Abstract
Widely used as anti-cancer and immunosuppressive agents, thiopurines have narrow therapeutic indices due to frequent toxicities, partly explained by TPMT genetic polymorphisms. Recent studies identified germline NUDT15 variation as another critical determinant of thiopurine intolerance, but the underlying molecular mechanisms and its clinical implications remain unknown. In 270 children enrolled in clinical trials for acute lymphoblastic leukemia in Guatemala, Singapore, and Japan, we identified 4 NUDT15 coding variants (p.Arg139Cys, p.Arg139His, p.Val18Ile, p.Val18_Val19insGlyVal) that resulted in 74.4%–100% loss of nucleotide diphosphatase activity. Loss-of-function NUDT15 diplotypes were consistently associated with thiopurine intolerance across three cohorts (P=0.021, 2.1×10−5, and 0.0054, respectively; meta-analysis P=4.45×10−8, allelic effect size=−11.5). Mechanistically, NUDT15 inactivated thiopurine metabolites and decreased its cytotoxicity in vitro, and patients with defective NUDT15 alleles showed excessive thiopurine active metabolites and toxicity. Taken together, our results indicate that a comprehensive pharmacogenetic model integrating NUDT15 variants may inform personalized thiopurine therapy.
Introduction
Thiopurines (mercaptopurine [MP], thioguanine [TG], and azathioprine) are widely used anti-cancer and immunosuppressive agents1–8. In acute lymphoblastic leukemia (ALL), prolonged daily exposure to MP is a major component of contemporary treatment regimens and indispensable for the cure of this disseminated malignancy7,9–13. However, MP can cause severe myelosuppression, resulting in frequent treatment disruptions, necessitating extensive supportive care, and increasing the risk of life-threatening infections6,13–22. Thiopurines are also commonly prescribed in patients with inflammatory bowel diseases (IBD, e.g., Crohn’s disease and ulcerative colitis), especially for their steroid-sparing potential and efficacy in remission maintenance4,6,14–17. Thiopurine treatment for IBD is associated with substantial hematopoietic toxicity, leading to discontinuation of therapy in up to 40% of patients and subsequent disease recurrence6,18–23. Therefore, the narrow therapeutic indices of thiopurines point to a strong need for applying evidence-based precision medicine approaches to the use of this class of medications.
As prodrugs, thiopurines are enzymatically converted to thioguanosine triphosphates (TGTP), through multiple sequential anabolic reactions. TGTP is further reduced to deoxy-thioguanosine triphosphate (TdGTP) that is incorporated into double strand DNA (DNA-TG) to trigger futile mismatch repair and eventually apoptosis1,24–28. The integration of thioguanine into DNA is widely accepted as one of the principal sites of action for thiopurine drugs and a major process responsible for their cytotoxic effects8,27–31. There are multiple pathways that negatively impact thiopurine effects: MP can be directly anabolized by thiopurine methyltransferase (TPMT) to the inactive methyl-mercaptopurine; thiopurine active metabolites can also be catabolized via dephosphorylation of thioguanine nucleotides (TGN)32,33. Therefore, thiopurine cytotoxicity is determined by the competition between the activation and inactivation pathways, components of which are affected by genetic variations. In particular, single nucleotide polymorphisms (SNPs) in the TPMT gene have been shown to cause loss of TPMT enzymatic activity, excessive levels of TGN, and predispose patients to thiopurine-related hematopoietic toxicity across multiple diseases23,34–38. In fact, TPMT-guided preemptive thiopurine dose adjustment has been shown to substantially reduce the risk of adverse effects without compromising therapeutic efficacy39–45 and is a prototype of precision medicine approaches to individualize drug therapy based on pharmacogenetics. However, substantial toxicity still occurs in some patients with normal TPMT activity, suggesting that additional variables, including other genetic variants, may contribute to the inter-patient variability in thiopurine metabolism.
Recent genome-wide association studies described a missense variant in the NUDT15 gene (rs116855232, referred to as the c.415C>T or the p.Arg139Cys variant hereafter) that is strongly associated with thiopurine-related myelosuppression in patients with IBD46 and in children with ALL47. Individuals homozygous for the risk allele at p.Arg139Cys were exquisitely sensitive to MP and tolerated only 8% of the standard dose, and this NUDT15 variant alone explained 22% of variance in MP tolerance47. Encoding a purported purine-specific nucleotide diphosphatase, NUDT15 is hypothesized to dephosphorylate the thiopurine active metabolites TGTP and TdGTP, thus preventing their incorporation into DNA and negatively affecting the desired cytotoxic effects of thiopurines. However, the exact mechanism by which NUDT15 regulates thiopurine metabolism has not been experimentally examined and the functional consequences of NUDT15 variants are unknown.
In this study, we systematically identified NUDT15 variants associated with thiopurine disposition and host toxicity, characterized their enzymatic properties, and comprehensively investigated the molecular pathways linking NUDT15 to thiopurine toxicity. Our results suggest that a comprehensive pharmacogenetic model integrating NUDT15 genetic variants may inform strategies to further individualize thiopurine therapy.
Results
NUDT15 variant discovery and function characterization
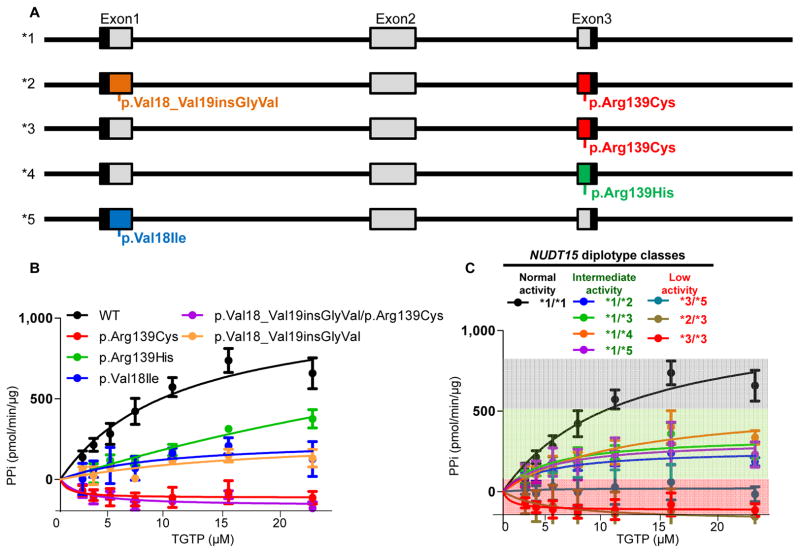
To systematically characterize NUDT15 polymorphisms associated with thiopurine toxicity, we sequenced all exonic regions of this gene in 3 cohorts including 270 children treated on frontline ALL clinical trials in Guatemala, Singapore, and Japan (Supplementary Table 1). In total, we identified 4 coding variants located in exons 1 and 3, all of which resulted in changes in amino acid sequence of the NUDT15 protein (Fig. 1A). The c.415C>T and c.416G>A variants were transition substitutions within the codon for amino acid residue Arg139, causing an arginine-to-cysteine and arginine-to-histidine change (p.Arg139Cys and p.Arg139His), respectively. The other 2 variants affected the Val18 residue: the c.52G>A variant resulted in a valine-to-isoleucine conversion (p.Val18Ile), and the c.36_37insGGAGTC led to an in-frame addition of a glycine and a valine residue (p.Val18_Val19insGlyVal). We inferred 5 NUDT15 haplotypes with distinct combinations of genotype at these 4 variants (referred to as haplotype *1–*5 hereafter, Fig. 1A). In particular, the p.Val18_Val19insGlyVal allele was in high linkage disequilibrium with the p.Arg139Cys allele, and the presence of both defined the common haplotype *2. Analyses of the phased 1,000 Genomes Project data identified similar haplotype patterns, and the prevalence of each haplotype varied substantially by ancestry (Supplementary Fig. 1).
