Abstract
Background
Accumulating evidence indicates that neuropathic pain is a neuro-immune disorder with enhanced activation of the immune system. Recent data provided proof that neuropathic pain patients exhibit increased numbers of immunosuppressive regulatory T cells (Tregs), which may represent an endogenous attempt to limit inflammation and to reduce pain levels. We here investigate the molecular mechanisms underlying these alterations.
Methods
Our experimental approach includes functional analyses of primary human T cells, 3′-UTR reporter assays, and expression analyses of neuropathic pain patients’ samples.
Results
We demonstrate that microRNAs (miRNAs) are involved in the differentiation of Tregs in neuropathic pain. We identify miR-124a and miR-155 as direct repressors of the histone deacetylase sirtuin1 (SIRT1) in primary human CD4+ cells. Targeting of SIRT1 by either specific siRNA or by these two miRNAs results in an increase of Foxp3 expression and, consecutively, of anti-inflammatory Tregs (siRNA: 1.7 ± 0.4; miR-124a: 1.5 ± 0.4; miR-155: 1.6 ± 0.4; p < 0.01). As compared to healthy volunteers, neuropathic pain patients exhibited an increased expression of miR-124a (2.5 ± 0.7, p < 0.05) and miR-155 (1.3 ± 0.3; p < 0.05) as well as a reduced expression of SIRT1 (0.5 ± 0.2; p < 0.01). Moreover, the expression of these two miRNAs was inversely correlated with SIRT1 transcript levels.
Conclusions
Our findings suggest that in neuropathic pain, enhanced targeting of SIRT1 by miR-124a and miR-155 induces a bias of CD4+ T cell differentiation towards Tregs, thereby limiting pain-evoking inflammation. Deciphering miRNA-target interactions that influence inflammatory pathways in neuropathic pain may contribute to the discovery of new roads towards pain amelioration.
Trial registration
German Clinical Trial Register DRKS00005954
Electronic supplementary material
The online version of this article (doi:10.1186/s12974-016-0712-6) contains supplementary material, which is available to authorized users.
Keywords: Neuropathic pain, miRNA, Regulatory T cells, Histone deacetylase sirtuin1, Analgesia
Background
Neuropathic pain is caused by impairment of somatosensory functions in both the peripheral and central nervous system [1]. It is often associated with spontaneous pain, dysesthesia, paraesthesia, and hyperalgesia (increased pain caused by painful stimuli) and allodynia (increased pain caused by non-painful stimuli) [2, 3]. The treatment of neuropathic pain is ambitious, and outcomes often are unsatisfactory [4]. Despite intensive analgesic treatment, significant attenuation of pain is only achieved in a limited number of patients [5].
There is emerging evidence that aberrant responses of the immune system substantially contribute to the development of neuropathic pain [6]. Immune cells respond to nerve injury by migration into the nervous system at the side of injury, thereby releasing mediators, which affect intercellular signaling [7]. Although the precise role of immune cells in neuropathic pain remains unclear, adoptive transfer of immune cells producing pro-inflammatory cytokines significantly increase pain sensitivity, whereas transfer of cells producing anti-inflammatory cytokines decrease pain sensitivity in nerve-injured rats [8]. Recent data investigating neuropathic pain in humans published by our group point into the same direction. We showed that patients exhibit altered ratios of peripheral T-helper cell subsets. Specifically, increased numbers of immunosuppressive regulatory T cells (Tregs) have been found [9, 10]. This could reflect an endogenous strategy to limit inflammation and to reduce pain levels in neuropathic pain, which is of interest with respect to future treatment approaches.
The current study aims to investigate the molecular mechanisms underlying these alterations. We focused on the expression of the histone deacetylase sirtuin1 (SIRT1), which is supposed to play a significant role in the development and function of Tregs [11, 12]. SIRT1 controls transcription factor forkheadbox-p3 (Foxp3), the master regulator of Treg differentiation [12]. Treatment with SIRT1 inhibitors increased Foxp3 gene expression with consecutive increase of Treg differentiation in mice [13]. We thus hypothesized that these mechanisms might also be involved in the Treg alterations observed in neuropathic pain patients.
Methods
Patients
Patients appearing with neuropathic pain in our Department of Pain Medicine were verified for fulfilling the inclusion criteria and asked about their agreement to participate in the study. Neuropathic pain was defined as “pain caused by a lesion or disease of the somatosensory nervous system” such as polyneuropathy, postherpetic neuralgia, or trigeminal neuralgia/neuropathy using the PainDETECT-questionnaire [14]. Additionally, quantitative sensory testing was performed to all patients, according to the protocol of the German Research Group on neuropathic pain [15]. Patients suffering from low back pain with radiculopathy (even if the radicular component was clearly predominant) or patients with autoimmune, chronic, inflammatory, neoplastic, or psychiatric diseases were excluded. None of the patients had been treated with corticosteroids or had received known immunomodulatory agents currently or in the past. Acute inflammation was excluded by determination of C-reactive protein (CRP), total- and differential leucocyte, and measurement of body temperature. Eleven patients fulfilled the inclusion criteria. Blood samples of these patients were obtained as well as from 9 healthy volunteers after written consents were obtained. Additionally, patients were asked to quote their average pain intensity using an 11-point numerical rating scale (NRS) with 0 representing “no pain” and 10 “worst pain imaginable”. For patients’ characteristics, see Table 1.
Table 1.
