Abstract
Background
Burkholderia mallei and B. pseudomallei are the causative agents of glanders and melioidosis, respectively, diseases with high morbidity and mortality rates. B. mallei and B. pseudomallei are closely related genetically; B. mallei evolved from an ancestral strain of B. pseudomallei by genome reduction and adaptation to an obligate intracellular lifestyle. Although these two bacteria cause different diseases, they share multiple virulence factors, including bacterial secretion systems, which represent key components of bacterial pathogenicity. Despite recent progress, the secretion system proteins for B. mallei and B. pseudomallei, their pathogenic mechanisms of action, and host factors are not well characterized.
Results
We previously developed a manually curated database, DBSecSys, of bacterial secretion system proteins for B. mallei. Here, we report an expansion of the database with corresponding information about B. pseudomallei. DBSecSys 2.0 contains comprehensive literature-based and computationally derived information about B. mallei ATCC 23344 and literature-based and computationally derived information about B. pseudomallei K96243. The database contains updated information for 163 B. mallei proteins from the previous database and 61 additional B. mallei proteins, and new information for 281 B. pseudomallei proteins associated with 5 secretion systems, their 1,633 human- and murine-interacting targets, and 2,400 host-B. mallei interactions and 2,286 host-B. pseudomallei interactions. The database also includes information about 13 pathogenic mechanisms of action for B. mallei and B. pseudomallei secretion system proteins inferred from the available literature or computationally. Additionally, DBSecSys 2.0 provides details about 82 virulence attenuation experiments for 52 B. mallei secretion system proteins and 98 virulence attenuation experiments for 61 B. pseudomallei secretion system proteins. We updated the Web interface and data access layer to speed-up users’ search of detailed information for orthologous proteins related to secretion systems of the two pathogens.
Conclusions
The updates of DBSecSys 2.0 provide unique capabilities to access comprehensive information about secretion systems of B. mallei and B. pseudomallei. They enable studies and comparisons of corresponding proteins of these two closely related pathogens and their host-interacting partners.
The database is available at http://dbsecsys.bhsai.org.
Keywords: Bacterial secretion system, Virulence factors, Pathogenic mechanisms of action, Host-pathogen interactions, Burkholderia mallei, Burkholderia pseudomallei
Background
Introduction
The highly infectious pathogen Burkholderia mallei is the causative agent of glanders, and its phylogenetically closest species, B. pseudomallei, is the causative agent of melioidosis. B. mallei evolved from an ancestral strain of B. pseudomallei by genome reduction and adaptation to an obligate intracellular lifestyle [1]. Given their considerable antibiotic resistance, ability to infect via aerosol exposure, and absence of vaccines, B. mallei and B. pseudomallei represent an emerging public health threat in their natural environment and as a potential bioterrorism threat. Although the two bacteria cause different diseases, they share multiple virulence factors, including bacterial secretion systems, which represent key components of bacterial pathogenicity [2, 3]. Recent research has provided new insights into B. mallei and B. pseudomallei pathogenicity [4, 5], but the identities of their secretion system proteins, their pathogenic mechanisms, and host factors are not completely characterized.
Several database systems provide general information about Burkholderia proteins [6–8], whereas more specific databases encompass information about single [9–12] or multiple [13, 14] bacterial secretion systems for many, but not all, Burkholderia species. In addition, existing databases of secretion systems for multiple pathogens provide general information about secretion system proteins of B. mallei and B. pseudomallei but lack specific information about protein functions and, especially, do not provide information about pathogenic mechanisms of action, experimental results, or host-interacting partners.
Our contribution
To catalogue in-depth information about secretion system proteins of B. mallei and their host factors, we previously developed the Database of Burkholderia mallei Secretion Systems (DBSecSys) [15]. Since its deployment in July 2014, over 500 users from 38 countries from around the world have accessed DBSecSys. The users’ interest motivated us to update the database and extend it with information about secretion system proteins for B. pseudomallei. This updated version, DBSecSys 2.0, provides not only annotation for B. mallei and B. pseudomallei proteins associated with secretion systems but also manually curated information about these proteins, their involvement in virulence, their host targets (proteins), and their mechanisms of action inferred from literature and host-pathogen interaction data. These features provide an ability to study and compare secretion systems of the two pathogens not only through characterization of their corresponding pathogen proteins but also through characterization of their host-interacting partners. These data and features are currently not available elsewhere, and they make DBSecSys 2.0 a unique resource for B. mallei and B. pseudomallei secretion system research.