Figure 1. NUDT15 genetic variants and their effects on nucleotide diphosphatase activity.
Four coding variants were identified representing 5 haplotypes (*1 to *5, panel A). Each variant NUDT15 was expressed in E. Coli and purified protein was subjected to diphosphatase activity measurement with TGTP as the substrate (panel B). Variant or wildtype proteins were combined to determine the level of NUDT15 activity in patients with different diplotypes (panel C). There were no significant differences in nucleotide diphosphatase activity between diplotypes within the intermediate group (green shade, P = 0.73) or within the low activity group (red shade, P = 0.19), as determined using Kruskal-wallis test. Center values (dots) represent mean of triplicates and error bars indicate standard deviation.
To define the functional consequences of NUDT15 polymorphisms, we characterized nucleotide diphosphatase activity for each of the 4 variant proteins (Fig. 1B). Wildtype NUDT15 efficiently converted the thiopurine active metabolite TGTP to the monophosphate thioguanosine nucleotide TGMP, with an apparent catalytic efficiency (Vmax/Km) of 107.9 ± 0.2. In contrast, 4 variant NUDT15 proteins showed 74.4% to 100% loss of enzymatic activity: Vmax/Km of 23.6 ± 0.9, 27.7 ± 1.1, and 14.9 ± 1.1 for the p.Arg139His, p.Val18Ile, p.Val18_Val19insGlyVal variants, respectively; enzymatic activity was not detectable for the p.Arg139Cys or the p.Val18_Val19insGlyVal/p.Arg139Cys variant protein. Similar differences in NUDT15 activity were observed when TdGTP was used as the substrate (Supplementary Fig. 2). While the p.Arg139Cys and p.Val18_Val19insGlyVal individually caused reduction of NUDT15 activity, the protein harboring both variants did not show further loss of function compared to the p.Arg139Cys variant alone (Fig. 1B), suggesting that the *2 haplotype is likely to have similar functional consequence as haplotypes with a single variant (e.g., *3). Thermostability assay showed substantially lower Tm values for all variant NUDT15 proteins (42.6 ± 0.1, 43.6 ± 0.2, 52.5 ± 0.3, 41.5 ± 0.1, and 54.0 ± 0.1 °C, for the p.Arg139Cys, p.Arg139His, p.Val18Ile, p.Val18_Val19insGlyVal/p.Arg139Cys and p.Val18_Val19insGlyVal, respectively), compared to the wildtype protein (56.3 ± 0.3 °C), consistent with perturbation of NUDT15 conformation by these substitutions (Supplementary Fig. 3).
Further, we tested different combinations of variant NUDT15 proteins to determine enzymatic activity for each of the 8 diplotypes observed in our clinical cohorts (Fig. 1C). Equimolar mixtures of the variant and wildtype NUDT15 proteins (equivalent of patients heterozygous at a single variant, e.g., *1/*3) showed intermediate activity, indicating a gene dosage effect. By comparison, the mixture of two different variant NUDT15 proteins (e.g., *3/*5) showed low activity that was comparable to a single variant with the same total protein concentration (e.g., *3/*3), suggesting that individuals with compound heterozygous genotypes are likely to have similar MP intolerance as those homozygous for a single NUDT15 variant. Therefore, we classified patients into 3 diplotypic groups (Fig. 1C): normal activity (*1/*1), intermediate activity (*1/*2, *1/*3, *1/*4 and *1/*5), and low activity (*2/*3, *3/*3 and *3/*5). There were no significant differences in enzymatic activity between diplotypes within the intermediate activity group (P = 0.73) or within the low activity group (P = 0.19).
Consistently in the Guatemalan, Singaporean, and Japanese cohorts (N = 159, 79, and 32, respectively), MP tolerance was highest in patients with the normal activity NUDT15 diplotype followed by those with intermediate activity, and lowest in patients with the diplotypes indicative of low NUDT15 activity (P = 0.021, 2.1 × 10−5, and 0.0054, respectively, Fig. 2). Of particular note, patients with *1/*2 diplotype (heterozygous at both the p.Arg139Cys and the p.Val18_Val19insGlyVal variants) showed similar degree of MP intolerance as the *1/*3 group (heterozygous at the p.Arg139Cys variant alone, Supplementary Fig. 4), corroborating our observation that single (p.Arg139Cys) and double (p.Val18_Val19insGlyVal/p.Arg139Cys) variant NUDT15 proteins exhibited similar enzymatic activity (Fig. 1B). Additionally, two patients with compound heterozygous NUDT15 genotype (*3/*5 and *2/*3) were exquisitely sensitive to MP, on the same scale as those homozygous for the variant allele (*3/*3), indicating cumulative effects of NUDT15 variants (Supplementary Fig. 4). A meta-analysis combining three cohorts indicated strong and consistent effects of NUDT15 variants on tolerated MP dosage reduction (P = 4.45 × 10−8, allelic effect size of −11.5 [95% confidence interval, −15.6 to −7.4], Fig. 2D) and we did not observe any statistically significant heterogeneity across cohorts (P = 0.34). Including patients with TPMT risk variants (rs1800462, rs1800460, and rs1142345) had minimal effects on the association of NUDT15 diplotype with tolerated MP dosage (meta-analysis P = 9.96 × 10−10 and 9.92 × 10−8 for before and after adjusting for TPMT variants respectively, Supplementary Fig. 5), indicating independent contributions of these two genes to MP metabolism and toxicity.
Figure 2. Association of NUDT15 diplotype with MP tolerance during ALL therapy in Guatemala, Singapore, and Japan.
Patients were classified as “normal”, “intermediate”, or “low” NUDT15 activity on the basis of their diplotype at 4 coding variants (panels A, B, and C for the Guatemalan, Singaporean, and Japanese cohorts, respectively). MP dosage was adjusted during maintenance therapy to avoid host toxicities and tolerated MP dosage was defined as the average over at least 14 daily dosages after at least 9 weeks of maintenance therapy. Cases with TPMT variants (rs1800462, rs1800460, and rs1142345) were excluded from the analysis. P value was calculated by using linear regression test, after adjusting for co-variates when applicable. In panels A–C, each box includes data between 25th and 75th percentiles, with horizontal line indicating median. Similar association analyses were performed to compare tolerated MP dosage between normal and intermediate NUDT15 groups (P = 0.04, 0.00049, and 0.0033 for Panels A, B, and C, respectively). Meta-analysis combining test statistics from three cohorts indicated consistent association (Forest plot, Panel D, P = 4.45 × 10−8), with no significant heterogeneity across cohorts (P = 0.34). Allelic effect size indicates the change in MP dosage for every copy of the NUDT15 risk allele. The length of each horizontal line represents the range of 95% confidence interval of allelic effect size with the tick indicating median (gray boxes are proportional to weights of each cohort used in meta-analysis). Dashed vertical line denotes allelic effect size in the meta-analysis with the lateral tips of diamond representing 95% confidence interval.
