Abstract
The Red Palm Weevil (RPW) Rhynchophorus ferrugineus (Olivier) is a voracious pest of palm species. In recent decades its range has expanded greatly, particularly impacting the date palm industry in the Middle East. This has led to conjecture regarding the origins of invasive RPW populations. For example, in parts of the Middle East, RPW is commonly referred to as the “Pakistani weevil” in the belief that it originated there. We sought evidence to support or refute this belief. First reports of RPW in Pakistan were from the Punjab region in 1918, but it is unknown whether it is native or invasive there. We estimated genetic variation across five populations of RPW from two provinces of Pakistan, using sequences of the mitochondrial cytochrome oxidase subunit I gene. Four haplotypes were detected; two (H1 and H5) were abundant, accounting for 88% of specimens across the sampled populations, and were previously known from the Middle East. The remaining haplotypes (H51 and H52) were newly detected (in global terms) and there was no geographic overlap in their distribution within Pakistan. Levels of haplotype diversity were much lower than those previously recorded in accepted parts of the native range of RPW, suggesting that the weevil may be invasive in Pakistan. The affinity of Pakistani haplotypes to those reported from India (and the geographical proximity of the two countries), make the latter a likely “native” source. With regards the validity of the name “Pakistani weevil”, we found little genetic evidence to justify it.
Keywords: Middle East, Pakistani weevil, Punjab Province, Khyber Pakhtunkhwa Province, COI
The Red Palm Weevil (RPW) Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae: Rhynchophorinae) has been recognized as a major economically important pest of palm species for more than a century. It has been found devastating over 40 different commercial and ornamental palm tree species, belonging to 23 different genera and 3 families (Faleiro et al. 2012, Giblin-Davis et al. 2013). These include date palm Phoenix dactylifera L. (Mukhtar et al. 2011), oil palm Elaeis guineensis Jaquin (Murphy and Briscoe 1999), coconut Cocos nucifera L.(Faleiro 2006), and Canary Island date palm P. canariensis Chabaud, (El-Mergawy and Al-Ajlan 2011). The larval stages of RPW typically reside within the trunk of an infested palm tree, destroying the vascular system and boring into the heart of the host (Wakil et al. 2015). Voracious feeding by these larvae may subsequently lead to tree collapse. In India, yield losses of 10–25% have been reported in coconut plantations (Murphy and Briscoe 1999). In the Arabian Peninsula (an area that accounts for 30% of global date production), RPW is estimated to damage up to 5% of the date palm plantations, resulting in losses of 5–20 million US dollars (El-Sabea et al. 2009). In Pakistan, production losses of 10–20% have been reported in different varieties of dates (Baloch et al. 1992).
Around the world, RPW is also variously referred to as the Asiatic palm weevil, coconut weevil, sago palm weevil, and red stripe weevil (although the actual specific identity of many reported populations, particularly those in SE Asia, is likely to be wrong; Rugman-Jones et al. 2013). Furthermore, because of the cryptic, internal nature of the beetle’s attack, and the resulting slow death of the palm tree, it has also been referred to as “the hidden enemy” and even date palm AIDS (Khamiss and Abdel Badeea 2013). In the Middle East, RPW is often referred to as the “Pakistani weevil” in the belief that it invaded the former from Pakistan. However, this is somewhat controversial nomenclature, since there is little empirical evidence supporting a causative link (Rugman-Jones et al. 2013). Furthermore, RPW is generally considered to be invasive in Pakistan, although it was first formerly reported in what are now the Multan, Muzaffargarh and Dera Ghazi Khan (D.G. Khan) Districts of the Pakistani province of Punjab, and the neighboring Indian state of Punjab, almost a century ago (Milne 1918). Wattanapongsiri (1966) has since defined the native range of RPW as an area stretching east from India throughout SE Asia (although its occurrence in Indonesia has recently been thrown into doubt; Rugman-Jones et al. 2013), and in his detailed revision of the genus Rhynchophorus, based on several extensive museum collections, he recorded only a single undated specimen from modern day Pakistan. However, it remains a possibility that RPW was “always” present in Pakistan.