Patient characteristics
| Item | Healthy | Neuropathic pain | p value |
|---|---|---|---|
| Numbers (n) | 9 | 11 | |
| Age | 36 ± 9 | 54 ± 12 | < 0.05 |
| Female | 55 % | 64 % | n.s. |
| BMI | 22.9 ± 2.9 | 25.3 ± 3.4 | n.s. |
| NRS (rest) | 0.0 ± 0.0 | 4.8 ± 2.3 | < 0.05 |
| NRS (motion) | 0.0 ± 0.0 | 7.6 ± 1.7 | < 0.05 |
| KAB | 1.5 ± 0.4 | 3.4 ± 0.7 | < 0.05 |
Results are expressed as mean ± standard deviation (SD)
BMI body mass index, NRS (rest/motion) numeric rating scale (0 to 10) of pain, 0: “no pain,” 10: “worst pain imaginable,” KAB questionnaire for self-perceived stress ranging (1–6), 1: “no stress,” 6: “maximum stress,” n.s. not significant
The prospective study protocol followed the principles of the Declaration of Helsinki and was reviewed and approved by the Ethics Committee of the LMU Munich and registered on German Clinical Trial Register (Registration Trial DRKS00005954). Patients included in this study have also been part of a recently published study by Luchting et al. [9] showing an anti-inflammatory T cell shift in patients suffering from neuropathic pain.
miRNA selection and target prediction
In the current manuscript, we focused on the evaluation of miR-124a and miR-155. These microRNAs (miRNAs) were selected as follows: We first sought to identify miRNAs that have been found to be differentially expressed in pain- and inflammation-related syndromes [16, 17]. Of these, only miR-124a and miR-155 were predicted to target SIRT1. These predictions were based on the established target prediction algorithm TargetScan [18].
RNA isolation and cDNA synthesis
Total RNA was isolated using either the RNAqueous® Micro Kit or the mirVana miRNA Isolation Kit followed by subsequent DNase treatment (Turbo DNase, Ambion) according to the manufacturer’s instructions. Quantity and purity of the isolated RNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific). Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using SuperScript III First Strand Synthesis System (Invitrogen), as per manufacturer’s instructions.
Quantitative RT-PCR
cDNA was synthesized from equal amounts of total RNA using Superscript III reverse transcriptase (Invitrogen) and oligo(dT) and random hexamer primers following the supplier’s instructions. Quantitative analyses of messenger RNA (mRNA) levels were performed in duplicates on a Light Cycler 480 (Roche Diagnostics) using either UPL probes and specific primers or specific single assays (Table 2, Roche Diagnostics, Penzberg). The cycling conditions comprised an initial denaturation phase at 95 °C for 5 min, followed by 45 cycles at 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 15 s. Data were normalized to the reference genes SDHA and TBP [19].
Table 2.
Primer sequences for real-time PCR
| Sequence/assay ID | |
|---|---|
| Foxp3 | Roche RealTime Ready Single Assay ID 113503 |
| SIRT1 | for 5′-TGT ACG ACG AAG ACG ACG AC-3′ (UPL probe #63) rev 5′-TTC ATC ACC GAA CAG AAG GTT-3′ (UPL probe #63) |
| TBP | for 5′-GAACATCATGGATCAGAACAACA-3′ (UPL probe #87) rev 5′-ATAGGGATTCCGGGAGTCAT-3′ (UPL probe #87) |
| SDHA | for 5′-GAGGCAGGGTTTAATACAGCA-3′ (UPL probe #80) rev 5′-CCAGTTGTCCTCCTCCATGT-3′ (UPL probe #80) |
Quantification of miRNA expression
Expression of miR-124a, miR-155, and U47 (endogenous control) was quantified using TaqMan miRNA assays (Applied Biosystems) following the manufacturer’s protocol. In brief, equal amounts of RNA (10 ng) were reverse transcribed using miRNA-specific stem-loop primers and the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR (RT-PCR) was performed in duplicate using LightCycler 480 Probes Master on the LightCycler 480 instrument applying the following cycling conditions: denaturing at 95 °C for 10 min, 45 cycles of 95 °C for 15 s, and 60 °C for 60 s. U47 RNA was used for normalization of miRNA expression data.
Western blot analysis
Thirty-five micrograms of total protein extracts was electrophoresed in an 8 % SDS–PAGE and subsequently electroblotted onto PVDF membranes. Non-specific binding sites on the membrane were blocked using 5 % non-fat dry milk in TBS-Tween. SIRT1 antibody (Cell Signaling Technology, Danvers, MA) was diluted in PBST supplemented with 1 % non-fat dry milk (dilution factor 1:2000). β-actine (Cell Signaling Technology, dilution factor 1:40,000) served as a loading control. Immunoreactive bands were visualized using horseradish peroxidase-labeled goat anti-mouse or goat anti-rabbit antibodies and the Signal Fire ECL Substrate (Cell Signaling Technology, Danvers, MA).
Purification of peripheral human CD4+ T cells
CD4+ T cells were isolated from peripheral blood mononuclear cells (PBMCs) by magnetic separation with Whole Blood CD4 MicroBeads (MACS Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. Viability and cell number were ascertained by ViCell analyzer (Beckman Coulter, Fullerton, CA).
Cell cultures and stimulation conditions
Primary CD4+ T cells were cultured in six-well plates in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10 % heat-inactivated fetal calf serum (Biochrom, Berlin, Germany), penicillin (100 IU/mL), streptomycin (100 μg/mL), sodium pyruvate, and l-glutamine (Gibco, Life Technologies, Darmstadt, Germany) at 37 °C in a humidified atmosphere of 5 % CO2 in air.
For differentiation into Tregs, CD4+ T cells (7 × 105/mL) were cultured under Treg differentiating conditions (anti-CD3/CD28 Dynabeads (Invitrogen, Carlsbad, Germany) for 36 h, rhIL-2 and TGF-ß for four additional days.) To evaluate the effect of miR-124a, miR-155, or siSIRT1 on Treg differentiation, CD4+ T cells were transfected with these miRNAs, siRNA, or negative control 6 h before stimulation of CD4+ T cells was initiated.
