following options is recommended, including zidovudine (AZT) +3TC+EFV, AZT +3TC+ nevirapine (NVP), or TDF +3TC (or FTC)+NVP.3 Although HAART can control the viral load to a clinically undetectable level in most HIV cases, a large hindrance for extended effective treatments is the emergence of drug resistance mutations. Patients acquiring or infected with drug-resistant mutant viruses have fewer treatment options and are at higher risk of morbidity and mortality, especially in developing countries where choices of HIV inhibitors are limited.4 Thus, identifying and characterizing the drug resistance mutations and their impacts on drug susceptibility as well as viral replication can help clinicians offer more effective treatment to HIV patients.
To date, most drug resistance data focused on HIV-1 subtype B. Specifically, 21 major NNRTI resistance mutations at nine sites have been reported by the HIV Drug Resistance Database (http://hivdb.stanford.edu/DR/NNRTIResiNote.html, last updated on March 2, 2014). However, subtype B is mainly prevalent in the Western world, accounting for only 11% of global HIV-1 infections,5 while non-B subtypes are playing more important roles in the whole pandemic. Limited data are available concerning the genetic mechanisms of drug resistance in non-B subtype viruses. In contrast, it has been observed that different HIV-1 subtypes may develop various mutations to certain HIV inhibitors in both in vitro and in vivo studies.6–11 Viral replication capacity (RC) also differs among various subtypes, which may be magnified under high selective pressure.12 Thus, it is important and valuable to evaluate the impacts of mutations in non-B subtypes on drug susceptibility and viral RC.
Circulating recombination form 08_BC (CRF08_BC) is one of the current predominant subtypes in China containing a recombinant reverse transcriptase (RT) gene derived from subtypes Thailand B and Indian C.13,14 More than 40% of HIV infections in Chinese blood donors were found to be the CRF08_BC subtype from 2007 to 2010.15 However, few data are available for the drug resistance mutations in this subtype in treatment-exposed patients. Previously, to select the potentially emerging drug resistance mutations during NVP treatment, we propagated a clinical isolate of CRF08_BC subtype (2007CNGX-HK) in human peripheral blood mononuclear cells with increasing NVP concentration for 40 passages (about 400 days).14 Several novel mutations (L228I, Y232H, and D404N) in RT were observed in that study to confer moderate-to-high-level NVP resistance either alone or in combination.14 In a later study, we demonstrated that D404N in the RT connection subdomain was an NNRTI resistance–related mutation that displayed cross-resistance to NVP, EFV, rilpivirine (RPV), and AZT.16
In this study, we extended our research to the other two novel mutations, L228I and Y232H. Both these mutations locate in the β12–β13 hairpin of the palm subdomain, which contains the residues 227–235 (FLWMGYELH) and forms the so-called primer grip in RT.17,18 The function of the primer grip is to maintain the primer terminus in correct orientation for nucleophilic attack on an incoming dNTP during DNA strand extension.17 Mutations in this region, such as F227A, G231A, Y232A, E233A, and H235A, have been reported to result in the loss of RNase H function.19 L234A was confirmed to affect the dimerization of p51 and p66 subunits.20 Of note, mutations at L228 and Y232 have been reported to be related to drug resistance in subtype B or C in many studies.18,19,21–24 It is very interesting to further identify whether these previously selected novel mutations (L228I and Y232H) affect drug susceptibility and viral replication in subtype CRF08_BC. This study intensively characterized L228I and Y232H either alone or in combination with the other associated mutations through site-directed mutagenesis based on an infectious clone of the CRF08_BC subtype.
Methods
Cells, plasmids, and drugs
The 293FT cells were obtained from the American Type Culture Collection. The TZM-bl cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme, Inc. Both 293FT and TZM-bl cells were cultured as described previously.25 HIV-1 CRF08_BC subtype wild-type (WT) infectious plasmid pBRGX was previously constructed.25 Drugs, including three NNRTIs [NVP, EFV, and etravirine (ETR)] and five NRTIs [AZT, abacavir (ABC), 3TC, FTC, and TDF], were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH.
Site-directed mutagenesis and virus production
Six mutation patterns, including L228I, Y188C, Y188C/L228I, A139V, Y232H, and A139V/Y232H, in RT were introduced into the pBRGX plasmid by site-directed mutagenesis as described previously.16 Multiple mutations were introduced into pBRGX one by one. The presence of desired mutations in the plasmids was confirmed by DNA sequencing. Mutation-specific primers and primers for amplifying RT in positive clone identification were designed and listed in Table 1.
Table 1.
Primer Sequences for Site-Directed Mutagenesis
| Amino acid mutation | Codon mutation | Genetic mutation in BRGX | Primer sequences (5′-3′) | Position in HXB2 |
|---|---|---|---|---|
| L228I-F | CTT-ATT | C3202A | CATTTATTTGGATGGGGTATGAACTCCATCCTGAC | 3226–3260 |
| L228I-R | CTT-ATT | C3202A | ATCCAAATAAATGGAGGTTCTTTCTGATGTTTCTTGTCTG | 3199–3238 |
| Y188C-F | TAT-TGT | A3083G | ACTTGTGTGTAGGATCTGACTTAGAAATAGGGCAGCAT | 3106–3143 |
| Y188C-R | TAT-TGT | A3083G | ATCCTACACACAAGTCATCCATGTATTGATAGATAACTATG | 3080–3120 |
| A139V-F | GCA-GTA | C2936T | TGAGGTACCAGGGATTAGATATCAGTACAATGTGCTTCCAC | 2960–3000 |
| A139V-R | GCA-GTA | C2936T | CCTGGTACCTCATTGTTTACACTAGGTATGGTAAATGCAG | 2932–2971 |
| Y232H-F | TAT-CAT | T3214C | ATGGGGCATGAACTCCATCCTGACAAATGGACAGTAC | 3237–3273 |
| Y232H-R | TAT-CAT | T3214C | AGTTCATGCCCCATCCAAAGAAATGGAGGTTC | 3219–3250 |
| Primers for RT amplification to sequence (5′-3′) | ||||
| HI1395 | ATGAAGAGGCTGCAGAATGGG | 1406–1426 | ||
| HI4635 | GGATTCTACTACTCCCTGACTTTGG | 4664–4688 | ||
Mutated nucleotides are underlined and in bold.