The objective of this study is to characterize genetic diversity within and between different RPW populations in Pakistan with the hope of identifying: (1) whether Pakistan forms part of the native range of RPW; or if not, (2) the most likely origin of Pakistani RPW populations; and finally, (3) whether there is evidence that Middle Eastern populations of RPW originated from Pakistan. We used sequences of the mitochondrial cytochrome oxidase I (COI) gene to investigate genetic variation in RPW from five different geographically isolated populations in the Punjab and Khyber Pakhtunkhwa (KPK) provinces of Pakistan, and also compared these with publically available sequences from RPW populations from around the world. If Pakistan is part of the native range, we expect to find relatively high levels of diversity (i.e., a large number of haplotypes; see Rugman-Jones et al. 2013). In contrast, relatively low levels of diversity are typical of invasive populations and by comparison with other populations, can be used to make inferences about their potential origins.
Materials and Methods
Specimen Collections
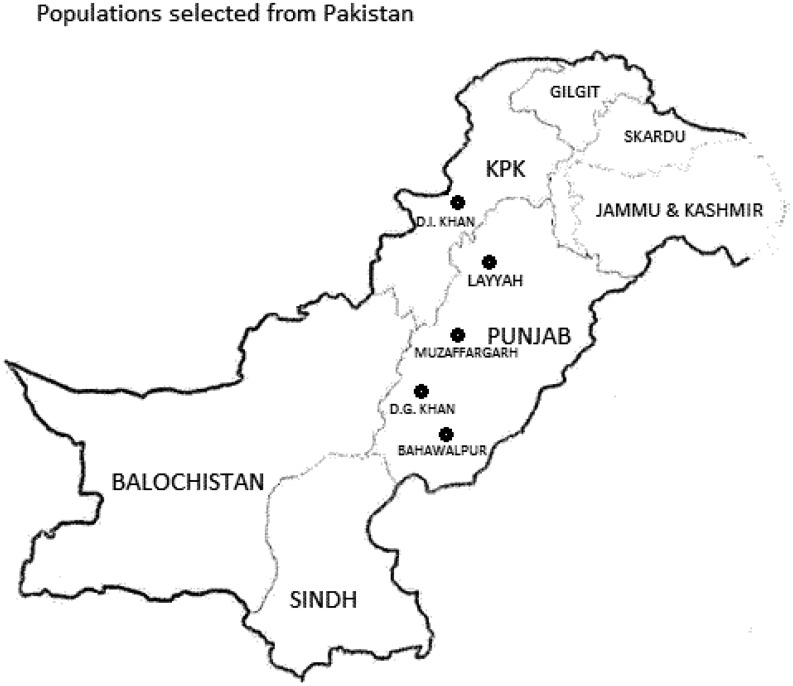
During February and March 2015, adult RPW were collected from five districts spread across the Punjab and KPK provinces of Pakistan (Table 1; Fig. 1). Live insects were collected from infested or fallen date palm plants, with the permission of the orchard’s owners or farmers, and permission from the Director of the Regional Agricultural Research Institute (RARI) (Bahawalpur). A total of 80 RPW adults were collected (Table 1), stored collectively in plastic jars containing 95% ethanol (one per location), and maintained at −20 °C in the Microbial Control Laboratory, University of Agriculture, Faisalabad. At the end of March 2015, the ethanol-preserved insects were transported to the laboratory of Dr. Richard Stouthamer (University of California Riverside [UCR]). During transport, the insects were not kept under controlled temperatures. At UCR, each weevil was transferred to an individual plastic vial, labeled, and kept in the freezer at −20 °C until processing.
Table 1.
Sampling information for RPW populations collected from date palm Phoenix dactylifera in Punjab and KPK provinces of Pakistan
| Population | Collection date | No. of specimens | Province | Geographical characteristics |
||
|---|---|---|---|---|---|---|
| Alt. (m) | Lat. | Long. | ||||
| Layyah | 27 Feb 2015 | 8 | Punjab | 143 | 30°58′N | 70°56′E |
| Bahawalpur | 14 Mar 2015 | 17 | Punjab | 252 | 29°59′N | 73°15′E |
| Dera Ghazi Khan | 10 Feb 2015 | 21 | Punjab | 150 | 29°57′N | 70°29′E |
| Muzaffargarh | 08 Mar 2015 | 18 | Punjab | 114 | 30°50′N | 71°54′E |
| Dera Ismail Khan | 01 Feb 2015 | 16 | KPK | 166 | 31°49′N | 70°52′E |
Fig. 1.
Map of collection sites in Punjab and KPK provinces of Pakistan.
