Abstract
Soil microbiota play a critical role in soil biogeochemical processes and have a profound effect on soil functions. Recent studies have revealed microbial co-occurrence patterns in soil microbial communities, yet the geographic pattern of topological features in soil microbial co-occurrence networks at the continental scale are largely unknown. Here, we investigated the shifts of topological features in co-occurrence networks inferred from soil microbiota along a continental scale in eastern China. Integrating archaeal, bacterial and fungal community datasets, we inferred a meta-community co-occurrence network and analyzed node-level and network-level topological shifts associated with five climatic regions. Both node-level and network-level topological features revealed geographic patterns wherein microorganisms in the northern regions had closer relationships but had a lower interaction influence than those in the southern regions. We further identified topological differences associated with taxonomic groups and demonstrated that co-occurrence patterns were random for archaea and non-random for bacteria and fungi. Given that microbial interactions may contribute to soil functions more than species diversity, this geographic shift of topological features provides new insight into studying microbial biogeographic patterns, their organization and impacts on soil-associated function.
Introduction
Soil microbiota play critical roles in a wide range of biogeochemical cycles and comprise the major pool of living biomass in soil ecosystems (Miltner et al., 2012; Xu et al., 2013). Increasing evidence indicates that a variety of soil factors can shape and be shaped by the microbiome, suggesting a promising avenue for increasing soil health via directed manipulation of the microbiome (Chaparro et al., 2012; Ellouze et al., 2013). Characterizing the capacity of the soil microbiota, its interaction with soil factors and its contribution to various soil biogeochemical processes therefore has the potential to provide important insights into soil functions. This would comprise a systems-level understanding of community function and structure (Fuhrman, 2009). To address this challenge, researchers have started mapping the soil microbiota (Hultman et al., 2015; Panke-Buisse et al., 2015), and the use of high-throughput sequencing analysis has allowed us to characterize the composition and functional attributes of soil microbial communities across broad spatial scales (Fierer and Jackson, 2006; Bates et al., 2011; Fierer et al., 2013). Additionally, recent studies have revealed co-occurrence patterns in soil microbial communities across a wide range of terrestrial ecosystems (Barberán et al., 2012; Fierer et al., 2012).
Network analyses-based approaches have recently been used to investigate co-occurrence patterns between microorganisms in complex environments ranging from the human gut to oceans and soils (Ruan et al., 2006; Fuhrman and Steele, 2008; Faust et al., 2012; Chow et al., 2013). Co-occurrence patterns are ubiquitous and particularly important in understanding microbial community structure, offering new insights into potential interaction networks, and revealing niche spaces shared by community members (Steele et al., 2011; Faust and Raes, 2012; Kara et al., 2013). Recent studies have explored large, complex microbial community datasets and have demonstrated previously unseen co-occurrence patterns, such as strong non-random associations, niche specialization (Faust et al., 2012), unexpected ecological relationships (Zhang et al., 2014), and deterministic processes at different taxonomic levels (Chaffron et al., 2010). Topology-based analysis of large networks has proven powerful for studying the characteristics of co-occurrence patterns at various taxonomic levels, and identifying keystone microbial groups in different soils (Lupatini et al., 2014). Here, we significantly advance this research by providing a comprehensive understanding of the topological shifts of soil bacterial, archaeal and fungal co-occurrence networks at a continental scale.
Eastern Asia represents an ideal continental scale system to explore a complete vegetation gradient from tropical forest to arctic tundra. Comparing the topological properties of the nodes associated with forest soil in different climatic regions and examining network-level topological features can provide us with insight into variations in the co-occurrence patterns along this successional climatic gradient. This approach helps contextualize microbial biogeography by taking into account the complex network of potential interactions among microbes in these environments. Specifically, we addressed the following questions: (i) Do the topological features of co-occurrence network vary between different climate regions? (ii) Do microorganisms from different kingdoms (bacteria, archaea, fungi) exhibit different co-occurrence patterns? (iii) What environmental factors correlate with variation in the topological features of interaction networks? To answer these questions, we performed ribosomal RNA amplicon sequencing analyses on natural, undisturbed forest soil microbiota spanning five successional climate regions and implemented co-occurrence network analysis to examine the topological feature dynamics across this continental scale. Our main objective was to characterize and better understand co-occurrence network patterns in soil microbial communities.