Construction and content
Database content
Table 1 shows the content of DBSecSys 2.0. Using the available literature on Burkholderia species and our experimental and computational work on B. mallei [16, 17], we compiled information for 204 B. mallei ATCC 23344 proteins and 281 B. pseudomallei K96243 proteins and their associated secretion systems, including 200 orthologous proteins. The majority of the proteins are associated with five secretion systems. In the updated database, we also included several recently identified secreted proteins that have been associated with virulence, but whose secretion mechanisms have not yet been determined (denoted as Undetermined Type in the database). We did not include information about contact-dependent interactions, which are involved in bacterial-bacterial interactions for B. pseudomallei [18], because the database focus is on secretion systems relevant for interactions with mammalian hosts.
Table 1.
Summary of the content of DBSecSys 2.0
| Number of B. mallei | |
|---|---|
| Proteins | 204 |
| Associated secretion systems | 5 |
| Virulence factors | 52 |
| Virulence attenuation experiments | 82 |
| Inferred mechanisms of action | 12 |
| Number of B. pseudomallei | |
| Proteins | 281 |
| Associated secretion systems | 5 |
| Virulence factors | 61 |
| Virulence attenuation experiments | 98 |
| Inferred mechanisms of action | 13 |
| Number of host-pathogen protein-protein interactions | |
| Human-B. mallei (experimental) | 569 |
| Murine-B. mallei (experimental) | 788 |
| Human-B. mallei (computational) | 608 |
| Murine-B. mallei (computational) | 435 |
| Human-B. pseudomallei (experimental) | 4 |
| Human-B. pseudomallei (computational) | 1,117 |
| Murine-B. pseudomallei (computational) | 1,165 |
Published experimental data served as the main source of information for 82 virulence attenuation experiments for 52 B. mallei secretion system proteins and 98 virulence attenuation experiments for 61 B. pseudomallei secretion system proteins. For a subset of these experiments, we transferred virulence attenuation between orthologous proteins of the two pathogens. In these cases, we provided evidence that the inferred virulence attenuation is based on homology and, thus, needs experimental verification.
Since the publication of the first version of the database DBSecSys, there were no new published protein-protein interactions (PPIs) between B. mallei and human and murine hosts. Thus, we imported PPIs between B. mallei and 1,633 host proteins (795 human and 838 murine) from the previous version. Experimental host-pathogen PPI data for B. pseudomallei are scarce. We found only 4 published PPIs between B. pseudomallei and the human host [19]. To provide a more comprehensive picture of the host-pathogen PPIs for B. pseudomallei, we mapped host-pathogen PPIs for B. mallei proteins in the database to their orthologous B. pseudomallei proteins. PPIs between B. mallei protein and host proteins were identified previously in yeast two-hybrid (Y2H) experiments, i.e., they were based on direct protein interactions. Thus, the transfer of PPIs to orthologous B. pseudomallei proteins can be justified. The database now contains 2,400 host-pathogen PPIs between B. mallei and these 2 hosts and 2,286 host-pathogen PPIs between B. pseudomallei and these hosts. As previously described [15], DBSecSys 2.0 also contains a set of 491 PPIs [20] among 357 human proteins and 36 PPIs [21, 22] among 47 murine proteins that were identified as interacting partners of B. mallei proteins in Y2H experiments [16].
Data sources
We identified secretion system proteins of B. mallei and B. pseudomallei using literature, experiments, orthology between B. mallei and B. pseudomallei proteins, and computational methods. For all experimental data and observations contained in DBSecSys 2.0, we provided links to their original sources and publications through PubMed.
Human-B. mallei and murine-B. mallei protein interaction data have been identified in Y2H experiments. As PPI detection experiments did not exhaustively screen for all possible host-B. mallei interactions, we used human-murine orthology information based on the HomoloGene [23] database to infer additional host-pathogen interactions, as previously described [15, 16]. Using these data, we transferred host-pathogen interaction data for B. pseudomallei to proteins that have orthologs to B. mallei proteins. We manually curated information about orthologous proteins between the two pathogens from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [24] and QuartetS_DB [25], including only bidirectional hits with a sequence identity larger than 95 %.
