Abstract
Background
The NAC (NAM, ATAF and CUC) transcriptional factors constitute a large family with more than 150 members in rice and some of them have been demonstrated to play crucial roles in plant abiotic stress response. Here, we report the characterization of a rice stress-responsive NAC gene, ONAC095, and the exploration of its function in drought and cold stress tolerance.
Results
Expression of ONAC095 was up-regulated by drought stress and abscisic acid (ABA) but down-regulated by cold stress. ONAC095 protein had transactivation activity and the C2 domain in C-terminal was found to be critical for transactivation activity. Transgenic rice lines with overexpression of ONAC095 (ONAC095-OE) and dominant chimeric repressor-mediated suppression of ONAC095 (ONAC095-SRDX) were generated. The ONAC095-OE plants showed comparable phenotype to wild type under drought and cold stress conditions. However, the ONAC095-SRDX plants displayed an improved drought tolerance but exhibited an attenuated cold tolerance. The ONAC095-SRDX plants had decreased water loss rate, increased proline and soluble sugar contents, and up-regulated expression of drought-responsive genes under drought condition, whereas the ONAC095-SRDX plants accumulated excess reactive oxygen species, increased malondialdehyde content and down-regulated expression of cold-responsive genes under cold condition. Furthermore, ONAC095-SRDX plants showed an increased ABA sensitivity, contained an elevated ABA level, and displayed altered expression of ABA biosynthetic and metabolic genes as well as some ABA signaling-related genes.
Conclusion
Functional analyses through dominant chimeric repressor-mediated suppression of ONAC095 demonstrate that ONAC095 plays opposite roles in drought and cold stress tolerance, acting as a negative regulator of drought response but as a positive regulator of cold response in rice.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0897-y) contains supplementary material, which is available to authorized users.
Keywords: Abscisic acid (ABA), Cold tolerance, Drought tolerance, NAC transcription factor, ONAC095, Rice (Oryza sativa L.)
Background
Environmental constraints such as drought, salt and extreme temperatures often affect adversely plant growth and development, which lead to great loss of productivity worldwide [1]. Extensive studies have revealed that plants can timely sense external signals and initiate effectively complicated signaling networks to respond to environmental stress by activating various cellular, physiological, biochemical and metabolic processes [2–4]. Abscisic acid (ABA), as a critical stress phytohormone, plays important roles in abiotic stress signaling networks, and the ABA-mediated stress signaling can be divided into ABA-dependent and ABA-independent pathways [4, 5]. A number of key genes that are involved in the ABA-dependent and ABA-independent stress pathways have been identified, including DRE-binding protein/C-repeat-binding factor (CBF), ABA-binding factor, MYC and MYB [4, 5]. In addition, stress-induced reactive oxygen species (ROS), including hydrogen peroxide (H2O2) and superoxide anion, are harmful by-products causing cellular oxidative damage at excess level [6], whereas ROS are also considered to play signaling roles in regulating abiotic stress response at proper cellular concentration [6–8].
Upon perception of environmental stimuli, stress-initiated signaling network often effectively and timely reprograms the expression of a large spectrum of stress-responsive genes [4, 9, 10]. For example, a total of 5866 genes (2145 up-regulated and 3721 down-regulated), accounting for ~18 % of the annotated genes in rice genome, were differentially expressed during drought stress in rice [11]. Such large proportion of differentially expressed genes during a specific abiotic stress response requires a synergistic action of different types of transcription factors (TFs) in both temporal and spatial manners. Genetic and molecular studies using knockout/knockdown mutants and/or overexpression lines have revealed that many families of TFs such as NAC, AP2/ERF, MYB, WRKY, bZIP, homeodomain, bHLH, NF-Y and CAMTA have members that play roles in abiotic stress response [12–17]. It was also suggested that some of the functionally characterized TF genes may have great potentials in improvement of abiotic stress tolerance in crop plants [18].
NAC proteins are plant-specific TFs [19] and constitute a large family with 151 members in rice [20–22]. The NAC TFs contain a highly conserved NAC domain at N-terminal, which determines DNA-binding activity, and a variable domain at C-terminal, which is responsible for transcription activity [19]. Beside the involvement in growth and development [23], the function of the NAC TFs in biotic and abiotic stress responses has been well documented in both model and crop plants [14, 15, 17, 24]. Transcriptional profiling revealed that a relative large portion of the Arabidopsis and rice NAC TF families exhibited differential expression patterns in response to various biotic and abiotic stresses [25–27]. Up to date, six rice NAC genes, e.g. ONAC048 (OsNAC6), ONAC048 (OsNAC111), ONAC122, ONAC131, ONAC054 (RIM1) and ONAC068 (OsNAC4), have been reported to be involved in pathogen defense response [28–31]. Meanwhile, at least 7 NAC genes including ONAC002 (SANC1/OsNAC9), ONAC048 (SNAC2/OsNAC6), ONAC009 (OsNAC5), ONAC122 (OsNAC10), ONAC045, ONAC058 (OsNAP) and ONAC022 have been shown to play roles in abiotic stress tolerance [32–42]. Overexpression of some of these NAC TF genes in transgenic rice improved significantly the drought and salinity tolerance and the ABA-mediated signaling pathway [32, 33, 35, 39–41], stomatal movement and root system [32, 35, 37–39] are involved in the improved abiotic stress tolerance in the transgenic plants.
In our previous study, a number of stress-responsive ONAC genes in rice response to biotic and abiotic stresses were identified through analysis of publicly available microarray data [26, 28]. In the present study, we performed a detailed functional analysis of ONAC095 in abiotic stress tolerance by overexpression and dominant chimeric repressor-mediated suppression of ONAC095 in transgenic rice. Our results revealed that dominant chimeric repressor-mediated suppression of ONAC095 function confers an improved drought tolerance but results in an attenuated cold tolerance in rice, demonstrating that ONAC095 plays opposite roles in drought and cold stress response.
Results
ONAC095 is a drought- and ABA-up-regulated but cold-down-regulated NAC gene
The ONAC095 gene (LOC_Os06g51070) encodes a 292 aa protein with a typical N-terminal NAC domain, which can be divided into 5 subdomains (A to E) [20] (Fig. 1a). Although the sequence outside the NAC domain is divergent, two conserved C1 and C2 domains [43] are present in the C-terminal region of the ONAC095 protein (Fig. 1a). ONAC095 is closely related to rice ONAC022 and Arabidopsis ANAC036, showing 62.4 % and 52.0 % of identity, respectively. The ONAC095 protein contains eight putative phosphorylation sites with probability of >90 %, including five serine (S) residues at positions of 8, 96, 181, 218 and 221 aa, two threonine (T) residues at positions of 99 and 199 aa, and one tyrosine (Y) residue at position of 286 aa (Fig. 1a). Bioinformatics analysis indicated that several stress-responsive cis-elements including 1 GCC box, 4 MYC recognition sites, 6 MYB recognition sites and 12 W-boxes are present in the promoter region (1.5 Kb upstream of the start codon) of the ONAC095 gene (Fig. 1b). We examined by quantitative real time-PCR (qRT-PCR) the responsiveness of ONAC095 to abiotic stress and ABA. Expression of ONAC095 in detached leaves was significantly and rapidly up-regulated within 3 hr by fast dehydration, giving 3.9 and 2.2 folds of increase at 1 and 3 hr after drought treatment, respectively (Fig. 1c). An increase of 2.1 folds in ONAC095 expression level was observed at 1 hr after treatment with 150 mM NaCl but the expression decreased rapidly to basal level at 3 hr after treatment (Fig. 1c). By contrast, the expression of ONAC095 was down-regulated gradually by cold (4 °c) treatment, showing ~5 folds of decrease at 12 and 24 hr after treatment (Fig. 1c). In heat (42 °c)-treated plants, the expression of ONAC095 increased within 12 hr and decreased to basal level at 24 hr after treatment (Fig. 1c). Significant induction of ONAC095 expression in ABA-treated plants was also observed, showing 3.3 folds of increase at 6 hr after treatment (Fig. 1c). These data suggest that ONAC095 is a drought- and ABA-up-regulated but cold-down-regulated stress-responsive rice NAC gene.
Fig. 1.