RT, reverse transcriptase.
Viruses were produced by transfection of constructed infectious plasmids into 293FT cells as described previously.25,26 Viral titer was detected by the measurement of p24 production using the Vironostika Kit (bioMérieux) according to the manufacturer's instructions and by detection of the 50% tissue culture infective dose (TCID50) using TZM-bl cells as described previously.27 Only those with p24 production higher than 50 mg/liter were subjected to phenotypic drug resistance assay and RC analysis.
Phenotypic susceptibility of virus variants
The susceptibility of virus variants to NNRTIs (NVP, EFV, and ETR) and NRTIs (AZT, ABC, 3TC, FTC, and TDF) was measured in a single-cycle cell culture–based phenotypic assay in TZM-bl cells as described previously.16 In brief, 1 × 104 cells were seeded on a 96-well flat-bottom plate in 100 μl of growth medium (Dulbecco's modified Eagle's medium +10% fetal bovine serum +1% penicillin/streptomycin). Next day, mutant and WT viruses equal to 200–400 blue focus forming unit in serially diluted drugs were added into each well of the plate in triplicate. The wells infected with equal amount of viruses but without drugs were taken as viral controls. Then, the plate was incubated at 37°C in a 5% CO2 incubator for 48 h. After the cells were fixed and stained, the number of blue cells was calculated using ImmunoSpot 3.2 software after imaged by ELIspot plate reader (Cellular Technology Limited). The 50% inhibitory concentration (IC50) values were determined by SigmaPlot 12.0 software (four-parameter logistic analysis). Viral susceptibility to drugs was expressed as the fold changes in IC50 of mutant virus compared to that of the WT reference virus. To compare the results in this study with those of previous work in the same criterion, the levels of resistance are divided into four groups based on fold changes as described previously,14,27 that is, high-level resistance (>20-folds), moderate-level resistance (4- to 20-folds), low-level resistance (2- to 4-folds), and susceptive (0- to 2-folds).
RC determination of virus variants
The RCs of WT and mutant viruses were evaluated in a noncompetitive RC assay as described previously.25,28–30 Briefly, 1 × 104 TZM-bl cells per well were seeded on a 96-well plate. Next day, virus stocks were 3- to 10-fold serially diluted and applied to the cells. Each virus was tested in triplicate for each concentration. After 48 h of incubation at 37°C in a 5% CO2 incubator, cells were fixed and stained. Virus-infected blue cells were counted as described above. RC was determined by normalizing the blue cell number to corresponding p24 production of each mutant virus. Final relative RC levels were expressed as a percentage of mutant viral RC with reference to that of the WT reference virus. RC of test virus (%) = (blue cell number/ng of p24test virus/blue cell number/ng of p24WT reference virus) × 100.
Molecular modeling of RT
To investigate whether any possible structural changes could be induced by the novel mutations, L228I and Y232H, the models of HIV-1 CRF08_BC RT (1–560 amino acids) were built by homology modeling with HIV-1 RT, NVP, and RNA/DNA hybrid crystal structure 4PUO31 using the server SWISS-MODEL32 with default parameters, in which WTs contain L228, A139, and Y232, while mutants contain 228I, 139V, and 232H. The models were further analyzed using Discovery Studio 4.1 Visualizer (Accelrys), and the Ramachandran plot was examined to ensure that the structures of the models are not in the unfavorable regions.
Statistical analysis
The data were evaluated for statistical significance by an unpaired Student's t-test using Prism software (version 5.0; GraphPad, Inc.). Values of p < .05 were considered significant. Results were given as mean ± standard deviation of triplicate independent experiments.
