Abstract
Chlamydia trachomatis serovars D-K are obligate intracellular bacteria that have tropism for the columnar epithelial cells of the genital tract. Chlamydia trachomatis infection has been reported to induce modifications in immune cell ligand expression on epithelial host cells. In this study, we used an in vitro infection model that resulted in a partial infection of C. trachomatis-exposed primary-like immortalized endocervical epithelial cells (A2EN). Using this model, we demonstrated that expression of the natural killer (NK) cell activating ligand, MHC class I-related protein A (MICA), was upregulated on C. trachomatis-infected, but not on noninfected bystander cells. MICA upregulation was concomitant with MHC class I downregulation and impacted the susceptibility of C. trachomatis-infected cells to NK cell activity. The specificity of MICA upregulation was reflected by a higher cytolytic activity of an NK cell line (NK92MI) against C. trachomatis-infected cells compared with uninfected control cells. Significantly, data also indicated that NK cells exerted a partial, but incomplete sterilizing effect on C. trachomatis as shown by the reduction in recoverable inclusion forming units (IFU) when cocultured with C. trachomatis-infected cells. Taken together, our data suggest that NK cells may play a significant role in the ability of the host to counter C. trachomatis infection.
Keywords: Chlamydia, cytolysis, endocervix, immune evasion, MICA, NK cell
Introduction
Genital infections with Chlamydia trachomatis serovars D-K are the most prevalent sexually transmitted bacterial infection (CDC, 2010). The propensity for these intracellular infections to remain relatively asymptomatic in women, combined with the ability of C. trachomatis to survive for extended periods in the genital tract, make this pathogen a major public health challenge. Although the microorganism is susceptible to antibiotics, asymptomatic patients typically go untreated. Infection that ascends into the upper tract can cause pelvic inflammatory disease that can eventually lead to tubal infertility, ectopic pregnancy, and chronic pelvic pain (Brunham & Rey-Ladino, 2005). Chlamydia trachomatis infection also enhances human immunodeficiency virus acquisition and shedding (Plummer et al., 1991; Ghys et al., 1997) and has been implicated as a cofactor in HPV-induced cervical neoplasia (reviewed in Paavonen, 2011] and possibly preterm labor (Baud et al., 2008). Co-evolution of C. trachomatis with its human host has driven the acquisition of several immune evasion strategies that likely contribute to the above and promote continued spread of disease (Brunham & Rey-Ladino, 2005).
Chlamydia trachomatis is an obligate intracellular pathogen and genital serovars have a tropism for columnar epithelial cells of the female and male genital tracts. When C. trachomatis is recognized by the host immune system, innate [natural killer (NK) cells (Tseng & Rank, 1998; Hook et al., 2004, 2005)]; innate-like [NK T (NKT) cells (Yang, 2007)] and adaptive [CD4+ (Ficarra et al., 2008) and CD8+ T cells (Igietseme et al., 1994; Roan & Starnbach, 2006; Ficarra et al., 2008; Igietseme et al., 2009)] immune constituents contribute to host cellular immune defense and/or host immune pathogenesis. To avert detection by CD8+ and CD4+ cells, genital serovars of C. trachomatis decrease epithelial cell surface expression of major histocompatibility (MHC) class I and class II antigen presenting molecules through the secretion of C. trachomatis Protease-like Activity Factor (CPAF), a chlamydia-encoded protein (Zhong et al., 1999, 2000, 2001; Shaw et al., 2002). CPAF is also involved in the degradation of CD1d, the host cell ligand for NKT cells, in penile genital epithelial cells (Kawana et al., 2007, 2008). While most experiments are conducted using supraphysiologic C. trachomatis exposure levels that insure high infection rates, these protocols do not reflect in vivo infection dynamics. We have recently reported the effects of C. trachomatis serovar D on endocervical epithelial cells in vitro using novel techniques that allow more physiologic partial infection of exposed cells and discrete assessment of infected and noninfected bystander cells within a mixed culture (Ibana et al., 2011a). These experiments revealed that cell surface expression of MHC class I products is decreased on both infected and noninfected, bystander cells and suggest that soluble and nonsoluble factors are involved in this downregulation (Ibana et al., 2011a). In this study, we use similar techniques to assess the effects of C. trachomatis infection on endocervical epithelial cell expression of the host cell-expressed NK cell activating ligand, MHC class I-related protein A (MICA; Brunham & Rekart, 2008).
Materials and methods
A2EN epithelial cell culture conditions and C. trachomatis infection
In all infection analyses, a primary-like immortalized endocervical epithelial cell line (A2EN) was utilized. A2EN was derived from primary epithelial cells grown out from an endocervical explant and which were immortalized by transduction with PA317/LXSN-16E6E7-conditioned medium as described previously (Herbst-Kralovetz et al., 2008). These cells were propagated in antibiotic-free keratinocyte serum-free medium (KSFM) supplemented with 30 μg mL−1 recombinant epidermal growth factor (rEGF), 0.1 ng mL−1 bovine pituitary extract (Invitrogen, Carlsbad, CA), and 0.4 mM CaCl2 (Sigma, St. Louis, MO); referred to herein as cKSFM. A2EN cells were grown under 2% O2 and 5% CO2 at 37 °C (Ficarra et al., 2008). Cells were infected with C. trachomatis serovar D (D/UW-3/Cx) in SPG (10 mM sodium phosphate [pH 7.2], 0.25 M sucrose, 5 mM L-glutamic acid) at a multiplicity of infection (MOI) of 1–3 to achieve infection rates of ~40–60% (Ibana et al., 2011a, b) for mixed cell analyses. For cytolytic assays, an MOI of 15 was used to achieve infection rates of 80–85% (Kawana et al., 2007). A mock-infected control and infections with UV-inactivated elementary bodies (UVEB) were included for each infection condition. UVEB were prepared by exposing purified EBs to UV-light (mineralight UVSL-25, at 115 volts, 60 cycles, 0.12 Amps) for 2 h at a 10 mm distance. UVEB were confirmed free of infectious chlamydial particles by infecting HeLa cells at an MOI of up to 100. Immediately after infection, SPG was removed and replaced with cKSFM. Cells for immunofluorescent staining were cultured in 12-well culture plates on coverslips. Cells for flow cytometric analyses were cultured in six-well culture plates and harvested using a mild cell detaching agent, Accutase (Innovative Cell Technologies, San Diego, CA), at the indicated times post infection (hours postinfection or hpi).