To account for the relationship among host proteins that interact with B. mallei and B. pseudomallei proteins, we used information from host PPI networks. For human PPIs, we used data from an experimentally detected PPI network compiled by Yu et al. [20], whereas for murine PPIs, we used data from an experimentally detected PPI network available in the BioGRID database (release 3.2.105) [21, 22].
We inferred pathogen mechanisms of action for proteins in the database using common terms from the available literature shown in Table 2. To better account for recent research about the two pathogens, the list of mechanisms of action was extended with 3 additional terms, cytotoxicity, intracellular survival, and regulation. For B. mallei proteins, we used mechanisms of action based on the literature, Y2H experiments, and additional computations, as previously described [15]. For B. pseudomallei proteins, we used the literature as the primary source of information about mechanisms of action. To provide additional information about possible mechanisms of action, we assigned all mechanisms of action bi-directionally between orthologous proteins of B. mallei and B. pseudomallei.
Table 2.
Description of pathogenic mechanisms of action included in DBSecSys 2.0
| Name | Pathogens use this mechanism to: |
|---|---|
| Actin cytoskeleton rearrangement | Subvert the host cell cytoskeleton to promote attachment to the host cell surface, internalization in the host cell, and prevent uptake by phagocytic cells. |
| Actin-based motility | Bind to host actin, triggering actin polymerization on the pathogens’ surface and producing a mechanical force that propels them through the host cell and facilitates cell-to-cell spread. |
| Adhesion | Attach to the host cell surface, promoting bacterial internalization in the host cell. |
| Apoptosis | Exert control on the processes that regulate apoptosis in the host. |
| Cytotoxicitya | Secrete toxins into the host cell. |
| Interference with signaling | Interfere with the host signaling cascade, promoting their internalization in the host cell and intracellular survival. |
| Interference with the immune response | Downregulate host inflammatory responses, promoting their internalization in the host cell and intracellular survival. |
| Intracellular survivala | Evade the host immune response and multiply in the host cell. |
| Invasion | Promote their ability to invade the host cell. |
| Multinucleated giant cell formation | Induce host cell fusion and multinucleated giant cell formation. |
| Phagosomal escape and evasion of autophagy | Ensure bacterial escape from endocytic vesicles as well as to evade autophagosomes, ensuring the pathogens’ intracellular survival and cell-to-cell spread. |
| Regulationa | Control secretion system activation and related mechanisms of pathogenicity. |
| Ubiquitination–deubiquitination | Interfere with host ubiquitination processes to attenuate the host immune response, to prevent their degradation, and to ensure their destruction when no longer required for establishing the infection. |
aMechanisms of action added in the updated database
We used the National Center for Biotechnology Information (NCBI) [23] and Uniprot [26] to annotate pathogen and host proteins, e.g., protein names and sequence information. For functional annotation of host proteins, we used Gene Ontology (GO) terms [27] and KEGG pathways [24].
Software architecture
In DBSecSys 2.0 database and Web interface, we used a three-tier architecture, which we developed for the previous version [15]: 1) a backend Oracle database server that stores the data contained in the DBSecSys 2.0 database, 2) the controller, and 3) the presentation tier. We used Java Platform, Enterprise Edition 7, JavaServer Faces 2, and ICEfaces 3 technologies. The presentation tier also includes JBrowse [28], D3.js [29], NVD3.js [30], and Cytoscape.js [31] JavaScript libraries that provide detailed interactive visualizations of genes on the reference sequence and of PPIs. For DBSecSys 2.0, we updated the data access layer to provide a more consistent and faster user experience. Some of the searches are 2 to 3 time faster than the previous version. We have also added a loading indicator to provide visual feedback for any long running searches. The DBSecSys 2.0 Web application is hosted on an Apache Tomcat Web server at http://dbsecsys.bhsai.org.