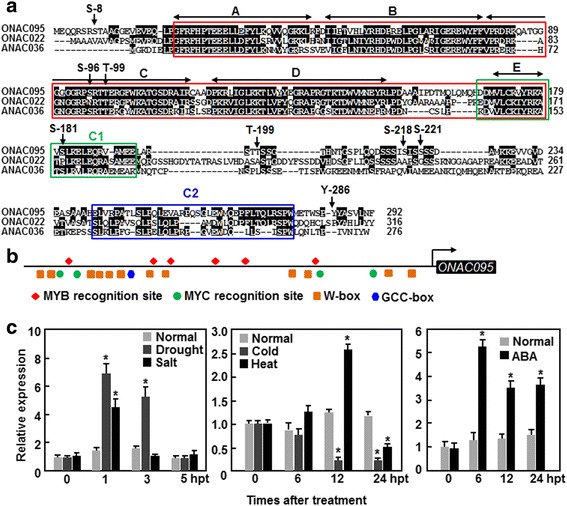
Structural feature of ONAC095, cis-elements in promoter of ONAC095 and stress-responsive expression of ONAC095. a Alignment of ONAC095 with rice ONAC022 and Arabidopsis ANAC036. The conserved NAC domain is boxed in red and the five highly conserved subdomains A to E are indicated by black arrowed lines. C1 and C2 domains are boxed with green and blue lines, respectively. Putative phosphorylation sites are indicated by arrows. S, serine; T, threonine; Y, tyrosine. b Distribution of major stress-related cis-elements in the promoter (1.5 Kb upstream of the start codon) of the ONAC095 gene. c Stress-responsive expression of ONAC095 by drought, salt, cold and heat stresses as well as by exogenous ABA. Drought stress was applied to two-week-old seedlings by transferring the seedlings to three layers filter papers for fast dehydration. Salt stress was applied to the seedlings by rooting the seedlings in 150 mM NaCl solution. Cold and heat stresses were applied by placing the seedlings in growth chambers with temperatures set at 4 °C or 42 °C, respectively. For ABA treatment, the seedlings were sprayed with 100 μM ABA or similar volume of solution as controls. Relative expression levels of ONAC095 were normalized by the transcript level of the Actin gene and the expression level was set as 1 at 0 hr after treatment. Data presented in c are the means ± SD from three independent experiments and columns with an asterisk indicate significant difference at p < 0.05 level between the treatments and normal controls
ONAC095 has transactivation activity that is determined by two conserved proline residues in the C-terminal C2 domain
To examine whether ONAC095 had transactivation activity, the full ONAC095 protein, a C-terminal-truncated N-terminal fragment ONAC095-N (lacking 152–292 aa at C-terminal) and an N-terminal-truncated C-terminal fragment ONAC095-C (lacking 1–151 aa at N-terminal) were each fused to the GAL4 DNA-binding domain in pBD vector (Fig. 2a). Yeasts harboring GAL4-ONAC095, ONAC095-N or ONAC095-C all grew on SD/Trp− medium (Fig. 2b). However, only yeasts harboring GAL4-ONAC095 or GAL4-ONAC095-C grew while yeasts carrying GAL4-ONAC095-N and empty pBD vector did not grow on SD/Trp−His− medium containing 4 mM 3-amino-1,2,4-triazole (3-AT) (Fig. 2b). Yeasts harboring GAL4-ONAC095 or GAL4-ONAC095-C showed significant β-galactosidase activity after addition of X-α-gal (Fig. 2b), indicating that ONAC095 had transactivation activity and the C-terminal is responsible for its transactivation activity. We then mapped the putative sequence responsible for transactivation activity in C-terminal by testing a series of truncated C-terminal constructs for their transactivation activity (Fig. 2a). Yeasts carrying GAL4-ONAC095-CΔ1 (lacking 259–292 aa from C-terminal) grew on SD/Trp−His− medium and displayed β-galactosidase activity (Fig. 2b). By contrast, yeasts carrying GAL4-ONAC095-CΔ2 (lacking 224–292 aa from C-terminal) or GAL4-ONAC095-CΔ3 (lacking 189–292 aa from C-terminal) did not grow on SD/Trp−His− medium and did not show β-galactosidase activity (Fig. 2b), suggesting that the specific sequence between 224–292 aa in C-terminal of ONAC095 is responsible for transactivation activity. To examine the possibility whether a part of the C2 domain may be the determinant responsible for the transactivation activity, we further tested the transactivation activity of truncated constructs GAL4-ONAC095-CΔC2 (lacking 242–292 aa from C-terminal), in which the C2 domain was fully deleted, and GAL4-ONAC095-C2, which spanned 242–278 aa containing the complete C2 domain (Fig. 2a). As shown in Fig. 2b, yeasts harboring GAL4-ONAC095-CΔC2 did not grow on SD/Trp−His− medium and did not show β-galactosidase activity while yeasts harboring GAL4-ONAC095-C2 grew on SD/Trp−His− medium and showed β-galactosidase activity, confirming that the C2 domain is responsible for transactivation activity of ONAC095. Because yeasts harboring GAL4-ONAC095-CΔ1 had transactivation activity, it is possible that the specific sequence for transactivation activity is located between 242–258 aa of ONAC095, a region containing five conserved amino acid residues in a consensus of xLxxPxxxxLPxLxxxx when aligned with ONAC022 and ANAC036 (Fig. 1a). To determine the importance of these five conserved residues in the transactivation activity, we constructed a series of mutated versions, ONAC095-C2-M1-5, in which the leucine (L) residues at 243, 251 and 254 aa and the proline (P) residues at 246 and 252 aa in 242–258 aa region were individually replaced with arginine (R) (Fig. 2c) and tested for their transactivation activity. As shown in Fig. 2d, yeasts harboring GAL4-ONAC095-C2-M2 or GAL4-ONAC095-C2-M4 did not grow on SD/Trp−His− medium and did not show β-galactosidase activity, whereas yeasts harboring GAL4-ONAC095-C2-M1, GAL4-ONAC095-C2-M3 or GAL4-ONAC095-C2-M5 did grow and show β-galactosidase activity, demonstrating that the conserved proline residues at 246 and 252 aa are critical and required for the transactivation activity of ONAC095.
Fig. 2.
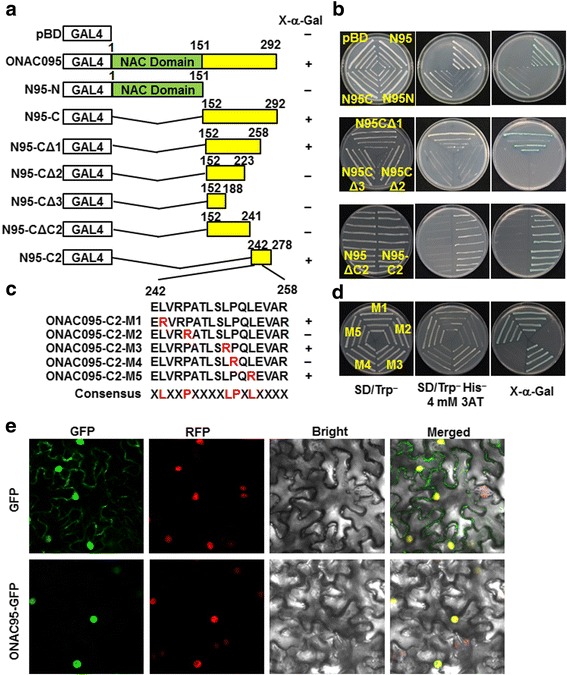
Transactivation activity and nuclear localization of ONAC095. a–d Transactivation activity and mapping of the specific sequence responsible for transactivation activity in ONAC095. a and c Diagrams showing different truncated constructs and mutated amino acid residues in C2 domain, respectively. b and d Transcription activity assay for truncated and mutated constructs of ONAC095 in yeast. Yeasts harboring different truncated and mutated constructs and empty vector were grown on the SD/Trp− plates or SD/Trp− His− with 4 mM 3-AT for 3 days at 30 °C. Transactivation activity was examined by the growth ability and production of blue pigment after addition of X-α-gal in the SD/Trp− His− plates for 1 day. e ONAC095 is localized in nucleus. Agrobacteria harboring pFGC-ONAC095 or pFGC-eGFP were infiltrated into leaves of N. benthamiana plants expressing a red nucleus marker protein RFP-H2B and leaf samples were collected at 24 hr after agroinfiltration. Microscopic examination was performed under a confocal laser scanning microscope in dark field for green fluorescence (left), red fluorescence (middle left), white field for cell morphology (middle right) and in combination (right), respectively
ONAC095 is a nucleus-localized protein
To examine the subcellular localization of ONAC095, the coding sequence of ONAC095 was fused in-frame with GFP at N-terminal in pFGC-EGFP vector and transiently expressed in leaves of Nicotiana benthamiana plants harboring a red nuclear marker RFP–H2B protein [44]. Microscopic observations of the agroinfiltrated N. benthamiana leaves collected at 24 hr after agroinfiltration revealed that the GFP:ONAC095 fusion was solely localized in nucleus, co-localized with the known nuclear marker RFP–H2B protein (Fig. 2e), whereas GFP alone distributed ubiquitously throughout the cell without specific compartmental localization (Fig. 2e). These data indicate that ONAC095 is a nucleus-localized protein.
Generation and characterization of ONAC095 overexpression and dominant chimeric repressor-mediated suppression transgenic lines
To explore the function of ONAC095 in abiotic stress tolerance, we generated transgenic rice lines with overexpression of ONAC095 or dominant chimeric repressor-mediated suppression of ONAC095 function. A maize ubiquitin promoter-driven overexpression construct ONAC095-OE was made by inserting the ONAC095 coding sequence into a modified binary vector PU1301 (Fig. 3a). Considering that functional redundancy often occurs in some of NAC TFs [31, 45], a dominant chimeric repressor-mediated suppression construct ONAC095-SRDX was also made by fusing the ONAC095 coding sequence at its C-terminal to a plant-specific transcriptional repression domain [46] (Fig. 3a). The ONAC095-OE and ONAC095-SRDX constructs were introduced into rice cv. Xiushui 134 calli through Agrobacterium-mediated transformation method. Eighteen independent ONAC095-OE lines and 21 independent ONAC095-SRDX lines were obtained. After screening phenotype and segregation of hygromycin (Hgr) resistance on 1/2 MS medium in T2 and T3 generations, two overexpression lines ONAC095-OE6 and ONAC095-OE12 and two dominant chimeric repressor-mediated suppression lines ONAC095-SRDX2 (S2) and ONAC095-SRDX3 (S3) were identified as single-copy homozygous transgenic lines. Southern blotting of genomic DNA probed with a fragment of the HptII gene confirmed that each of these selected ONAC095-OE and ONAC095-SRDX lines contained a single copy of the transgenic construct (Fig. 3b). qRT-PCR analysis revealed that the transcript levels of ONAC095 in T3 generation plants of ONAC095-OE6 and ONAC095-OE12 lines were ~11 and ~57 times higher than that in wild-type (WT) plants, respectively, whereas the transcript levels of ONAC095-SRDX in T3 generation plants of ONAC095-SRDX2 and ONAC095-SRDX3 lines were ~14 and ~18 times higher over that in WT, respectively (Fig. 3c). Considering that ONAC022 is closely related to ONAC095 [42], we also examined whether altered expression of ONAC095 in transgenic plants affected the expression of ONAC022. qRT-PCR data showed that the expression level of ONAC022 in ONAC095-OE and ONAC095-SRDX plants was comparable to that in WT (Fig. 3d), indicating that altered expression of ONAC095 does not affect the expression of ONAC022 in transgenic rice. We did not observe any difference in plant height and root length between ONAC095-OE and WT plants grown in greenhouse (Fig. 3e–g). However, we noticed that ONAC095-SRDX plants showed growth retardation (Fig. 3e), leading to 11–15 % of reduction in plant height (Fig. 3f), as compared to WT. The root lengths and 1000-grain weights from ONAC095-OE and ONAC095-SRDX plants grown in greenhouse were comparable to WT (Fig. 3g and h). Thus, it is likely that dominant chimeric repressor-mediated suppression of ONAC095 function has a negative impact on rice growth and development.