Immunofluorescent staining
Mock-infected, UVEB-infected and C. trachomatis-infected cells grown on coverslips were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde, then washed and permeabilized with 0.5% saponin. Cells adhered to the coverslips were blocked with Background Sniper blocking reagent (Biocare Medical, Concord, CA) to inhibit nonspecific staining. To visualize the chlamydial inclusion bodies, C. trachomatis were stained using Meriflour antichlamydial-LPS conjugated to fluorescein isothiocyanate (FITC; Fisher Scientific, Pittsburgh, PA). DAPI (Invitrogen) was used to stain nucleic acids. Stained cells were fixed with Prolong Gold antifade reagent (Invitrogen). Inclusion forming units (IFU) were assessed as previously described by Shirey et al. (2006).
Flow cytometric analyses of MHC class I and MICA expression on infected and uninfected cells
Mock-infected and UVEB-infected A2EN cells and A2EN cells infected with C. trachomatis at a MOI of 2 were harvested, fixed, surface stained with anti-MHC class I–PE (eBiosciences, San Diego, CA) or anti-MICA-PE (BD Biosciences, San Jose, CA), permeabilized using Perm/fix reagent (BD Biosciences) and intracellularly stained with antichlamydial-LPS-FITC (Accurate, Westbury, NY). Cells were analyzed by flow cytometry. Noninfected cells were delineated from C. trachomatis-infected cells in C. trachomatis-infected cultures using Flowjo software (Tree Star, Ashland, OR) by setting the threshold at the baseline fluorescent intensity of unlabeled, mock-infected controls as detected on FL1 (FITC) fluorescence. Infected cells from C. trachomatis-exposed cultures were separated from noninfected bystander cells by setting the gating tool on the population of cells with fluorescence intensity above the threshold. After primary separation of C. trachomatis-infected cells and noninfected bystander cells, MICA and MHC class I expression on noninfected bystander and C. trachomatis-infected cells were determined in the FL2 channel (PE) and were quantified by assessing the median fluorescence intensity (MFI) emitted in the FL2 channel by the gated cell population.
Interexperimental variations in MFI absolute values owing to voltage setting differences between independent experiments were corrected using data transformation. Briefly, absolute MFI data from three to six independent experiments were expressed relative to mock-infected MFI from the same experiment [relative MFI (RMFI) = mock MFI/experimental MFI]. To assess for the effects of C. trachomatis infection on MHC class I and MICA expression relative to the mock-infected control, ‘delta MFI’ was calculated using the formula: ‘delta MFI’ = 1 − RMFI for each experiment. Because Mock RMFI = 1, mock ‘delta MFI’ = 0. ‘Delta MFI’ data points therefore represent the degree of change in absolute MFI comparing experiment-specific C. trachomatis-infected cell populations to its corresponding mock-infected control. A value 0 indicates no change in MHC class I or MICA; negative values indicate a downregulation and positive values indicate an upregulation of the surface ligand expression.
NK cell culture conditions and phenotypic characterization
NK92MI (ATCC, Manassas, VA), an interleukin 2 (IL-2) independent NK cell line was utilized in in vitro cytolytic assays. NK92MI cells were propagated in Alpha-Minimum Essential Medium without ribonucleosides and deoxyribonucleosides (Invitrogen), supplemented with 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid (Sigma), 12.5% horse serum (ATCC), and 12.5% fetal bovine serum (Invitrogen). To assess the expression of MHC class I receptor (KIR) and MICA receptor (NKG2D) on this cell line, NK92MI cells were stained with anti-NKG2D-APC (BD Pharmingen) and anti-KIR-FITC (AbD Serotec) and analyzed by flow cytometry. To compare the cytolytic granule expression of NK92MI with that of peripheral blood mononuclear cell-derived NK cells, both groups of cells were surface stained with anti-CD3-PerCP Cy5.5 (BD Pharmingen) and anti-CD56-APC (BD Biosciences) antibodies. Following surface staining, the cells were permeabilized using perm/fix reagent (BD Biosciences) and intracellularly stained with antigranzyme-PE (Cell Sciences) and antiperforin-FITC (Abcam) antibodies. Perforin and granzyme expression in CD3-CD56+ gated NK cells were assessed using the FlowJo software (TreeStar).
Epithelial cell/NK cell coculture
The endocervical epithelial cell line, A2EN was used as experimental target cells. Infection of A2EN with C. trachomatis serovar D was performed as previously described by Kawana et al. (2007). Chlamydia trachomatis-exposed cells were subsequently cultured for 34 or 42 hpi. Cocultures were established by adding NK92MI cells to the infected A2EN at 34 hpi or 42 hpi. NK92MI cells were cocultured with A2EN cells at ratios of 10 : 1, 5 : 1 and 2.5 : 1 (effector-to-target ratios), for an additional 4 h following the 34 and 42 hpi time points. In a matched C. trachomatis-infected A2EN-NK92MI coculture, 2 μg of neutralizing anti-MICA antibody (AbD Serotec) was added to the culture medium with NK92MI. For the assessment of cytolysis, 50 μL aliquots of cell culture supernatants were collected at the end of the four-hour incubation of the A2EN-NK92MI coculture. For IFU determinations, cell culture supernatants and cell lysates were collected in SPG at the end of coculture incubations. Paired, mock-infected and UVEB-infected A2EN cultures were included in each experimental condition as C. trachomatis infection negative controls. K562 (ATCC), a human erythroleukemia line, was utilized as a control target for NK92MI.