Results
L228I conferred low-level resistance to ETR and enhanced viral resistance to NVP in the context of Y188C
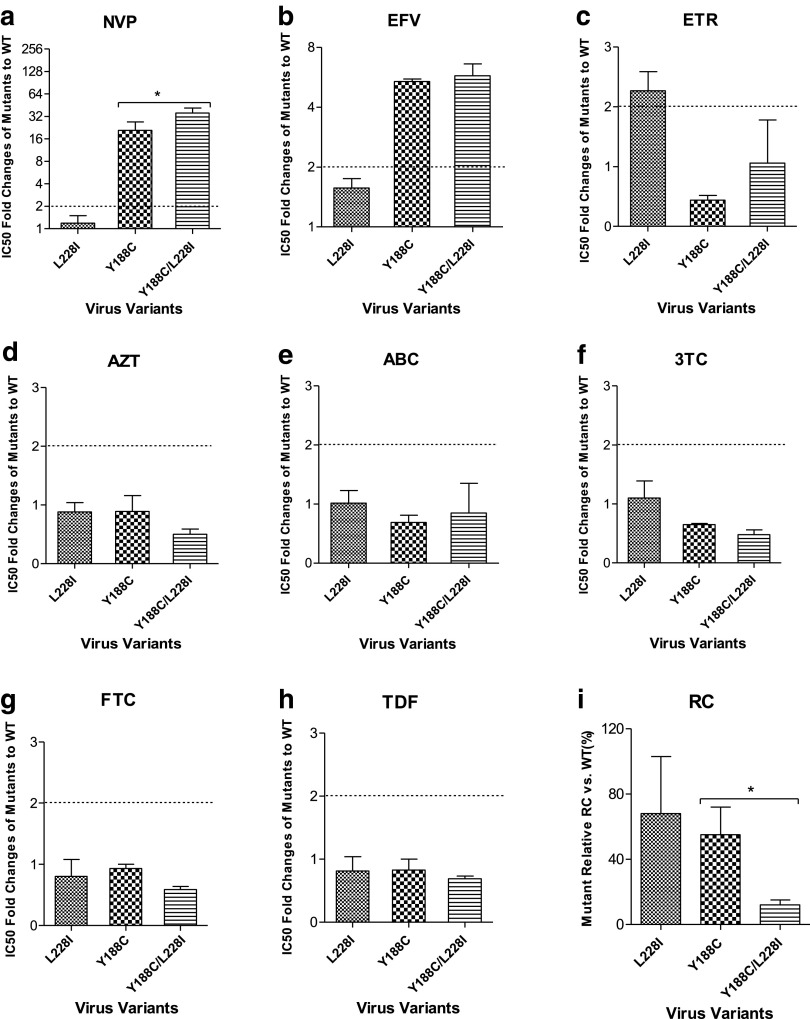
To define the exact phenotypic effect of a single mutation L228I on NVP and other RT inhibitors, susceptibility assays using TZM-bl cells were performed. Our results showed that a single L228I mutation failed to display a substantial effect on viral susceptibility to the first-generation NNRTIs, NVP (Fig. 1a) or EFV (Fig. 1b) (<2-fold), but exerted a low-level resistance to the second-generation NNRTI, ETR (2.27-fold) (Fig. 1c). Viruses with the primary mutation of Y188C conferred a high-level resistance to NVP (20.86-fold) (Fig. 1a) and a moderate-level resistance to EFV (5.38-fold) (Fig. 1b). Interestingly, the addition of L228I to Y188C significantly increased viral resistance to NVP (35.47-fold) compared to that of Y188C alone (20.86-fold) (p = .0444) (Fig. 1a), indicating that L228I could further reduce viral susceptibility to NVP and EFV in the context of Y188C. The viruses above containing mutations Y188C, L228I, and Y188C/L228I were susceptible to all the current clinically available NRTIs (AZT, ABC, 3TC, FTC, and TDF) (<2-fold) (Fig. 1d–h).
FIG. 1.
Phenotypic analysis and viral RC of L228I-associated virus variants. Compared with WT virus, the fold changes of IC50 of L228I-associated virus variants against NNRTIs NVP (a), EFV (b), and ETR (c), as well as NRTIs AZT (d), ABC (e), 3TC (f), FTC (g), and TDF (h), were determined by phenotypic assay. Viral RCs of these variants were tested by a single-cycle cell culture assay (i). In phenotypic assay, TZM-bl cells were infected with virus variants and treated with serially diluted RT inhibitors. In RC detection assay, TZM-bl cells were infected with serially diluted virus variants without inhibitors. The dashed line is labeled at twofolds of IC50 ratio to indicate the drug resistance standard. 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; IC50, 50% inhibitory concentration; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; NVP, nevirapine; RC, replication capacity; RT, reverse transcriptase; TDF, tenofovir; WT, wild type.
To evaluate the effect of the mutation L228I on viral replication, we performed a single-cycle cell culture assay. Compared to the WT virus, L228I retained a 68% RC of WT, which was close to that of Y188C (55% of WT). Y188C/L228I significantly reduced viral RC to 12% of WT compared to that of single Y188C (55%, p = .0475) (Fig. 1i). The results suggested that L228I might impair viral replication in the presence of Y188C.
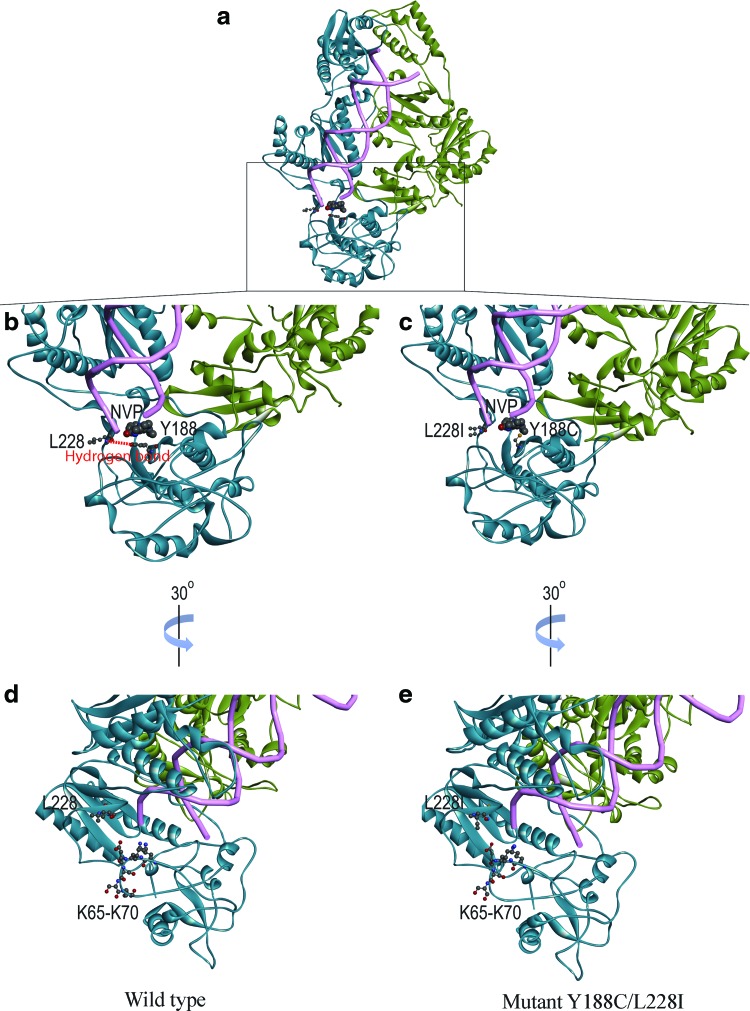