Utility and Discussion
Users can browse and query cross-linked data in DBSecSys 2.0 through a Web-based graphical user interface, which supports five functionalities: query for 1) pathogen protein or host protein annotation, 2) pathogen proteins associated with a secretion system, 3) pathogen proteins associated with the pathogen’s mechanism of action, 4) host-pathogen interactions, and 5) experimentally screened virulence factors. We provided details of these functionalities for a single organism in the previous version of the database, which have been updated in the current User’s Guide. Users can download query results in a tab-delimited format and host-pathogen PPIs in the Proteomics Standards Initiative Molecular Interactions (PSI-MI) Tab 2.5 format [32]. DBSecSys 2.0 also provides links to external resources for detailed information about individual proteins (NCBI, Uniprot, GO, and KEGG), as well as links to relevant publications in PubMed.
Importantly, the new database content and design enable comparative analysis between organisms. We describe two such applications below.
Application 1: search for orthologous proteins in B. mallei and B. pseudomallei
Users can study individual (pathogen or host) proteins on the “Proteins” page of the Web interface. Users can search for association of a pathogen protein with a secretion system using standard protein/gene identifiers, such as Locus Tag, Name, or Uniprot ID. When the ortholog of the query protein is present in the database, DBSecSys 2.0 provides a link to it. For example, a search by Name for the virulence factor TssM [19, 33] returns links to orthologous proteins in the two organisms. Users can click one of the links, e.g., the link to B. mallei, to display the protein page with detailed annotation information about this protein, including its Locus Tag BMAA0729, evidence about its association with the type 2 secretion system (T2SS), virulence attenuation experiments, inferred mechanisms of action, and host-pathogen interactions. Selecting the box labeled “Pathogen Ortholog Comparison” and clicking the link for BMAA0729, the database returns information about TssM’s orthologous protein in B. pseudomallei, with Locus Tag BPSS1512, on the lower half of the screen. This enables direct comparison of annotations for the two orthologs. The information about orthologs is very similar because of the database curation strategy to transfer virulence information, mechanisms of action, and host-pathogen interactions between two orthologous proteins. Comparison between secretions systems of these orthologous proteins provides additional insights.
Application 2: comparison of related secretion systems in B. mallei and B. pseudomallei
Users can study pathogen proteins associated with a specific secretion system on the “Secretion Systems” page of the Web interface. Continuing the previous example, users can compare secretion systems for two TssM proteins by clicking the link to T2SS and selecting the link “Compare Burkholderia mallei (strain ATCC 23344) and Burkholderia pseudomallei (strain K96243)” from the drop-down list “Select Pathogen.” The lists of proteins in the corresponding secretion systems of the two pathogens are displayed on the same page and enable direct comparison between individual proteins. In addition, users can open the link to the genome browser, which depicts the positions of these proteins on the genomes of the two organisms, including linear genome maps and schematic circular chromosomes. Positioning the mouse over the marked red bands on the circular chromosomes shows individual proteins or secretion system clusters. Clicking at the positions on chromosome 2 of each organism corresponding to TssM, BMAA0729 in B. mallei and BPSS512 in B. pseudomallei, shows their positions on the corresponding linear genome maps. In addition, by adjusting the coverage of the genomic regions, users can visualize the position of this T2SS protein relative to the adjacent type 6 secretion system (T6SS) proteins in B. mallei and B. pseudomallei, as shown in Fig. 1. Users can inspect the syntenic regions for the two pathogens to visualize orthologous pairs. For examples, the nearest neighbor of TssM is TssN, which is shown at Locus Tags BMAA0728 in B. mallei and BPSS1513 in B. pseudomallei. Previously, we found that both transposon insertion and deletion of this gene in B. mallei fully attenuate virulence [16, 34]. Because of its importance for virulence and its regulation by the same mechanisms as cluster 1 T6SS [35–37], an important virulence factor, we putatively assigned TssN to T6SS.
Fig. 1.