NK cell cytolytic assays
The cytolytic activity of NK cells was assessed using Cyto-Tox 96 (Promega, Madison, WI), a nonradioactive assay based on the release of lactate dehydrogenase. Supernatants collected from the 4 h cell cocultures were added to pyruvate substrate and diaphorase. The formation of colored products was quantified spectrophotometrically at 490 nm. K562 cells were used as a positive control for NK cell cytolytic activity. In each experiment, controls for target spontaneous release, target maximum release, volume correction, culture medium background and effector cell spontaneous release were included. Cytotoxicity was determined as follows:
Assessment of IFUs from NK cell/epithelial cell cocultures
To assess the infectivity of C. trachomatis particles in cultures of infected A2EN cells alone and infected A2EN cells cocultured with NK cells, aliquots from pooled cell culture supernatants and cell lysates were collected from 34 to 42 hpi cultures after exposure to culture medium alone or culture medium with NK92MI for 4 h. HeLa cells plated to confluence on a coverslip of known area were infected with dilutions of cell lysates and supernatants from infected A2EN cells. Infected HeLa cells were fixed, permeabilized, stained with Chlamydial-LPS-FITC, and counterstained with DAPI. DAPI/FITC fluorescence from five randomly selected fields per coverslip was visualized using a 20× objective and a Zeiss AxioObserver microscope outfitted with a Zeiss AxioCam MRm. Images were acquired using Zeiss AxioVision software version 4.6, and the area of each image was calculated using the AxioVision’s scale calibration. Acquired RGB images were processed using the open-source ImageJ derivative, Fiji (http://fiji.sc/wiki/index.php/Fiji) as follows. Images were split into red (discarded), blue and green channels to separate signals from cell nuclei (DAPI), and inclusions (Chlamydial-LPS-FITC). The images in each channel were converted to 8-bit gray-scale and thresholded automatically using the intermodes method to create binary 1-bit images. Binary images were subjected to water shedding to separate the majority of overlapping nuclei and overlapping inclusions. Finally, Fiji’s ‘Analyze Particles’ function was used to enumerate nuclei and inclusions. Circularity was set at 0.3–1.0 during particle analysis. IFUs were then calculated using the formula:
Statistical analyses
Statistical analyses were performed using the Prism software (graphpad). Two-tailed Student’s T-tests were employed to test for significant differences between experimental conditions. A P-value of < 0.05 was considered significant.
Results
Chlamydia trachomatis differentially modulates MICA ligand expression on directly infected and noninfected bystander cells
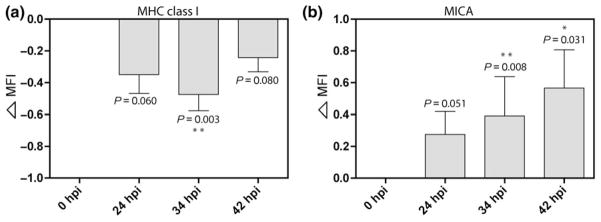
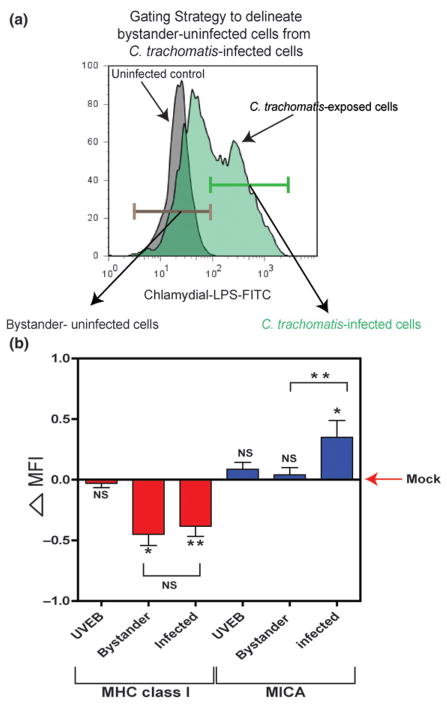
Using standard infection conditions, the cell surface expression of MHC class I and of MICA were analyzed by flow cytometry approximately 6–24 h prior to completion of one C. trachomatis serovar D developmental cycle (Fig. 1). As predicted, MHC class I expression decreased beginning at 24 hpi, with a significant decrease observed at 34 hpi. Intriguingly, MHC class I downregulation was less significant toward the later stage of the C. trachomatis developmental cycle, 42 hpi (Fig. 1a). In contrast, cell surface expression of MICA increased slightly at 24 hpi and continued to increase through 42 h hpi (Fig. 1b). Using methods that infect only a subpopulation of A2EN cells in culture and that allow the host protein response to infection (Fig. 2), we analyzed the change in MHC class I and MICA expression in bystander-noninfected cells and C. trachomatis-infected cells. These two cell populations were delineated by gating based on Chlamydial-LPS positivity (Fig. 2a). We found that C. trachomatis exposure increased the cell surface expression of MICA in infected cells through 38 hpi but had no effect on bystander-noninfected cells (Fig. 2b). In contrast to MHC class I alterations, which affect noninfected bystander cells and C. trachomatis-infected cells (Ibana et al., 2011a), MICA expression on noninfected bystander cells in C. trachomatis-exposed cultures was unaffected. Further, we also demonstrated that active C. trachomatis infection is required for changes in ligand expression to occur, as these phenomena were not observed when cells were exposed to UV-inactivated EBs (Fig. 2b). These data clearly indicate distinct kinetics and effects of C. trachomatis on MHC class I and MICA and suggest that cytokines and/or chemokines released by infected host cells do not influence MICA expression on neighboring cells.
Fig. 1.
Alterations in MHC class I and MICA expression on A2EN endocervical epithelial cells after Chlamydia trachomatis infection. Flow cytometric assessment of MHC class I and MICA expression on mock-infected cells and C. trachomatis-infected A2EN cells at 24, 34, and 42 hours postinfection (hpi). Chlamydia trachomatis-infected cells were gated based on Chlamydial-LPS-FITC positivity. Data shown are representative of means and standard deviations of ‘delta MFI’ from four independent experiments. (a) MHC class I expression of C. trachomatis-infected cells was significantly downregulated at 34 hpi (**P < 0.01). (b) MICA expression progressively increased over time postinfection. Statistical analyses were performed using Student’s t-test; P-values are included to demonstrate the trends toward significant differences in MHC class I and MICA expression of A2EN cells at different times post-C. trachomatis infection when compared to the mock-infected control. The formula for calculating ‘delta MFI’ is described in materials and methods; a delta MFI of 0.5 represents a 100% increase in the absolute MFI value relative to mock-infected cell ligand expression.
Fig. 2.
Differential MHC class I and MICA expression on Chlamydia trachomatis-infected and uninfected, bystander A2EN endocervical epithelial cells. The expression of MHC class I and MICA were assessed in different populations of A2EN cells 38 hpi. (a) Uninfected bystander cells were delineated from C. trachomatis-infected cells based on Chlamydial-LPS-FITC positivity. Uninfected bystander cells were identified by setting the gate below the threshold of basal-FITC signals from uninfected cells (Chlamydia-negative, mock-infected control). Chlamydia trachomatis-infected cells were identified by gating for cells above the threshold FITC signal of the control. (b) The degree of change in MHC class I and MICA expression on A2EN exposed to UV-irradiated elementary bodies (UVEB) and those from bystander-uninfected cells and C. trachomatis-infected cells from an A2EN cell culture exposed to infectious EBs are presented as delta MFI values. The horizontal line represents the delta MFI of mock-infected cells and the degree of difference in MHC class I and MICA expression between the different A2EN cell groups and the mock-infected control is illustrated by the diversion from mock delta MFI = 0. Negative delta MFI values indicate a decrease in MHC class I expression, while positive delta MFI values indicate an increase in MICA expression; ‘delta MFI’ = 0.5 represents a 100% upregulation of the absolute MFI value (see Materials and methods for ‘delta MFI’ calculation). Statistical analyses were performed using Student’s t-test. Asterisks indicate a significant difference relative to the mock-infected control (*P < 0.5; **P < 0.01). Differences in MHC class I and MICA expression between bystander cells and C. trachomatis-infected cells were also tested. Delta MFIs shown in the graph are the means and standard deviations from three to six independent experiments.