Fig. 3.
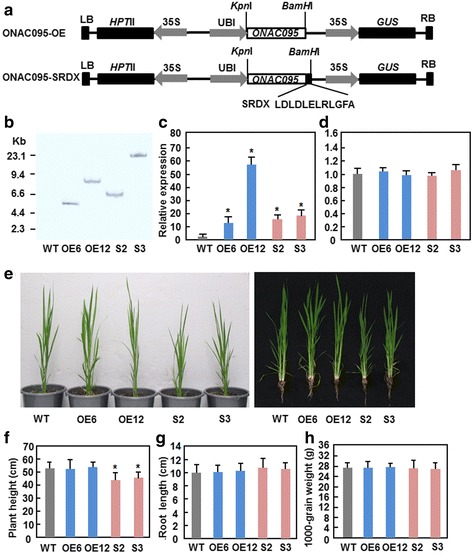
Characterization of ONAC095-OE and ONAC095-SRDX transgenic rice lines and their growth phenotypes. a Schematic diagrams showing the overexpression ONAC095-OE and the dominant chimeric repressor-mediated suppression ONAC095-SRDX constructs used for transformation. HptII, hygromycin phosphotransferase II; LB, left border; RB, right border; Ubi, maize ubiquitin promoter; 35S, CaMV 35S promoter; GUS, β-glucuronidase. b Confirmation of single-copy transgenic lines by Southern blot analysis. Fifty micrograms of genomic DNA were digested with EcoRI and probed with a DIG-labeled fragment of the HptII gene. c Transcript levels of ONAC095 and ONAC095-SRDX in ONAC095-OE and ONAC095-SRDX transgenic lines. Leaf samples from two-week-old seedlings were used for analysis of the transcript levels by qRT-PCR. d Transcript levels of ONAC022 in ONAC095-OE and ONAC095-SRDX transgenic lines. Leaf samples from two-week-old seedlings were used for analysis of the transcript levels by qRT-PCR. e Growth phenotype of two-month-old ONAC095-OE and ONAC095-SRDX plants grown under normal watered condition in greenhouse. f and g Plant height and root length of two-month-old ONAC095-OE and ONAC095-SRDX plants grown under normal watered condition in greenhouse. h Weights of 1000-grain from ONAC095-OE and ONAC095-SRDX plants grown under normal watered condition in greenhouse. Data presented (c, d, f, g) and (h) are the mean ± SD from three independent experiments and columns with an asterisk indicate significant difference at p ≤ 0.05 level between WT and OE/SRDX lines. WT, wild type; OE6, ONAC095-OE6; OE12, ONAC095-OE12; S2, ONAC095-SRDX2; S3, ONAC095-SRDX3
Dominant chimeric repressor-mediated suppression of ONAC095 function confers an improved drought tolerance
We first explored the involvement of ONAC095 in drought tolerance by phenotyping ONAC095-OE and ONAC095-SRDX plants under drought condition and comparing with WT. In our repeated drought stress experiments, drought symptom, represented by rolled leaves and wilted plants, in ONAC095-OE lines at 20 days after drought treatment and at 7 days after recovery of watering was indistinguishable from WT (Fig. 4a), indicating that overexpression of ONAC095 in transgenic rice does not affect the drought tolerance. By contrast, drought symptom in ONAC095-SRDX plants at 20 days after drought treatment and at 7 days after recovery of watering was markedly less severe than WT (Fig. 4a). At 7 days after recovery of watering, the survival rate of ONAC095-SRDX plants was ~30 % higher than WT (Fig. 4b). To explore the possible mechanism responsible for the improved drought stress tolerance in ONAC095-SRDX plants, we analyzed and compared some stress-related physiological and biochemical changes and the expression of several selected drought stress-responsive genes between ONAC095-SRDX and WT plants grown under normally watered and/or drought stressed conditions. The rate of water loss, as calculated from the relative water content (RWC), in detached leaves of ONAC095-SRDX plants decreased by 9–15 %, as compared with WT, at 2 and 3 hr after detachment (Fig. 4c). Under normally watered condition, the contents of proline and soluble sugars in ONAC095-SRDX plants were comparable to those in WT (Fig. 4d and e). However, the contents of proline and soluble sugars in ONAC095-SRDX and WT plants at 10 days under drought stressed condition were increased significantly as compared to those in plants grown under normally watered condition (Fig. 4d and e). Further, the increase in contents of proline and soluble sugars in ONAC095-SRDX plants was much evident than those in WT under drought stressed condition, resulting in increase of 30–43 % for proline content and 28–31 % for soluble sugar content, respectively (Fig. 4d and e). Similarly, the expression levels of OsPP2C28, a member of the PP2C family known to be involved in abiotic stress response [47], OsbZIP23 and OsAP37, two stress-responsive TF genes [48, 49], and OsRAB21, OsRAB16B and OsERD1 (a homolog of Arabidopsis AtERD1), three stress-related genes [50, 51], in ONAC095-SRDX and WT plants grown under normally watered condition were comparable (Fig. 4f). Under drought stressed condition, the expression of these genes was significantly up-regulated in both ONAC095-SRDX and WT plants compared to those in plants grown under normally watered condition; however, the expression levels in ONAC095-SRDX plants showed a further increase over those in WT (Fig. 4f). Together, these data indicate that dominant chimeric repressor-mediated suppression of ONAC095 function in ONAC095-SRDX plants confers an improved drought stress tolerance that may be resulted from reduced transpiration rate, increased contents of stress-related metabolites, and up-regulated expression of drought-responsive genes.
Fig. 4.
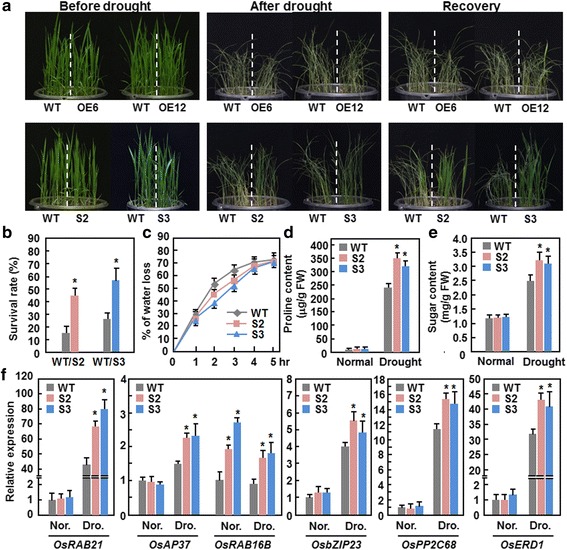
Dominant chimeric repressor-mediated suppression of ONAC095 function conferred an improved drought tolerance in ONAC095-SRDX plants. a Growth performance of ONAC095-OE, ONAC095-SRDX and WT plants at different stages during a drought stress experiment. ONAC095-OE and ONAC095-SRDX plants were grown in same barrels with WT and were subjected to drought stress treatment by withholding water for 20 days. Drought stressed plants were recovered for another 7 days by normally watering. b–e Comparison of survival rate, water loss rate, contents of proline and soluble sugars between ONAC095-SRDX and WT plants with or without drought stress treatment. Plants with >20 % green leaves were considered to be survivals (b). Rates of water loss in detached leaves of 4-week-old ONAC095-SRDX and WT plants were measured at indicated time points over a period of 5 hr after detachment (c). Leaf samples form 4-week-old ONAC095-SRDX and WT plants grown under normally watered and drought-stressed (at 6 days after water withholding) conditions were collected and measured for contents of proline (d) and soluble sugars (e). f Expression of drought stress-related genes in ONAC095-SRDX and WT plants before and after drought stress treatment. Leaf samples were collected from normally watered and drought stressed plants for 15 days. Relative expression levels were normalized by the transcript level of the Actin gene as an internal control and the expression level of each gene of interest in WT plants under normal condition was set as 1. Nor., normally watered condition; Dro., drought stressed condition. Data presented in (b–f) are the means ± SD from three independent experiments and columns with an asterisk indicate significant difference at p < 0.05 level between WT and SRDX lines. WT, wild type; OE6, ONAC095-OE6; OE12, ONAC095-OE12; S2, ONAC095-SRDX2; S3, ONAC095-SRDX3
Dominant chimeric repressor-mediated suppression of ONAC095 function attenuates cold stress tolerance
The fact that the expression of ONAC095 was down-regulated by cold stress led us to examine whether ONAC095 plays a role in cold stress tolerance by phenotyping ONAC095-OE and ONAC095-SRDX plants under cold stress condition and comparing with WT. In repeated cold stress experiments, ONAC095-OE plants displayed indistinguishable cold stress symptoms such as rolled leaves and wilted plants from those of WT at 5 days after cold (4 °C) treatment and at 7 days after recovery (Fig. 5a), indicating that overexpression of ONAC095 in transgenic rice does not affect the cold tolerance. By contrast, the ONAC095-SRDX plants showed more severe cold stress symptoms at 7 days after recovery from cold stress than those of WT (Fig. 5a). At 7 days after recovery from cold stress, <10 % of the ONAC095-SRDX plants survived while ~90 % of WT survived (Fig. 5b). To elucidate the possible mechanism responsible for the attenuated cold stress tolerance in the ONAC095-SRDX plants, we analyzed and compared some stress-related physiological changes, e.g. malondialdehyde (MDA) content, electrolyte leakage and chlorophyll content, and the expression changes of several selected cold-responsive genes between ONAC095-SRDX and WT plants grown under unstressed or cold stressed conditions. Under unstressed condition, MDA content, relative electrolyte leakage and chlorophyll content in ONAC095-SRDX plants were comparable to those in WT (Fig. 5c–e). However, MDA content and relative electrolyte leakage in ONAC095-SRDX and WT plants at 1 day under cold stressed condition were increased significantly but chlorophyll content was decreased markedly in ONAC095-SRDX and WT plants as compared to those in plants grown under unstressed condition (Fig. 5c–e). Further, increase in MDA content and relative electrolyte leakage and decrease in chlorophyll content were more evident in ONAC095-SRDX plants than those in WT under cold stressed condition, resulting in increase of 0.9–1.5 folds for MDA content and of 4.3–4.7 folds for relative electrolyte leakage and decrease of 25–30 % for chlorophyll content, respectively (Fig. 5c–e). Under unstressed condition, the expression level of OsICE1 [52] in ONAC095-SRDX plants was comparable to that in WT but the expression levels of OsWRKY76 [53] in ONAC095-SRDX plants were significantly lower than that in WT (Fig. 5f). Under cold stressed condition, the expression levels of OsWRKY76 and OsICE1 were significantly up-regulated in both ONAC095-SRDX and WT plants compared to those in plants grown under unstressed condition; however, the cold-induced expression levels in ONAC095-SRDX plants were lower than those in WT (Fig. 5f). Collectively, these data indicate that dominant chimeric repressor-mediated suppression of ONAC095 function in ONAC095-SRDX plants attenuates the cold stress tolerance that may be resulted from increased MDA content and down-regulated expression of cold-responsive genes.