DNA Extraction and Amplification
Each specimen was extracted using the protocol detailed in Rugman-Jones et al. (2013). Specifically, a small piece (2–5 mm3) of muscle tissue was dissected from a single tibia using flame-sterilized scissors and forceps, and allowed to air dry for 1 min on sterile filter paper. The tissue was then transferred to a sterile 0.6 ml microcentrifuge tube and ground up in 6 µl proteinase-K (>600 mAU/ml; Qiagen, Valencia, CA) using a glass pestle. To this was added 120 µl of a 5% (w/v) suspension of Chelex 100 resin (Bio-Rad Laboratories, Hercules, CA) and the reaction was incubated at 55 °C for 1 h followed by 10 min at 99 °C. After this the tubes were centrifuged for 4 min at 14,000 rpm to pellet the Chelex. Subsequently, 80 µl of supernatant was carefully transferred to a new microcentrifuge tube. Polymerase chain reaction (PCR) was used to amplify a section of the mitochondrial gene (mtDNA) cytochrome oxidase subunit 1 (COI) from each specimen. PCR was performed in 25 µl reactions containing 2 µl of DNA template, ddH2O, 1× ThermoPol PCR Buffer (New England BioLabs, Ipswich, MA), an additional 1 mM MgCl2, 400 µM dUTP, 200 µM each dATP, dCTP and dGTP, 10 µg BSA (NEB), 1 U Taq polymerase (NEB), and 0.2 µM of each PCR primer. Initial reactions utilized the primers C1-J-1718 and C1-N-2329 (Simon et al. 1994). Reactions were performed in a Mastercycler ep gradient S thermocycler (Eppendorf North America Inc., New York, NY) programmed for an initial denaturing step of 2 min at 94 °C; followed by five cycles of 30 s at 94 °C, 1 min 30 s at 45 °C, and 1 min at 72 °C; followed by a further 35 cycles of 30 s at 94 °C, 1 min 30 s at 51 °C, and 1 min at 72 °C; and, a final extension of 5 min at 72 °C. Amplification was verified by standard agarose gel electrophoresis, and samples that failed to amplify were subject to two further attempted amplifications, first using the primer set BRON and SIMON (El-Mergawy et al. 2011), and if that failed, pairing BRON with C1-N-2329 (see Rugman-Jones et al. 2013). The integrity of the DNA extracted from any specimen that still failed to yield a COI amplicon was tested in a fourth PCR, this time targeting the 28S rRNA region with the primers (28sF3633 and 28sR4076) and protocol detailed in Rugman-Jones et al. (2013).
Cleaning and Sequencing
PCR products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA), and sequenced in both directions at the Institute for Integrative Genome Biology core instrumentation facility (UCR). Sequences were aligned in MAFFT v7.293 (http://mafft.cbrc.jp/alignment/server/) using the G-INS-i strategy, then trimmed to 528 bp (to match the extensive dataset used by Rugman-Jones et al. 2013) using BioEdit version 7.0.9.0 (Hall 1999), and finally translated using the EMBOSS-Transeq website (http://www.ebi.ac.uk/Tools/emboss/transeq/index.html) to confirm the absence of nuclear pseudogenes (Song et al. 2008). All sequences were deposited in GenBank (Benson et al. 2008); accession numbers KU696489-537.
Genetic Analyses
Sequences of the COI gene generated in this study were collapsed into haplotypes, and the number and nature of polymorphic sites was characterized, using DnaSP v5.10.01 (Librado and Rozas 2009). Genetic variation within each Pakistani population (Table 1), was characterized by calculating the number of COI haplotypes and haplotype diversity (Hd; the probability that two randomly sampled haplotypes are different) using DnaSp; and the average number of nucleotide differences in pairwise comparisons among COI sequences (k) using MEGA version 6 (Tamura et al. 2013). Standard error of k was computed by bootstrapping with 1,000 replicates. Since most estimators of population differentiation can be highly unreliable when using a single locus and relatively small sample sizes, genetic variation between topographical populations was also investigated simply by obtaining population-pairwise estimates of k, again in MEGA. In order to put variation within Pakistan into a global context, we then combined our sequences with those from three earlier studies (El-Mergawy et al. 2011, Rugman-Jones et al. 2013, Wang et al. 2015; GenBank accessions GU581319-GU581628, KF311358-KF311740, and KF413063-KF413073, respectively). All sequences were trimmed to a uniform length, resulting in a matrix of 539 sequences, each 528 bp long. Sequences were again collapsed into haplotypes using DnaSP v5.10.01 and a “global” haplotype network was constructed using the statistical parsimony method of Templeton et al. (1992) in the software program TCS, version 1.21 (Clement et al. 2000).