Changes in host NK ligand expression after C. trachomatis exposure impacts epithelial cell susceptibility to NK lysis
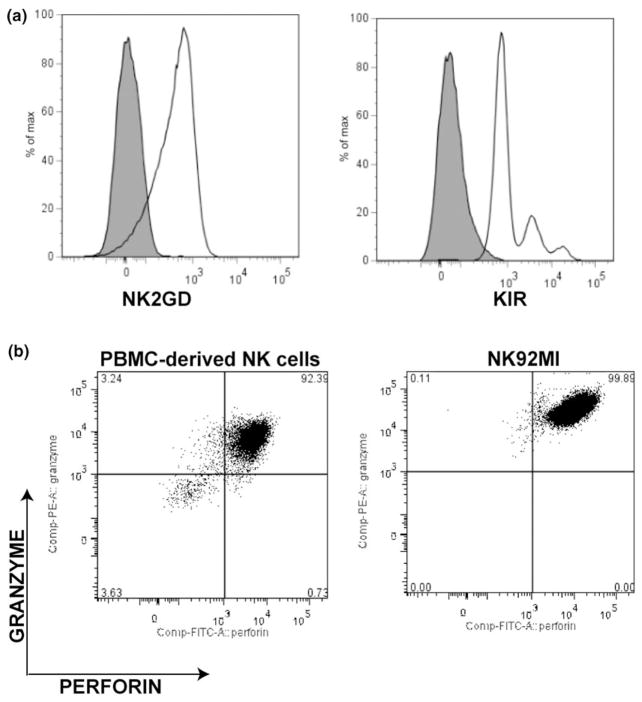
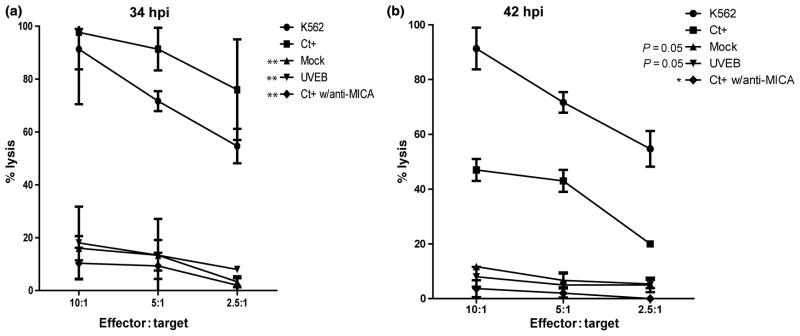
To assess the physiological consequences of C. trachomatis serovar D-mediated MHC class I and MICA modulation, mock-infected, UVEB-infected, and C. trachomatis-infected A2EN cells were exposed to NK92MI cells in coculture experiments. NK92MI expresses NK2GD and KIR –receptors for MICA and MHC class I, respectively, (Fig. 3a). Similar to NK cells derived from peripheral blood mononuclear cells, these cells also contain the intracellular cytolytic granule proteins perforin and granzyme (Fig. 3b). Morphologic assessment of C. trachomatis-infected and mock-infected cocultures revealed that the majority of mock-infected cells retain normal A2EN monolayer morphology over 4 h of exposure (data not shown), while infected cells reveal morphologic evidence of cell lysis, including membrane blebbing (Video S1, Supporting information). Quantification of LDH release confirmed a significant increase in A2EN cell lysis among infected cells at 34 hpi when compared to mock-infected control (P < 0.01), suggesting that C. trachomatis infection enhances the susceptibility of infected endocervical epithelial cell to NK cell cytolytic activity (Fig. 4a). Pertinent to these observations, addition of a neutralizing anti-MICA antibody significantly decreased NK92MI lytic activity against C. trachomatis-infected cultures (P < 0.01). This indicates that the enhanced C. trachomatis-infected cell lysis by NK cells was dependent on MICA. Furthermore, no significant increase in susceptibility to NK cell lysis was observed in A2EN cells infected with UV-inactivated Chlamydial elementary bodies, supporting previous data that active C. trachomatis infection is required for the modulation of NK ligand expression to increase NK cell lysis.
Fig. 3.
NK cell line (NK92MI) phenotypic assessment. (a) NK92MI expression of the NK cell receptor for MHC class I, NK2GD, and the NK cell receptor for MICA, KIR. (b) A comparison of NK92MI perforin and granzyme expression with PBMC-derived NK cells. Receptor and cytolytic granule expressions were assessed by flow cytometry. Data revealed that NK92MI expresses NK cell receptors for MHC class I and MICA and is equipped with the necessary cytolytic molecules to mount a granule-mediated cytolysis of target cells. Results are representative of three profiling experiments.
Fig. 4.
Lysis of A2EN endocervical epithelial cells by NK cells in co-culture. (a) Percent lysis of Chlamydia trachomatis-infected A2EN cells at 34 hpi (b) Percent lysis of C. trachomatis-infected A2EN at 42 hpi. Percent lysis of C. trachomatis-infected cells at 34 and 42 hpi were assessed after 4 h incubation with NK92MI at 10 : 1, 5 : 2 and 2.5 : 1 effector-to-target ratios by measuring LDH release. The percent lysis of C. trachomatis-infected cells was compared against percent lyses of mock-infected control, UVEB-infected cells, and anti-MICA-treated cultures. Differences in percent lyses of C. trachomatis-infected culture and control cultures were assessed using Students t-testing. Asterisks indicate statistical significance (*P < 0.05; **P < 0.01). Data shown are means and standard deviations of percent lysis from two experiments performed in triplicate.