To investigate the possible mechanism behind the effects of the novel mutation L228I, the three-dimensional RT model of HIV-1 subtype CRF08_BC was built by homology modeling with HIV-1 RT, NVP, and the RNA/DNA hybrid crystal structure (4PUO) through the server SWISS-MODEL as described previously.16 As shown in Figure 2a, the whole structure of WT CRF08_BC subtype RT is a heterodimer of p51 (green) and p66 (blue) subunits, binding an RNA/DNA hybrid (Fig. 2a). The orientation of L228 is in close proximity to the binding pocket of NVP but not interacting with NVP (Fig. 2b). At the same time, a hydrogen bond is formed between L228 and Y188 in the WT RT (Fig. 2b). When L228I and Y188C are introduced, the hydrogen bond will be abolished (Fig. 2c), possibly disrupting the core structure of p66 subunit close to the binding site of NVP. Intriguingly, we also observed that the residue L228 is located at the entrance of the RNA/DNA hybrid together with the hydrophilic residues K65–K70 (Fig. 2d). The mutation L228I with larger side chain might slightly block this entrance site and inhibit the reverse transcription (Fig. 2e).
FIG. 2.
RT models of WT and mutant Y188C/L228I. The RT models of WT and mutant are shown in ribbon diagram. Subunits of p66 and p51 are colored as blue and green, respectively (a). The residues at 228, 188 (b, c), and K65–K70 (d, e) in p66 subunit are highlighted. Their atoms are labeled in ball-and-stick format. The hydrogen bond between L228 and Y188 is shown in red dotted line (b). The DNA/RNA hybrid is shown in tube format in pink, while the NNRTI NVP is shown in CPK format (a). The figures were produced using DS Visualizer (Accelrys). Color images available online at www.liebertpub.com/aid
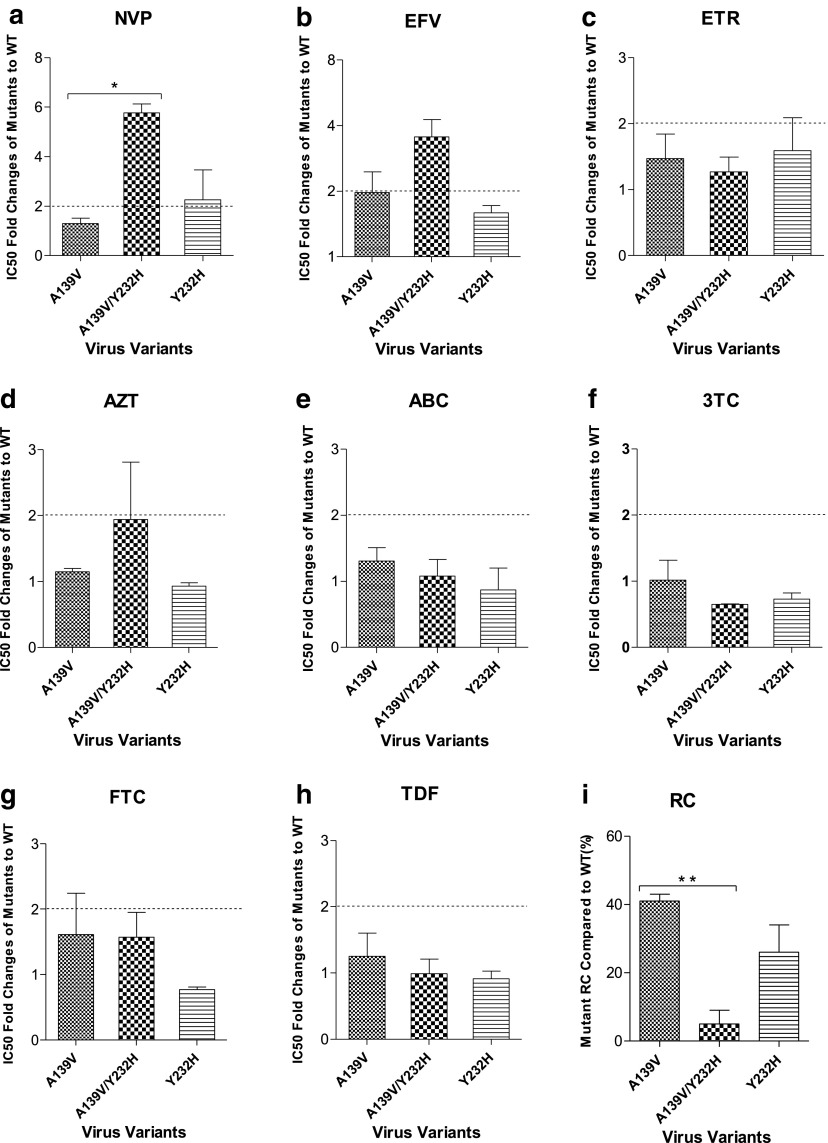
Previously, Y232H was selected together with A139V, and the combination of A139V–Y232H was suggested to exhibit a high-level resistance to NVP.14 Here, the mutant viruses containing Y232H and/or A139V were produced to detect the viral susceptibility to RT inhibitors. As shown in Figure 3, no resistance to NVP, EFV, or ETR was observed in virus containing a single mutation A139V, while Y232H alone induced a low-level resistance to NVP (2.26-fold). Of note, the combination of A139V and Y232H conferred a moderate-level resistance to NVP (5.76-fold), which was significantly higher than that of A139V alone (p = .0045) (Fig. 3a) and a low-level resistance to EFV (3.55-fold) (Fig. 3b). No resistance to ETR (Fig. 3c) and any of the NRTIs (AZT, ABC, 3TC, FTC, and TDF) (Fig. 3d–h) was detected in either single or the combinatorial pattern of A139V and Y232H. In parallel, viral RCs of A139V and Y232H were also detected (Fig. 3i). The single mutation A139V displayed 41% RC of WT, while Y232H exhibited 26% RC of WT. The combination of A139V–Y232H dramatically reduced viral RC to 5% of WT, which was significantly lower than that of single A139V (p = .0088).