Materials and methods
Soil sampling
We collected three soil samples from a 100 × 100 m2 plot in natural forestry communities at 110 sites across eastern China (distances ranging from 0.7 to 3671.8 km) using a uniform sampling protocol (Supplementary Figure S1). Each soil sample was combined with five soil cores that were taken at a depth ranging from 0 to 15 cm. We removed loose debris from the forest floor and combined each set of five soil cores as one soil sample, giving three biological replicates per plot. All soil samples were transported to our laboratory on ice. Coarse roots and stones were removed, and a subset of the soil was air-dried for analysis of edaphic properties. Based on regional climates and geographic distribution, these sites were categorized into five climatic regions in accordance with the Köppen−Geiger climate classification system (http://en.wikipedia.org/wiki/Köppen_climate_classification/). These included the ‘south region' comprising tropical wet and dry climates (Aw) and two warm temperate climates (Cfa and Cwa), and the ‘north region' comprising warm summer continental climates (Dwb) and hot summer continental climates (Dwa). We obtained mean annual air temperatures and mean annual precipitation values from the WorldClim database (www.worldclim.org). Soil collection protocols and methods for investigating edaphic and environmental properties are described in Supplementary Information.
Ribosomal RNA (rRNA) amplicon sequencing and processing
DNA was extracted from soil samples using the MP FastDNA SPIN Kit for soil (MP Biomedicals, Solon, OH, USA), as per the manufacturer's instructions. Equal amounts (200 ng) of DNA extract from the three replicates were pooled to form a composite DNA sample. DNA purity and concentrations were analyzed with a NanoDrop spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). Isolated DNA was stored at −20 °C for microbial diversity and sequence analyses. We performed 16 S rRNA gene amplification for archaea and bacteria and 18 S rRNA gene amplification for fungi using the microbial tag-encoded FLX amplicon pyrosequencing (TEFAP) procedures described earlier (Sun et al., 2011). A region of the 16 S rRNA genes for archaea (V3−V5 regions) and bacteria (V1−V3 regions) were amplified by primer pairs, A340F90 (GYGCASCAGKCGMGAAW)/A806R96 (GGACATCVSGGGTATCTAAT) and Gray28F (GAGTTTGATCNTGGCTCAG)/Gray519R (GTNTTACNGCGGCKGCTG), respectively. A region of the fungal 18 S rRNA gene was amplified by primer pair, funSSUF (TGGAGGGCAAGTCTGGTG)/funSSUR (TCGGCATAGTTTATGGTTAAG). We used negative (for DNA extraction and PCR) and positive controls throughout the experiment. The amplicons were subjected to 454 pyrosequencing on 11 plates by using the GS-FLX+ implemented by Research and Testing Laboratory (Lubbock, TX, USA).
All pyrosequencing data processing, including sequence quality control, operational taxonomic unit (OTU)-based analysis, taxonomy analysis and diversity indices calculation, was performed using the Mothur software V 1.35.1 (Quast et al., 2013). Briefly, sequences were sorted by barcodes into archaea, bacteria, and fungi. Sequences with barcode ambiguities, those less than 200 bp in length and with average quality scores <25 were culled. Quality-filtered sequences were aligned to the SILVA database release 111 and chimeras were de novo detected and removed by using the UCHIME modules in Mothur. Each unique sequence was considered as an individual OTU and was classified at a 50% confidence threshold within the SILVA database release 111 (Quast et al., 2013). The OTU matrices were rarefied to 300, 3000 and 2000 sequences per sample for archaea, bacteria and fungi, respectively. Following rarefaction, a total of 83 samples were further analyzed by network analyses.
Network construction
To reduce rare OTUs in the data set, we removed OTUs with relative abundances less than 0.01% of the total number of archaeal, bacteria, and fungal sequences, respectively. The co-occurrence network was inferred based on the Spearman correlation matrix constructed with the WGCNA package (Langfelder and Horvath, 2012). The nodes in this network represent OTUs and the edges that connect these nodes represent correlations between OTUs. We adjusted all P-values for multiple testing using the Benjamini and Hochberg false discovery rate (FDR) controlling procedure (Benjamini et al., 2006), as implemented in the multtest R package. The direct correlation dependencies were distinguished using the network deconvolution method (Feizi et al., 2013). Based on correlation coefficients and FDR-adjusted P-values for correlation, we constructed co-occurrence networks. The cutoff of FDR-adjusted P-values was 0.001. The cutoff of correlation coefficients was determined as 0.78 through random matrix theory-based methods (Luo et al., 2006). Network properties were calculated with the igraph package. We generated network images with Gephi (http://gephi.github.io/). All samples were divided into groups by climatic region. The impact of each sample group on the Spearman correlation value of each edge in the network was assessed by dividing the omission score (OS) (Spearman correlation value without these samples) by the absolute original Spearman score (Lima Mendez et al., 2015). To account for group size, the OS was computed repeatedly for random, same-size sample sets. Nonparametric P-values were calculated as the number of times random OSs were smaller than the sample group OS, divided by the number of random OSs (500 for each taxon pair). Edges were classified as region-specific when the ratios of OSs to absolute original scores were below one and adjusted P-values were below 0.05.