NUDT15 regulates thiopurine activation and cytotoxicity
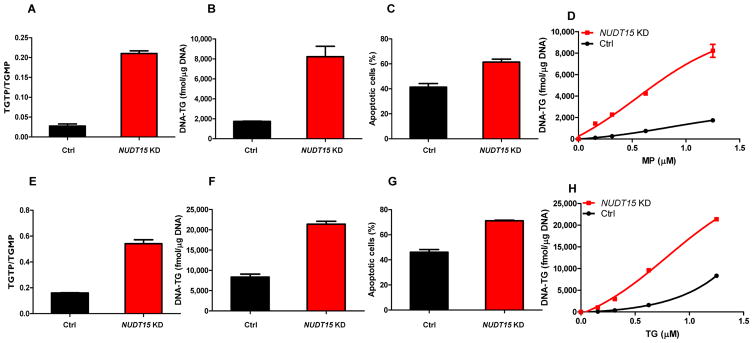
Because NUDT15 converts TGTP to TGMP (also TdGTP to TdGMP), we hypothesized that NUDT15 prevents the incorporation of these thiopurine metabolites into DNA (DNA-TG) and negatively regulates thiopurine activation and consequently its cytotoxicity. To recapitulate the loss of NUDT15 activity resulting from genetic variation, we first established stable NUDT15 knockdown in a human lymphoid cell line (Supplementary Fig. 6). In the control cells transduced with scramble shRNA, MP was extensively metabolized and the ratio of TGTP to TGMP was 0.028 (± 0.0052), with TGTP representing only 2.7% (± 0.5%) of total intracellular thioguanine nucleotides (TGN). When NUDT15 expression was downregulated, the level of TGTP increased significantly with a 7.5-times higher TGTP to TGMP ratio (0.21 [± 0.0067]) and 6.2-times higher %TGTP in TGN (16.8% [± 0.3%]) compared to control cells, indicating a significant shift in equilibrium toward TGTP (Fig. 3A). Consequently, DNA-TG was markedly higher in NUDT15 knockdown cells compared to control (8,219.9 ± 853.6 fmol/μg DNA and 1,734.7 ± 25.7 fmol/μg DNA, respectively, Fig. 3B), leading to a significant increase in MP-induced apoptosis in these cells (Fig. 3C, Supplementary Fig. 7A). With increasing concentrations of MP, there was concomitant elevation of intracellular DNA-TG, and the rate at which MP was incorporated into DNA (i.e., DNA-TG) was substantially greater in the NUDT15 knockdown cells compared to control cells (Fig. 3D). Similarly, when we repeated these experiments with TG or azathioprine, NUDT15 knockdown cells consistently showed increased levels of active metabolites (e.g., TGTP and DNA-TG), and higher susceptibility to TG- or azathioprine-induced apoptosis (Fig. 3E–H, Supplementary Figs. 7B and 8). Together, these results indicate that NUDT15 inactivates thiopurine metabolites and is directly related to their cytotoxic effects, thus providing a clear biological mechanism explaining the susceptibility to thiopurine host toxicity in patients inheriting loss-of-activity variants in NUDT15.
Figure 3. Effects of NUDT15 on thiopurine metabolism and cytotoxicity.
NUDT15 knockdown (NUDT15 KD, red) cells were established by lentiviral transduction of NUDT15-specific shRNA, and control cells (black) were transduced with non-targeted vectors. Thiopurine metabolites (cytosolic TGMP and TGTP, and DNA-incorporated thioguanine, [DNA-TG]) were analyzed after treatment with 1.25 μM of MP (A and B) or TG (E and F) for 48 hours. DNA-TG was measured in cells exposed to increasing concentrations of MP (D) and TG (H). Cytotoxicity was determined by MTT assay following 72-hour incubation with MP (C, 1.25 μM) or TG (G, 1.25 μM). Mean values are plotted in each panel with error bars indicating standard deviation from triplicates.
NUDT15 variants and thiopurine metabolism in vivo
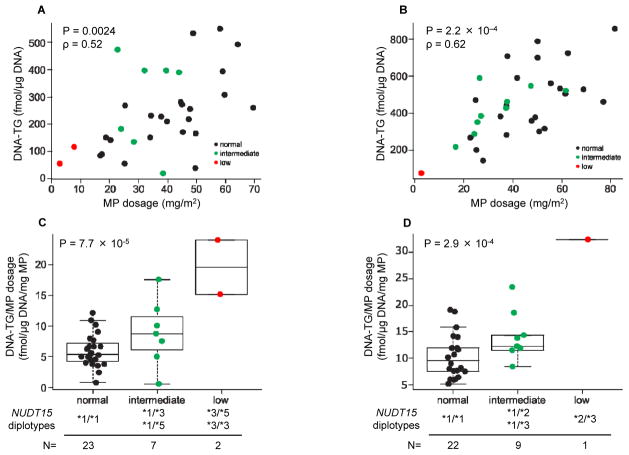
Building upon the in vitro results, we next sought to determine how inherited NUDT15 deficiency influences thiopurine activation in patients by directly monitoring DNA-TG levels in white blood cells from children with ALL receiving daily MP treatment. In 32 patients in the Singaporean cohort, DNA-TG levels strongly correlated with the actual MP dosage delivered (P = 0.0024, Fig. 4A). The positive correlation between MP dosage and DNA-TG was also validated in the Japanese cohort (N = 32, P = 2.2 × 10−4, Fig. 4B). The ratio of DNA-TG to MP dosage (i.e., the amount of DNA-TG converted from every unit MP dose) varied significantly by NUDT15 genotype. In the Singaporean cohort (Fig. 4C and Supplementary Fig. 9A), the average DNA-TG to MP dosage ratio was 6.0 ± 2.7 fmol/μg DNA/mg MP in children with wildtype NUDT15 (*1/*1). In contrast, those with intermediate activity NUDT15 diplotypes (*1/*3 and *1/*5) achieved 8.8 (± 5.3 fmol/μg DNA) of DNA-TG for every mg of MP and this ratio was highest in patients with homozygous or compound heterozygous NUDT15 diplotypes (19.6 ± 6.3 fmol/μg DNA/mg MP, P = 7.7 × 10−5). Similarly in the Japanese cohort (Fig. 4D and Supplementary Fig. 9B), the DNA-TG to MP dosage ratio was 9.6 ± 4.1, 12.3 ± 4.5, and 32.4 fmol/μg DNA/mg MP for children with normal, intermediate, and low activity NUDT15 diplotypes, respectively (P = 2.9 × 10−4). A meta-analysis combining the Singaporean and Japanese cohorts indicated strong and consistent association of NUDT15 diplotype with normalized DNA-TG (P = 2.2 × 10−8) and we did not observe significant heterogeneity between cohorts (P = 0.63).