FIG. 3.
Phenotypic analysis and viral RC of Y232H-associated virus variants. Compared with WT virus, the fold changes of IC50 of Y232H-associated virus variants against NNRTIs NVP (a), EFV (b), and ETR (c), as well as NRTIs AZT (d), ABC (e), 3TC (f), FTC (g), and TDF (h), were determined by phenotypic assay, while viral RCs of these variants were tested by a single-cycle cell culture assay (i). In phenotypic assay, TZM-bl cells were infected with virus variants and treated with serially diluted RT inhibitors. In RC detection assay, TZM-bl cells were infected with serially diluted virus variants without inhibitors. *p < .05, **p < .01. The dashed line is labeled at twofolds of IC50 ratio to indicate the drug resistance standard.
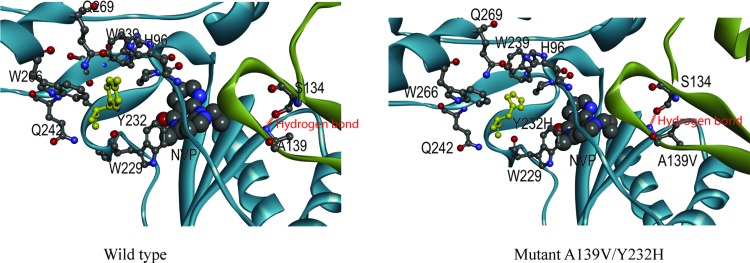
To further investigate the possible mechanism behind the reduced drug susceptibility and viral RC by A139V and Y232H, the RT model of subtype CRF08_BC was also built based on 4PUO as prescribed previously.16 As shown in Figure 4, Y232 in p66 subunit near the NVP binding pocket interacts with the adjacent amino acids W266, Q269, W239, H96, W229, and Q242. When Y232 is substituted into 232H, the primary interactions between Y232 and W266, Q269, W239, H96, and W229 will be abolished, which might induce structural change and disruption in p66 subunit. In addition, residue A139 in p51 subunit locates in a β7–β8 loop region, which is in the RT dimerization interface and close to the NVP binding pocket. A hydrogen bond is formed between S134 and A139. The substitution of alanine to valine at residue 139 might influence the hydrogen bond by increasing the hydrophobicity of the side chain as well as the core structure of the loop. Therefore, the combination of A139V–Y232H might cause significant conformational change to the RT structure than each of them alone.
FIG. 4.
RT models of WT and mutant A139V/Y232H. The models of WT and mutant are shown in ribbon diagram. Subunits of p66 and p51 are colored as blue and green, respectively. The residues at 232 and its vicinal sites in p66 subunit, as well as A139 and S134 in p51 subunit, are highlighted. Their atoms are labeled in ball-and-stick format. Y232 is colored in yellow. The hydrogen bond between A(V)139 and S134 is shown in red dotted line. The DNA/RNA hybrid is shown in tube format in pink, and the NNRTI NVP is shown in CPK format. The figures were produced using DS Visualizer (Accelrys). Color images available online at www.liebertpub.com/aid
Discussion
In the previous study,14 we selected two novel mutations, L228I and Y232H, in HIV-1 CRF08_BC subtype RT under increasing NVP pressure. The results suggested that these two mutations conferred moderate-to-high-level resistance to NVP in combination with other mutations. To further identify the effects of these mutations on viral susceptibility and RC, this study constructed six mutant viruses containing mutations Y188C, L228I, Y188C/L228I, A139V, Y232H, and A139V/Y232H based on an HIV-1 CRF08_BC subtype infectious clone. We found that L228I alone conferred a low-level resistance to ETR. The combinations of Y188C/L228I and A139V/Y232H enabled the virus to be cross-resistant to both NVP and EFV accompanied with severe RC loss. Modeling study suggested that the copresence of L228I and Y188C or A139V and Y232H might lead to significant conformational changes in RT due to abolished hydrogen bonding or amino acid interactions, resulting in drug resistance and reduced viral RC.