Results
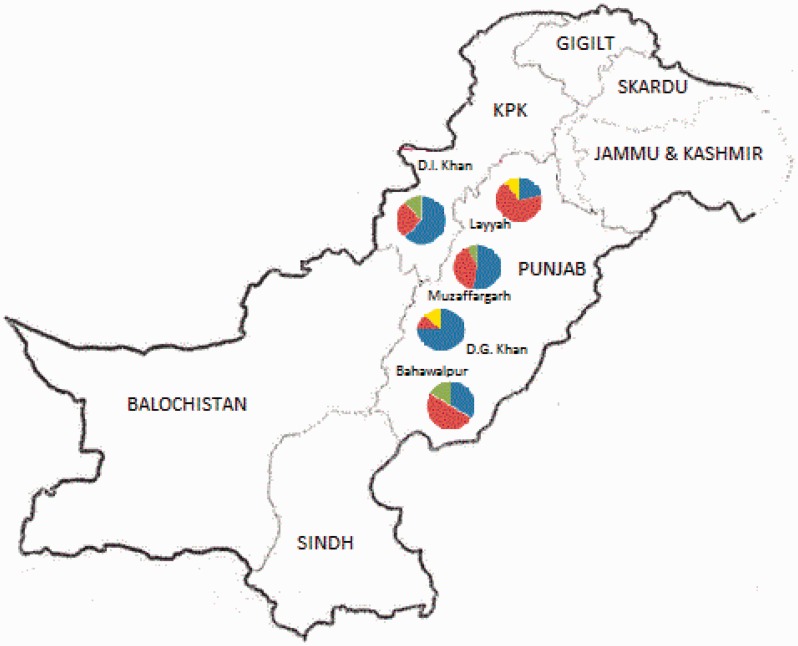
Using various combinations of four PCR primers, sequences of the COI gene were successfully obtained from 50 of the 80 RPW specimens collected from the Punjab and KPK provinces of Pakistan. For the remaining 30 “failed” specimens, attempts to amplify the highly conserved 28S rRNA also failed, suggesting that our extractions from those specimens had yielded no amplifiable DNA. Among the 50 specimens successfully sequenced, we found four haplotypes. The four haplotypes were very closely related, with only three polymorphic nucleotides (positions 63, 156, and 174) (see GenBank accessions). All substitutions were synonymous. Just two haplotypes accounted for the majority (88%) of the specimens and corresponded to haplotypes H1 (n = 20) and H5 (n = 24) previously encountered by El-Mergawy et al. (2011) and Rugman-Jones et al. (2013). These haplotypes were common in all five Pakistani populations (Table 2; Fig. 2). The remaining two haplotypes had restricted, and non-overlapping distributions, being found in three and two populations, and our “global” haplotype analysis revealed that neither had been encountered in the earlier studies of El-Mergawy et al. (2011), Rugman-Jones et al. (2013), or Wang et al. (2015). Hereafter they are referred to as H51 (n = 4) and H52 (n = 2), respectively.
Table 2.
Genetic characterization of five RPW populations from the Punjab and KPK provinces of Pakistan based on a 528-bp section of the mitochondrial COI gene
| Population | No. of specimens | No. of haplotypes | Haplotypes | Haplotype diversity (Hd) |
|---|---|---|---|---|
| Layyah | 9 | 3 | H1, H5, H52 | 0.556 |
| Bahawalpur | 11 | 3 | H1, H5, H51 | 0.691 |
| Dera Ghazi Khan | 8 | 3 | H1, H5, H52 | 0.464 |
| Muzaffargarh | 15 | 3 | H1, H5, H51 | 0.590 |
| Dera Ismail Khan | 7 | 3 | H1, H5, H51 | 0.668 |
Haplotypes H1 and H5 are numbered in accordance with El-Mergawy et al. (2011) and Rugman-Jones et al. (2013).
Fig. 2.
Distribution of mitochondrial haplotypes across five populations of RPW from the Punjab and KPK provinces of Pakistan. Haplotypes H1 and H5 are numbered in accordance with El-Mergawy et al. (2011) and Rugman-Jones et al. (2013).