Figure 4. NUDT15 variants and MP metabolism in children during ALL therapy.
DNA-TG levels were analyzed in Singaporean (A and C) and Japanese (B and D) cohorts. Sixty-three and 44 samples were successfully measured from 32 cases with wildtype TPMT in Singapore and Japanese cohorts, respectively. An average DNA-TG level was estimated for each patient. The associations between MP dosage and DNA-TG were evaluated by two-sided Spearman rank test (A and B). DNA-TG level was normalized based on actual MP dosage (the average of 14 days) prior to metabolite measurements and then correlated with NUDT15 diplotype (as normal, intermediate, or low NUDT15 activity) using a linear regression model (two-sided, C and D). Similar association analyses were also performed to evaluate the difference in normalized DNA-TG between normal and intermediate NUDT15 groups (P = 0.14 and 0.039 for Panels C and D, respectively). Meta-analysis combining test statistics from two cohorts indicated consistent association of NUDT15 diplotype with normalized DNA-TG (P = 2.2 × 10−8), without significant heterogeneity across cohorts (P = 0.63). Each box includes data between 25th and 75th percentiles, with horizontal line indicating median.
Discussion
The current study has comprehensively characterized the effects of inherited variants of NUDT15 on the metabolism of thiopurine and on the clinical tolerance of thiopurine toxicity. These novel findings are important for multiple reasons: First, these NUDT15 variants are highly penetrant and confer exquisite sensitivity to thiopurines (homozygous carriers can tolerate < 10% of standard dosage), with effect sizes comparable to TPMT variants that are clinically implemented to guide thiopurine dose reduction46,47. In fact, in a multiethnic cohort of children with ALL, the NUDT15 variant p.Arg139Cys alone explained 22% of variance in MP tolerance47. Secondly, TPMT variants are generally rare among Asian populations48 and thus of limited relevance for guiding thiopurine dosing in these race groups. In contrast, NUDT15 genetic variation is substantially over-represented in Asians and is their predominant genetic cause for thiopurine toxicity46,47. The unequivocal evidence linking NUDT15 p.Arg139Cys variant to thiopurine toxicity (particularly in Asians46,47,49,50) strongly indicates its potential clinical relevance, and also raises the question of whether (or what) clinical action is warranted for these at-risk patients.
A critical barrier to integrating NUDT15 genotypes into thiopurine dosing algorithm has been the paucity of data to establish the pharmacologic basis for dose reduction for patients with NUDT15 risk alleles. Addressing this knowledge gap, our results from a variety of laboratory model systems and in patients collectively indicate that NUDT15 deficiency directly resulted in excessive levels of thiopurine active metabolites (TGTP and DNA-TG) and increased host toxicity. Therefore, reducing thiopurine doses for patients who carry the NUDT15 variants would likely tailor their exposure to a level of TGTP and DNA-TG that is similar to wildtype patients receiving standard thiopurine doses. This is a highly plausible strategy for utilizing NUDT15 genotype to individualize thiopurine therapy to mitigate toxicity, as it is the same principle used for TPMT-based dose adjustments already implemented clinically51. In fact, in 285 children with newly diagnosed ALL, leukemic cells with intermediate activity NUDT15 diplotypes were also significantly more sensitive to TG than those with wildtype diplotype (P = 0.03, Supplementary Fig. 10). Therefore, genotype-guided thiopurine dose reductions in ALL patients inheriting NUDT15 risk alleles would likely minimize side effects without compromising antileukemic efficacy. Future clinical studies will be needed to precisely define the optimal dosage of thiopurines in patients with different NUDT15 diplotypes. The strength of the association between NUDT15 variants and tolerated MP dosage also differed slightly among three ALL cohorts (strongest in Singaporeans and weakest in Guatemalans, Fig. 2). Among these three populations, Guatemalans had the lowest tolerated MP dosage even with wildtype NUDT15 and had smaller differences in MP tolerance across NUDT15 genotype groups (although still significant). It is possible that other novel genetic variants associated with thiopurine toxicity exist uniquely in Guatemalans, potentially contributing to their overall sensitivity to this class of drugs. These hypotheses should be examined in future studies.
Our targeted sequencing of NUDT15 identified 4 coding variants, all of which influenced NUDT15 activity and were linked to thiopurine toxicity (Figs. 1 and 2). Intriguingly, 2 of these variants both affect residue Arg139 that is located in the α helix a2 at the base of the substrate binding pocket of NUDT1552. It has been hypothesized that the substitution of arginine with cysteine (p.Arg139Cys) at this position might introduce a disulfide bond and thus structural perturbation that interferes with TGTP binding52. In contrast, the arginine-to-histidine change caused by the p.Arg139His variant might lead to a reduced eletrophilicity and compromise substrate interaction. The other 2 NUDT15 variants locate at residue Val18 within the β sheet at the N-terminus of the protein, and it is unclear how the substitution and insertion at this position structurally hinder NUDT15 function. It is also possible that coding variants can affect NUDT15 protein synthesis and/or degradation and thus indirectly influence enzymatic activity, as we have shown for TPMT variants53. However, when ectopically expressed in HEK293T cells, the NUDT15 variants identified herein showed relatively high protein stability over time in vitro, comparable to wildtype NUDT15 (Supplementary Fig. 11); it will be important to document similar lack of differences in patients with these NUDT15 genotypes.
It was originally believed that NUDT15 functions as a sanitizer to remove damaged nucleotides, e.g., oxo-dGTP generated from radical oxygen species54. However, this was challenged by more recent data showing a strong preference of NUDT15 for thiopurine metabolites over oxo-dGTP52. NUDT15 is also shown to efficiently hydrolyze dGTP with potential importance in purine nucleotide homeostasis52, but its exact physiological functions remain unknown. It is unclear whether loss-of-function NUDT15 variants confer susceptibility to any diseases, although there is no evidence in the literature to suggest that this is the case. It is plausible that NUDT15 deficiency has limited effects on endogenous nucleotide metabolism under normal physiological conditions because of compensation by other NUDT proteins with overlapping enzymatic functions52,55. In contrast, the prominent role of NUDT15 in thiopurine inactivation signifies potential importance of the NUDT family of enzymes in the metabolism of other nucleotide analog drugs, which are widely used for diverse human diseases (especially in antiviral therapy)56. Therefore, comprehensive identification of pharmacogenetic variants in NUDT genes may have broad clinical relevance.