At codon 228, the most commonly reported mutation was L228H/R. It was suggested to be associated with treatment containing both NRTIs22 and NNRTIs21,33. Another study reported that L228H/R was associated with the accumulation of both type I and type II thymidine analogue mutation resistance pathways in both subtypes B and C.34 Recently, L228R was also identified in RT of HIV-1 subtype CRF_BC strains isolated from NVP-containing HAART treatment–experienced patients.35 However, it was completely absent from those treatment-naive patients in China.35 That study also found that L228R did not show any significant resistance effect (<2-fold) to NNRTIs, including NVP, EFV, delavirdine (DLV), and ETR.35 To our knowledge, only one study observed L228I in the context of V75M and F227L in one Iranian HIV-1–positive patient who failed to antiretroviral therapy.36 No further investigation on L228I was reported thereafter. In this study, although viruses with a single L228I mutation remained susceptive to NVP and EFV, the combinations of Y188C–L228I exhibited a high level of cross-resistance to these two first-line HAART drugs, which was in agreement with our previous results.14 No significant resistance to any NRTIs (AZT, ABC, 3TC, TDF, and FTC) was detected by the L228I-containing viruses. Our results suggested that L228I might be a novel accessory NNRTI resistance–related mutation in subtype CRF08_BC. If L228I and Y188C occur at the same time during the genotypic resistance test before the treatment starts, the first-generation NNRTIs, including NVP and EFV, should be avoided to be prescribed. Additionally, we also assessed the drug susceptibility to NNRTIs of Y188C and found that Y188C conferred a high-level resistance to NVP and a moderate-level resistance to EFV, which was consistent with previous reports from our and others' studies.37–39
Modeling study showed that a hydrogen bond was formed between L228 and Y188 in WT RT. When only a single mutation L228I emerged, it would not affect this hydrogen bond. However, when Y188C and L228I were introduced, the hydrogen bond would be abolished. Since Y188 is in the NNRTI binding pocket,40 the abolished hydrogen bond might affect the NNRTI binding. This might be helpful to understand why in this study a single mutation L228I did not induce significant drug resistance to NVP and EFV but Y188C could confer high- or moderate-level resistance to these two NNRTIs.
To determine the impacts of Y188C and L228I on the viral RC, a single-cycle cell culture assay was performed in this study. Results showed that the single mutation Y188C or L228I slightly impaired viral RC. The copresence of Y188C and L228I conferred significant lower RC than the single mutation Y188C, which indicated that there was a synergistic effect between L228I and Y188C on reducing viral RC. Modeling study showed that L228 locates in the primer grip and the lager side chain of L228I might block the entrance site of RNA/DNA hybrid during reverse transcription, which might also lead to impaired viral replication.
In addition to L228I, our previous study also selected another novel mutation Y232H in the primer grip region, which exhibited a high-level resistance to NVP together with A139V.14 In this study, the phenotypic results showed that a single Y232H mutation only showed a low-level resistance to ETR but kept susceptive to NVP and EFV. A single mutation A139V also did not impact viral susceptibility to NVP and EFV, which is consistent with the results of another study in subtype B infection.41 Nevertheless, the combination of A139V and Y232H displayed moderate and low levels of resistance to NVP and EFV, respectively. This observation suggested that there was a synergistic drug resistance effect between A139V and Y232H. The first-line HAART regimen should be considered to include ETR instead of NVP or EFV in the context of the combinational pattern of A139V and Y232H. Of note, the resistance level to NVP induced by A139V/Y232H in this study was a little lower than that suggested previously.14 This might be associated with the cumulative effect of other additional mutations since in previous study A139V and Y232H were present together with three other mutations (Y181C, H221Y, and E396G).
Furthermore, we also found that both Y232H and A139V had substantial negative impacts on viral replication. This might be associated with their spatial positions. Modeling study suggested that when Y232 in the p66 subunit was substituted into 232H, the previous interactions between Y232 and its vicinal residues in the palm and thumb subdomains might be abolished. This might induce further conformational change to p66 subunit, leading to reduced viral RC. Besides, it has been reported that the β7–β8 loop (codons 132–140) in p51 subunit of RT is important for RT dimerization,41–43 and the integrity of the loop is maintained by hydrogen bonding between the side chains of S134 and T139. In this study, the RT model also showed a hydrogen bond between A139 and S134 in the p51 subunit. However, the substitution of alanine to valine (A139V) at codon 139 would influence the hydrogen bond by increasing the hydrophobicity of the side chain as well as the core structure of the loop. Therefore, we hypothesize that RT dimerization might also be influenced by the mutation A139V, resulting in impaired RT activity. This may explain why A139V could diminish part of viral RC. When both Y232H and A139V are induced simultaneously, the RT configuration changes would be enhanced and result in more severe impairment to viral RC.
In addition, other substitutions at codon 139, such as T139K/R, were also reported to be associated with RTI treatment in clinical samples of subtype B patients,23,35,44 but they did not appear in our previous selection study. This might be related to the subtype-specific codon differences. As shown in Table 2, the WT amino acid at codon 139 in HIV-1 RT is different in subtype CRF08_BC (Ala, A) from subtype B (Thr, T). It is supposed to be easier for Ala (GCA) to be mutated into Val (GTA) than Thr (ACA) since A139V needs one base pair change (C-T), while T139V requires two (AC-GT). Similarly, it should be easier for Thr (T, ACA) to be substituted by Lys (K, AAA) and Arg (R, AGA) for the same reason. That might be helpful to explain why we selected 139V but not 139K/R in subtype CRF08_BC.
Table 2.
Mutations at 139, 228, and 232 in Subtype B and CRF08_BC
| Subtype B | Subtype CRF08_BC | |||
|---|---|---|---|---|
| Site in RT | Amino acid | Codon | Amino acid | Codon |
| Wild type | ||||
| 139 | Thr (T) | ACA | Ala (A) | GCA |
| 228 | Leu (L) | CTT | Leu (L) | CTT |
| 232 | Tyr (Y) | TAT | Tyr (Y) | TAT |
| Mutants | ||||
| 139 | Lys (K) | AAA | Val (V) | GTA |
| Arg (R) | AGA | |||
| Ile (I) | ATA | |||
| 228 | Arg (R) | CGT | Ile (I) | ATT |
| His (H) | CAT | |||
| 232 | His (H) | CAT | His (H) | CAT |
CRF08_BC, circulating recombination form 08_BC.