Interestingly, the differences in lysis of C. trachomatis-infected A2EN vs. mock-infected, UVEB-exposed and anti-MICA-treated targets are markedly greater at 34 hpi than at 42 hpi (Fig. 4). These data indicate that there is a significant decrease in the efficiency of lysis of C. trachomatis-infected A2EN cells at later time points postinfection (42 hpi) when compared to earlier stage infection (34 hpi) and suggest that the temporal modulation of MHC class I downregulation may impact the susceptibility of C. trachomatis-infected cells to NK cell lysis.
NK-mediated cell lysis may exert a limited sterilizing effect on chlamydial growth
Infected host cell lysis could result in the release of infectious or noninfectious chlamydial particles. To evaluate the infectivity of the C. trachomatis released from NK cell-exposed infected cells, pooled A2EN cell lysates and culture supernatants from C. trachomatis-infected cells cocultured with NK cells were compared with those cultured for the same period of time postinfection but in the absence of NK cells. The marked decrease in recoverable IFU from cells cocultured with NK92MI cells (Fig. 5; Fig. S1) suggests that these effector cells exert some degree of sterilizing effect on C. trachomatis-infected endocervical cells and that host NK cells could decrease the infectious burden during C. trachomatis infection. Surprisingly, however, we note that although efficient lysis of C. trachomatis-infected cells was observed at 34 hpi, the observed decrease in IFU recovered was only twofold. These data suggest that C. trachomatis may be equipped with some form of escape mechanisms despite NK cell-mediated lysis of its host cells.
Fig. 5.
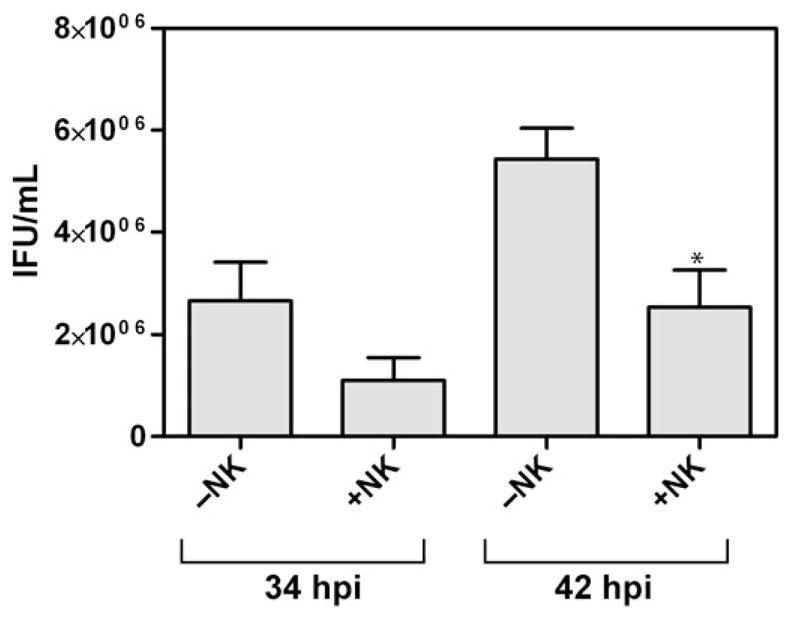
Infectious particles recovered from Chlamydia trachomatis-infected cells cultured in the presence and absence of NK cells. Quantification of IFUs in pooled cell lysates and culture supernatants derived from 34 to 42 hpi C. trachomatis-infected cultures collected after 4-h incubation with or without NK cells. A twofold decrease in IFU was observed in the presence of NK cells. Graph represents mean and standard deviation based on four sets of experiments.
Discussion
Infectious pathogens evade innate and adaptive host immune detection through modulation of host responses. Successful pathogens, including C. trachomatis, exert overlapping and redundant mechanisms that often include alterations in those host ligands that mediate interactions with innate and adaptive immune cells (Tortorella et al., 2000). While well-orchestrated, pathogen protective strategies would promote evasion of antigen nonspecific innate immunity and antigen-specific adaptive responses, co-evolution of pathogen and host enable a balance between pathogen evasion and host protection. For C. trachomatis, we and others have shown that host cell MHC class I, Class II, and CD1d are degraded in infected cells relatively late in the pathogen’s developmental cycle (Zhong et al., 1999; : Zhong et al., 2000; : Zhong et al., 2001; Kawana et al., 2007, 2008). This occurs well after the initiation of chemokine/cytokine secretion by C. trachomatis-infected epithelial cells, which usually does not begin until 20–24 h after infection (Rasmussen et al., 1997). The latter delay may allow a window for unfettered pathogen growth and development. We have recently demonstrated that downregulation of cell surface expression of MHC class I in C. trachomatis-infected A2EN cells can be seen on infected cells and on bystander, noninfected cells in culture (Ibana et al., 2011a), which may further protect C. trachomatis pathogens from antigen-specific clearance. By harnessing our capability to assess the host epithelial cell response to C. trachomatis in both bystander-noninfected cells and C. trachomatis-infected cells, we now show that the effects on MHC class I and on MICA kinetically occur in tandem, beginning prior to 24 hpi and lasting until late in the developmental cycle. Unlike its effects on MHC class I, the effects of C. trachomatis on MICA expression include an upregulation of expression, effects that are significantly more prolonged (still rising at 42 hpi) and effects that are limited to infected cells. The onset of increases in MICA correlate with the reported timing of increased pro-inflammatory cytokine and chemokine secretion in C. trachomatis-infected cells in vitro (Rasmussen et al., 1997). Still, the fact that increases in MICA are seen only on infected cells but not on uninfected bystanders in the same culture suggests that soluble mediators are not sufficient for these effects.
Chlamydia trachomatis infection mediates MHC class I downregulation through direct mechanisms involving the degradation of the transcription factor, RFX5, by chlamydia protease-like activity factor (Zhong et al., 2000). We have previously demonstrated that ‘soluble factors’ could also mediate the downregulation of MHC class I (Ibana et al., 2011a). The downregulation of MHC class I by cytokines, including IL-10 (Caspar-Bauguil et al., 2000) and CXCL12 (Wang et al., 2008) has been demonstrated in other culture models, supporting our previous observation that MHC class I downregulation occurs indirectly in the bystander-noninfected cells present in C. trachomatis-infected A2EN cells (Ibana et al., 2011a). Cytokine-mediated induction of dendritic cell MICA transcription by IFNα has been reported (Jinushi et al., 2003), but the overall effects of cytokines on MICA expression appear to be quite pleiotropic with varying effects depending on cell type and environment (reviewed in Champsaur & Lanier, 2010). In the present study, we observed that MICA is upregulated only in infected cells, demonstrating that the mechanisms underlying C. trachomatis-associated changes in MICA differ from those altering expression of MHC class I and suggesting C. trachomatis infection does not promote the production of soluble MICA-inducing mediators in our culture system.