Our results from comprehensive in vitro and in vivo studies strongly indicate that NUDT15-related thiopurine toxicity follows an additive genetic mode of inheritance, with the severity of the phenotype proportional to the cumulative number of risk alleles in NUDT15. Patients homozygous or compound heterozygous for NUDT15 risk alleles experience especially excessive MP toxicity, compared to those with intermediate or normal activity diplotypes (Fig. 2). This is not unexpected because homozygous variant NUDT15 protein is enzymatically inactive (Fig. 1C) and complete loss of this important metabolizing enzyme results in disproportionally high levels of active metabolite and thus toxicity, as also seen in patients with inherited TPMT deficiency12,35. The interpretation of NUDT15 genotype becomes more complicated when a patient carries multiple functional variants. For example, if two variants are on the same haplotype, the patient is likely to have intermediate NUDT15 activity because the remaining wildtype copy is still functional (e.g., *1/*2). In contrast, if each of these two variants affects a different copy of NUDT15 (compound heterozygous, e.g., *3/*5), enzymatic activity is completely lost in these patients similar to subjects with homozygous variant genotype (e.g., *3/*3). These two scenarios indicate distinct levels of thiopurine tolerance and can be distinguished only through haplotype-based analyses. Using phased sequencing data from the 1,000 Genomes Project, we inferred NUDT15 diplotypes in major race/ethnic groups worldwide (Supplementary Fig. 1) and experimentally determined the enzymatic activity of each haplotype. NUDT15 deficiency (carrying low or intermediate activity diplotypes) was most common in East Asians (22.6%) and also prevalent in South Asians (evenly distributed in India, Pakistan, Bangladesh, and Sri Lanka at 13.6%) and Native American populations (e.g., 21.2% in Peruvians and 12.5% in Mexicans). In fact, in our Japanese cohort, approximately 1 in every 3 individuals carried risk variant(s) in the NUDT15 gene, a frequency that is even higher than that of TPMT deficiency (e.g., 1 in every 12 Europeans or 1 in every 8 Africans)51. We have also identified rare NUDT15 variants in the Broad Institute Exome Aggregation Consortium dataset including whole exome seq of 60,706 individuals (Supplementary Table 2), although the function of these variants and their effects on thiopurine metabolism are unknown.
As previously described, cases with both variants of NUDT15 and TPMT variants were significantly less tolerant to MP tolerance than those with risk alleles in one of these two genes47. Therefore, integration of NUDT15 in thiopurine dosing algorithm may have a major implication for Asian populations, whereas TPMT variants are most informative for thiopurine toxicity in Europeans and Africans. A polygenic dosing algorithm that incorporates both NUDT15 and TPMT variants would potentially provide a robust approach to personalize thiopurine therapy in major racial and ethnic groups represented in diverse populations worldwide.
Online Methods
Patients and thiopurine therapy
A total of 270 children treated on frontline ALL clinical trials in Guatemala, Singapore, and Japan were included in this study: 266 had newly-diagnosed ALL and 4 cases were diagnosed with mixed phenotype acute leukemia or lymphoma (Supplementary Table 1). The Guatemalan cohort comprised 159 patients (self-identified as Native American or Hispanic/mestizo) from the LLAG-0707 ALL protocol57 at the Unidad Nacional de Oncología Pediátrica, Guatemala City, Guatemala. This protocol included daily MP treatment during the maintenance phase with a planned MP dosage of 50–75 mg/m2 per day with provisions for dose adjustments based on the degree of myelosuppresion (target white blood cell count between 1.5–3.0 × 109/L) or infections. The Singaporean cohort included 79 children treated on the MaSpore ALL 2003 and MaSpore ALL 2010 protocols58 at National University Hospital, Singapore. Planned daily MP dosage was 50 mg/m2 and 75 mg/m2 for the standard/intermediate and high-risk arms respectively, and clinically titrated to a target white blood cell count between 2.0–4.0 × 109/L. The Japanese cohort consisted of 32 children treated on the Japanese Pediatric Leukemia/Lymphoma Study Group (JPLSG) ALL-B12 protocol (UMIN000009339) for newly-diagnosed ALL at 13 hospitals and oncology centers in Japan. The standard MP dosage during maintenance phase was 50 mg/m2 per day and was adjusted to a target white blood cell count between 2.0–3.0 × 109/L. In all 3 cohorts, MP dosage was considered to be stable after at least 9 weeks of daily MP dosing during maintenance therapy (with appropriate dose titration), following which the average of daily dosage over at least 14 days was used to define MP tolerance (i.e., tolerated MP dosage).
Patients were selected on the basis of the availability of germline DNA and availability of MP dosing and tolerance history. Germline DNA was extracted from peripheral blood obtained during clinical remission. This study was approved by the respective institutional review boards, and informed consent was obtained from parents, guardians, and/or patients, as appropriate.
NUDT15 sequencing and TPMT genotyping
Coding regions (exons 1, 2 and 3) of the NUDT15 gene were first amplified by polymerase chain reaction (PCR) from germline DNA, followed by Sanger sequencing (primer sequences are provided in Supplementary Table 3). Sequence alignment and comparison were performed using the CLC Genomics Workbench (CLC Bio, Qiagen) to identify genetic variants. NUDT15 variant genotype was coded as 0, 1, or 2 to indicate the number of variant alleles, and sequencing data were analyzed by using PHASE59 to infer haplotypes. TPMT variants (rs1800462, rs1800460, and rs1142345) were manually genotyped as described previously60 for all patients, and individuals with variant genotypes (N = 20 and 2 in the Guatemalan and Singaporean cohorts, respectively) were excluded from subsequent analyses unless otherwise indicated.
NUDT15 purification, enzymatic activity measurement, and thermostability assay
Human NUDT15 cDNA (accession number BC133017) was cloned into the pCold II expression vector (TaKaRa Bio) with an N-terminal His tag, as previously described54. Missense and insertion variants (p.Arg139Cys, p.Arg139His, p.Val18Ile, p.Val18_Val19insGlyVal, and p.Val18_Val19insGlyVal/p.Arg139Cys) were introduced by site-directed mutagenesis using the QuikChange II XL kit (Agilent Technologies). Wildtype or variant NUDT15 protein was ectopically expressed in E. coli BL21 with IPTG induction (0.5 mM, 24 hours at 15 °C), and purified by affinity chromatography using the TALON metal affinity resin (Clontech). Purified proteins were concentrated with an Amicon Ultra 0.5 ml centrifugal filter (Sigma-Aldrich) in 10 mM pH 8.0 Tris-HCl buffer containing 0.1 mM EDTA, 5 mM DTT and 10% glycerol. Protein purity was determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Coomassie Blue staining (Supplementary Fig. 12), and the molecular weight of each NUDT15 protein was confirmed by Time-of-Flight mass spectrometry using the Waters LCT Premier XE MS (Supplementary Table 4).
NUDT15 diphosphatase activity was measured by quantifying the release of pyrophosphate using the PiPer Pyrophosphate Assay Kit (Life Technologies). Each NUDT15 protein (100 ng) was incubated with varying concentrations of TGTP (1.88 μM to 21.4 μM) or TdGTP (0.99 μM to 35.7 μM) at 37 °C for 10 minutes before heat inactivation (99 °C for 20 minutes). Free pyrophosphate was then quantified immediately by fluorescence spectrometry according to manufacturers’ instructions, and Km and Vmax values were determined by Michaelis-Menten kinetics methods in Prism (GraphPad). Each experiment was done in triplicates and repeated at least three times.
To determine protein thermostability, 1 μg of wildtype or variant NUDT15 protein was incubated with 10× Sypro®Orange (Molecular Probes), and the mixture was heated from 20 °C to 95 °C in increments of 0.2 °C in the Quant Studio 12K Flex real time PCR system (Applied Biosystems). Fluorescence changes were monitored with a charged-couple device camera, with excitation and emission wavelengths at 490 nm and 575 nm, respectively. Tm values, the temperature midpoint for the protein unfolding transition, were calculated based on the Boltzmann model in Prism (GraphPad).