Flow cytometric staining and analysis
For identification and quantification of Tregs, multicolor flow cytometry was used after surface staining of peripheral blood mononuclear cells with specific antibodies. These antibodies include anti-human CD4 and Foxp3. To quantify the number of Tregs after transfection of CD4+ T cells and incubation under Treg skewing conditions, Tregs were identified by surface staining with anti-human CD4+ and intracellular staining with FoxP3 antibody (Biolegend, San Diego, CA, USA). The amount of Tregs was expressed as a ratio of CD4+Foxp3+ T cells as a percentage of CD4+ T cells. Tregs in patients and healthy volunteers were identified after surface staining of PBMCs with monoclonal antibodies specific for anti-human CD4, CD25, and CD127 and intracellular staining with an anti-human Foxp3 antibody. CD4+CD25highCD127lowFoxp3+ cells were defined as Tregs.
Cloning and mutagenesis of vector constructs
The psiCHECK-2 Target Expression Vector (Promega, Madison, WI, USA) was used for generation of 3′-untranslated region (3′-UTR) reporter constructs as described before [20]. Briefly, the 3′-UTR of SIRT1 containing the predicted target sites of miRNA-124a and miRNA-155 were amplified by PCR from human genomic DNA (100 ng) with the primers given in Table 3 (synthesized by Metabion, Martinsried, Germany). Cycling conditions were as follows: 95 °C for 3 min denaturing; 30 cycles of 95 °C for 30 s, 61.2 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 5 min. PCR products were cloned into the PmeI and XhoI restriction sites of the psiCHECK-2 plasmid. Site-directed mutagenesis [20] of the putative miR-124a or the three miR-155 binding sites was performed using the QuickChange Lightning Mutagenesis Kit (Stratagene) with the primers given in Table 4. All plasmids were verified by sequence analysis (MWG Biotech, Ebersberg, Germany).
Table 3.
Primer sequences for the 3′-UTR of SIRT1
| Primer | Sequence | Restriction site |
|---|---|---|
| SIRT1-3′UTR for | 5′-CTCGAGCTGTGAAACAGGAAGTAACAGACA-3′ | XhoI |
| SIRT1-3′UTR rev | 5′-GTTTAAA CTGGCAGTAATGGTCCTAGCTG-3′ | PmeI |
Restriction enzymes and the cutting sides of these enzymes are italicized
Table 4.
Primers for mutagenesis
| Primer | Sequence | Position |
|---|---|---|
| SIRT mut 124a for | 5′-TATTTAAAAGCTTAGCCTGGATTAAAACTAGAGATCAACTTTCTCAGA-3′ | 1211–1217 |
| SIRT mut 124a rev | 5′-GCTGAGAAAGTTGATCTCTAGTTTTAATCCAGGCTAAGCTTTTAAATA-3′ | |
| SIRT mut 155_1 for | 5′-CAGGAATTGTTCCACCAGGGTTAGGAACTTTAGCATGTC-3′ | 36–42 |
| SIRT mut 155_1 rev | 5′-GACATGCTAAAGTTCCTAACCCTGGTGGAACAATTCCTG-3′ | |
| SIRT mut 155_2 for | 5′-TTGATCTTTTCCACAAGGGTTAAACTGCCAAAATGTG-3′ | 929–935 |
| SIRT mut 155_2 rev | 5′-CACATTTTGGCAGTTTAACCCTTGTGGAAAAGATCAA-3′ | |
| SIRT mut 155_3 for | 5′-GAAATTGCACAGTAAGGGTTTATTTTTCAGACCATT-3′ | 1408–1414 |
| SIRT mut 155_3 rev | 5′-AATGGTCTGAAAAATAAACCCTTACTGTGCAATTTC-3′ |
Sequenzes of the mutagenesis are italicized
Cell transfections and luciferase assay
Cell transfections were performed by electroporation using the Neon™ transfection system (Invitrogen, Life Technologies, Darmstadt, Germany). CD4+ T cells were transfected with 50 nM pre-miR-124a, pre-miR-155, or negative control. For luciferase assay, HEK-293 cells were co-transfected with 1 μg of psiCheck-2 dual luciferase reporter plasmids containing the 3′-UTR of SIRT1 and either pre-miR-124a, pre-miR-155, or negative control (Ambion, Austin, TX, USA) at a final concentration of 50 nM. HEK-293 cells (European Collection of Cell Cultures) were grown in Dulbecco’s modified Eagle medium (DMEM—Lonza, Walkersville, MD) supplemented with 10 % heat-inactivated fetal bovine serum (FBS), 1 % penicillin/streptomycin/glutamine, and 1 % NEAA at 37 °C in a humidified atmosphere of 5 % CO2 in air. Forty-eight hours after transfection, cells were lysed and analyzed for firefly and renilla luciferase activity using the Dual-Glo-Luciferase Assay System (Promega), and Renilla luciferase activities were normalized to Firefly activities. All experiments were performed in triplicates.
Statistical analyses
All statistical analyses were performed using SigmaStat 12.0 (Systat Software, Chicago, USA). Every statistical analysis was started with testing for normal distribution using the Shapiro-Wilk Test. Further analyses were performed with Student’s t test for all data with normal distribution and the nonparametric Mann-Whitney rank-sum test for all data without normal distribution. Values are expressed as mean ± standard deviation (SD). p values <0.05 were considered as statistically significant.
Results
SIRT1 mRNA expression is decreased in neuropathic pain
First, to confirm our previous findings, we determined the Foxp3/CD4+ cell ratio in neuropathic pain patients as compared to healthy volunteers. As shown in Fig. 1a, neuropathic pain patients exhibited significantly elevated Foxp3/CD4+ ratios (1.6 ± 0.9 in neuropathic pain vs. 0.8 ± 0.5 in healthy controls; p < 0.05). Noteworthy, number of Tregs was not correlated with age (Additional file 1: Figure S1A). Next, we investigated the mRNA expression of SIRT1 in CD4+ T cells obtained from patients suffering from neuropathic pain as compared to healthy volunteers. As shown in Fig. 1b, neuropathic pain patients exhibited a markedly reduced SIRT1 mRNA expression (0.5 ± 0.2 in neuropathic pain vs. 1.0 ± 0.4 in healthy controls; p < 0.01, Fig. 1b).
Fig. 1.