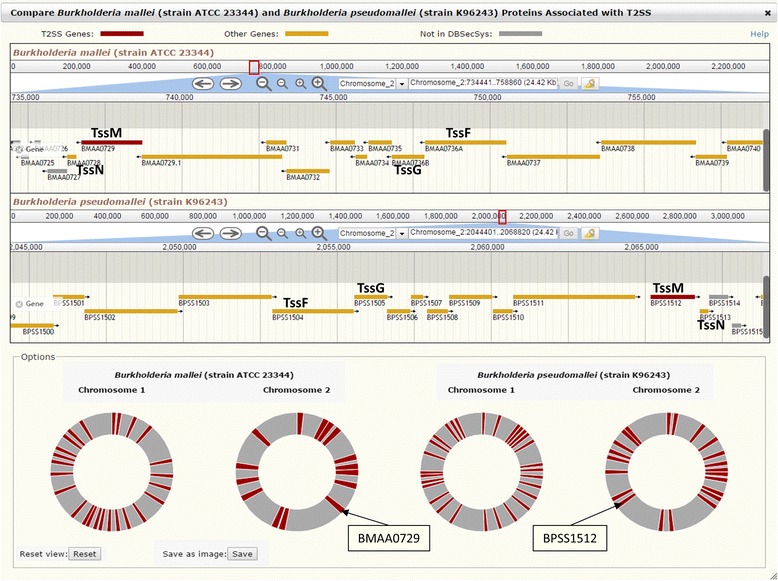
Comparison of related secretion systems in Burkholderia mallei and B. pseudomallei. The genome browser shows the location of the orthologous proteins TssM, TssN, TssF, and TssG and their association with related secretion systems on the bacterial reference sequences for B. mallei strain ATCC 23344 and B. pseudomallei strain K96243. Type 2 secretion system (T2SS) and type 6 secretion system (T6SS) proteins are depicted in red and yellow, respectively
Another interesting protein is TssF, with Locus Tag BPSS1504 in B. pseudomallei, which was recently identified as an important virulence factor for B. pseudomallei [38]. Neither NCBI nor KEGG provided a B. mallei ortholog for this protein, because the corresponding genomic region was originally annotated as a pseudogene [39]. However, experimental evidence indicated that this region in B. mallei codes for two proteins, designated as BMAA0736A and BMAA0736B [35]. Based on the annotation from the Pathosystems Resource Integration Center (PATRIC) [6], these two proteins are orthologs to TssF and TssG, respectively, in B. pseudomallei, and their protein sequences in B. mallei have the same length and 99 % sequence identity to the corresponding B. pseudomallei orthologs. We included these sequences in DBSecSys 2.0, providing for a more comprehensive comparison of cluster 1 T6SS for the two pathogens.
Database updates
The database content will be expanded with information for additional pathogens and will be updated periodically. Before each update, the current state of the database will be frozen and archived, and can be requested by contacting dbsecsys@bhsai.org.
Conclusions
We previously developed DBSecSys, a database of B. mallei secretion system proteins. The database organization provided capability to store and retrieve comprehensive information about pathogen proteins associated with secretion systems, such as their annotation, involvement in virulence, host protein targets, and mechanisms of action inferred from the literature and host-pathogen interaction data. In DBSecSys 2.0, we improved data querying and retrieval, enabling comparison of secretion systems for multiple pathogens. In addition, we expanded the database with new information about B. pseudomallei and updated the information about B. mallei. These updates provide unique capabilities to access comprehensive information about the secretion systems of B. mallei and B. pseudomallei. Importantly, they enable studies and comparisons of corresponding proteins of these two closely related pathogens and their host-interacting partners.
Acknowledgments
None.
Funding
This work was supported by the Defense Threat Reduction Agency (project CBS.MEDBIO.02.10.BH.021) and by the U.S. Medical Research and Materiel Command (Ft. Detrick, MD) as part of the U.S. Army’s Network Science Initiative.
Availability of data and materials
The Web-enabled DBSecSys 2.0 database is publicly accessible at http://dbsecsys.bhsai.org. The DBSecSys 2.0 Web interface has been tested in the following Web browsers: Google Chrome (version 48), Microsoft Internet Explorer (version 11), and Mozilla Firefox (version 31). The “User’s Guide” page of the DBSecSys 2.0 Web interface includes a step-by-step description of all DBSecSys 2.0 features. For additional questions about the database, please contact dbsecsys@bhsai.org.