To assess NUDT15 protein stability in vitro, variant or wildtype NUDT15 cDNA with a N-terminal FLAG tag on the pcDNA3.1 backbone (Invitrogen) was transiently expressed in HEK293T cells (American Type Culture Collection, ATCC) using polyethylenimine reagent (Polysciences). Cycloheximide (50 μg/ml) was added 48 hours following transfection, and NUDT15 protein levels were monitored after 0, 24, and 48 hours by Western blot with β-actin as the loading control (anti-FLAG antibody, #2368, Cell Signaling).
NUDT15 knockdown and thiopurine metabolism in vitro
Lentiviral vectors containing shRNAs specific for human NUDT15 (TRCN0000050311) or scramble sequence (SHC016) were purchased from Sigma-Aldrich. Viral particles were prepared using the calcium chloride method in HEK293T cells and stable knockdown was established in the human lymphoid cell line Nalm6 (German Collection of Microorganisms and Cell Cultures, DSMZ) by lentiviral transduction of shRNA and puromycin selection, following procedures described previously61. Cell lines were authenticated by karyotyping and cytogenetics and tested negative for mycoplasma contamination. The level of NUDT15 knockdown was determined by real-time quantitative PCR with human tubulin as the internal control (Supplementary Fig. 6A and primer sequences are provided in Supplementary Table 3). NUDT15 expression was also determined at the protein level by Western blot with β-actin as the loading control (anti-NUDT15 antibody, ab122521, Abcam, Supplementary Fig. 6B).
To determine thiopurine metabolism in vitro, 5 × 106 NUDT15 knockdown or control cells were treated with varying concentrations of MP or TG at 37 °C for 48 hours. Cytosolic thiopurine metabolites (e.g., TGTP, TGDP, and TGMP) were analyzed with liquid chromatography-tandem mass spectrometry (LC-MS/MS) according to a previously reported method62. In parallel, aliquots of MP-, TG-, or azathioprine-treated cells were processed to extract genomic DNA, and DNA-TG was measured using LC-MS/MS as described previously63. MP, TG, and azathioprine cytotoxicity was determined for NUDT15 knockdown and control cells by using the colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay at the end of the 72-hour drug exposure. In vitro experiment were done in triplicates and repeated at least three times.
Thiopurine metabolism in children with ALL
To investigate in vivo thiopurine metabolism, DNA incorporation of TdGTP (i.e., the level of DNA-TG) was longitudinally monitored during the maintenance therapy phase in a subset of children enrolled on the MaSpore ALL 2010 and JPLSG ALL-B12 protocols. In the Singaporean cohort (N = 32), peripheral blood was collected every 3 months during maintenance, with a maximum of 3, 4, and 5 time points for those on the high, intermediate, and standard risk arms, respectively. In the Japanese cohort (N = 32), peripheral blood collection was scheduled at the 13th and 41st week of maintenance therapy. For both cohorts, genomic DNA was extracted from leukocytes within 72 hours of blood draw and DNA-TG was quantified using LC-MS/MS as described previously63, and detailed MP dosing history was recorded for 14 days prior to each sample collection.
To examine the effects of NUDT15 variants on leukemic cell sensitivity to thiopurine, we also evaluated TG LC50 (drug concentration that is lethal to 50% of cells) in 285 children with newly-diagnosed ALL. Primary ALL cells were exposed to increasing concentrations of TG for 96 hours, after which cell viability was determined using MTT assay to estimate LC50, as described previously64.
Statistical analyses
Each patient was assigned a NUDT15 “genetic score” of 0, 1, or 2 indicating low, intermediate, or normal NUDT15 activity, on the basis of his/her NUDT15 genetic diplotype and experimentally determined enzymatic activity for each diplotype (Fig. 1C). Within each ALL cohort, we first evaluated the association of patient clinical features with tolerated MP dosage, using the Wilcoxon rank test (gender), the Kruskal-Wallis test (ancestry and leukemia type), or the Spearman rank test (age as a continuous variable, Supplementary Table 1). The correlation between NUDT15 genetic score and tolerated MP dosage (an average value for each patient if there were multiple measurements) was then tested in Guatemalan, Singaporean, and Japanese cohorts separately, using the linear regression model. Age and gender were included as covariates in regression model for the Singaporean cohort because they were related to MP dosage. A similar statistical model was applied to determine the association of NUDT15 genetic score and the ratio of DNA-TG to MP dosage (DNA-TG/MP dosage) in the Singaporean and Japanese cohorts. Correlation between tolerated MP dosage and DNA-TG was determined by using the Spearman rank test. Kruskal-wallis test was used to evaluate the differences in nucleotide diphosphatase activity, tolerated MP dosage, or normalized DNA-TG among diplotypes within the intermediate activity group and among those within the low activity group. Meta-analyses were also performed combining test statistics of each cohort using random effect model based on inverse variant method65 with ancestry, age, and gender as covariates, and inter-cohort heterogeneity was tested using the Q-statistic. All statistical tests were two-sided and chosen as appropriate according to data distribution, and the threshold for statistical significance was defined as P < 0.05. R (version 3.0) was used for all analyses unless indicated otherwise.
Supplementary Material
Acknowledgments
The authors thank the patients and parents who participated in the clinical trials included in this study, Dr. Hidemi Toyoda at Mie University for his assistance in processing the JPLSG samples, and Dr. Colton Smith at St. Jude Children’s Research Hospital for querying the 1,000 Genomes Project data. This work was supported by the National Institutes of Health (CA021765 and GM115279), the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, the Order of St. Francis Foundation, the V Foundation for Cancer Research, and the Danish Childhood Cancer Foundation. The JPLSG ALL-B12 study is supported by the Japanese Ministry of Health and the Ma-Spore ALL studies are supported by National Medical Research Council (Singapore), Children’s Cancer Foundation, and Viva Foundation for Children with Cancer. JJY is an American Society of Hematology Scholar, TM is supported by the Mie Prefecture Study Abroad Scholarship, Mie, Japan, UH and MS are supported by the Robert Bosch Foundation, Stuttgart, Germany, KH is supported by the Pediatric Oncology Education Program grant (CA23944), and TI is supported by Alex’s Lemonade Stand Foundation’s POST program.
Footnotes
Author Contributions
Supervised research: J.J.Y.; Conceived and designed the experiments: T.M., R.N., H.H., K.S., A.E.J.Y, W.E.E. and J.J.Y.; Performed the experiments: T.M., R.N., V.P.A., X.Z., T.N.L, K.H., J.N., K.K., U.H., R.M., L.L., C.R.N., T.I., Z.C., E.C., C.J., Y.L. and M.S.; Performed statistical analysis: V.P.A., W.Y., ; Analyzed the data: T.M., R.N., V.P.A., W.Y., J.N., U.H., R.M., L.L., T.I., C.J., Y.L. M.S., H.H., K.S. and J.J.Y.; Contributed to reagents/materials/analysis tools: F.A., K.K., Y.K., M.K., K.K., C.R.N., S.K., Z.C., E.C., D.B., H.I., C.H.P., M.V.R., A.M., H.H., K.S. and A.E.J.Y.; Wrote the paper: T.M., W.E.E. and J.J.Y.