Fig. 5.
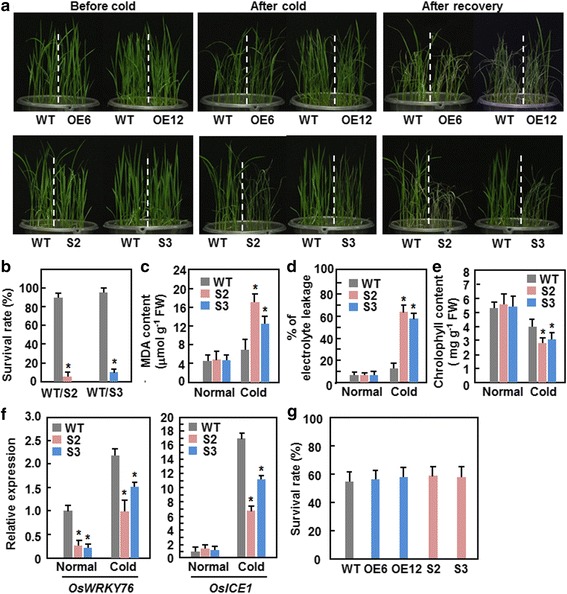
Dominant chimeric repressor-mediated suppression of ONAC095 function in ONAC095-SRDX plants attenuated cold stress tolerance but did not affect heat stress tolerance. a Growth performance of ONAC095-OE, ONAC095-SRDX and WT plants at different stages during a cold stress experiment. ONAC095-OE and ONAC095-SRDX plants were grown in same barrels with WT and were subjected to cold stress treatment in a growth chamber with temperature at 4 °C for 5 days for ONAC095-OE/WT plants or for 1 day for ONAC095-SRDX/WT plants. The cold stressed plants were recovered under normal condition for 7 days. b Survival rates of the cold stress-treated plants. c–e Comparison of MDA content (c), electrolyte leakage (d) and chlorophyll content (e) in leaves of ONAC095-SRDX and WT plants with or without cold treatment. f Expression of cold stress-related genes in ONAC095-SRDX and WT plants before and after 1 day of cold treatment. Relative expression levels were normalized by the transcript level of the Actin gene as an internal control and the expression level of the tested genes in WT plants under normal condition was set as 1. g Survival rates of the heat stress-treated plants. Three-week-old WT, ONAC095-OE and ONAC095-SRDX plants were treated with heat stress in a growth chamber with temperature of 45 °C for 5 days and survivals were recorded at 7 days after recovering under normal condition. Data presented in (b–g) are the means ± SD from three independent experiments and columns with an asterisk indicate significant difference at p < 0.05 level between WT and SRDX lines. WT, wild type; OE6, ONAC095-OE6; OE12, ONAC095-OE12; S2, ONAC095-SRDX2; S3, ONAC095-SRDX3
We also examined whether altered expression of ONAC095 affected the thermotolerance in rice by phenotyping ONAC095-OE and ONAC095-SRDX plants under heat stress condition and comparing with WT. In repeated experiments, both the ONAC095-OE and ONAC095-SRDX plants displayed indistinguishable phenotype from WT and similar survival rate to WT after heat stress treatment (Fig. 5g), indicating that overexpression of ONAC095 or dominant chimeric repressor-mediated suppression of ONAC095 function does not affect the thermotolerance in transgenic rice.
Dominant chimeric repressor-mediated suppression of ONAC095 function accelerates cold stress-induced ROS accumulation
Considering that ROS is often linked to oxidative damage during abiotic stress response, we analyzed and compared the generation and accumulation of ROS between ONAC095-SRDX and WT plants grown under unstressed and cold stressed conditions to explore the involvement of ROS in attenuated cold stress tolerance in ONAC095-SRDX plants. In situ detection of ROS by 3,3’-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining and quantitative measurement of H2O2 revealed that no significant difference in accumulation of H2O2 and superoxide anion in leaf tissues was observed between ONAC095-SRDX and WT plants grown under unstressed condition (Fig. 6a–d). At 1 days after cold (4 °C) treatment, significant accumulation of H2O2 and superoxide anion in leaves of both ONAC095-SRDX and WT plants was detected (Fig. 6a–c); however, the accumulation levels of H2O2 and superoxide anion in leaves of ONAC095-SRDX plants were more evident as compared to those in WT (Fig. 6a and b) or higher than those in WT (Fig. 6c). The levels of H2O2 in leaves of the ONAC095-SRDX and WT plants increased under drought stress condition; however, the accumulation levels of H2O2 in leaves of ONAC095-SRDX plants were comparable to that in WT (Fig. 6d). In unstressed plants, the activity of superoxide dismutase (SOD) and catalase (CAT) in ONAC095-SRDX plants was lower than that in WT (Fig. 6e and f); the decrease of SOD and CAT activity in ONAC095-SRDX plants was more evident at 1 day after cold stress treatment, leading to 30 % of decrease in activity, as compared to those in WT (Fig. 6e and f). The expression levels of OsRbohA, OsRbohG and OsRbohH genes, encoding for NADPH oxidases involved in ROS generation [7, 54], were comparable between ONAC095-SRDX and WT plants (Fig. 6g). At 1 day after cold treatment, the expression levels of OsRbohA, OsRbohG and OsRbohH in leaves of ONAC095-SRDX plants increased significantly, showing 0.6–1.0, 0.5–0.6 and 9.2–9.4 folds of increases, respectively, as compared to those in WT (Fig. 6g). Taken together, these data indicate that an abnormal generation and accumulation of ROS, due to up-regulated expression of OsRboh genes and reduced activity of antioxidant enzymes, occurs in ONAC095-SRDX plants upon cold stress, and the accumulated ROS may then attenuate the cold stress tolerance via cellular oxidative damage.
Fig. 6.
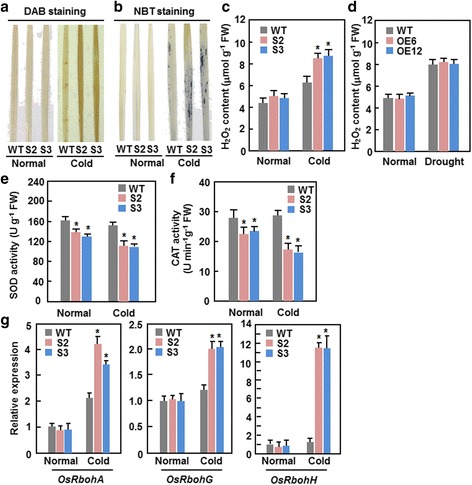
Dominant chimeric repressor-mediated suppression of ONAC095 function accelerated cold-induced ROS accumulation in ONAC095-SRDX plants. Three-week-old ONAC095-SRDX and WT plants were subjected to cold stress treatment by moving into a growth chamber with temperature of 4 °C for 1 day and leaf samples from cold stressed and unstressed plants were collected for different analyses. a and b In situ detection of H2O2 and superoxide anion in leaves by DAB and NBT staining, respectively. c and d Quantification of H2O2 level in leaves of ONAC095-SRDX and WT plants with or without cold (c) or drought (d) stress treatment. e and f Activity of SOD and CAT in leaves of ONAC095-SRDX and WT plants with or without cold stress treatment. g Expression of selected OsRboh genes in ONAC095-SRDX and WT plants with or without cold stress treatment. Relative expression levels were normalized by the transcript level of the Actin gene as an internal control and the expression level of the tested genes in WT plants under normal condition was set as 1. Data presented in (c–g) are the means ± SD from three independent experiments and columns with an asterisk indicate significant differences at p < 0.05 level between WT and OE/SRDX lines. WT, wild type; OE6, ONAC095-OE6; OE12, ONAC095-OE12; S2, ONAC095-SRDX2; S3, ONAC095-SRDX3
Dominant chimeric repressor-mediated suppression of ONAC095 function increases ABA sensitivity
The fact that expression of ONAC095 was induced by ABA led us to explore whether altered expression of ONAC095 affected ABA sensitivity in ONAC095-OE and ONAC095-SRDX plants. We first examined the ABA sensitivity of ONAC095-OE and ONAC095-SRDX lines and compared with WT by analyzing seed germination and seedling growth in presence of ABA. In the absence of ABA, seeds of ONAC095-OE, ONAC095-SRDX and WT lines germinated normally and no difference was observed among ONAC095-OE, ONAC095-SRDX and WT (Fig. 7a and b). In the presence of 5 μM ABA, however, germination of ONAC095-OE seeds was comparable to WT but germination of ONAC095-SRDX seeds was significantly inhibited, showing by 25–30 % of decrease, in comparison to WT (Fig. 7a and b). Similarly, growth of ONAC095-OE and ONAC095-SRDX seedlings in the absence of ABA were similar to WT (Fig. 7c); however, in the presence of 5 μM ABA, growth of ONAC095-OE seedlings was comparable to WT but growth of ONAC095-SRDX seedlings was significantly inhibited as compared with WT (Fig. 7c–f). Weight of single seedling, length of shoot and root of ONAC095-SRDX seedlings were decreased by 30–40 % as compared with WT in the presence of 5 μM ABA (Fig. 7d–f). These data indicate that overexpression of ONAC095 does not affect the ABA sensitivity in ONAC095-OE lines but dominant chimeric repressor-mediated suppression of ONAC095 function in ONAC095-SRDX lines results in increased ABA sensitivity. We next examined whether altered expression of ONAC095 affected the endogenous ABA level and ABA-mediated signaling in ONAC095-OE and ONAC095-SRDX lines. As shown in Fig. 7g, the endogenous ABA level in ONAC095-SRDX plants was significantly higher than that in WT, leading to 33–42 % of increase; however, no significant difference in ABA content was detected between the ONAC095-OE and WT plants. Accordingly, the expression of ABA biosynthetic gens OsNCED4 and OsNCED5 was up-regulated but the expression of an ABA metabolic gene OsABA8OX39 was down-regulated in ONAC095-SRDX plants grown under normal condition (Fig. 7h). Furthermore, the expression levels of OsPP2C30 and OsPP2C49, two PP2Cs involved in ABA signaling [47], in ONAC095-SRDX plants were also significantly up-regulated as compared to those in WT (Fig. 7h). Together, these results indicate that dominant chimeric repressor-mediated suppression of ONAC095 function affects the endogenous ABA level through regulation of the expression of ABA biosynthetic and metabolic genes and thereby modulates an activated ABA signaling in ONAC095-SRDX plants.