Topological feature analysis
We calculated topological features (Supplementary Table S1) for each node in the network with the igraph package (Csardi and Nepusz, 2006). This feature set included betweenness centrality (the number of shortest paths going through a node), closeness centrality (the number of steps required to access all other nodes from a given node), transitivity (the probability that the adjacent nodes of a node are connected, also called the clustering coefficient) and degree (the number of adjacent edges). The betweenness centrality feature was used to measure the centrality of each node in the network. Nodes were further classified as peripheral, intermediate or central by ranking all nodes according to centrality, partitioning this ranked list into three equally populated bins, which were termed ‘centrality tiers' (Greenblum et al., 2012). Nodes with high degree (>100) and low betweenness centrality values (<5000) are recognized as keystone species in co-occurrence networks.
Statistical analyses
The Spearman's rank correlation test was used to examine the correlation between abundance and each topological feature. To test for differences in topological features between climatic regions, we used the Wilcoxon rank-sum test in R. The correlation coefficients across all node-level topological features supported by the igraph package were calculated, and a feature set without any pairwise correlations >0.95 was selected for further analysis. We generated sub-networks for each soil sample from meta-community networks by preserving OTUs presented in each site using subgraph functions in igraph packages. Network-level topological features provided in igraph packages were calculated for each sub-network. We grouped each sub-network by sampling location and used Wilcoxon rank-sum test to determine the different network-level topological features between climatic regions. We then predicted the spatial distribution of these topological features based on Krige interpolation using the function autoKrige in automap packages (Hiemstra et al., 2009). The correlation coefficients between network-level topological features and environmental factors were calculated. The importance of environmental factors (geographic factors, climatic factors and soil properties) for network-level topological features was estimated with multiple regression on distance matrices (MRM) in ecodist packages. The Euclidean distance matrices for environmental factors and network-level topological features standardized with decostand of vegan package were used in MRM models. To test the relationship between network-level topological features and environmental factors, we further compared the first component of principal component analysis for network-level topological features with soil pH or the first principal component analysis components of soil carbon, iron and nitrogen parameters, respectively.
Results
Data sets
We analyzed 1 502 091 Roche 454 FLX-derived rRNA gene amplicon reads (SRR2177920) from 110 soil samples collected from natural, undisturbed forests across eastern China (Supplementary Figure S1). The majority of archaeal sequences belonged to the phyla Crenarchaeota (92.3%) and Euryarchaeota (7.1%). Bacterial sequences primarily comprised phyla (and sub-phyla), Alphaproteobacteria (27.8%), Actinobacteria (11.5%), Acidobacteria (6.8%) and Betaproteobacteria (5.1%). The most abundant fungal phyla were the Ascomycota (73.7%), Mucoromycota (15.5%) and the Basidiomycota (6.3%). Of the 110 soil samples, 83 were selected after filtering. These sites were located in five climatic regions represented by 10 samples in tropical wet and dry climates (Aw), 46 in warm temperate climates (Cfa and Cwa), 21 in hot summer continental climates (Dwa) and 6 in warm summer continental climates (Dwb). These soil samples comprised 1810 archaeal, 648 bacterial and 1370 fungal OTUs with relative abundance greater than 0.01%.