Competing Financial Interests Statement
The authors have no competing financial interest to disclose.
URLs
Japanese Pediatric Leukemia/Lymphoma Study Group ALL-B12 (UMIN000009339), http://www.umin.ac.jp; R version 3.0, http://www.r-project.org; 1,000 Genomes Project, http://www.1000genomes.org; the Broad Institute Exome Aggregation Consortium, http://exac.broadinstitute.org
References
- 1.Karran P, Attard N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nat Rev Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29:551–65. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vora A, et al. Treatment reduction for children and young adults with low-risk acute lymphoblastic leukaemia defined by minimal residual disease (UKALL 2003): a randomised controlled trial. Lancet Oncol. 2013;14:199–209. doi: 10.1016/S1470-2045(12)70600-9. [DOI] [PubMed] [Google Scholar]
- 4.Reinisch W, et al. Azathioprine versus mesalazine for prevention of postoperative clinical recurrence in patients with Crohn’s disease with endoscopic recurrence: efficacy and safety results of a randomised, double-blind, double-dummy, multicentre trial. Gut. 2010;59:752–9. doi: 10.1136/gut.2009.194159. [DOI] [PubMed] [Google Scholar]
- 5.Maltzman JS, Koretzky GA. Azathioprine: old drug, new actions. J Clin Invest. 2003;111:1122–4. doi: 10.1172/JCI18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg R, Irving PM. Toxicity and response to thiopurines in patients with inflammatory bowel disease. Expert Rev Gastroenterol Hepatol. 2015:1–10. doi: 10.1586/17474124.2015.1039987. [DOI] [PubMed] [Google Scholar]
- 7.Koren G, et al. Systemic exposure to mercaptopurine as a prognostic factor in acute lymphocytic leukemia in children. N Engl J Med. 1990;323:17–21. doi: 10.1056/NEJM199007053230104. [DOI] [PubMed] [Google Scholar]
- 8.Elion GB. The purine path to chemotherapy. Science. 1989;244:41–7. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- 9.Duration and intensity of maintenance chemotherapy in acute lymphoblastic leukaemia: overview of 42 trials involving 12 000 randomised children. Lancet. 1996;347:1783–8. doi: 10.1016/s0140-6736(96)91615-3. [DOI] [PubMed] [Google Scholar]
- 10.Lennard L, Lilleyman JS. Variable mercaptopurine metabolism and treatment outcome in childhood lymphoblastic leukemia. J Clin Oncol. 1989;7:1816–23. doi: 10.1200/JCO.1989.7.12.1816. [DOI] [PubMed] [Google Scholar]
- 11.Lilleyman JS, Lennard L. Mercaptopurine metabolism and risk of relapse in childhood lymphoblastic leukaemia. Lancet. 1994;343:1188–90. doi: 10.1016/s0140-6736(94)92400-7. [DOI] [PubMed] [Google Scholar]
- 12.Relling MV, Hancock ML, Boyett JM, Pui CH, Evans WE. Prognostic importance of 6-mercaptopurine dose intensity in acute lymphoblastic leukemia. Blood. 1999;93:2817–23. [PubMed] [Google Scholar]
- 13.Schmiegelow K, Nielsen SN, Frandsen TL, Nersting J. Mercaptopurine/Methotrexate maintenance therapy of childhood acute lymphoblastic leukemia: clinical facts and fiction. J Pediatr Hematol Oncol. 2014;36:503–17. doi: 10.1097/MPH.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teml A, Schaeffeler E, Herrlinger KR, Klotz U, Schwab M. Thiopurine treatment in inflammatory bowel disease: clinical pharmacology and implication of pharmacogenetically guided dosing. Clin Pharmacokinet. 2007;46:187–208. doi: 10.2165/00003088-200746030-00001. [DOI] [PubMed] [Google Scholar]
- 15.Candy S, et al. A controlled double blind study of azathioprine in the management of Crohn’s disease. Gut. 1995;37:674–8. doi: 10.1136/gut.37.5.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser AG, Orchard TR, Jewell DP. The efficacy of azathioprine for the treatment of inflammatory bowel disease: a 30 year review. Gut. 2002;50:485–9. doi: 10.1136/gut.50.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanauer SB, et al. Postoperative maintenance of Crohn’s disease remission with 6-mercaptopurine, mesalamine, or placebo: a 2-year trial. Gastroenterology. 2004;127:723–9. doi: 10.1053/j.gastro.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Connell WR, Kamm MA, Ritchie JK, Lennard-Jones JE. Bone marrow toxicity caused by azathioprine in inflammatory bowel disease: 27 years of experience. Gut. 1993;34:1081–5. doi: 10.1136/gut.34.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Boer NK, van Bodegraven AA, Jharap B, de Graaf P, Mulder CJ. Drug Insight: pharmacology and toxicity of thiopurine therapy in patients with IBD. Nat Clin Pract Gastroenterol Hepatol. 2007;4:686–94. doi: 10.1038/ncpgasthep1000. [DOI] [PubMed] [Google Scholar]
- 20.de Jong DJ, Goullet M, Naber TH. Side effects of azathioprine in patients with Crohn’s disease. Eur J Gastroenterol Hepatol. 2004;16:207–12. doi: 10.1097/00042737-200402000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Hindorf U, Lindqvist M, Hildebrand H, Fagerberg U, Almer S. Adverse events leading to modification of therapy in a large cohort of patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2006;24:331–42. doi: 10.1111/j.1365-2036.2006.02977.x. [DOI] [PubMed] [Google Scholar]
- 22.Posthuma EF, et al. Fatal infectious mononucleosis: a severe complication in the treatment of Crohn’s disease with azathioprine. Gut. 1995;36:311–3. doi: 10.1136/gut.36.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwab M, et al. Azathioprine therapy and adverse drug reactions in patients with inflammatory bowel disease: impact of thiopurine S-methyltransferase polymorphism. Pharmacogenetics. 2002;12:429–36. doi: 10.1097/00008571-200208000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Fotoohi AK, Coulthard SA, Albertioni F. Thiopurines: factors influencing toxicity and response. Biochem Pharmacol. 2010;79:1211–20. doi: 10.1016/j.bcp.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Hedeland RL, et al. DNA incorporation of 6-thioguanine nucleotides during maintenance therapy of childhood acute lymphoblastic leukaemia and non-Hodgkin lymphoma. Cancer Chemother Pharmacol. 2010;66:485–91. doi: 10.1007/s00280-009-1184-5. [DOI] [PubMed] [Google Scholar]
- 26.Ebbesen MS, et al. Incorporation of 6-thioguanine nucleotides into DNA during maintenance therapy of childhood acute lymphoblastic leukemia-the influence of thiopurine methyltransferase genotypes. J Clin Pharmacol. 2013;53:670–4. doi: 10.1002/jcph.81. [DOI] [PubMed] [Google Scholar]
- 27.Diouf B, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat Med. 2011;17:1298–303. doi: 10.1038/nm.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krynetskaia NF, et al. Msh2 deficiency attenuates but does not abolish thiopurine hematopoietic toxicity in msh2−/− mice. Mol Pharmacol. 2003;64:456–65. doi: 10.1124/mol.64.2.456. [DOI] [PubMed] [Google Scholar]
- 29.Swann PF, et al. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–11. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 30.Tidd DM, Paterson AR. Distinction between inhibition of purine nucleotide synthesis and the delayed cytotoxic reaction of 6-mercaptopurine. Cancer Res. 1974;34:733–7. [PubMed] [Google Scholar]