Authors’ contributions
V.M., N.Z., D.DS., and A.W. performed the data curation. V.M. and K. K. designed the database schema. V.M., K.K., and J.R. designed the Web interface. K.K. developed the front- and back-end infrastructure. V.M., N.Z., A.W., and J.R. conceived the work. N.Z., V.M., K.K., and J.R. wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or the U.S. Department of Defense. This paper has been approved for public release with unlimited distribution.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- DBSecSys
Database of Burkholderia mallei and Burkholderia pseudomallei secretion systems
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- NCBI
National Center for Biotechnology Information
- PATRIC
Pathosystems resource integration center
- PPI
Protein-protein interaction
- Y2H
Yeast two-hybrid
- T2SS
Type 2 secretion system
- T6SS
Type 6 secretion system
Contributor Information
Vesna Memišević, Email: vmemisevic@bhsai.org.
Kamal Kumar, Email: kkumar@bhsai.org.
Nela Zavaljevski, Email: nzavaljevski@bhsai.org.
David DeShazer, Email: david.deshazer.civ@mail.mil.
Anders Wallqvist, Email: sven.a.wallqvist.civ@mail.mil.
Jaques Reifman, Email: jaques.reifman.civ@mail.mil.
References
- 1.Losada L, Ronning CM, DeShazer D, Woods D, Fedorova N, Kim HS, Shabalina SA, Pearson TR, Brinkac L, Tan P, et al. Continuing evolution of Burkholderia mallei through genome reduction and large-scale rearrangements. Genome Biol Evol. 2010;2:102–116. doi: 10.1093/gbe/evq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allwood EM, Devenish RJ, Prescott M, Adler B, Boyce JD. Strategies for intracellular survival of Burkholderia pseudomallei. Front Microbiol. 2011;2:170. doi: 10.3389/fmicb.2011.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willcocks SJ, Denman CC, Atkins HS, Wren BW. Intracellular replication of the well-armed pathogen Burkholderia pseudomallei. Curr Opin Microbiol. 2016;29:94–103. doi: 10.1016/j.mib.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Hatcher CL, Muruato LA, Torres AG. Recent Advances in Burkholderia mallei and B. pseudomallei Research. Curr Trop Med Rep. 2015;2(2):62–69. doi: 10.1007/s40475-015-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stone JK, DeShazer D, Brett PJ, Burtnick MN. Melioidosis: molecular aspects of pathogenesis. Expert Rev Anti Infect Ther. 2014;12(12):1487–1499. doi: 10.1586/14787210.2014.970634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42(Database issue):D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brittnacher MJ, Fong C, Hayden HS, Jacobs MA, Radey M, Rohmer L. PGAT: a multistrain analysis resource for microbial genomes. Bioinformatics. 2011;27(17):2429–2430. doi: 10.1093/bioinformatics/btr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24(23):2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi D, Liu L, Tai C, Deng Z, Rajakumar K, Ou HY. SecReT4: A web-based bacterial type IV secretion system resource. Nucleic Acids Res. 2013;41:D660–D665. doi: 10.1093/nar/gks1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Souza RC, del Rosario Quispe Saji G, Costa MO, Netto DS, Lima NC, Klein CC, Vasconcelos AT, Nicolas MF. AtlasT4SS: A curated database for type IV secretion systems. BMC Microbiol. 2012;12:172. doi: 10.1186/1471-2180-12-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Huang H, Sun M, Zhang Q, Guo D. T3DB: an integrated database for bacterial type III secretion system. BMC Bioinformatics. 2012;13:66. doi: 10.1186/1471-2105-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Yao Y, Xu HH, Hao L, Deng Z, Rajakumar K, Ou HY. SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ Microbiol. 2015;17(7):2196–2202. doi: 10.1111/1462-2920.12794. [DOI] [PubMed] [Google Scholar]
- 13.Bleves S, Dunger I, Walter MC, Frangoulidis D, Kastenmuller G, Voulhoux R, Ruepp A. HoPaCI-DB: host-Pseudomonas and Coxiella interaction database. Nucleic Acids Res. 2014;42(Database issue):D671–D676. doi: 10.1093/nar/gkt925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res. 2016;44(D1):D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memišević V, Kumar K, Cheng L, Zavaljevski N, DeShazer D, Wallqvist A, Reifman J. DBSecSys: A database of Burkholderia mallei secretion systems. BMC Bioinformatics. 2014;15(1):244. doi: 10.1186/1471-2105-15-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Memišević V, Zavaljevski N, Pieper R, Rajagopala SV, Kwon K, Townsend K, Yu C, Yu X, Deshazer D, Reifman J, et al. Novel Burkholderia mallei virulence factors linked to specific host-pathogen protein interactions. Mol Cell Proteomics. 