Meta-community co-occurrence network
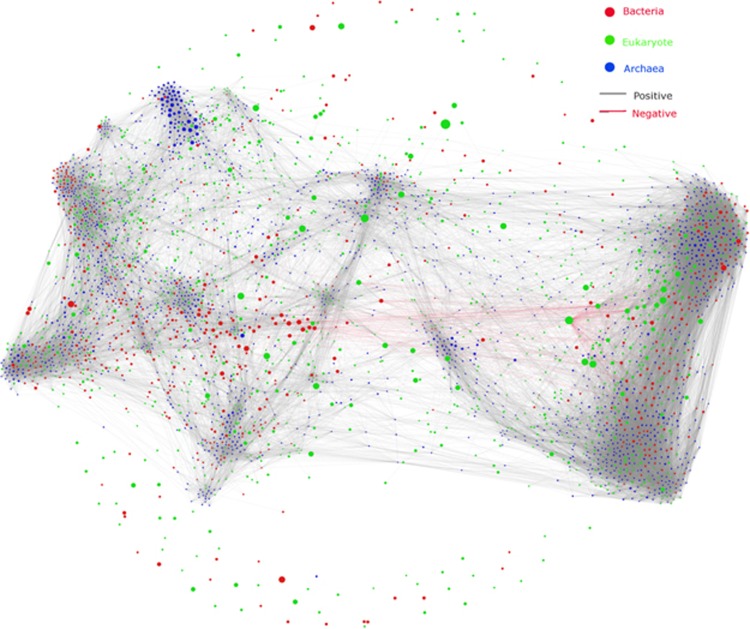
We inferred a meta-community co-occurrence network based on correlation relationships and P-values for correlations adjusted with FDR (Benjamini et al., 2006). The edges arising from indirect interactions in this network were recognized by a deconvolution procedure (Feizi et al., 2013). This generated a meta-community co-occurrence network capturing 66 443 associations among 3828 microbial OTUs (Figure 1). In total, 92.2% of the edges were identified as global, and only 7.8% of edges were region-specific (with region-specific OTUs), including 752 edges in Aw, 519 edges in Cfa, 1049 edges in Cwa, 2809 edges in Dwa and 634 edges in Dwb. The global network roughly followed a scale-free degree distribution (Supplementary Figure S2), meaning that most OTUs had low-degree values, and only a few hub nodes had high-degree values. To determine the difference of degree distribution for nodes from archaea, bacteria and fungi, we classified edges into nine groups (Supplementary Figure S3), each representing edges that linked nodes between different kingdoms. The degree distribution for edges between archaeal nodes was represented by a binomial distribution with a maximum abundance at approximately 20, which may indicate a random structure of networks and a random co-occurrence pattern. The degrees for bacteria and fungi were distributed according to power-law distributions, which indicated a scale-free network structure and a non-random co-occurrence pattern. We have not further analyzed degree distribution patterns at the phylum level because the numbers of nodes for most of the phyla were too small to generate reliable degree-abundance plots.
Figure 1.
The co-occurrence network interactions of soil bacteria, archaea and fungi. The connection stands for a strong (Spearman's ρ>0.78) and significant (P-value<0.001) correlation. The nodes represented unique sequences in the data sets. The size of each node is proportional to the relative abundance.
Betweenness centrality of climatic region-associated OTUs
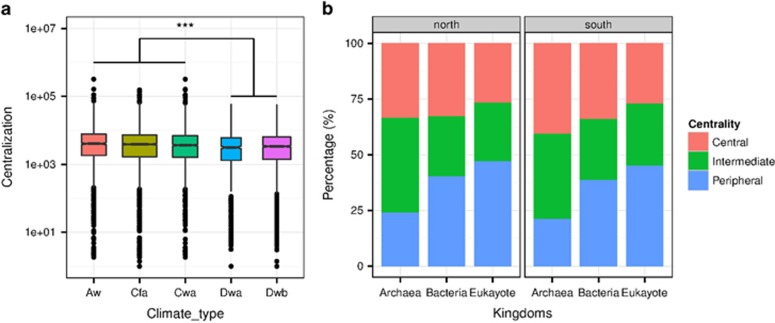
Using the meta-community co-occurrence network outlined above, we examined whether OTUs associated with a specific climatic region exhibited unique node-level topological features. We firstly focused on betweenness centrality, which measures the number of shortest paths going through a given node, as a proxy for the location of this node in relation to other nodes. High betweenness centrality values indicate a core location of this node in the network, whereas low betweenness centrality values indicate a more peripheral location.
Significantly lower betweenness centrality scores were observed for OTUs associated with Dwa and Dwb regions than those associated with Aw, Cfa and Cwa regions (P=8.1 × 10−5, Wilcoxon rank-sum test, Figure 2a). This suggests that the soil microbes from the southern regions were more often located in core, central positions within the network than those from the northern regions. By partitioning the OTUs into three kingdoms, we found significantly higher centrality scores for archaeal OTUs as compared to bacterial and fungal OTUs (P<2.2 × 10−16, and Wilcoxon rank-sum test, Supplementary Figure S4). The betweenness centrality scores were significantly lower for archaeal OTUs associated with Dwa and Dwb regions than for archaeal OTUs associated with Aw, Cfa and Cwa regions (P=6.4 × 10−6, Wilcoxon rank-sum test). However, the betweenness centrality scores for bacterial or fungal OTUs were not significantly different across the different climatic regions (P=0.41 and 0.20, respectively, Wilcoxon rank-sum test). We partitioned the OTUs into three centrality-based tiers and found an overrepresentation of archaeal OTUs in the central and intermediate tiers (Figure 2b). In soil samples from Aw, Cfa and Cwa regions, 78.5% of the archaeal OTUs were classified into the central and intermediate tiers, compared with 61.1% of the bacterial and 54.7% of the fungal OTUs. Similarly, in soil samples from Dwa and Dwb regions, 75.6% of the archaeal OTUs were classified into central and intermediate tiers, compared with 60.3% of bacterial and 52.5% of fungal OTUs.