- 31.Krynetski EY, Krynetskaia NF, Bianchi ME, Evans WE. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70, ERp60, and glyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res. 2003;63:100–6. [PubMed] [Google Scholar]
- 32.Tzoneva G, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–71. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer JA, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45:290–4. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Relling MV, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–8. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 35.Lennard L, Lilleyman JS, Van Loon J, Weinshilboum RM. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–9. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 36.Evans WE, et al. Preponderance of thiopurine S-methyltransferase deficiency and heterozygosity among patients intolerant to mercaptopurine or azathioprine. J Clin Oncol. 2001;19:2293–301. doi: 10.1200/JCO.2001.19.8.2293. [DOI] [PubMed] [Google Scholar]
- 37.Gardiner SJ, Gearry RB, Begg EJ, Zhang M, Barclay ML. Thiopurine dose in intermediate and normal metabolizers of thiopurine methyltransferase may differ three-fold. Clin Gastroenterol Hepatol. 2008;6:654–60. doi: 10.1016/j.cgh.2008.02.032. quiz 604. [DOI] [PubMed] [Google Scholar]
- 38.Regueiro M, Mardini H. Determination of thiopurine methyltransferase genotype or phenotype optimizes initial dosing of azathioprine for the treatment of Crohn’s disease. J Clin Gastroenterol. 2002;35:240–4. doi: 10.1097/00004836-200209000-00008. [DOI] [PubMed] [Google Scholar]
- 39.Yates CR, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126:608–14. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 40.Krynetski EY, et al. A single point mutation leading to loss of catalytic activity in human thiopurine S-methyltransferase. Proc Natl Acad Sci U S A. 1995;92:949–53. doi: 10.1073/pnas.92.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tai HL, et al. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 42.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–4. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 43.Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol. 2010;11:507–9. doi: 10.1016/S1470-2045(10)70097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arico M, et al. The seventh international childhood acute lymphoblastic leukemia workshop report: Palermo, Italy, January 29–30, 2005. Leukemia. 2005;19:1145–52. doi: 10.1038/sj.leu.2403783. [DOI] [PubMed] [Google Scholar]
- 45.Schmiegelow K, et al. Thiopurine methyltransferase activity is related to the risk of relapse of childhood acute lymphoblastic leukemia: results from the NOPHO ALL-92 study. Leukemia. 2009;23:557–64. doi: 10.1038/leu.2008.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang SK, et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet. 2014;46:1017–20. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang JJ, et al. Inherited NUDT15 Variant Is a Genetic Determinant of Mercaptopurine Intolerance in Children With Acute Lymphoblastic Leukemia. J Clin Oncol. 2015;33:1235–42. doi: 10.1200/JCO.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kham SK, et al. Thiopurine S-methyltransferase activity in three major Asian populations: a population-based study in Singapore. Eur J Clin Pharmacol. 2008;64:373–9. doi: 10.1007/s00228-007-0426-x. [DOI] [PubMed] [Google Scholar]
- 49.Kakuta Y, et al. NUDT15 R139C causes thiopurine-induced early severe hair loss and leukopenia in Japanese patients with IBD. Pharmacogenomics J. 2015 doi: 10.1038/tpj.2015.43. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka Y, et al. Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br J Haematol. 2015 doi: 10.1111/bjh.13518. [DOI] [PubMed] [Google Scholar]
- 51.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89:387–91. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter M, et al. Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2. Nat Commun. 2015;6:7871. doi: 10.1038/ncomms8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tai HL, Krynetski EY, Schuetz EG, Yanishevski Y, Evans WE. Enhanced proteolysis of thiopurine S-methyltransferase (TPMT) encoded by mutant alleles in humans (TPMT*3A, TPMT*2): mechanisms for the genetic polymorphism of TPMT activity. Proc Natl Acad Sci U S A. 1997;94:6444–9. doi: 10.1073/pnas.94.12.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takagi Y, et al. Human MTH3 (NUDT18) protein hydrolyzes oxidized forms of guanosine and deoxyguanosine diphosphates: comparison with MTH1 and MTH2. J Biol Chem. 2012;287:21541–9. doi: 10.1074/jbc.M112.363010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLennan AG, Cartwright JL, Gasmi L. The human NUDT family of nucleotide hydrolases. Enzymes of diverse substrate specificity. Adv Exp Med Biol. 2000;486:115–8. doi: 10.1007/0-306-46843-3_23. [DOI] [PubMed] [Google Scholar]
- 56.Jordheim LP, Durantel D, Zoulim F, Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov. 2013;12:447–64. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- 57.Antillón-Klussmann FVP, Garrido C, Castellanos M, De Alarcon P, Ribeiro R. Treatment for acute lymphoblastic leukemia in limited income country: the experience of the Unidad Nacional de Oncologia Pediatrica (UNOP) of Guatemala. Pediatr Blood Cancer. 2010;55:861. [Google Scholar]
- 58.Yeoh AE, et al. Minimal residual disease-guided treatment deintensification for children with acute lymphoblastic leukemia: results from the Malaysia-Singapore acute lymphoblastic leukemia 2003 study. J Clin Oncol. 2012;30:2384–92. doi: 10.1200/JCO.2011.40.5936. [DOI] [PubMed] [Google Scholar]
- 59.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang JJ, Bhojwani D. Thiopurine S-methyltransferase pharmacogenetics in childhood acute lymphoblastic leukemia. Methods Mol Biol. 2013;999:273–84. doi: 10.1007/978-1-62703-357-2_20. [DOI] [PubMed] [Google Scholar]
- 61.Xu H, et al. Inherited Coding Variants at the CDKN2A Locus Influence Susceptibility to Acute Lymphoblastic Leukemia in Children. Nature Communications. 2015 doi: 10.1038/ncomms8553. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofmann U, et al. Simultaneous quantification of eleven thiopurine nucleotides by liquid chromatography-tandem mass spectrometry. Anal Chem. 2012;84:1294–301. doi: 10.1021/ac2031699. [DOI] [PubMed] [Google Scholar]
- 63.Jacobsen JH, Schmiegelow K, Nersting J. Liquid chromatography-tandem mass spectrometry quantification of 6-thioguanine in DNA using endogenous guanine as internal standard. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;881–882:115–8. doi: 10.1016/j.jchromb.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 64.Holleman A, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–42. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 65.Higgins JP, Thompson SG, Spiegelhalter DJ. A re-evaluation of random-effects meta-analysis. J R Stat Soc Ser A Stat Soc. 2009;172:137–159. doi: 10.1111/j.1467-985X.2008.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.