Fig. 7.
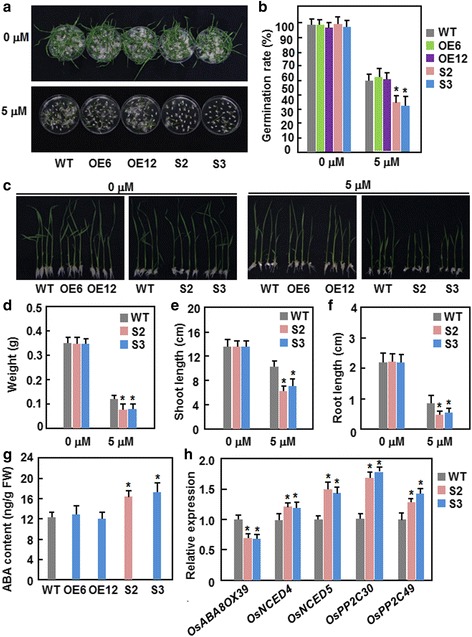
Dominant chimeric repressor-mediated suppression of ONAC095 function increased ABA sensitivity in ONAC095-SRDX lines. a and b Germination performance and rates of ONAC095-OE, ONAC095-SRDX and WT seeds on 1/2 MS medium supplemented with or without 5 μM ABA. c–f Growth performance of ONAC095-OE, ONAC095-SRDX and WT seedlings grown on 1/2 MS medium with or without 5 μM ABA. Weight of single seedling (d) and length of shoot (e) and root (f) were measured at 6 days after germination. g ABA content in two-week-old ONAC095-OE, ONAC095-SRDX and WT plants grown under normally watered condition. h Expression levels of ABA biosynthesis- and metabolism-related and ABA signaling-related genes in ONAC095-SRDX and WT plants. Relative expression levels were normalized by the transcript level of the Actin gene as an internal control and the expression level of the tested genes in WT plants under normal condition was set as 1. Data presented in (b) and (d–h) are the means ± SD from three independent experiments and columns with an asterisk indicate significant difference at p < 0.05 level between WT and OE/SRDX lines. WT, wild type; OE6, ONAC095-OE6; OE12, ONAC095-OE12; S2, ONAC095-SRDX2; S3, ONAC095-SRDX3
Discussion
NAC TFs constitute a large family with 151 members in rice [20–22]; however, only a limited number of NAC TFs have been functionally characterized so far. In the present study, we demonstrated through functional analyses using overexpression and dominant chimeric repressor-mediated suppression transgenic lines that ONAC095 plays opposite roles in drought and cold stress tolerance in rice. Furthermore, biochemical studies revealed that the C2 domain in C-terminal and two proline residues in C2 domain are critical for transactivation activity of ONAC095. Our functional and biochemical studies demonstrate that ONAC095 functions as a dual regulator of abiotic stress response in rice.
NAC TFs consist of a conserved DNA-binding NAC domain, which is responsible for the oligomerization into dimeric proteins [19], and a more divergent C-terminal region, which functions as a transcription regulatory domain [55]. Most of the previously identified NAC TFs were reported to act as transcription activators, although a few of NAC TFs were found to be transcriptional repressors [56, 57]. We showed that ONAC095 is a transcriptional activator, similar to its closely related homologous ONAC022 that can bind specifically to a canonical NAC TF recognition cis-element in vitro and act as a transcriptional activator [42]. We also found that the C-terminal of ONAC095 is responsible for its transactivation activity (Fig. 2), consistent with a common concept that NAC TFs have a C-terminal transcriptional activation domain [32, 35, 41, 42]. The ONAC095 protein, similar to its closely related homologous rice ONAC022 [42] and Arabidopsis ANAC036 [43], contains two unique conserved C1 and C2 domains (Fig. 1a), which are not present in other NAC TFs [43]. The C1 domain, consisting of the putative NAC subdomain E and its immediately downstream sequence, was proposed to be involved in DNA-binding ability [43]. In our study, however, the truncated constructs ONAC095-CΔ2 (spanning 152–223 aa) and ONAC095-CΔC2 (spanning 152–241 aa), which harbor the full C1 domain, abolished the transactivation activity (Fig. 2a and b), indicating that the C1 domain is required for transactivation activity. Our biochemical analysis of transactivation activity with a series of truncated constructs of the C-terminal of ONAC095 further identified the possible sequence region and/or critical residues that are responsible for transactivation activity in ONAC095. The truncated construct ONAC095-CΔ1 (spanning 152–258 aa), containing a part of the C2 domain, had transactivation activity but the truncated construct ONAC095-CΔC2 (spanning 152–241 aa), in which the full C2 domain was deleted, did not (Fig. 2a and b), implying that a determinant for transactivation activity exists between 242–258 aa in C-terminal. This is further supported by the observation that the C2 domain itself had transactivation activity (Fig. 2a and b). It was also found that the transactivation activity of SNAC1, which does not have a C2 domain, is located between 242–271 aa [32]. Further mutation analysis revealed that the conserved proline residues at 246 and 252 aa are critical and required for transactivation activity of ONAC095 (Fig. 2c and d). This is consistent with the observation that a single amino acid residue at 270 aa determines the transactivation activity in SNAC1 [32].
Although expression of ONAC095 was induced by drought stress as well as by ABA (Fig. 1c), dominant chimeric repressor-mediated suppression of ONAC095 function in ONAC095-SRDX plants led to higher survival ratio and better growth performance under drought stress condition (Fig. 4), demonstrating that ONAC095 acts as a negative regulator of drought tolerance in rice. This differs from previously reported rice NAC TFs that play positive roles in improving drought stress tolerance [32–42]. The mechanisms responsible for the improved drought tolerance in ONAC095-SRDX plants can be explained by several physiological, biochemical and molecular changes. Firstly, accumulation of compatible solutes such as soluble sugars and free proline is a common phenomenon in response to abiotic stress [58]. Higher levels of free proline and soluble sugars under drought stress condition (Fig. 4d and e) may partially account for the improved drought and salt tolerance. Secondly, ABA plays a critical role in regulating abiotic stress response in plants [4, 5]. Generally, high level of endogenous ABA may strengthen and/or accelerate stress response and thus correlate with improved abiotic stress tolerance [59, 60]. The ONAC095-SRDX plants contained an elevated level of endogenous ABA (Fig. 7g), which may be resulted from up-regulated expression of ABA biosynthetic genes such as OsNCEDs [61] and down-regulated expression of ABA metabolic gene like OsABA8OX39 [60] (Fig. 7h), and exhibited an increased ABA sensitivity (Fig. 7a–f). Thirdly, an increased endogenous ABA level in plants generally initiates ABA-mediated signaling pathway that regulates the expression of many stress-responsive genes [62]. The expression of some early ABA signaling and regulatory genes such as OsPP2C30/49/68 [47], OsbZIP23 [48] and OsAP37 [49] and late ABA-responsive genes such as OsRAB16B and OsRAB21 [50], was significantly up-regulated in ONAC095-SRDX plants (Figs. 4f and 7h), implying an activated ABA-mediated signaling pathway in the transgenic plants. These data suggest that dominant chimeric repressor-mediated suppression of ONAC095 function in ONAC095-SRDX plants may modify an ABA-mediated signaling pathway to regulate the expression of stress-responsive genes and thereby confer an improved drought tolerance.
Expression of ONAC095 was down-regulated by cold stress (Fig. 1c) and the ONAC095-SRDX plants showed severely wilted appearance, lower survival rate and higher percentage of electrolyte leakage after cold treatment (Fig. 5), indicating that dominant chimeric repressor-mediated suppression of ONAC095 function led to an attenuated cold tolerance. The ONAC095-SRDX plants accumulated more H2O2 and superoxide anion under cold stress (Fig. 6a–c), implying that dominant chimeric repressor-mediated suppression of ONAC095 function may lose the ability to control the balance between the cold-induced ROS generation and scavenging ability. This is supported by the observations that expression of 3 OsRboh genes, coding for NADPH oxidases involved in ROS generation [7, 54], were up-regulated while the activities of SOD and CAT, two ROS scavenging enzymes, were decreased in ONAC095-SRDX plants (Fig. 6e–g). Excessive accumulation of ROS often causes the production of MDA, which is toxic to plant cells at high level [8]. After cold stress, ONAC095-SRDX plants had significantly higher MDA level (Fig. 5c). Thus, it is reasonable that abnormal ROS accumulation resulted from imbalanced generation and scavenging may be one of the mechanisms that cause attenuated clod tolerance in ONAC095-SRDX plants. On the other hand, it is generally accepted that plant response to cold stress, like that to drought stress, often requires both ABA-dependent and ABA-independent signaling pathways [63]. The CBF-mediated signaling pathway is believed to be an ABA-independent signaling pathway [64]. ONAC095-SRDX plants contained an elevated ABA level (Fig. 7g) and showed down-regulated expression of OsICE1 (Fig. 5f), an upstream TF that regulates the expression of CBF genes [52]. These results indicate that an ABA-independent rather than ABA-dependent signaling is involved in ONAC095-regulated cold response in ONAC095-SRDX plants, probably partially through the CBF signaling pathway.