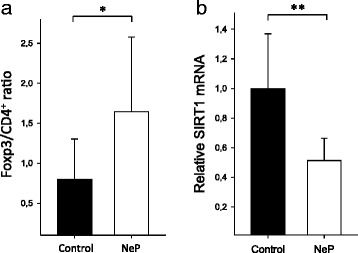
SIRT1 is down-regulated in CD4+ T cells of patients with neuropathic pain. a T cells obtained from patients with neuropathic pain were characterized by surface staining with monoclonal antibodies specific for anti-human CD4, CD25, and CD127 and intracellular staining with an anti-human Foxp3 antibody followed by flow cytometric analysis. The Foxp3/CD4+ ratio found in neuropathic pain patients (NeP, n = 11) and in healthy controls (control, n = 9) is shown. b Total RNA was extracted from purified CD4+ T cells of patients with neuropathic pain (NeP, n = 11) as well as from CD4+ T cells of healthy volunteers (control, n = 9). Relative expression of SIRT1 was quantified by quantitative PCR (qPCR) using SDHA and TBP as reference genes. The results indicate fold reduction of SIRT1 in patients with neuropathic pain vs. healthy volunteers. Data are given as means ± SD; *p < 0.05, **p < 0.01
Knockdown of SIRT1 in human CD4+ T cells induces Treg differentiation in vitro
SIRT1 is known as an important negative regulator of Foxp3 expression in murine T cells. To gain insight into its functions in human T lymphocytes, we next analyzed the impact of SIRT1 knockdown in primary human CD4+ T cells on Foxp3 expression and Treg differentiation. Transfection of human CD4+ T cells with SIRT1 siRNA significantly reduced both SIRT1 mRNA and protein expression (mRNA 0.5 ± 0.1, n = 6; p < 0.01, Fig. 2a) as compared to normal control (NC). After incubation of transfected and stimulated cells under Treg skewing conditions for 4 days, an increase of Foxp3 mRNA expression by approximately 30 % was found (1.3 ± 0.1, n = 3; p < 0.01, Fig. 2b). Accordingly, Treg differentiation was clearly enhanced (1.7 ± 0.4, n = 3; p < 0.01, Fig. 2c). These findings imply that a decreased SIRT1 expression may significantly contribute to the increase of Treg cells in neuropathic pain.
Fig. 2.
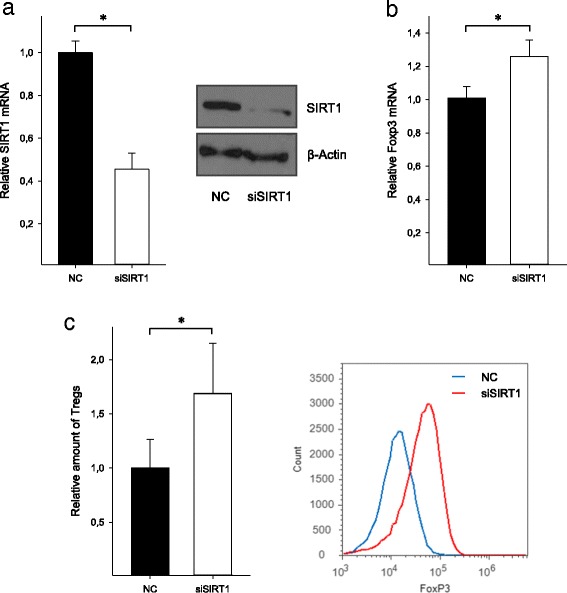
SIRT1 is a negative regulator of Foxp3 expression in human T cells. SIRT1 knockdown in primary human CD4+ T cells of healthy volunteers was performed by specific siRNA, and Foxp3 mRNA expression and Treg differentiation was analyzed after incubation under Treg skewing conditions for 4 days. a Successful transfection of siRNA was confirmed by qPCR (left panel) and Western Blot analysis (right panel). One blot is representative of n = 3. b Relative Foxp3 mRNA expression after transfection and Treg differentiation as measured by qPCR. c Treg subpopulation as determined by FACS analysis (left panel), a histogram representative of n = 6 individual experiments performed in duplicates is shown in the right panel. Data are given as means ± SD; *p < 0.01, n = 6
miR-124a and miR-155 are potential candidates of SIRT1 regulation in neuropathic pain
We next hypothesized that regulation by specific miRNAs may influence SIRT1 expression and may thus account for the observed alterations of SIRT1 mRNA expression in neuropathic pain. To this end, we combined results of published microarray data in patients with chronic pain syndromes with target prediction in silico. These analyses revealed miR-124a and miR-155 as potential candidates involved in the regulation of SIRT1 in neuropathic pain. Target prediction tools suggested three specific binding sites for miR-155 and a single specific binding side for miR-124a with high probability within the 3′-UTR of the SIRT1 transcript (Fig. 3a).
Fig. 3.