In conclusion, we identified and characterized two novel accessory NNRTI resistance–associated mutations L228I and Y232H in the RT of HIV-1 subtype CRF08_BC virus. Attention should be paid to these novel mutations in the genotypic resistance test beside those reported mutations in subtype B. Overall, our study contributed by filling critical gaps in the global profile of HIV-1–induced drug resistance in non-B subtype. The results provide valuable information for drug resistance surveillance, HIV-1 genotypic resistance testing, and the treatment modulation of HIV-1–infected individuals in China.
Acknowledgments
We are highly appreciative of the resources made available by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. These include TZM-bl from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme, Inc., as well as the following RT inhibitors, including AZT, 3TC, ABC, FTC, TDF, NVP, EFV, and ETR. This work was supported by a grant from the AIDS Trust Fund of Hong Kong Special Administrative Region Government of the People's Republic of China (MSS 140R).
Author Contributions
X.M.Z. and B.J.Z. conceived and designed the experiments. X.M.Z., Q.Z., H.W., and K.Z. performed the experiments. T.C.K.L. and X.L. constructed RT modeling study and analyzed the results. X.M.Z., H.C., J.Z., Z.W.C., D.Y.J., and B.J.Z. analyzed results and wrote the article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hammer SM, Squires KE, Hughes MD, et al. : A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med 1997;337:725–733 [DOI] [PubMed] [Google Scholar]
- 2.Gulick RM, Mellors JW, Havlir D, et al. : Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med 1997;337:734–739 [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization: Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach. WHO Press, Geneva, Switzerland, 2013 [PubMed] [Google Scholar]
- 4.Tang MW, Shafer RW: HIV-1 antiretroviral resistance: Scientific principles and clinical applications. Drugs 2012;72:e1–e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delviks-Frankenberry KA, Lengruber RB, Santos AF, et al. : Connection subdomain mutations in HIV-1 subtype-C treatment-experienced patients enhance NRTI and NNRTI drug resistance. Virology 2013;435:433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holguin A, Ramirez de Arellano E, Rivas P, Soriano V: Efficacy of antiretroviral therapy in individuals infected with HIV-1 non-B subtypes. AIDS Rev 2006;8:98–107 [PubMed] [Google Scholar]
- 7.Brenner B, Turner D, Oliveira M, et al. : A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 2003;17:F1–F5 [DOI] [PubMed] [Google Scholar]
- 8.Caride E, Brindeiro R, Hertogs K, et al. : Drug-resistant reverse transcriptase genotyping and phenotyping of B and non-B subtypes (F and A) of human immunodeficiency virus type I found in Brazilian patients failing HAART. Virology 2000;275:107–115 [DOI] [PubMed] [Google Scholar]
- 9.Caride E, Hertogs K, Larder B, et al. : Genotypic and phenotypic evidence of different drug-resistance mutation patterns between B and non-B subtype isolates of human immunodeficiency virus type 1 found in Brazilian patients failing HAART. Virus Genes 2001;23:193–202 [DOI] [PubMed] [Google Scholar]
- 10.Grossman Z, Istomin V, Averbuch D, et al. : Genetic variation at NNRTI resistance-associated positions in patients infected with HIV-1 subtype C. AIDS 2004;18:909–915 [DOI] [PubMed] [Google Scholar]
- 11.Lai MT, Lu M, Felock PJ, et al. : Distinct mutation pathways of non-subtype B HIV-1 during in vitro resistance selection with nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 2010;54:4812–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spira S, Wainberg MA, Loemba H, Turner D, Brenner BG: Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J Antimicrob Chemother 2003;51:229–240 [DOI] [PubMed] [Google Scholar]
- 13.Ouyang Y, Shao Y, Ma L: HIV-1 CRF_BC recombinants infection in China: Molecular epidemic and characterizations. Curr HIV Res 2012;10:151–161 [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Zhang XM, Zhang HJ, et al. : In vitro selection of HIV-1 CRF08_BC variants resistant to reverse transcriptase inhibitors. AIDS Res Hum Retroviruses 2015;31:260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng P, Wang J, Huang Y, et al. : The human immunodeficiency virus-1 genotype diversity and drug resistance mutations profile of volunteer blood donors from Chinese blood centers. Transfusion 2012;52:1041–1049 [DOI] [PubMed] [Google Scholar]
- 16.Zhang XM, Wu H, Zhang Q, et al. : A novel mutation, D404N, in the connection subdomain of reverse transcriptase of HIV-1 CRF08_BC subtype confers cross-resistance to NNRTIs. J Antimicrob Chemother 2015;70:1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cases-Gonzalez CE, Menendez-Arias L: Increased G → A transition frequencies displayed by primer grip mutants of human immunodeficiency virus type 1 reverse transcriptase. J Virol 2004;78:1012–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisniewski M, Palaniappan C, Fu Z, Le Grice SF, Fay P, Bambara RA: Mutations in the primer grip region of HIV reverse transcriptase can increase replication fidelity. J Biol Chem 1999;274:28175–28184 [DOI] [PubMed] [Google Scholar]
- 19.Palaniappan C, Wisniewski M, Jacques PS, Le Grice SF, Fay PJ, Bambara RA: Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem 1997;272:11157–11164 [DOI] [PubMed] [Google Scholar]
- 20.Ghosh M, Jacques PS, Rodgers DW, Ottman M, Darlix JL, Le Grice SF: Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 1996;35:8553–8562 [DOI] [PubMed] [Google Scholar]