Figure 2.
Betweenness centralization associated with different climatic regions (a) and percentage of bacterial, archaeal and fungal nodes with different centralization (b). ***P<0.001.
Linking climatic region-associated OTUs to additional node-level topological features
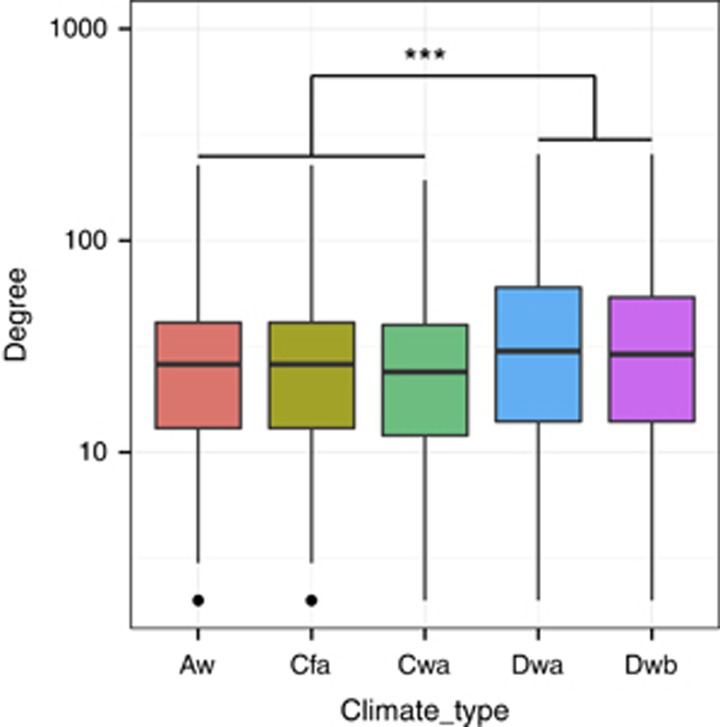
We next examined a number of additional node-level topological measures for each OTU in the meta-community co-occurrence network, including degree and closeness. In contrast to betweenness centrality, these measures are more local in nature, taking into account only the immediate neighborhood of OTUs, and hence capturing a different aspect of network topological features. Degrees of OTUs associated with Dwa and Dwb regions were significantly higher than those associated with Aw, Cfa and Cwa regions (P<2.2 × 10−16, Wilcoxon rank-sum test, Figure 3). Partitioning the set of OTUs in the network into archaeal, bacterial and fungal OTUs, we found that the degrees differed in different climatic regions. Specifically, archaeal OTUs associated with Dwa and Dwb regions had a significantly higher degree compared to archaeal OTUs associated with Aw, Cfa and Cwa regions (P=2.8 × 10−6, Wilcoxon rank-sum test, Supplementary Figure S5). Bacterial OTUs associated with Dwa and Dwb regions had a marginally lower degree than OTUs associated with Aw, Cfa and Cwa regions (P=0.05, Wilcoxon rank-sum test, Supplementary Figure S5). The degrees of fungal OTUs associated with the different climatic regions were not significantly different. Closeness followed a similar trend but no significant differences were observed across the climatic regions for either archaeal, bacterial or fungal OTUs (Supplementary Figure S6).
Figure 3.
Node degree values associated with different climatic regions. ***P<0.001.
We further assessed the relationships between degrees and relative abundances of OTUs in the three domains (Supplementary Figure S7). The number of edges between archaeal nodes increased with relative abundance (R=0.151, P=9.9 × 10−11, Spearman's rank correlation test). Conversely, the degrees associated with bacterial and fungal nodes decreased with increasing relative abundance (R=−0.369 and −0.501, respectively, P<2.2 × 10−16, Spearman's rank correlation test). The degree of abundant OTUs is expected to be high when the co-occurrence pattern is random. Therefore, the co-occurrence pattern was expected to be random for archaeal nodes and non-random for bacterial and fungal nodes.