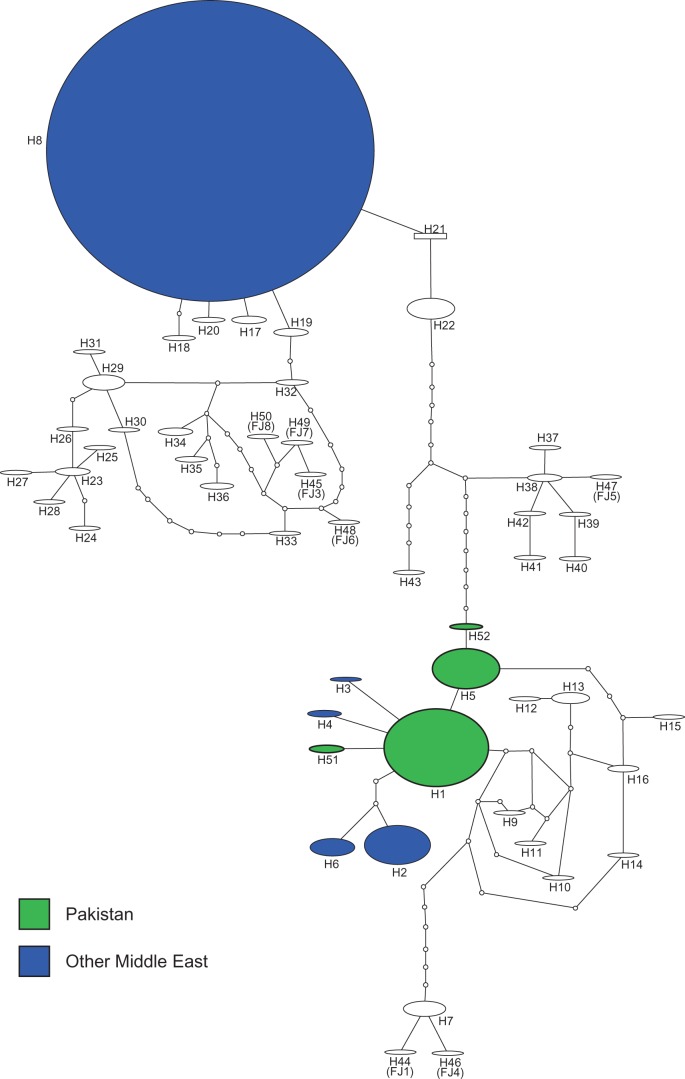
Given the very similar nature of the four Pakistani haplotypes, and the prevalence of just two of those haplotypes across the five Pakistani RPW populations, estimates of genetic variation within and between populations were very low (k was <1 in all comparisons; Table 3). In global terms, the haplotypes detected in our Pakistani samples were most similar to native haplotypes from other parts of the Indian sub-continent (Fig. 3), although divergence was ∼1% (k between Pakistani haplotypes and H9–16 from Goa and Sri-Lanka [Rugman-Jones et al. 2013] was 5.88). Outside of the native range, the most common Pakistani haplotypes (H1 and H5) were also invasive in the United Arab Emirates, Oman, and Syria (Fig. 3).
Table 3.
Variation in a 528-bp segment of the cytochrome oxidase subunit I (COI) region of mitochondrial DNA (mtDNA) of R. ferrugineus
| Population | Layyah | Bahawalpur | Dera Ghazi Khan | Muzaffargarh | Dera Ismail Khan |
|---|---|---|---|---|---|
| Layyah | 0.72 [0.53] | – | – | – | – |
| Bahawalpur | 0.74 [0.48] | 0.84 [0.58] | – | – | – |
| Dera Ghazi Khan | 0.83 [0.63] | 0.91 [0.61] | 0.50 [0.34] | – | – |
| Muzaffargarh | 0.69 [0.50] | 0.73 [0.53] | 0.67 [0.47] | 0.67 [0.53] | – |
| Dera Ismail Khan | 0.78 [0.53] | 0.79 [0.55] | 0.71 [0.46] | 0.69 [0.50] | 0.85 [0.61] |
MEGA version 6 was used to compute average number of pairwise nucleotide differences (k) within (diagonal element) and between (below diagonal) populations in the Punjab and KPK provinces of Pakistan. Estimates of standard error [in brackets] were obtained using a bootstrap procedure with 1000 replicates.