Given that plants are frequently exposed to a multitude of environmental stresses complicated molecular mechanisms, both coordinately and separately, that ensure general adaptive responses are essential for plants to survive under stress conditions. It has been shown that cross-talks at different levels between cold and drought signaling pathways occur in plant cells upon perception of environmental cues [4]. Our results in the present study show that dominant chimeric repressor-mediated suppression of ONAC095 function improves drought tolerance but attenuates cold tolerance in transgenic rice plants (Figs. 4 and 5), demonstrating that ONAC095 acts as a negative regulator of drought response but functions as a positive regulator of cold response. Such opposite roles in cold and drought response indicate that ONAC095 may be involved in the negative cross-talk between cold and drought responses. Similar observations were also reported for rice OsGH3-2 and Arabidopsis AtATL78, which play opposite roles in drought and cold stress tolerance [65, 66]. On the other hand, it is well known that plants operate specific signaling pathways in response to cold and drought stresses, which differentially activate distinct subsets of stress-related genes [9, 64]. In this regard, it is likely that ONAC095, as a transcriptional activator, may regulate different target genes upon perception of drought and cold stress signals and thereby exert its differential functions in drought and cold stress response. Further comparative analysis of gene expression profiling between ONAC095-SRDX and WT plants under drought and cold stress conditions will provide insights into the ONAC095-regulated signaling pathways.
ONAC095 is closely related to rice ONAC022 and Arabidopsis ANAC036 [42]; however, the function and action mode of ONAC095 differ from ONAC022 and ANAC036. Overexpression of ONAC022 or ANAC036 in transgenic plants resulted in a dwarf phenotype [42, 43], while dominant chimeric repressor-mediated suppression of ONAC095 function led to a stunted growth phenotype (Fig. 3e and f), indicating that ONAC095 is required for normal growth and development in rice. On the other hand, the ONAC022-overexpressing rice plants contained an elevated ABA level and conferred an improved drought and salt tolerance [42] while the ONAC095-suppressed rice plants showed an increased drought stress tolerance and ABA level (Figs. 4a, b and Fig. 7g). Thus, it is likely that ONAC095 and ONAC022, two closely related NAC TFs, have distinguishable biological functions in abiotic stress response through distinct mechanisms.
ONAC095 was induced by dehydration but repressed by cold stress (Fig. 1c). It is reasonable to speculate that the ONAC095-OE plants with increased expression level of ONAC095 (Fig. 3c) would mimic “tolerance” to drought stress and should “increase” drought stress tolerance, while they would be more “sensitivity” to cold stress and should “decrease” cold stress tolerance. In contrast, the ONAC095-SRDX plants, in which the ONAC095 function was suppressed by a dominant chimeric repressor, would mimic “sensitivity” to drought stress and should “decrease” drought stress tolerance, while they would also mimic “tolerance” to cold stress and should “increase” cold stress tolerance. Our experimental data indicate that, whereas the ONAC095-SRDX plants did exhibit clear phenotype under drought and cold stress, the ONAC095-OE plants did not show any altered response to drought and cold stress (Figs. 4a and 5a). One possibility is that ONAC095 is necessary but not sufficient to its function in growth/development and abiotic stress response. Alternatively, post-translational modification such as phosphorylation may be required for the function of the ONAC095 protein. This is partially supported by the presence of several putative phosphorylation sites with probability of >90 % in ONAC095 protein (Fig. 1a). If this is the case, it is then reasonable that simply overexpression of the ONAC095 gene in transgenic rice should not be enough to confer an altered phenotype. Recently, it was reported that transgenic rice plants overexpressing a phosphomimicking mutated OsWRKY53 showed further-enhanced disease resistance than the native OsWRKY53-overexpressing rice plants [67]. Thus, detailed biochemical assays are required to examine whether ONAC095 can be phosphorylated, and if so, to determine the putative phosphorylation sites. Once the phosphorylation feature is established, creating transgenic lines with overexpression of phosphomimicking mutated version of ONAC095 and examining the phenotype under abiotic stress including drought stress will clarify whether post-translational phosphorylation is required for the function of ONAC095. Additionally, it is also possible that the choice of the constitutive ubiquitin promoter to control the expression of ONAC095 in overexpression transgenic lines led to malfunction of ONAC095, thereby resulting in indistinguishable phenotype in ONAC095-OE plants from WT plants under drought and cold stress. Surprisingly, neither the ONAC095-OE nor the ONAC095-SRDX plants exhibited altered response to heat stress (Fig. 5g), although expression of ONAC095 was significantly induced by heat stress (Fig. 1c).
Expression of ONAC095 was affected differentially in response to drought, salt, cold and heat stress (Fig. 1c); however, no typical abiotic stress-related cis-element like DREB or ABRE was identified in the ONAC095 promoter (Fig. 1b). One possibility is that the promoter of ONAC095 may contain unidentified novel abiotic stress- and/or ABA-related cis-elements that drive its expression in response to abiotic stress. Similar observations were reported for some stress-responsive rice ONAC genes. For example, although there are no predicted stress-related cis-elements in promoters of the OsNAC4, OsNAC5 and OsNAC6 genes, all these three ONAC genes respond to abiotic stress [35]. Alternatively, the responsiveness of ONAC095 to abiotic stress is modulated by other TFs via the stress-related cis-elements like W-box and GCC-box in the promotor region of ONAC095 (Fig. 1b). In this regard, ONAC095 may function during relatively late stage in the stress response network [10]. Further detailed analysis of the ONAC095 promoter and its cis-elements will provide new insights into the regulation mechanism of ONAC095 expression during stress response.
Because ONAC095 is a transcriptional activator rather than a transcriptional repressor, it is unlikely that ONAC095 directly activates the expression of the ABA- and drought-related genes in ONAC095-SRDX plants; instead, it may suppress the expression of negative regulators for these genes during drought stress. By contrast, ONAC095 may directly regulate its target genes to modulate basal cold stress tolerance as dominant chimeric repressor-mediated suppression of ONAC095 function in ONAC095-SRDX plants led to down-regulation of some cold-responsive genes. On these regards, it is likely that ONAC095 may regulate different sets of genes that act separately in drought and cold tolerance.
Conclusion
ONAC095 is a transcriptional activator and the C2 domain in C-terminal and two proline residues in C2 domain are critical for transactivation activity of ONAC095. Functional analyses of the dominant chimeric repressor-mediated suppression transgenic lines demonstrate that ONAC095 acts as a negative regulator of drought response but as a positive regulator of cold response in rice. Further RNA-seq analysis of the ONAC095 regulon and chromatin immunoprecipitation-based identification of downstream target genes will provide new insights into how ONAC095 differentially regulates the drought and cold tolerance in rice. Although ONAC095 plays opposite roles in drought and cold stress tolerance, our knowledge that dominant chimeric repressor-mediated suppression of ONAC095 function improves drought tolerance can be used to generate drought-tolerant rice germplasms or materials for potentially application in temperate regions.
Methods
Plant materials, growth conditions and treatments
Rice (Oryza sativa L.) cv. Yuanfengzao (provided by Professor Rongyao Chai, Zhejiang Academy of Agricultural Sciences, Hangzhou, China) was used for analyses of gene expression in response to abiotic stress and ABA treatment while cv. Xiushui 134 (provided by Professor Rongyao Chai, Zhejiang Academy of Agricultural Sciences, Hangzhou, China), which was established for genetic transformation with high frequency in our lab [42], for generation of transgenic lines and phenotype analyses. Seeds were pre-germinated in water for 2 days and the germinated seeds were then planted into a soil mix. All rice plants were grown in a growth room with a cycle of 26 °C 14 hr light (>3000 lux)/22 °C 10 hr dark or in a greenhouse with natural sunlight. For ABA treatment, two-week-old seedlings were sprayed with 100 μM ABA in 0.1 % ethanol solution or with 0.1 % ethanol solution as controls. Drought treatment was applied by placing two-week-old seedlings into three layers of filter papers for fast dehydration and salt treatment was given by rooting the seedlings in a 150 mM NaCl solution. For extreme temperature stress treatments, seedlings were transferred to a growth chamber with temperature at 4 °C for cold treatment or a growth chamber with temperature at 42 °C for heat treatment. Samples were collected at different time points after treatment and stored at −80 °C until use.
Cloning and bioinformatics analysis of ONAC095
Coding sequence of ONAC095 was amplified with primers of ONAC095-F and ONAC095-R (Additional file 1: Table S1) designed based on the predicted cDNA in Rice Genome Annotation database and cloned into pMD19-T vector, yielding plasmid pMD19-ONAC095. Multiple sequence alignment was performed using ClustalW program in the LaserGene software [68]. The promoter sequence (1500 bp upstream from the transcription start site) of the ONAC095 gene was searched for putative cis-elements at the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [69]. Putative phosphorylation sites were searched at the NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/) [70].