Such distinct topological features may additionally be used to highlight keystone species in co-occurrence networks. Specifically, nodes with high degree (>100) and low betweenness centrality values (<5000) are recognized as keystone species in co-occurrence networks (Berry and Widder, 2014). A large fraction of these keystone species were unclassified archaea related to the phylum Thaumarchaeota (59 OTUs, whose relative abundance ranged from 0.010 to 0.227%). Major keystone bacterial species included members of the phylum Actinobacteria (16 OTUs, with relative abundances ranging from 0.012 to 0.028%), comprising the orders Gaiellales (8 OTUs), Rubrobacteriales (2 OTUs), Solirubrobacteriales (2 OTUs), Acidobacteriales (1 OTUs) and Corynebacteriales (1 OTUs), and the phylum Proteobacteria (6 OTUs, relative abundance ranged from 0.010 to 0.014%), comprising the order Rhizobiales (5 OTUs) and the uncultured bacterium GR-WP33-30 (1 OTU). Fungal keystone species included members of sub-phylum Pezizomycotina (6 OTUs, relative abundance ranging from 0.013 to 0.023%) and an unclassified Fungal OTU (relative abundance 0.011%).
Network-level topological features changed with climatic regions
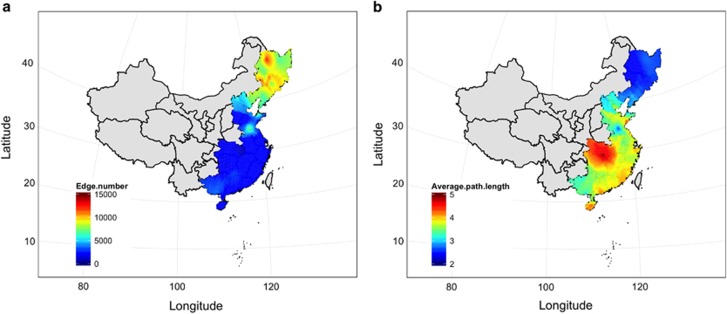
We generated sub-networks for each soil sample by keeping OTUs associated with specific samples and all edges among them in the meta-community co-occurrence network. A number of network-level topological features were calculated for sub-networks and separated into three clusters based on hierarchical cluster analysis on the dissimilarities of those features (Supplementary Figure S8). The first cluster included cluster number, diameter, degree assortativity, betweenness centralization and average path length. The second cluster included transitivity and node number. The third cluster included degree centralization, average nearest-neighbor degree, density, edge number and closeness centralization. To extend our results beyond the 83 soils directly assayed, we predicted spatial distribution maps of network-level topological features using a kriging interpolation method (Heimstra et al., 2009). The predicted spatial patterns showed that the edge numbers (Figure 4a), similar to other topological features in this cluster (Supplementary Figure S8), such as density (Supplementary Figure S9), degree centralization (Supplementary Figure S10) and average nearest neighbor degree (Supplementary Figure S11), were higher in the northern regions (Dwa and Dwb) than those found in the southern regions (Aw, Cwa and Cfa) (P<5.3 × 10-9, Wilcoxon rank-sum test). These results indicated that the network in the northern regions was more connected than the network in the southern regions. In contrast, average path lengths of sub-networks for soils were lower in the northern regions compared to those in the southern regions (Dwa and Dwb) (P=4.0 × 10−9, Wilcoxon rank-sum test, Figure 4b). This small world feature suggests a closer relationship in the northern regions. Patterns in topological features, such as degree of assortativity (Supplementary Figure S12), betweenness centralization (Supplementary Figure S13) and cluster number (Supplementary Figure S14), were similar to patterns observed for average path lengths (P<4.2 × 10−7, Wilcoxon rank-sum test). However, the patterns in node numbers and transitivity of sub-networks showed no significant differences across the climatic regions (P=0.15 and 0.99, respectively). The Wilcoxon rank-sum test showed that the size of the microbial community was not significantly different between the northern and the southern regions (Supplementary Figures S15 and S16).
Figure 4.
The spatial distribution of edge numbers (a) and average path lengths (b).