Fig. 3.
Relationships between four Pakistani COI haplotypes and 48 others occurring around the world. Haplotype network constructed from 539 COI sequences (each 528 bp long) generated by the present study and three earlier studies (see text). Each haplotype is represented by an oval, or a rectangle for that with the highest outgroup probability (H21, from Thailand). Size of each haplotype is indicative of the number of specimens sharing that haplotype. H1–43 are numbered according to El-Mergawy et al. (2011) and Rugman-Jones et al. (2013); H44–50 correspond to additional haplotypes from Wang et al. (2015; therein referred to as FJ1 and FJ3–FJ8); and, H51–52 are new to this study.
Discussion
In the Middle East, RPW is often referred to as the “Pakistani weevil” in the belief that it invaded from Pakistan. However, strong empirical evidence to justify this belief has not been forthcoming (e.g., Rugman-Jones et al. 2013). Furthermore, there is some doubt as to the actual status (invasive or native) of RPW in Pakistan, although it has typically been considered an invasive pest there. In this study, we found four haplotypes across 50 specimens, from five sampled populations located in the Pakistani provinces of Punjab and KPK. Of these haplotypes, two (H1 and H5) were common in all the populations sampled. The other two were relatively rare, and non-overlapping, with one (H51; Fig. 3) represented by only four specimens [two from Bahawalpur and one from Muzaffargarh and Dera Ismail Khan (D.I. Khan)], and the other (H52; Fig. 3) by two specimens (one from Layyah and another from Dera Ghazi Khan). These rare haplotypes have not been detected outside of Pakistan (El-Mergawy et al. 2011, Rugman-Jones et al. 2013, Wang et al. 2015). In global terms, the four Pakistani haplotypes were most similar to native haplotypes in the remainder of the Indian sub-continent (Fig. 3), and H1 and H5 were also common in invasive populations in parts of the Middle East (UAE, Oman, and Syria).
Low levels of genetic diversity are atypical of RPW populations across its described native range, but very characteristic of invasive populations of this species around the globe (see Rugman-Jones et al. 2013). Levels of genetic diversity detected in our study lay somewhere between the two extremes. If we consider RPW to be an invasive, this suggests that the RPW populations in the Punjab and KPK provinces of Pakistan have resulted from: a) the influx of a large number of weevils during a single invasion event; and/or b) multiple invasions from one or more sources. The genetic similarity between our Pakistani haplotypes and those previously reported from the Indian state of Goa suggests that India would have been the most likely source of any invasion. This is also the most likely scenario in a biogeographical sense. Commercial cultivation of dates in India is focused largely in the western states of Gujarat, Rajasthan and Punjab, adjacent to the border with Pakistan, and (to a lesser extent) the southernmost states of Tamil Nadu and Kerala. RPW is a strong flyer and it is easy to imagine that Pakistan may have been invaded by weevils from one or more of these Indian states. However, convincing support for such a hypothesis will require much wider sampling of India, which sadly remains a genetic “black hole” since the country does not allow researchers to collect and export specimens, due to Indian claims of intellectual property rights over genetic resources.
In contrast to the invasive “argument”, our results could also be interpreted as evidence that the Punjab and KPK provinces of Pakistan actually fall within the native range of the weevil. At least three known hosts of RPW are native to Pakistan [Nannorhops ritchiana (Griffith) Aitchison, Phoenix loureiroi Kunth and P. sylvestris Roxburgh (Champion et al. 1960, Mughal 1992, Malumphy and Moran 2007, Malik 2015)], and the date palm Phoenix dactylifera has been cultivated in the Sindh province of Pakistan for more than a thousand years (http://edu.par.com.pk/wiki/dates/). Furthermore, the presence of RPW in the Punjab province of Pakistan, and what is now the neighboring Indian state of Punjab, was first documented almost a century ago (Milne 1918). Therefore, we must consider the possibility that small populations of RPW have always been present in Pakistani Punjab, but have simply gone ignored, or unnoticed, because of the relative isolation of the region, and/or because their economic impact was (at that time) not significant. However, it is perhaps surprising that Wattanapongsiri (1966), in his revision of the genus Rhynchophorus, considered only a single R. ferrugineus specimen from anywhere in Pakistan (a specimen from the Kalat District of the modern Balochistan province, held in the Bavarian State Collection of Zoology, Munich). Prior to the Partition of India in 1947, the two “Punjabs” were considered a single province under the governance of the British Raj, and British collectors described vast numbers of insects from the entire Indian sub-continent (including Pakistan), depositing the bulk of their specimens at the British Museum of Natural History, London. Had RPW been abundant at that time, it seems unlikely that such a conspicuous insect would have escaped collection. However, despite having access to the BMNH collections (among many others), Wattanapongsiri (1966) included only the single “Balochistan” specimen, in his work. Unfortunately, that specimen was without a collection date, and so sheds little further light on the history of RPW in Pakistan.