Transactivation activity and subcellular localization assays
For analysis of the transactivation activity, the coding sequence and the truncated and mutated sequences of ONAC095 were obtained by PCR with different pairs of gene-specific primers (Additional file 1: Table S1) and cloned into pBD at EcoRI/BamHI sites [71]. The recombinant plasmids and pBD empty vector were transformed into yeast strain AH109. The transformed yeasts were plated on SD/Trp− medium or SD/Trp− His− medium containing 4 mM 3-AT and incubated for 3 days at 30 °C. Transactivation activity was assessed according to the growth status and production of blue pigment after addition of X-α-gal (5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside) on SD/Trp− His− medium. For analysis of the subcellular localization, the coding sequence of ONAC095 was amplified using primers of ONAC095GFP-F and ONAC095GFP-R (Additional file 1: Table S1) and cloned into pFGC-EGFP at BamHI/XbaI sites [72], yielding plasmid pFGC-GFP-ONAC095. Agrobacteria harboring pFGC-GFP-ONAC095 or pFGC-EGFP were infiltrated separately into leaves of N. benthamiana plants expressing a nuclear marker RFP–H2B protein [44] (provided by Dr. Michael Goodin, Department of Plant Pathology, University of Kentucky, USA). The agroinfiltrated leaves were collected at 2 days after agroinfiltration and GFP fluorescence signals were detected under a Zeiss LSM 510 Meta confocal laser scanning microscope (Oberkochen, Germany) using a 500–530 nm emission filter [73].
Binary vector construction, rice transformation and characterization of the transgenic lines
To construct the overexpression vector, the coding sequence of ONAC095 was amplified with primers of ONAC095OE-F and ONAC095OE-R (Additional file 1: Table S1) and cloned into a modified pCAMBIA1301 vector PU1301 under the control of the maize ubiquitin promoter [74], yielding PU1301-ONAC095-OE. To construct the chimeric suppression vector, the ONAC095 coding sequence without the stop codon was amplified using the forward primer ONAC095OE-F and the reverse primer ONAC095SRDX-R, which contains a synthetic SRDX (LDLDLELRLGFA) coding sequence fused at the C-terminus [46], and cloned into PU1301, yielding PU1301-ONAC095-SRDX. The resulting constructs PU1301-ONAC095-OE and PU1301-ONAC095-SRDX were introduced into calli of rice cv. Xiushui134 through standard Agrobacterium-mediated transformation protocol [75]. Putative single-copy ONAC095-OE and ONAC095-SRDX transgenic lines were screened according to a 3:1 segregation of HgrR : HgrS by planting seeds of T2 generation on 1/2 MS medium containing 50 μg/L Hgr. Homozygous single-copy ONAC095-OE and ONAC095-SRDX transgenic lines were selected based on phenotype of 100 % HgrR for seeds of T3 generation on 1/2 MS medium containing 50 μg/L Hgr. To confirm these single-copy transgenic lines, genomic DNA was extracted using the CTAB procedure [76] and 50 μg of genomic DNA was digested with EcoRI. After separation by electrophoresis on a 0.8 % agarose gel, DNAs in gel were transferred by capillary action onto a Hybond-N+ nylon membrane (Amersham Biosciences, Little Chalfont, UK) and hybridized with a 589 bp HptII probe labelled with DIG by the random priming method using a DIG High Prime DNA Labeling and Detection kit (Roche Diagnostics, Shanghai, China). Detection of DIG signals was performed according to the manufacturer’s recommendation.
Phenotype analyses for abiotic stress tolerance and ABA sensitivity
For drought stress assay, three-week-old ONAC095-OE and ONAC095-SRDX plants were grown with WT plants in same barrels and were subjected to drought stress by withholding water for 20 days, followed by recovery with normal water supply for another 7 days [42]. For cold stress assay, three-week-old ONAC095-OE and ONAC095-SRDX plants were grown with WT plants in same barrels and then transferred into a growth chamber with temperature at 4 °C with a cycle of 16 hr light/8 hr dark for 5 days for ONAC095-OE/WT plants and for 1 day for ONAC095-SRDX/WT plants, followed by transferring to the growth room with normal condition for recovery. For heat tolerance assay, three-week-old ONAC095-OE and ONAC095-SRDX plants were grown with WT plants in same barrels and were transferred into a growth chamber with temperature at 45 °C with a cycle of 16 hr light /8 hr dark for 5 days. After heat treatment, the plants were recovered at 28 °C for 7 days [77]. Plants with >20 % green leaves were considered to be survivals, and the others were considered to be dead plants. Survival rates were calculated as the percentage of survivals in the total plants used in the experiments. In abiotic stress assays, eight plants for each of the transgenic and WT lines were included in a single replicate and four replicates were set for each of the experiments. For ABA sensitivity assay, 60 seeds were plated on 1/2 MS medium with or without 5 μM ABA under 28 °C/25 °C (day/night) with a 12 hr photoperiod. Seed germination was recorded at 6 days after plating and weight of single seedling and length of shoot and root were measured at 10 days after germination [42].
Physiological and biochemical measurements
Samples for physiological and biochemical measurements except the RWC assay were collected from the drought and cold stress assays. RWC in detached leaves was measured according to a previously reported method [78]. Briefly, fully expanded leaves of three-week-old ONAC095-SRDX and WT plants were detached to record the leaf fresh weight (WF), turgid leaf weight (WT) and dry weights (WD), and RWC was calculated from the equation RWC (%) = (WF − WD)/(WT − WD) × 100 %. Electrolyte leakage was measured following a modified method [38]. Measurement of chlorophyll content was performed as described previously [79] using 0.5 g of leaf samples and the chlorophyll content was calculated according to the formula Chl (A + B) = 5.24 × A664 + 22.24 × A648. Quantification of MDA content was performed following a previously described protocol [38] using 0.2 g leaf samples. Free proline was determined using colorimetric method [80] with 0.5 g leaf sample and total soluble sugars was measured as previously described [81] using anthrone reagent with 0.5 g leaf sample. Measurement of H2O2 was followed by a previously described protocol [82] using trichloroacetic acid reagent with 0.5 g leaf sample. Quantification of ABA was performed by a HPLC-Triple quadrupole liquid chromatography-mass spectrometry system (Model 1290/6460, Aglient Technologies, Santa Clara, CA) according to a previously described method [83]. Activity of SOD and CAT was determined spectrophotometrically according to previously described methods [84]. In situ detection of H2O2 and superoxide anion in leaf tissues was performed by DAB staining [85] and NBT staining [86], respectively.
qRT-PCR analysis of gene expression
Total RNA was extracted using TRIzol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instruction. First-strand cDNA was synthesized from 1 μg of total RNA with SuperScript III Kit (Invitrogen, Shanghai, China) according to the manufacturer’s instruction. qRT-PCR reaction contained 12.5 μL SYBR premix Ex TaqTM (TaKaRa, Dalian, China), 1 μL cDNA sample and 10 μM each primer in a final volume of 25 μL and was performed on a CFX96 Real-time System (Bio-Rad, Hercules, CA, USA). A rice Actin gene (accession number KC140129) was used as an internal control to normalize the data and relative expression levels of genes of interest were calculated using the 2-ΔΔCT method. Gene-specific primers used in qRT-PCR are listed in Additional file 1: Table S1.
Statistical analysis
All experiments were repeated independently for at least three times and data are shown as mean ± SD of three independent experiments. Data were subjected to statistical analysis according to the Student’s t-test and the probability values of p < 0.05 were considered as significant difference.
Acknowledgements
We are grateful to Dr. Shiping Wang (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, China) for providing the binary vector, Professor Rongyao Chai (Zhejiang Academy of Agricultural Sciences, Hangzhou, China) for seeds of rice cultivars and Dr. Michael Goodin (Department of Plant Pathology, University of Kentucky, USA) for providing the H2B-RFP N. benthamiana line.
Funding
This study was supported by National Natural Science Foundation (No. 31272028), National Transgenic Major Project of China (No. 2014ZX08009-003-001 and No. 2016YFD0100601), and Research Fund for the Doctoral Program of Higher Education of China (20120101110070).
Availability of data and materials
The data sets supporting the findings of this article are included within the article.
Author’s contributions
FS conceived the study. LH and FS designed the experiments. LH, YH, HZ and DL performed the experiments. LH and FS analyzed the data. FS drafted the manuscript, and all authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- ABA
Abscisic acid
- CAT
Catalase
- DAB
3, 3’-diaminobenzidine
- MDA
Malondialdehyde
- NBT
Nitroblue tetrazolium
- qRT-PCR
Quantitative reverse transcription PCR
- ROS
Reactive oxygen species
- SOD
Superoxide dismutase
- TF
Transcription factor
Additional file
Table S1. Primers used in this study for different purposes. (DOC 76 kb)
Contributor Information
Lei Huang, Email: leihero2008@163.com.
Yongbo Hong, Email: yongbohong@126.com.
Huijuan Zhang, Email: zhanghj82@zju.edu.cn.
Dayong Li, Email: dyli@zju.edu.cn.
Fengming Song, Email: fmsong@zju.edu.cn.