- 21.Saracino A, Monno L, Scudeller L, et al. : Impact of unreported HIV-1 reverse transcriptase mutations on phenotypic resistance to nucleoside and non-nucleoside inhibitors. J Med Virol 2006;78:9–17 [DOI] [PubMed] [Google Scholar]
- 22.Gonzales MJ, Wu TD, Taylor J, et al. : Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS 2003;17:791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahriar R, Rhee SY, Liu TF, et al. : Nonpolymorphic human immunodeficiency virus type 1 protease and reverse transcriptase treatment-selected mutations. Antimicrob Agents Chemother 2009;53:4869–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wohrl BM, Krebs R, Thrall SH, Le Grice SF, Scheidig AJ, Goody RS: Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J Biol Chem 1997;272:17581–17587 [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Zhang X, Wu H, et al. : Parental LTRs are important in a construct of a stable and efficient replication-competent infectious molecular clone of HIV-1 CRF08_BC. PLoS One 2012;7:e31233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang HJ, Wang YX, Wu H, Jin DY, Wen YM, Zheng BJ: The y271 and i274 amino acids in reverse transcriptase of human immunodeficiency virus-1 are critical to protein stability. PLoS One 2009;4:e6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Zhang HJ, Zhang XM, et al. : Identification of drug resistant mutations in HIV-1 CRF07_BC variants selected by nevirapine in vitro. PLoS One 2012;7:e44333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asahchop EL, Wainberg MA, Oliveira M, et al. : Distinct resistance patterns to etravirine and rilpivirine in viruses containing nonnucleoside reverse transcriptase inhibitor mutations at baseline. AIDS 2013;27:879–887 [DOI] [PubMed] [Google Scholar]
- 29.Xu HT, Quan Y, Schader SM, Oliveira M, Bar-Magen T, Wainberg MA: The M230L nonnucleoside reverse transcriptase inhibitor resistance mutation in HIV-1 reverse transcriptase impairs enzymatic function and viral replicative capacity. Antimicrob Agents Chemother 2010;54:2401–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta S, Fransen S, Paxinos EE, Stawiski E, Huang W, Petropoulos CJ: Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: Assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance. Antimicrob Agents Chemother 2010;54:1973–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das K, Martinez SE, Bandwar RP, Arnold E: Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: Insights into requirements for RNase H cleavage. Nucleic Acids Res 2014;42:8125–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold K, Bordoli L, Kopp J, Schwede T: The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006;22:195–201 [DOI] [PubMed] [Google Scholar]
- 33.Ceccherini-Silberstein F, Svicher V, Sing T, et al. : Characterization and structural analysis of novel mutations in human immunodeficiency virus type 1 reverse transcriptase involved in the regulation of resistance to nonnucleoside inhibitors. J Virol 2007;81:11507–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koning FA, Castro H, Dunn D, et al. : Subtype-specific differences in the development of accessory mutations associated with high-level resistance to HIV-1 nucleoside reverse transcriptase inhibitors. J Antimicrob Chemother 2013;68:1220–1236 [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Li Z, Xing H, et al. : Identification of the critical sites of NNRTI-resistance in reverse transcriptase of HIV-1 CRF_BC strains. PLoS One 2014;9:e93804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahman MK, Ayoub K, Mahboodi F: Genotypic correlation of a virologic response to lamivudine, stavudine and nevirapine in patients for whom combination therapy had failed. Iranian J Clin Infect Dis 2008;3:215–219 [Google Scholar]
- 37.Johnson VA, Brun-Vezinet F, Clotet B, et al. : Update of the drug resistance mutations in HIV-1: 2007. Top HIV Med 2007;15:119–125 [PubMed] [Google Scholar]
- 38.Tambuyzer L, Azijn H, Rimsky LT, et al. : Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir Ther 2009;14:103–109 [PubMed] [Google Scholar]
- 39.Wu H, Zhang X, Zhang H, et al. : In vitro selection of HIV-1 CRF08_BC variants resistant to reverse transcriptase inhibitors. AIDS Res Hum Retroviruses 2015;31:260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sluis-Cremer N, Temiz NA, Bahar I: Conformational changes in HIV-1 reverse transcriptase induced by nonnucleoside reverse transcriptase inhibitor binding. Curr HIV Res 2004;2:323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nissley DV, Radzio J, Ambrose Z, et al. : Characterization of novel non-nucleoside reverse transcriptase (RT) inhibitor resistance mutations at residues 132 and 135 in the 51 kDa subunit of HIV-1 RT. Biochem J 2007;404:151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auwerx J, Rodriguez-Barrios F, Ceccherini-Silberstein F, et al. : The role of Thr139 in the human immunodeficiency virus type 1 reverse transcriptase sensitivity to (+)-calanolide A. Mol Pharmacol 2005;68:652–659 [DOI] [PubMed] [Google Scholar]
- 43.Balzarini J, Auwerx J, Rodriguez-Barrios F, et al. : The amino acid Asn136 in HIV-1 reverse transcriptase (RT) maintains efficient association of both RT subunits and enables the rational design of novel RT inhibitors. Mol Pharmacol 2005;68:49–60 [DOI] [PubMed] [Google Scholar]
- 44.Ceccherini-Silberstein F, Gago F, Santoro M, et al. : High sequence conservation of human immunodeficiency virus type 1 reverse transcriptase under drug pressure despite the continuous appearance of mutations. J Virol 2005;79:10718–10729 [DOI] [PMC free article] [PubMed] [Google Scholar]