MICA was first described as cell stress-induced protein in the gastrointestinal epithelium (Groh et al., 1996). Increased MICA expression has been observed during both viral (cytomegalovirus) and bacterial (M. tuberculosis) infections (Groh et al., 2001; Das et al., 2001). Our observation that upregulation of MICA was limited to C. trachomatis-infected cells may indicate that this induction is via infection-derived stress or danger signals that are absent in noninfected bystander cells. Currently, the exact mechanism underlying the induction of MICA expression during viral and bacterial infection is not completely understood. Interestingly, a recent study suggested that human microRNAs can regulate MICA expression, allowing the maintenance of MICA protein expression at a particular threshold while facilitating acute upregulation of MICA during cellular stress (Stern-Ginossar et al., 2008). If C. trachomatis infection induces MICA expression by interfering with the host microRNA-mediated control pathways, this may explain why MICA induction does not occur on uninfected bystander cells. The latter effect would protect the host from unwarranted NK cell activation. While we speculate here on the involvement of microRNA-mediated control of MICA expression, we do not discount other potential mechanisms involving MICA upregulation during C. trachomatis infection. To this point, our observations certainly call for further studies on how C. trachomatis may facilitate direct and indirect control of host ligand expressions, as this may be significant in furthering our understanding of the impact of this bacterium on a variety of host cellular immune responses, including cytolytic CD8 T cells and NK cells.
The cytolytic CD8 T cell is a key mediator in the control of many intracellular microbial infections. However, the protective role of CD8 T cells against C. trachomatis infection is not clear, as numerous reports based on mouse models of C. trachomatis infection suggest that CD4 T cells are central to protective immunity against this bacterium. Nevertheless, it has also been shown that adoptive transfer of Chlamydia-specific CD8 T cells to MoPn-infected mice results in the resolution of infection (Igietseme et al., 1994). In vitro, it has also been demonstrated that a Chlamydia-specific-CD8 T cell clone exhibits cytolytic activity against C. trachomatis-infected human epithelial cells in coculture experiments (Kim et al., 1999). Furthermore, differing from mouse models (Su and Caldwell, 1995), a significant CD8 T cell infiltrate is observed in the human endocervix during C. trachomatis infection (Ficarra et al., 2008). If one accepts the possibility that CD8 T cells may play some role in protective immunity against C. trachomatis infections in humans, when viewed from the perspective of the pathogen, our results suggest that decreased MHC expression on infected and neighboring noninfected cells may be advantageous to chlamydial survival in vivo, widening the time frame for unfettered growth within the infected cell and possibly for spread of the infection. However, from the perspective of the host response to infection, a decrease in MHC expression in conjunction with the increase in MICA expression on infected cells may be, through NK cell-mediated cytolysis, the pathogen’s death knell.
While MHC downregulation could be utilized by C. trachomatis to evade host CD4+ and CD+8 T cell responses, MICA upregulation in combination with MHC class I downregulation is associated with enhanced susceptibility of intracellular microorganisms to NK cell activity (Bauer et al., 1999). The role of NK cells in the early response to genital chlamydial infection has been implicated in murine studies that demonstrate that depletion of NK cells results in exacerbation of chlamydial pathogenesis (Tseng & Rank, 1998). Our in vitro data also indicate that C. trachomatis infection renders A2EN endocervical epithelial cells susceptible to NK cell lysis. This finding is similar to observations reported by others (Hook et al., 2004) using infected SiHa cervical epithelial cells and NK cells derived from human peripheral blood mononuclear cells. In this study, we extended Hook et al.’s (2004) work by examining the temporal modulation of both MHC class I and MICA during the course of C. trachomatis infection of an immortalized primary endocervical epithelial cell (A2EN). Our data suggest that NK cells lyse C. trachomatis-infected cells more efficiently at 34 hpi, when secondary differentiation to infectious EB is at an early stage, compared with a later stage (42 hpi). The increased activity of NK cells toward early stage C. trachomatis-infected cells may be beneficial to the host by reducing the levels of infectious EBs that can be released. We also investigated the effect of NK-mediated lysis of C. trachomatis-infected cells on the level of recoverable IFUs. Curiously, although we observed that the recoverable IFUs decreased in the presence of NK cells, the magnitude of this decrease was smaller than effects on cytolysis efficiency.
NK cytolytic activity is primarily mediated by perforin, a pore-forming protein that acts as a channel for entry of granzymes (Reviewed in Lieberman, 2003), both of which are expressed in the NK cell line used here. Granzymes induce apoptosis in target cells, consistent with the membrane blebbing and cytolysis we observed when C. trachomatis-infected A2EN cells were exposed to the NK cell line (NK92MI). Therefore, while NK lysis may deprive C. trachomatis of its intracellular niche, we hypothesize that C. trachomatis may be equipped with a mechanism to survive or escape NK cell-mediated host cell lysis. Thus, we believe that our data warrants further investigation on the impact of NK cell activity on C. trachomatis, as this may reveal novel survival mechanisms used by this bacterium against host innate immune response. This capacity of Chlamydia is reminiscent of recent observations made with the sexually transmitted pathogen Neisseria gonorrheae, which is able to escape/suppress the effects of neutrophil-associated oxidative bursts (Johnson & Criss, 2011).
Interestingly, while our data and that of Hook et al. (2004) demonstrate increased susceptibility of C. trachomatis-infected cells to NK cell lysis, Mavoungou et al. (1999) have demonstrated that NK cells purified from the peripheral blood of C. trachomatis-infected patients have reduced IFNγ release and lytic capacity. These patients included those with genital and nongenital C. trachomatis serovars. Discrepancies among existing human studies on the role of NK cells in clearing C. trachomatis may reflect heterogeneity among NK cell receptors and their host-expressed ligands. Gene polymorphism in the site encoding the human activating NK cell receptor, NKG2D, has been shown to influence NK cell activity and susceptibility to some infectious diseases (Ma et al., 2010). Polymorphisms in human MICA have also been reported and may alter susceptibility to NK cell lysis (Ahmad et al., 2002; Karacki et al., 2004; Tosh et al., 2006). In light of the recent findings by Mei et al. (2009) that C. trachomatis-specific IgG antibodies were associated with tubal pathology but that a specific MICA allele (MICA*800) was negatively correlated with the presence of these antibodies in infertile women, we believe that specific MICA allele restriction may have an impact of susceptibility of the human host to C. trachomatis infection and in the development of disease. Therefore, while our data indicate that C. trachomatis infection may generally induce susceptibility to NK cell activity, we hypothesize that an individual’s NK2GD and MICA allelic composition may modify the degree of protection conferred by NK cells. Thus, in some individuals, a specific NKG2D and MICA allelic composition may facilitate C. trachomatis’ escape from the NK cell-mediated immune response more efficiently than other alleles. Such possibilities may explain why C. trachomatis infection remains an endocervical infection is some women but establishes acute ascending infection in others. They may also provide insight into why infection may be spontaneously cleared in several weeks or months in some individuals but remain for highly extended periods of time in others (Morre et al., 2002; Molano et al., 2005; Brunham & Rekart, 2008).