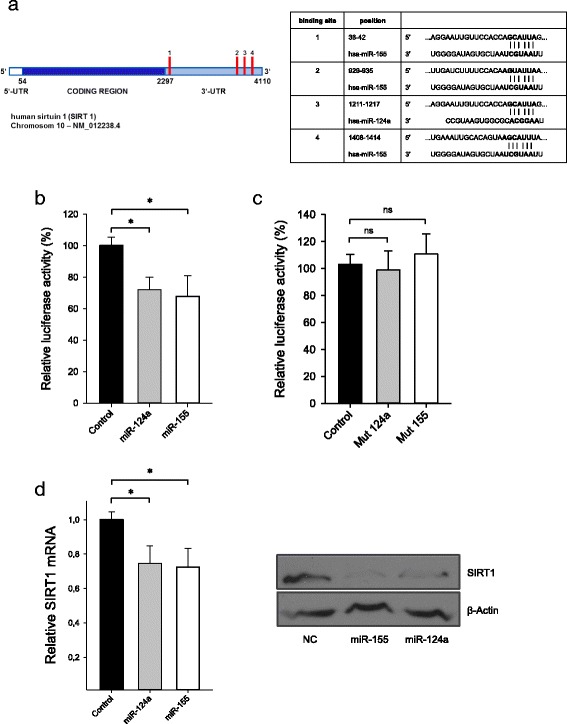
SIRT1 is a novel miR-124a and miR-155 target. Depiction of the genomic structure of the human SIRT1 gene on chromosome 10 and location of the putative miR-124a and miR-155 binding sites within its 3′-UTR (a). Target prediction algorithm identified a putative miR-124a binding site and three putative miR-155 binding sites, indicated by the red bars. Positions and seed sequences of the putative binding sites are listed in the adjacent table (a). A reporter vector containing the SIRT1 3′-UTR was co-transfected with pre-miR-124a or pre-miR-155 into HEK-293 cells, and hRLuc reporter activity was determined relative to a vector construct containing the SIRT1 3′-UTR co-transfected with pre-miR-scrambled control (b). Control constructs lacking either the miR-124a (Mut 124a) or the miR-155 binding sites (Mut 155) were generated by site-directed mutagenesis. Both mutant vectors were co-transfected with the respective miRNA or with scrambled control into HEK-293 cells, and hRLuc reporter activity was determined; luciferase activity relative to scrambled control is given. Data are means ± SD; ns not significant, *p < 0.01, n = 8. c CD4+ T cells of healthy donors were transiently transfected with miR-124a, miR-155, or scrambled control, respectively, and stimulated with anti-CD3/CD28 Dynabeads for 36 h. Relative SIRT1 mRNA was detected by qPCR, n = 6, *p < 0,01 (d, left panel), and SIRT1 protein expression was determined by Western Blot analysis (d, right panel). One blot is representative of n = 3
SIRT1 expression is directly regulated by miR-124a and miR-155
To provide an experimental proof of a direct interaction between miR-124a and/or miR-155 with the SIRT1 3′-UTR, we performed luciferase reporter assay on a psiCheck-2 plasmid containing a Renilla luciferase gene upstream of the SIRT1 3′-UTR. HEK293 cells were transiently co-transfected with the reporter vector construct and either pre-miR-124a or pre-miR-155 or NC, and luciferase activity was measured. As shown in Fig. 3b, reporter activity was significantly reduced by both miRNAs (miR-124a 72 ± 7 %, miR-155 68 ± 13 %, n = 8; p < 0.01), as compared to NC.
Site-directed mutagenesis of either the miR-124a or the three miR-155 binding sites within the 3′-UTR of SIRT1 strongly diminished the inhibitory effect of the respective miRNA (Fig. 3c). These data demonstrate that both miRNAs regulate SIRT1 expression by direct targeting of specific binding sites within the 3′-UTR of SIRT1.
Next, we validated the impact of miR-124a and miR-155 on the expression of SIRT1. We assessed SIRT1 mRNA levels after transfection of human CD4+ T cells with either miR-124a or miR-155 mimics or with negative control. As depicted in Fig. 3d, SIRT1 mRNA (miR-124a 0.75 ± 0.1, miR-155 0.72 ± 0.1; n = 6; p < 0.01) and protein expression significantly decreased after transient transfection of both miRNAs as compared to control.
Taken together, we provide evidence that SIRT1 mRNA expression in primary human T cells is directly regulated by miR-124a and miR-155.
miR-124a and miR-155 control SIRT expression in neuropathic pain
We next determined the expression of miR-124a and miR-155 in CD4+ T cells obtained from patients with neuropathic pain and from healthy volunteers. Expression of both miRNAs was significantly higher in patients with neuropathic pain as compared to healthy volunteers (miR-124 2.5 ± 0.7, p < 0.05, Fig. 2a, miR-155 1.3 ± 0.3; p < 0.05, Fig. 4a). Correlation analyses in human CD4+ T cells revealed for both miRNAs a significant inverse correlation with SIRT1 transcript levels (miR-124a: r = −0.75, p < 0.001, n = 20, miR-155: r = −0.6, p = 0.006, n = 20, Fig. 4b, c), which strongly points to an important role of both miRNAs as regulators of SIRT1 in vivo. There was no significant correlation between age and either miR-124a, miR-155, or SIRT1 mRNA expression (Additional file 1: Figure S1B-D).
Fig. 4.
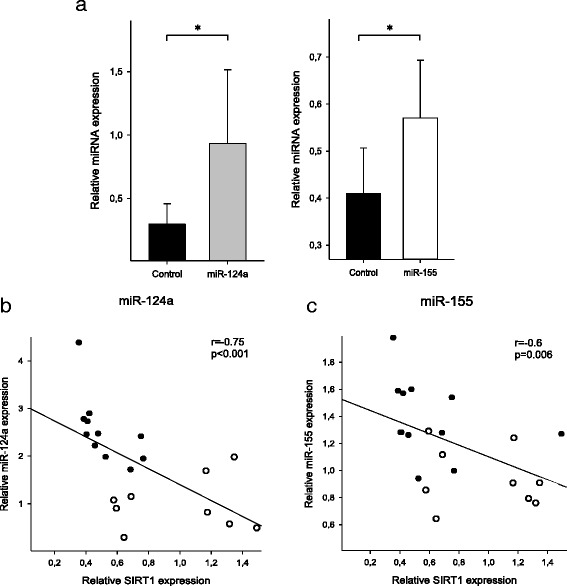
miR-124a and miR-155 are upregulated in CD4+ T cells of patients with neuropathic pain. a Total RNA was extracted from purified CD4+ T cells of patients suffering from neuropathic pain (n = 11) as well as from those of healthy volunteers (n = 9, control). Relative expression of endogenous miR-124a and miR-155 was quantified by qPCR using U47 RNA as normalizing control. Expression of miR-124a (a, left panel) and miR-155 (a, right panel) in patients with neuropathic pain as compared to healthy volunteers is shown. Data are given as means ± SD; *p < 0.05. Correlation of miR-124a expression (b) and miR-155 expression (c) and SIRT1 transcript levels in human CD4+ T cells
miR-124a and miR-155 increase Treg differentiation
To investigate the impact of both miRNAs on Treg differentiation, we transfected human CD4+ T cells with either pre-miR-124a or pre-miR-155 followed by culturing under Treg skewing conditions for 4 days. As shown in Fig. 5a, Foxp3 mRNA expression was significantly increased in miRNA-transfected cells as compared to controls (miR-124a 1.5 ± 0.4; n = 6; p < 0.01; miR-155 1.5 ± 0.4; n = 6; p < 0.01; Fig. 5a). Additionally, expression of the Treg signature molecules EOS, CTLA4, and IL2RA [21] was also elevated (Additional file 1: Figure S2). Accordingly, an enhancement of Treg differentiation was found (miR-124a 1.5 ± 0.4; n = 6; p < 0.01; miR-155 1.6 ± 0.4; n = 6; p < 0.01; Fig. 5b, c). In accordance with these in vitro findings, we found a significant correlation between both miR-124a and miR-155 expression and Foxp3 mRNA expression in human CD4+ T cells (Additional file 1: Figure S3). These findings demonstrate an impact of miR-124a and miR-155 on Treg differentiation via targeting of SIRT1.