Whether native or invasive, RPW has certainly been present in Pakistan for some time. The recent “rise” of RPW in the Punjab and KPK provinces has likely been exacerbated by anthropogenic movement of date palm germplasm from the neighboring provinces of Sindh and Baluchistan where date palms have been cultivated for centuries, and/or the rise of date cultivation in neighboring Indian states. Again, this is difficult to substantiate without sampling of those areas, and that should be a priority for further genetic work.
It has been claimed by some Middle Eastern countries that RPW originally crossed into Arabia in ornamental plants imported from Pakistan in 1985 (DAWN News 2003). While our data cannot completely refute this hypothesis it cannot fully support it either. Although both of the abundant Pakistani haplotypes detected in our study have been recorded in the Middle Eastern countries of UAE and Oman (and H1 only also in Syria), a third haplotype H8, has not been detected in Pakistan, but was found to be widespread in Saudi Arabia (El-Mergawy et al. 2011, Rugman-Jones et al. 2013). Indeed, El-Mergawy et al. (2011) detected three further haplotypes from Oman and UAE. If the Middle East was invaded solely by RPW from Pakistan it is hard to explain why there are additional haplotypes in the Middle East that have not been detected in Pakistan. One answer, originally put forward by Rugman-Jones et al. (2013), is that whether or not the Middle East has been invaded from Pakistan, it has also been invaded from somewhere else in the native range of RPW (most likely Thailand). It should also be noted that RPW-like damage was recorded in Iraq around the same time RPW was first recorded in the Punjab, although no specimens were collected to confirm this (Buxton 1920). There is currently no genetic data available for Iraqi populations of RPW, but given its relative proximity to the Middle East, it is possible that the latter (and indeed Pakistan) were invaded by RPW from Iraq. Intensive sampling of Iraq should be a priority for future genetic work.
Acknowledgments
We are indebted to Higher Education Commission, Islamabad (Pakistan) to provide funds for conducting this research at University of California, Riverside, USA and this publication is the part of research (2Av1-263) for PhD degree program.
References Cited
- Baloch H. B., Rustamani M. A., Khuhro R. D., Talpur M. A., Hussain T. 1992. Incidence and abundance of date palm weevil in different cultivars of date palm. Proc. Pak. Cong. Zool. 12: 445–447. [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. 2008. GenBank. Nucleic Acids Res. 36: 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton P. A. 1920. Insect pests of dates and the date palm in Mesopotamia and elsewhere. Bull. Entomol. Res. 11: 287–304. [Google Scholar]
- Champion H. G., Seth S. K., Khattak G. M. 1960. Manual of silviculture for Pakistan. Pakistan Forest Institute, Peshawar. [Google Scholar]
- Clement M., Posada D., Crandall K. A. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1660. [DOI] [PubMed] [Google Scholar]
- DAWN News 2003. Insect pests ravage red date palm trees. (http://www.dawn.com/news/89129/insect-pests-ravage-red-date-palm-trees (accessed 27 May 2016).