Supplementary Material
Acknowledgments
This work was supported by NIH grants U19AI061972 and AI095859 and by the Louisiana Vaccine Center and the South Louisiana Institute for Infectious Disease Research sponsored by the Louisiana Board of Regents. We thank Connie Porretta for technical assistance with flow cytometric experiments and Dr. Tim Foster for insightful comments with respect to data presentation.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Video S1. Time-lapse cinematography of a NK cell killing a C. trachomatis-infected endocervical epithelial cell.
Fig. S1. An example of the automated counting method described in the Materials and methods section.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ahmad T, Marshall SE, Mulcahy-Hawes K, et al. High resolution MIC genotyping: design and application to the investigation of inflammatory bowel disease susceptibility. Tissue Antigens. 2002;60:164–179. doi: 10.1034/j.1399-0039.2002.600207.x. [DOI] [PubMed] [Google Scholar]
- Baud D, Regan L, Greub G. Emerging role of Chlamydia and Chlamydia-like organisms in adverse pregnancy outcomes. Curr Opin Infect Dis. 2008;21:70–76. doi: 10.1097/QCO.0b013e3282f3e6a5. [DOI] [PubMed] [Google Scholar]
- Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- Brunham RC, Rekart ML. The arrested immunity hypothesis and the epidemiology of chlamydia control. Sex Transm Dis. 2008;35:53–54. doi: 10.1097/OLQ.0b013e31815e41a3. [DOI] [PubMed] [Google Scholar]
- Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- Caspar-Bauguil S, Puissant B, Nazzal D, Lefevre J-C, Thomsen M, Salvayre R, Benoist H. Chlamydia pneumonia induces interleukin-10 production that down-regulates major histocompatibility complex class I expression. J Infect Dis. 2000;182:1394–1401. doi: 10.1086/315856. [DOI] [PubMed] [Google Scholar]
- CDC. 2009 Sexually transmitted disease surveillance. 2010 from http://www.cdc.gov/std/stats09/chlamydia.htm.
- Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das H, Groh V, Kiujl C, Sugita M, Morita CT, Spies T, Bukowski JF. MICA engagement by human Vgamma2Vdelta2 T cells enhances their antigen-dependent effector function. Immunity. 2001;15:83–93. doi: 10.1016/s1074-7613(01)00168-6. [DOI] [PubMed] [Google Scholar]
- Ficarra M, Ibana JS, Poretta C, et al. A distinct cellular profile is seen in the human endocervix during Chlamydia trachomatis infection. Am J Reprod Immunol. 2008;60:415–425. doi: 10.1111/j.1600-0897.2008.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghys PD, Fransen K, Diallo MO, et al. The associations between cervicovaginal HIV shedding, sexually transmitted diseases and immunosuppression in female sex workers in Abidjan, Cote d’Ivoire. AIDS. 1997;11:F85–F93. doi: 10.1097/00002030-199712000-00001. [DOI] [PubMed] [Google Scholar]
- Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. P Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement of MIC induced on virus infected cells. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- Herbst-Kralovetz MM, Quayle AJ, Ficarra M, Greene S, Rose WA, 2nd, Chesson R, Spagnuolo RA, Pyles RB. Quantification and comparison of toll-like receptor expression and responsiveness in primary and immortalized human female lower genital tract epithelia. Am J Reprod Immunol. 2008;59:212–224. doi: 10.1111/j.1600-0897.2007.00566.x. [DOI] [PubMed] [Google Scholar]
- Hook CE, Telyatnikova N, Goodall JC, Braud VM, Carmichael AJ, Wills MR, Gaston JS. Effects of Chlamydia trachomatis infection on the expression of natural killer (NK) cell ligands and susceptibility to NK cell lysis. Clin Exp Immunol. 2004;138:54–60. doi: 10.1111/j.1365-2249.2004.02596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook CE, Matyszak MK, Gaston JS. Infection of epithelial and dendritic cells by Chlamydia trachomatis results in IL-18 and IL-12 production, leading to interferon-gamma production by human natural killer cells. FEMS Immunol Med Microbiol. 2005;45:113–120. doi: 10.1016/j.femsim.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Ibana JA, Schust DJ, Sugimoto J, Nagamatsu T, Greene SJ, Quayle AJ. Chlamydia trachomatis immune evasion via downregulation of MHC class I surface expression involves direct and indirect mechanisms. Infect Dis Obstet Gynecol. 2011a doi: 10.1155/2011/420905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibana JA, Belland RJ, Zea AH, Schust DJ, Nagamatsu T, AbdelRahman YM, Tate DJ, Beatty WL, Aiyar AA, Quayle AJ. Inhibition of indoleamine 2,3-dioxygenase activity by levo-1-methyl tryptophan blocks gamma interferon-induced Chlamydia trachomatis persistence in human epithelial cells. Infect Immun. 2011b;79:4425–4437. doi: 10.1128/IAI.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igietseme JU, Magee DM, Williams DM, Rank RG. Role for CD8+ T cells in antichlamydial immunity defined by Chlamydia-specific T-lymphocyte clones. Infect Immun. 1994;62:5195–5197. doi: 10.1128/iai.62.11.5195-5197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igietseme JU, He Q, Joseph K, Eko FO, Lyn D, Ananaba G, Campbell A, Bandea C, Black CM. Role of T lymphocytes in the pathogenesis of Chlamydia disease. J Infect Dis. 2009;200:926–934. doi: 10.1086/605411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinushi M, Takehara T, Kanto T, Tatsumi T, Groh V, Spies T, Miyagi T, Suzuki T, Sasaki T, Hayashi N. Critical role for MHC class I-related chain A and B expression on IFN-alpha-stimulated dendritic cells in NK cell activation: impairment of chronic hepatitis C virus infection. J Immunol. 2003;170:1249–1256. doi: 10.4049/jimmunol.170.3.1249. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Criss AK. Resistance of Neisseria gonorrheae to neutrophils. Front Microbiol. 2011;2:1–12. doi: 10.3389/fmicb.2011.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karacki PS, Gao X, Thio CL, et al. MICA and recovery from hepatitis C virus and hepatitis B virus infections. Genes Immun. 2004;5:261–266. doi: 10.1038/sj.gene.6364065. [DOI] [PubMed] [Google Scholar]