Fig. 5.
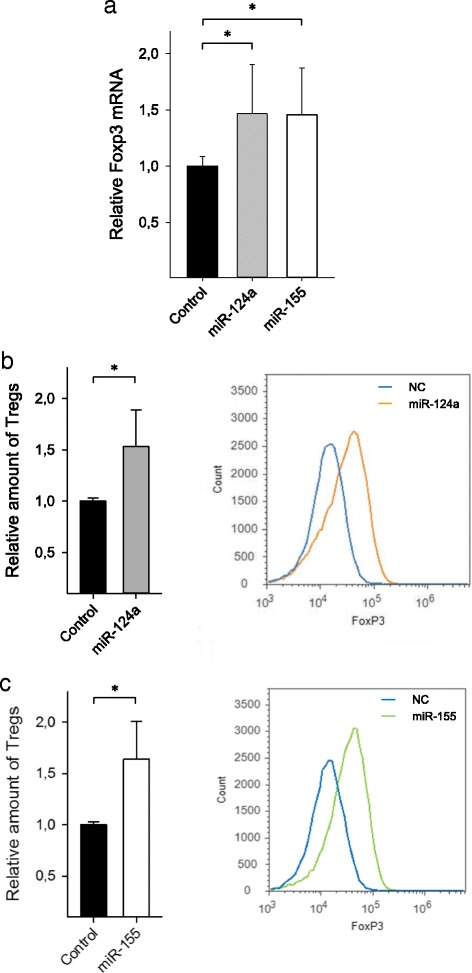
miR-124a and miR-155 enhance Treg differentiation. Human CD4+ T cells were transfected with either pre-miR-124a, pre-miR-155, or scrambled control, followed by culturing under Treg skewing conditions for 4 days. Relative Foxp3 mRNA expression was detected by qPCR (a). The number of Tregs was determined by FACS analysis (b, c). Additionally, representative histograms of the FACS analysis are shown in b and c (right panels). Data are given as means ± SD; *p < 0.01, n = 6
Discussion
The pathophysiology of neuropathic pain is not fully understood. Recent studies have established proof that aberrant responses of the adaptive immune system substantially contribute to the development of this clinical disorder. Underlying mechanisms, however, are largely unknown. In this study, we show an involvement of miRNAs in the regulation of inflammatory processes in neuropathic pain. We identify miR-124a and miR-155 as direct repressors of the deacetylase SIRT1. Targeting of SIRT1 by these miRNAs results in an increase of Foxp3 expression and, consecutively, of anti-inflammatory Tregs. We here show that in patients suffering from neuropathic pain as compared to healthy volunteers, an increased expression of miR-124a and miR-155 inhibits SIRT1 expression, which enhances CD4+ T cell differentiation towards Tregs.
Peripheral nerve injury leads to the release of factors that recruit and activate immune cells from the circulation. These cells secrete pro-inflammatory mediators that contribute to the development of pain symptoms. In particular, the T cell response is considered an important contributor to the development of neuropathic pain. In animal models of peripheral nerve injury, pain sensitivity of T cell deficient animals was significantly attenuated, which could be restored by adoptive transfer of pro-inflammatory cytokine producing Th1 cells [8]. On the other hand, expansion of Tregs, which limit immune responses of pro-inflammatory T cells, led to a significant reduction of pain hypersensitivity [22] while depletion of Tregs promoted pain hypersensitivity by inducing altered systemic concentrations of cytokines in mice [6]. The latter findings point towards a possible role of Tregs in the limitation of pain promoting inflammatory responses. In a very recently published study, we reported an increase of the Treg subpopulation in the peripheral blood of patients suffering from neuropathic pain, which also points into that direction. The pathways leading to the observed Treg induction, however, have not been addressed yet.
We here suggest a decreased expression of the histone-deacetylase SIRT1 as a possible underlying mechanism. SIRT1 is known to control Treg differentiation and function (i) by promoting Foxp3 gene expression and (ii) by Foxp3 lysine ε-aminodeacetylation leading to ubiquitination and proteasomal degradation. Here, we show that targeting of SIRT1 by specific siRNA promotes Treg differentiation of human CD4+ T cells in vitro. Similar results have been found in a recently published study, which reported an increased differentiation of naive T cells to Tregs after treatment with SIRT1 inhibitors in mice. Our findings strongly suggest that a decrease of SIRT1 expression contributes to the observed increase of Treg cells in neuropathic pain patients.