2013;12(11):3036–3051. doi: 10.1074/mcp.M113.029041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memišević V, Zavaljevski N, Rajagopala SV, Kwon K, Pieper R, DeShazer D, Reifman J, Wallqvist A. Mining host-pathogen protein interactions to characterize Burkholderia mallei infectivity mechanisms. PLoS Comput Biol. 2015;11(3) doi: 10.1371/journal.pcbi.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koskiniemi S, Garza-Sanchez F, Edman N, Chaudhuri S, Poole SJ, Manoil C, Hayes CS, Low DA. Genetic analysis of the CDI pathway from Burkholderia pseudomallei 1026b. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan KS, Chen Y, Lim YC, Tan GY, Liu Y, Lim YT, Macary P, Gan YH. Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J Immunol. 2010;184(9):5160–5171. doi: 10.4049/jimmunol.0902663. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Wallqvist A, Reifman J. Inferring high-confidence human protein-protein interactions. BMC Bioinformatics. 2012;13(1):79. doi: 10.1186/1471-2105-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O’Donnell L, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40(Database issue):D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C, Desai V, Cheng L, Reifman J. QuartetS-DB: a large-scale orthology database for prokaryotes and eukaryotes inferred by evolutionary evidence. BMC Bioinformatics. 2012;13:143. doi: 10.1186/1471-2105-13-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium U. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41(Database issue):D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19(9):1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bostock M, Ogievetsky V, Heer J. D(3): Data-Driven Documents. IEEE Trans Vis Comput Graph. 2011;17(12):2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 30.NVD3.js [http://nvd3.org]
- 31.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: An interactive web-based network browser. Bioinformatics. 2010;26(18):2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon--level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanks J, Burtnick MN, Brett PJ, Waag DM, Spurgers KB, Ribot WJ, Schell MA, Panchal RG, Gherardini FC, Wilkinson KD, et al. Burkholderia mallei tssM encodes a putative deubiquitinase that is secreted and expressed inside infected RAW 264.7 murine macrophages. Infect Immun. 2009;77(4):1636–1648. doi: 10.1128/IAI.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozue JAC S, Amemiya K, Chua J, Cote CK, Toothman RG, Dankmeyer JR, Klimko CP, Wilhelmsen CL, Raymond JW, Zavaljevski N, Reifman J, Wallqvist A. Phenotypic characterization of a novel virulence-factor deletion strain of Burkholderia mallei that provides partial protection against inhalational glanders in mice. Front Cell and Infect Microbiol. 2016;6:21. doi: 10.3389/fcimb.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrazek J, Nierman WC, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64(6):1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 36.Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, et al. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun. 2011;79(4):1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Schroder I, French CT, Jaroszewicz A, Yee XJ, Teh BE, Toesca IJ, Miller JF, Gan YH. Characterization and analysis of the Burkholderia pseudomallei BsaN virulence regulon. BMC Microbiol. 2014;14:206. doi: 10.1186/s12866-014-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopf V, Gohler A, Eske-Pogodda K, Bast A, Steinmetz I, Breitbach K. BPSS1504, a cluster 1 type VI secretion gene, is involved in intracellular survival and virulence of Burkholderia pseudomallei. Infect Immun. 2014;82(5):2006–2015. doi: 10.1128/IAI.01544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, et al. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci U S A. 2004;101(39):14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Web-enabled DBSecSys 2.0 database is publicly accessible at http://dbsecsys.bhsai.org. The DBSecSys 2.0 Web interface has been tested in the following Web browsers: Google Chrome (version 48), Microsoft Internet Explorer (version 11), and Mozilla Firefox (version 31). The “User’s Guide” page of the DBSecSys 2.0 Web interface includes a step-by-step description of all DBSecSys 2.0 features. For additional questions about the database, please contact dbsecsys@bhsai.org.