- Kawana K, Quayle AJ, Ficarra M, et al. CD1d degradation in Chlamydia trachomatis-infected epithelial cells is the result of both cellular and chlamydial proteasomal activity. J Biol Chem. 2007;282:7368–7375. doi: 10.1074/jbc.M610754200. [DOI] [PubMed] [Google Scholar]
- Kawana K, Matsumoto J, Miura S, et al. Expression of CD1d and ligand-induced cytokine production are tissue specific in mucosal epithelia of the human lower reproductive tract. Infect Immun. 2008;76:3011–3018. doi: 10.1128/IAI.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-K, Angevine M, Demick K, et al. Induction of Class I-restricted CD8+ CTLs specific for the major outer membrane protein of Chlamydia trachomatis in human genital tract infections. J Immunol. 1999;162:6855–6866. [PubMed] [Google Scholar]
- Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- Ma J, Guo X, Wu X, Li J, Zhu X, Li Z, Pan L, Li T, Li H, Liu Y. Association of NKG2D genetic polymorphism with susceptibility to chronic hepatitis B in a Han Chinese population. J Med Virol. 2010;82:1501–1507. doi: 10.1002/jmv.21855. [DOI] [PubMed] [Google Scholar]
- Mavoungou E, Poaty-Mavoungou V, Toure FS, Sall A, Delicat A, Yaba P, Mandeme Y, Nabias R, Lansoud-Soukate J. Impairment of natural killer cell activity in Chlamydia trachomatis infected individuals. Trop Med Int Health. 1999;4:719–727. doi: 10.1046/j.1365-3156.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- Mei B, Luo Q, Du K, Huo Z, Wang F, Yu P. Association of MICA gene polymorphisms with Chlamydia trachomatis infection and related tubal pathology in infertile women. Hum Reprod. 2009;24:3090–3095. doi: 10.1093/humrep/dep339. [DOI] [PubMed] [Google Scholar]
- Molano M, Meijer CJ, Weiderpass E, Arslan A, Posso H, Franceschi S, Ronderos M, Munoz N, van den Brule AJ. The natural course of Chlamydia trachomatis infection in asymptomatic Colombian women: a 5-year follow-up study. J Infect Dis. 2005;191:907–916. doi: 10.1086/428287. [DOI] [PubMed] [Google Scholar]
- Morre SA, van den Brule AJ, Rozendaal L, Boeke AJ, Voorhorst FJ, de Blok S, Meijer CJ. The natural course of asymptomatic Chlamydia trachomatis infections: 45% clearance and no development of clinical PID after one-year follow-up. Int J STD AIDS. 2002;13(Suppl 2):12–18. doi: 10.1258/095646202762226092. [DOI] [PubMed] [Google Scholar]
- Paavonen J. Chlamydia trachomatis infections of the female genital tract: state of the art. Ann Med. 2011 doi: 10.3109/07853890.2010.546365. [DOI] [PubMed] [Google Scholar]
- Plummer FA, Simonsen JN, Cameron DW, et al. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163:233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- Rasmussen SJ, Eckmann L, Quayle AJ, Shen L, Zhang YX, Anderson DJ, Fierer J, Stephens RS, Kagnoff MF. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Invest. 1997;99:77–87. doi: 10.1172/JCI119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roan NR, Starnbach MN. Antigen-specific CD8+ T cells respond to Chlamydia trachomatis in the genital mucosa. J Immunol. 2006;177:7974–7979. doi: 10.4049/jimmunol.177.11.7974. [DOI] [PubMed] [Google Scholar]
- Shaw AC, Vandahl BB, Larsen MR, Roepstorff P, Gevaert K, Vandekerckhove J, Christiansen G, Birkelund S. Characterization of a secreted Chlamydia protease. Cell Microbiol. 2002;4:411–424. doi: 10.1046/j.1462-5822.2002.00200.x. [DOI] [PubMed] [Google Scholar]
- Shirey KA, Jung JY, Carlin JM. Up-regulation of gamma interferon receptor expression due to Chlamydiatoll-like receptor interaction does not enhance signal transducer and activator of transcription 1 signaling. Infect Immun. 2006;74:6877–6884. doi: 10.1128/IAI.00505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Gur C, Biton M, Horowitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Su H, Caldwell HD. CD4+ T cells play a significant role in adoptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect Immun. 1995;63:3302–3308. doi: 10.1128/iai.63.9.3302-3308.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- Tosh K, Ravikumar M, Bell JT, Meisner S, Hill AV, Pitchappan R. Variation in MICA and MICB genes and enhanced susceptibility to paucibacillary leprosy in South India. Hum Mol Genet. 2006;15:2880–2887. doi: 10.1093/hmg/ddl229. [DOI] [PubMed] [Google Scholar]
- Tseng CT, Rank RG. Role of NK cells in early host response to chlamydial genital infection. Infect Immun. 1998;66:5867–5875. doi: 10.1128/iai.66.12.5867-5875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang L, Qiao A, Watson K, Zhang J, Fan GH. Activation of CXCR4 triggers ubiquitination and downregulation of major histocompatibility complex class I (MHC-I) on epithelioid carcinoma HeLa cells. J Biol Chem. 2008;283:3951–3959. doi: 10.1074/jbc.M706848200. [DOI] [PubMed] [Google Scholar]
- Yang X. Natural killer T (NKT) cell subsets in chlamydial infections. Adv Exp Med Biol. 2007;601:243–246. doi: 10.1007/978-0-387-72005-0_25. [DOI] [PubMed] [Google Scholar]
- Zhong G, Fan T, Liu L. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med. 1999;189:1931–1938. doi: 10.1084/jem.189.12.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Liu L, Fan T, Fan P, Ji H. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J Exp Med. 2000;191:1525–1534. doi: 10.1084/jem.191.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.