SIRT1 is subject to regulation on a transcriptional and posttranscriptional level [23, 24]. Particularly in tumors and endothelial cells, miRNAs have been shown to influence SIRT1 expression (e.g., miR-29c [23], miR-141 [23], miR-200 [24], miR-204 [25]). Based on the assumption that alterations of miRNA profiles might also be involved in the regulation of SIRT1 in neuropathic pain, we focused on miRNAs as potential suppressors of SIRT1 in this context. We identified miRNA-124a and miRNA-155 as potential candidates binding to the SIRT1-3′-UTR with high probability in silico. miR-155 is expressed in multiple types of immune cells and has been proposed to affect a wide range of immunological processes under physiologic conditions as well as in the course of immune responses [26–31]. In mice studies, it has been shown that the expression of miR-155 in Treg cells is required to maintain normal Treg numbers and function, which was in part attributed to miR-155-mediated SOCS1 repression [32, 33]. In a rat model of neuropathic pain, inhibition of miR-155 was shown to reduce cytokine production of microglial cells via SOCS1 repression, thereby attenuating pain symptoms [34].
miR-124a is predominantly expressed in the central nervous system (CNS). There, it displays specific temporal and spatial expression profiles in various cell types and affects a variety of biological functions. Dysregulation of miR-124 has been linked to several pathologic conditions of the CNS, such as brain tumors, neurodegeneration, epilepsy, and neuroimmune disorders. Furthermore, miR-124a is involved in macrophage polarization, which impacts a variety of diseases. For example, in animal models of pain, intrathecal application of miR-124a resulted in a decrease of pro-inflammatory cytokines secreted by microglia/macrophages, which led to a reduction of persistent hyperalgesia [35, 36].
Roads of miRNA regulation are redundant and highly dependent on the cellular and physiological context. Here, we reveal a new function of miR-124a and miR-155 in T cells in neuropathic pain: Our experiments show that both miRNAs suppress SIRT1 mRNA expression by direct targeting of specific binding sites. Accordingly, overexpression of miR-124a and miR-155 in human CD4+ T cells in vitro suppressed SIRT1 and, in accordance with our in vitro results obtained by transfection of SIRT1 siRNA, induced a bias towards Treg differentiation.
Clinical data also support this hypothesis: In T cells of neuropathic pain patients, we detected an increased expression of miR-124a and miR-155. Moreover, the expression of these two miRNAs was inversely correlated with SIRT1 transcript levels, which strongly supports the hypothesis that the Treg shift observed in neuropathic pain, indeed, is at least partially driven by a miRNA-mediated mechanism.
Conclusions
Increasing peripheral Treg numbers may be an endogenous attempt to limit inflammation, thus reducing pain levels in neuropathic pain. We here demonstrate that lymphocytic miRNAs significantly contribute to these adaptive processes. Deciphering miRNA-target interactions that influence inflammatory pathways in neuropathic pain may thus help to develop new approaches of pain amelioration.
Acknowledgements
The authors are grateful to G. Groeger, J. Rink, and S. Alijagic for the excellent technical assistance.
Funding
The study was partly funded by the Hella-Langer Stiftung, Germany (to SCA). The funding institution had no role in the data collection, analysis, interpretation, patient recruitment, or any other aspect pertinent to the study.
Availability of data and materials
All material used in this manuscript will be made available to researchers subject to confidentiality.
Authors’ contributions
JH, BL, and MH performed the experiments; JH, BL, LCH, and SK wrote the manuscript; BL and SCA recruited the patients and prepared the blood samples; all authors analyzed the data; JH, BL, and SK designed the experiments. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The prospective study protocol followed the principles of the Declaration of Helsinki and was reviewed and approved by the Ethics Committee of the LMU Munich (ethical approval number: 331–10) and registered on German Clinical Trial Register (Registration Trial DRKS00005954).
Abbreviations
- Tregs
Regulatory T cells
- SIRT1
Histone deacetylase sirtuin1
- Foxp3
Transcription factor forkheadbox-p3
- NeP
Neuropathic pain
- CRP
C-reactive protein
- NRS
Numerical rating scale
- PBMCs
Peripheral blood mononuclear cells
- qPCR
Quantitative real-time PCR (RT-PCR)
- FACS
Fluorescent-activated cell sorting
- KAB
Kurzfragebogen zur aktuellen Beanspruchung
- SDHA
Succinate dehydrogenase complex subunit A
- TBP
TATA box binding protein
- SD
Standard deviation
- CNS
Central nervous system
- BMI
Body mass index
Additional file
Correlation analysis of age and (A) Treg numbers, (B) SIRT1 mRNA, (C) miRNA-124 expression, and (D) miR-155 expression. Analyses revealed no significant correlations. Black dots: Neuropathic pain patients, white dots: Healthy volunteers. Figure S2 Human CD4 + T cells were transfected with either pre-miR-124a, pre-miR-155, or scrambled control, followed by culturing under Treg skewing conditions for 4 days. Relative mRNA expression the Treg signature molecules EOS, CTLA4, and IL2RA was detected by qPCR; *p < 0.05, **p < 0.01, n = 5. Figure S3. Correlation analysis of either miR-124a (A) or miR-155 (B) and Foxp3 mRNA expression. Black dots: Neuropathic pain patients, white dots: Healthy volunteers. (PPTX 227 kb)
Contributor Information
Jens Heyn, Email: Jens.Heyn@med.uni-muenchen.de.
Benjamin Luchting, Email: Benjamin.Luchting@med.uni-muenchen.de.
Ludwig C. Hinske, Email: Ludwig.Hinske@med.uni-muenchen.de
Max Hübner, Email: Max.Huebner@med.uni-muenchen.de.
Shahnaz C. Azad, Email: Shahnaz.Azad@med.uni-muenchen.de
Simone Kreth, Phone: +49-894400-76423, Email: Simone.Kreth@med.uni-muenchen.de.