Linking network-level topological features to edaphic properties
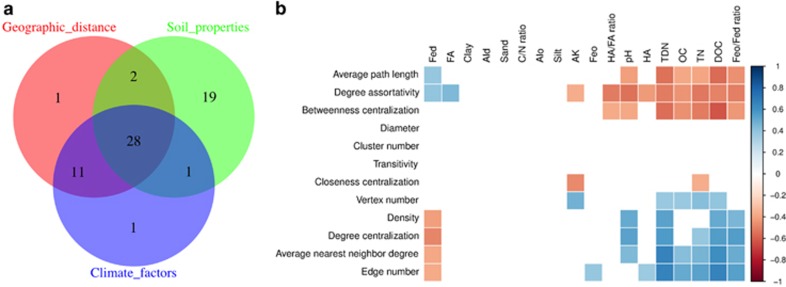
We used multiple regression with distance matrices (MRM) to estimate the contribution of different factors including geographic distances between sampling sites, regional climate factors (mean annual air temperature and mean annual precipitation) and soil properties to network-level topological features (Figure 5a). Soil properties contributed the largest partial regression coefficient (R2=0.48, P<0.0001), while geographic distance and regional climate factors contributed smaller, but significant, partial regression coefficients (R2=0.41 and 0.42, respectively, P<0.001). Soil properties together with geographic distance and regional climate factors explained 28% (P<0.001) of the network-level topological feature variation, and separately explained 19% of the variation (P<0.001). Since geographic distance and regional climate factors were highly correlated with each other (R=−0.75 to 0.97, P<0.0001, using Spearman's rank correlation test, Supplementary Figure S17), their combined effect explained 39% of the variation (P<0.001).
Figure 5.
The importance of geographic distance, climatic factors and soil properties for network-level topological features (a), and correlation between soil properties and network-level topological features (b). The R2 values were estimated with the MRM models. The correlation matrix keeps correlation with P<0.05.
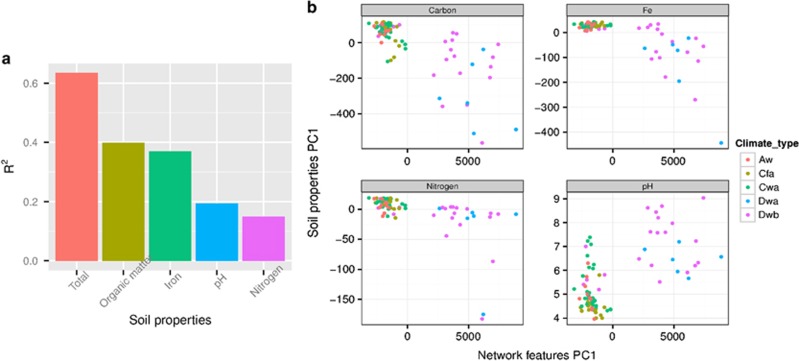
We then examined the correlations among network topological features and environmental factors in soils (Figure 5b, Supplementary Figures S18–S27). Edge number, average nearest-neighbor degree, degree centralization and network density were positively correlated with total dissolved nitrogen (R>0.53, P<0.001, Spearman's rank correlation test), dissolved organic carbon (R>0.50, P<0.001, Spearman's rank correlation test) and acid oxalate, soluble Fe (Feo)/free Fe oxides (Fed) ratio (R>0.45, P<0.001, Spearman's rank correlation test), and were negatively correlated with Fed (R<−0.38, P<0.001, Spearman's rank correlation test). Average path length and degree assortativity had negative correlations with soil pH (R<−0.38, P<0.001, Spearman's rank correlation test), total dissolved nitrogen (R<−0.52, P<0.001, Spearman's rank correlation test), dissolved organic carbon (R<−0.49, P<0.001, Spearman's rank correlation test) and Feo/Fed ratio (R<−0.44, P<0.001, Spearman's rank correlation test), and were positively correlated with Fed (R>0.31, P<0.001, Spearman's rank correlation test). Node numbers were positively correlated with potassium in soils (R=0.47, P<0.001, Spearman's rank correlation test). The contribution of soil organic matter and iron to the topological features of co-occurrence networks, estimated with MRM, was twice that of soil nitrogen and pH (Figure 6a). To identify the relationships between network-level topological features and soil properties, we compared the first principal components of topological features (99.9% of variance) with soil iron (95.6% of variance), carbon (98.9% of variance), nitrogen (97.5% of variance) and soil pH (Figure 6b). Variations of topological features and soil carbon, nitrogen and iron were smaller in the southern regions (Aw, Cwb and Cfb) as compared to those observed for the northern regions (Dwa and Dwb). The variation of topological features was correlated with soil iron, carbon and nitrogen.
Figure 6.
The contribution of soil organic matter, iron, nitrogen and pH to network-level topological features (a) and the relationships between the first component of network-level topological features and edaphic property groups (b).