- El-Mergawy R.A.A.M., Nasr M. I., Abdallah N., Silvain J. F. 2011. Mitochondrial genetic variation and invasion history of red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), in Middle-East and Mediterranean Basin. Int. J. Agric. Biol. 13: 631–637. [Google Scholar]
- El-Mergawy R.A.A.M., Al-Ajlan A. M. 2011. Red palm weevil, Rhynchophorus ferrugineus (Olivier): economic importance, biology, biogeography and integrated pest management. J. Agric. Sci. Technol. 1: 1–23. [Google Scholar]
- El-Sabea A.M.R., Faleiro J. R., Abo-El-Saad M. M. 2009. The threat of red palm weevil Rhynchophorus ferrugineus to date plantations of the Gulf region in the Middle-East: an economic perspective. Outlooks Pest Manag. 20: 131–134. [Google Scholar]
- Faleiro J. R. 2006. A review of the issues and management of the red palm weevil Rhynchophorus ferrugineus (Coleoptera: Rhynchophoridae) in coconut and date palm during the last one hundred years. Int. J. Trop. Insect Sci. 26: 135–154. [Google Scholar]
- Faleiro J. R., Ben Abdullah A., El-Bellaj M., Al-Ajlan A. M., Oihabi A. 2012. Threat of red palm weevil, Rhynchophorus ferrugineus (Olivier) to date palm plantations in North Africa. Arab J. Plant Prot. 30: 274–280. [Google Scholar]
- Giblin-Davis R. M., Faleiro J. R., Jacas J. A., Peña J. E., Vidyasagar P.S.P.V. 2013. Coleoptera: biology and management of the red palm weevil, Rhynchophorus ferrugineusI, pp. 1–34. In Peña J. E. (ed.), Potential invasive pests of agricultural crop species. CABI, Wallingford. [Google Scholar]
- Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Khamiss O., Abdel Badeea A. 2013. Initiation, characterization and karyotyping of a new cell line from red palm weevil Rhynchophorus ferrugineus adapted at 27 °C. In AFPP – Palm Pest Mediterranean Conference 16, 17 and 18 January 2013, Nice, France. (http://www.fredon-corse.com/standalone/4/F155xZqGY3a55K97Z4NE03nG.pdf) (accessed 27 May 2016).
- Librado P., Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Malik K. A. 2015. Flora of Pakistan. (http://www.efloras.org/florataxon.aspx?flora_id=5&taxon_id=20427) (accessed 27 May 2016).
- Malumphy C., Moran H. 2007. Red palm weevil Rhynchophorus ferrugineus. Plant Pest Notice Cent. Sci. Lab. 50: 1–3. [Google Scholar]
- Milne D. 1918. The date palm and its cultivation in the Punjab. The Punjab Government, Lyallpur, 153 pp.
- Mughal M. S. 1992. Spotlight on species: Nannorrhops ritchieana. Pak. J. Forest. 42: 162–166. [Google Scholar]
- Mukhtar M., Rasool K. G., Parrella M. P., Sheikh Q. I., Pain A., Lopez-Llorca L. V., Aldryhim Y. N., Mankin R. W., Aldawood A. S. 2011. New initiatives for management of red palm weevil threats to historical Arabian data palms. Fla. Entomol. 94: 733–736. [Google Scholar]
- Murphy S. T., Briscoe B. R. 1999. The red palm weevil as an alien invasive: biology and the prospects for biological control as a component of IPM. Biocontrol News Info. 20: 35–46. [Google Scholar]
- Rugman-Jones P. F., Hoddle C. D., Hoddle M. S., Stouthamer R. 2013. The lesser of two weevils: molecular-genetics of pest palm weevil populations confirm Rhynchophorus vulneraturs (Panzer 1798) as a valid species distinct from R. ferrugineus (Olivier 1790), and reveal the global extent of both. PLoS One. 8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Frati F., Beckenbach A., Crespi B., Liu H., Flook P. K. 1994. Evolution, weighting and phylogentic utility of mitochondrial gene sequence and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87: 651–701. [Google Scholar]
- Song H., Buhay J. E., Whiting M. F., Crandall K. A. 2008. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA. 105: 13486–13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher, G., Peterson, D., Filipski, A., Kumar B. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evo. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A. R., Crandall K. A., Sing C. F. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data III. Cladogram estimation. Genetics. 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil W., Faleiro J. R., Miller T. A. 2015. Sustainable Pest Management in Date Palm: Current Status and Emerging Challenges. Springer International Publishing AG, Switzerland. p. 429. [Google Scholar]
- Wang G., Zhang X., Hou Y., Tang B. 2015. Analysis of the population genetic structure of Rhynchophorus ferrugineus in Fujian, China, revealed by microsatellite loci and mitochondrial COI sequences. Entomol. Exp. Appl. 155: 28–38. [Google Scholar]
- Wattanapongsiri A. 1966. A revision of the genera Rhynchophorus and Dynamis (Coleoptera: Curculionidae). Dept. Agric. Sci. Bull. 1: 1–328. [Google Scholar]