References
- 1.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 2.Mehrotra R, Bhalothia P, Bansal P, Basantani MK, Bharti V, Mehrotra S. Abscisic acid and abiotic stress tolerance - different tiers of regulation. J Plant Physiol. 2014;171(7):486–96. doi: 10.1016/j.jplph.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Osakabe Y, Osakabe K, Shinozaki K, Tran LS. Response of plants to water stress. Front Plant Sci. 2014;5:86. doi: 10.3389/fpls.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58(2):221–7. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–9. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33(4):453–67. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol. 2011;14(6):691–9. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 8.You J, Chan Z. ROS regulation during abiotic stress responses in crop plants. Front Plant Sci. 2015;6:1092. doi: 10.3389/fpls.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–73. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 2014;5:170. doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zong W, Zhong X, You J, Xiong L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol Biol. 2013;81(1–2):175–88. doi: 10.1007/s11103-012-9990-2. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Song Y, Li S, Zhang L, Zou C, Yu D. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta. 2012;1819(2):120–8. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17(6):369–81. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol. 2013;4:248. doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castilhos G, Lazzarotto F, Spagnolo-Fonini L, Bodanese-Zanettini MH, Margis-Pinheiro M. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Sci. 2014;223:1–7. doi: 10.1016/j.plantsci.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci. 2015;6:902. doi: 10.3389/fpls.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran LS, Nishiyama R, Yamaguchi-Shinozaki K, Shinozaki K. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops. 2010;1(1):32–9. doi: 10.4161/gmcr.1.1.10569. [DOI] [PubMed] [Google Scholar]
- 19.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10(6):239–47. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics. 2008;280(6):547–63. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 22.Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465(1–2):30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Bollhoner B, Prestele J, Tuominen H. Xylem cell death: emerging understanding of regulation and function. J Exp Bot. 2012;63(3):1081–94. doi: 10.1093/jxb/err438. [DOI] [PubMed] [Google Scholar]
- 24.Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Deyholos MK. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006;6:25. doi: 10.1186/1471-2229-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Huang L, Hong Y, Zhang H, Song F, Li D. Comprehensive analysis suggests overlapping expression of rice ONAC transcription factors in abiotic and biotic stress responses. Int J Mol Sci. 2015;16(2):4306–26. doi: 10.3390/ijms16024306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuruzzaman M, Sharoni AM, Satoh K, Karim MR, Harikrishna JA, Shimizu T, et al. NAC transcription factor family genes are differentially expressed in rice during infections with Rice dwarf virus, Rice black-streaked dwarf virus, Rice grassy stunt virus, Rice ragged stunt virus, and Rice transitory yellowing virus. Front Plant Sci. 2015;6:676. doi: 10.3389/fpls.2015.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun L, Zhang H, Li D, Huang L, Hong Y, Ding XS, et al. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol Biol. 2013;81(1–2):41–56. doi: 10.1007/s11103-012-9981-3. [DOI] [PubMed] [Google Scholar]
- 29.Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, Omura T. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to Rice dwarf virus. Plant J. 2009;57(4):615–25. doi: 10.1111/j.1365-313X.2008.03712.x. [DOI] [PubMed] [Google Scholar]
- 30.Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che F-S. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 2009;28(7):926–36. doi: 10.1038/emboj.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokotani N, Tsuchida-Mayama T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Kaku H, Minami E, Nishizawa Y. OsNAC111, a blast disease-responsive transcription factor in rice, positively regulates the expression of defense-related genes. Mol Plant-Microbe Interact. 2014;27(10):1027–34. doi: 10.1094/MPMI-03-14-0065-R. [DOI] [PubMed] [Google Scholar]
- 32.Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A. 2006;103(35):12987–92. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2008;67(1–2):169–81. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- 34.Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun. 2009;379(4):985–9. doi: 10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
- 35.Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, et al. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics. 2010;284(3):173–83. doi: 10.1007/s00438-010-0557-0. [DOI] [PubMed] [Google Scholar]
- 36.Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153(1):185–97. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW, et al. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J. 2013;11(1):101–14. doi: 10.1111/pbi.12011. [DOI] [PubMed] [Google Scholar]
- 38.Song SY, Chen Y, Chen J, Dai XY, Zhang WH. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta. 2011;234(2):331–45. doi: 10.1007/s00425-011-1403-2. [DOI] [PubMed] [Google Scholar]
- 39.Redillas MC, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, et al. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J. 2012;10(7):792–805. doi: 10.1111/j.1467-7652.2012.00697.x. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Wang Y, Lv B, Li J, Luo L, Lu S, et al. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014;55(3):604–19. doi: 10.1093/pcp/pct204. [DOI] [PubMed] [Google Scholar]
- 41.Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci U S A. 2014;111(27):10013–8. doi: 10.1073/pnas.1321568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong Y, Zhang H, Huang L, Li D, Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front Plant Sci. 2016;7:4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato H, Motomura T, Komeda Y, Saito T, Kato A. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J Plant Physiol. 2010;167(7):571–7. doi: 10.1016/j.jplph.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Chakrabarty R, Banerjee R, Chung SM, Farman M, Citovsky V, Hogenhout SA, et al. PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant-Microbe Interact. 2007;20(7):740–50. doi: 10.1094/MPMI-20-7-0740. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Basnayake BMVS, Zhang H, Li G, Li W, Virk N, Mengiste T, Song F. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol Plant-Microbe Interact. 2009;22(10):1227–38. doi: 10.1094/MPMI-22-10-1227. [DOI] [PubMed] [Google Scholar]
- 46.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34(5):733–9. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 47.Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics. 2010;11:435. doi: 10.1186/1471-2164-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang Y, Tang N, Du H, Ye H, Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008;148(4):1938–52. doi: 10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK. Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 2009;150(3):1368–79. doi: 10.1104/pp.109.137554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono A, Izawa T, Chua NH, Shimamoto K. The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol. 1996;112(2):483–91. doi: 10.1104/pp.112.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–98. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17(8):1043–54. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot. 2013;64(16):5085–97. doi: 10.1093/jxb/ert298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell. 2007;19(12):4022–34. doi: 10.1105/tpc.107.055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J. 2010;426(2):183–96. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell. 2010;22(4):1249–63. doi: 10.1105/tpc.108.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HS, Park BO, Yoo JH, Jung MS, Lee SM, Han HJ, et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J Biol Chem. 2007;282(50):36292–302. doi: 10.1074/jbc.M705217200. [DOI] [PubMed] [Google Scholar]
- 58.Liu J, Zhu JK. Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 1997;114(2):591–6. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du H, Wang N, Cui F, Li X, Xiao J, Xiong L. Characterization of the β-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 2010;154(3):1304–18. doi: 10.1104/pp.110.163741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai S, Jiang G, Ye N, Chu Z, Xu X, Zhang J, Zhu G. A Kkey ABA catabolic gene, OsABA8ox3, is involved in drought stress resistance in rice. Plos One. 2015;10(2):e0116646. doi: 10.1371/journal.pone.0116646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor IB, Burbidge A, Thompson AJ. Control of abscisic acid synthesis. J Exp Bot. 2000;51(350):1563–74. doi: 10.1093/jexbot/51.350.1563. [DOI] [PubMed] [Google Scholar]
- 62.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6(5):410–7. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 64.Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12(10):444–51. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Du H, Wu N, Fu J, Wang S, Li X, Xiao J, Xiong L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot. 2012;63(18):6467–80. doi: 10.1093/jxb/ers300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim SJ, Kim WT. Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 2013;587(16):2584–90. doi: 10.1016/j.febslet.2013.06.038. [DOI] [PubMed] [Google Scholar]
- 67.Chujo T, Miyamoto K, Ogawa S, Masuda Y, Shimizu T, Kishi-Kaboshi M, Takahashi A, Nishizawa Y, Minami E, Nojiri H, Yamane H, Okada K. 2014 Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS One. 2014;9:e98737. doi: 10.1371/journal.pone.0098737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burland TG. DNASTAR’s lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 69.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blom N, Gammeltoft S, Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294(5):1351–62. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 71.Louvet O, Doignon F, Crouzet M. Stable DNA-binding yeast vector allowing high-bait expression for use in the two-hybrid system. Biotechniques. 1997;23(5):816–20. doi: 10.2144/97235bm11. [DOI] [PubMed] [Google Scholar]
- 72.Li Z, Jayasankar S, Gray DJ. Expression of a bifunctional green fluorescent protein (GFP) fusion marker under the control of three constitutive promoters and enhanced derivatives in transgenic grape (Vitis vinifera) Plant Sci. 2001;160(5):877–87. doi: 10.1016/S0168-9452(01)00336-3. [DOI] [PubMed] [Google Scholar]
- 73.Anderson KI, Sanderson J, Gerwig S, Peychl J. A new configuration of the Zeiss LSM 510 for simultaneous optical separation of green and red fluorescent protein pairs. Cytometry A. 2006;69(8):920–9. doi: 10.1002/cyto.a.20323. [DOI] [PubMed] [Google Scholar]
- 74.Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S. The expression pattern of a rice disease resistance gene Xa3/Xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics. 2007;177(1):523–33. doi: 10.1534/genetics.107.075176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47(6):969–76. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- 76.Luo H, Song F, Zheng Z. Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J Exp Bot. 2005;56(420):2673–82. doi: 10.1093/jxb/eri260. [DOI] [PubMed] [Google Scholar]
- 77.Li XM, Chao DY, Wu Y, Huang X, Chen K, Cui LG, Su L, Ye WW, Chen H, Chen HC, Dong NQ, Guo T, Shi M, Feng Q, Zhang P, Han B, Shan JX, Gao JP, Lin HX. Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat Genet. 2015;47(7):827–33. doi: 10.1038/ng.3305. [DOI] [PubMed] [Google Scholar]
- 78.Schonfeld MA, Johnson RC, Carver BF. Water relations in winter wheat as drought resistance indicator. Crop Sci. 1988;28:526–31. doi: 10.2135/cropsci1988.0011183X002800030021x. [DOI] [Google Scholar]
- 79.Lichtenthaler HK. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;18:350–82. doi: 10.1016/0076-6879(87)48036-1. [DOI] [Google Scholar]
- 80.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39(1):205–7. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 81.Bailey RW. The reaction of pentoses with anthrone. Biochem J. 1958;68(4):669–72. doi: 10.1042/bj0680669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alexieva V, Sergiev I, Mapelli S, Karanov E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant, Cell & Environ. 2001;24(12):1337–44. doi: 10.1046/j.1365-3040.2001.00778.x. [DOI] [Google Scholar]
- 83.Fu J, Chu J, Sun X, Wang J, Yan C. Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci. 2012;28(11):1081–7. doi: 10.2116/analsci.28.1081. [DOI] [PubMed] [Google Scholar]
- 84.Mittova V, Volokita M, Guy M, Tal M. Activities of SOD and the ascorbate-glutathione cycle enzymes in subcellular compartments in leaves and roots of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol Plant. 2000;110(1):42–51. doi: 10.1034/j.1399-3054.2000.110106.x. [DOI] [Google Scholar]
- 85.Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 1997;11(6):1187–94. doi: 10.1046/j.1365-313X.1997.11061187.x. [DOI] [Google Scholar]
- 86.Doke N. Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol Plant Pathol. 1983;23(3):345–57. doi: 10.1016/0048-4059(83)90019-X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the findings of this article are included within the article.