References
- 1.Baron R. Mechanisms of disease: neuropathic pain—a clinical perspective. Nat Clin Pract Neurol. 2006;2:95–106. doi: 10.1038/ncpneuro0113. [DOI] [PubMed] [Google Scholar]
- 2.Bouhassira D, Lanteri-Minet M, Attal N, Laurent B, Touboul C. Prevalence of chronic pain with neuropathic characteristics in the general population. Pain. 2008;136:380–7. doi: 10.1016/j.pain.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Scholz J, Woolf CJ. The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci. 2007;10:1361–8. doi: 10.1038/nn1992. [DOI] [PubMed] [Google Scholar]
- 4.van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155:654–62. doi: 10.1016/j.pain.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Freeman R, Baron R, Bouhassira D, Cabrera J, Emir B. Sensory profiles of patients with neuropathic pain based on the neuropathic pain symptoms and signs. Pain. 2014;155:367–76. doi: 10.1016/j.pain.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Lees JG, Duffy SS, Perera CJ, Moalem-Taylor G. Depletion of Foxp3+ regulatory T cells increases severity of mechanical allodynia and significantly alters systemic cytokine levels following peripheral nerve injury. Cytokine. 2015;71:207–14. doi: 10.1016/j.cyto.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Sommer C, Kress M. Recent findings on how proinflammatory cytokines cause pain: peripheral mechanisms in inflammatory and neuropathic hyperalgesia. Neurosci Lett. 2004;361:184–7. doi: 10.1016/j.neulet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Moalem G, Xu K, Yu L. T lymphocytes play a role in neuropathic pain following peripheral nerve injury in rats. Neuroscience. 2004;129:767–77. doi: 10.1016/j.neuroscience.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 9.Luchting B, Rachinger-Adam B, Heyn J, Hinske LC, Kreth S, Azad SC. Anti-inflammatory T-cell shift in neuropathic pain. J Neuroinflammation. 2015;12:12. doi: 10.1186/s12974-014-0225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luchting B, Rachinger-Adam B, Zeitler J, Egenberger L, Mohnle P, Kreth S, Azad SC. Disrupted TH17/Treg balance in patients with chronic low back pain. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akimova T, Xiao H, Liu Y, Bhatti TR, Jiao J, Eruslanov E, Singhal S, Wang L, Han R, Zacharia K, et al. Targeting sirtuin-1 alleviates experimental autoimmune colitis by induction of Foxp3+ T-regulatory cells. Mucosal Immunol. 2014;7:1209–20. doi: 10.1038/mi.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beier UH, Akimova T, Liu Y, Wang L, Hancock WW. Histone/protein deacetylases control Foxp3 expression and the heat shock response of T-regulatory cells. Curr Opin Immunol. 2011;23:670–8. doi: 10.1016/j.coi.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beier UH, Wang L, Bhatti TR, Liu Y, Han R, Ge G, Hancock WW. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol Cell Biol. 2011;31:1022–9. doi: 10.1128/MCB.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freynhagen R, Baron R, Gockel U, Tolle TR. painDETECT: a new screening questionnaire to identify neuropathic components in patients with back pain. Curr Med Res Opin. 2006;22:1911–20. doi: 10.1185/030079906X132488. [DOI] [PubMed] [Google Scholar]
- 15.Maier C, Baron R, Tolle TR, Binder A, Birbaumer N, Birklein F, Gierthmuhlen J, Flor H, Geber C, Huge V, et al. Quantitative sensory testing in the German Research Network on Neuropathic Pain (DFNS): somatosensory abnormalities in 1236 patients with different neuropathic pain syndromes. Pain. 2010;150:439–50. doi: 10.1016/j.pain.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK. MicroRNA modulation in complex regional pain syndrome. J Transl Med. 2011;9:195. doi: 10.1186/1479-5876-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schomberg D, Ahmed M, Miranpuri G, Olson J, Resnick DK. Neuropathic pain: role of inflammation, immune response, and ion channel activity in central injury mechanisms. Ann Neurosci. 2012;19:125–32. doi: 10.5214/ans.0972.7531.190309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 19.Ledderose C, Heyn J, Limbeck E, Kreth S. Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res Notes. 2011;4:427. doi: 10.1186/1756-0500-4-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyn J, Hinske LC, Ledderose C, Limbeck E, Kreth S. Experimental miRNA target validation. Methods Mol Biol. 2013;936:83–90. doi: 10.1007/978-1-62703-083-0_7. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt A, Eriksson M, Shang MM, Weyd H, Tegnér J. Comparative analysis of protocols to induce human CD4 + Foxp3+ regulatory T cells by combinations of IL-2, TGF-beta, retinoic acid, rapamycin and butyrate. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Austin PJ, Kim CF, Perera CJ, Moalem-Taylor G. Regulatory T cells attenuate neuropathic pain following peripheral nerve injury and experimental autoimmune neuritis. Pain. 2012;153:1916–31. doi: 10.1016/j.pain.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Bae HJ, Noh JH, Kim JK, Eun JW, Jung KH, Kim MG, Chang YG, Shen Q, Kim SJ, Park WS, et al. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. 2014;33:2557–67. doi: 10.1038/onc.2013.216. [DOI] [PubMed] [Google Scholar]
- 24.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286:25992–6002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Wang X, Chen P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer. 2013;13:290. doi: 10.1186/1471-2407-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin B, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, Boudousquie C, Utzschneider DT, Escobar TM, Perret R, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–53. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–59. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zawislak CL, Beaulieu AM, Loeb GB, Karo J, Canner D, Bezman NA, Lanier LL, Rudensky AY, Sun JC. Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc Natl Acad Sci U S A. 2013;110:6967–72. doi: 10.1073/pnas.1304410110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, Liao YH. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan Y, Yang J, Xiang K, Tan Q, Guo Q. Suppression of microRNA-155 attenuates neuropathic pain by regulating SOCS1 signalling pathway. Neurochem Res. 2015;40:550–60. doi: 10.1007/s11064-014-1500-2. [DOI] [PubMed] [Google Scholar]
- 35.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willemen HL, Huo XJ, Mao-Ying QL, Zijlstra J, Heijnen CJ, Kavelaars A. MicroRNA-124 as a novel treatment for persistent hyperalgesia. J Neuroinflammation. 2012;9:143. doi: 10.1186/1742-2094-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All material used in this manuscript will be made available to researchers subject to confidentiality.