Discussion
We have performed a co-occurrence network-based analysis using integrated datasets of archaeal, bacterial and fungal OTUs to delineate the geographic patterns of topological features along a climatic gradient across eastern China. The results from this study show that both node-level and network-level topological features are different between the northern (Dwa and Dwb) and the southern regions (Aw, Cfa and Cwa). OTUs typifying the northern regions had lower betweenness centrality values and higher degree values as compared to OTUs typifying the southern regions. As the topology of the network could reflect interactions between microorganisms, the betweenness centrality represents the importance of the control potential that an individual OTU exerts over the interactions of other OTUs in that network. OTUs with low betweenness centrality values represent microorganisms that are located away from the core of the network, compared to other OTUs (Greenblum et al., 2012). Such species are likely to have low influence on other interactions in the community. Degree value is a local quantification feature that informs us about the number of direct co-occurrence interactions for a specific OTU (Greenblum et al., 2012). Our results suggest that microorganisms in forest soils from the northern regions have stronger relationships but have a lower influence compared to microorganisms from the southern regions. This tendency is also supported by the spatial patterns of network-level topological features that cause the tendency of degree values to be higher in the northern regions while network betweenness centrality values tend to be lower in these regions. The geographic pattern of microbial communities has been widely reported. Likewise, our results provide evidence for the geographic pattern of co-occurrence relationships in microbial communities. One explanation for this topological differentiation is the niche differentiation in soil environments occurring as a result of high variations in water-energy conditions between the northern and southern regions of eastern China (Zheng et al., 2013). The high precipitation conditions in the southern regions could make the soil habitats more homogeneous. The weak niche differentiation possibly results in stronger interactions between soil microorganisms (Faust and Raes, 2012a). In contrast, low precipitation in the northern regions may lead to significant niche differentiation, which avoids competition and enables microorganisms to co-exist within communities for extended periods of time. Meanwhile, this niche differentiation likely inhibits the interactions between different species in the northern regions. Another potential explanation for the topological shifts between the northern and southern regions is the evolutionary history of microbial communities. Keystone nodes in co-occurrence networks tend to have high degrees and low betweenness centrality values (Berry and Widder, 2014). Keystone species represented by OTUs in co-occurrence networks were identified in the northern regions. According to the growth processes of a scale-free network, keystone nodes are commonly recognized as initiating components in networks (Barabási, 2009). This suggests that keystone lineages in microbial co-occurrence networks have a longer evolutionary history.
Our results also demonstrated that topological features vary between archaea, bacteria and fungi. In addition, the three investigated kingdoms tend to have different co-occurrence patterns. Archaeal degrees followed a binomial distribution, whereas bacterial and fungal degrees followed power-law distributions. The unexpected distinction between these kingdoms may be indicative of some differences in underlying interaction patterns. The power-law distribution pattern for networks is not surprising, as the degree of distribution in many real-world networks such as the internet (Adamic and Huberman, 2000), social networks (Barabasi et al., 2002) and biological networks (Bergman and Siegal, 2003) follows power-law distributions. Recent studies on microbial co-occurrence networks showed power-law distributions with 90−97% identity classifications for 16 S rRNA OTUs (Chaffron et al., 2010; Barberán et al., 2012; Faust et al., 2012). However, the universal primers used in these studies underrepresent archaeal sequences, thus severely underestimating the presence and contribution of archaea in global co-occurrence networks. To explore archaeal diversity comprehensively, we sequenced a partial fragment of the archaeal 16 S rRNA gene using archaea-specific primers. The binomial distribution of archaeal degrees indicates that the archaeal interaction is structured as a random network following the Erdos−Renyi model (Newman, 2003), in which the presence or absence of edges is a random process. One proposed explanation for the binomial distribution of archaeal edge degrees is neutral processes, meaning that all interactions between archaea are equally likely. Indeed the network density of the Archaea-specific network is lower than for the respective sub-networks for bacteria and fungi. Accordingly, an increase in species abundance may result in a degree increase of individual species in a co-occurrence network, as suggested by the positive correlation between degree values and abundance of archaeal OTUs. A recent archaeal biogeography study showed that the diversity patterns of soil archaea are mainly influenced by stochastic processes (Zheng et al., 2013). This study revealed that the contribution of neutral processes is more important than deterministic factors for soil archaea. Conversely, the negative correlation between degree values and relative abundance for bacterial and fungal OTUs indicates non-random interactions. If the links were random, OTUs with higher relative abundances are more likely to interact with other OTUs, and the degree values for OTUs would increase with an increase in their relative abundance. However, the negative correlation between degree value and relative abundance suggests that the degree is not determined by abundance, and therefore indicates a non-random pattern.
The centrality values were higher for archaeal OTUs than for bacterial and fungal OTUs in both the northern and southern regions. Given the random pattern in the archaeal co-occurrence network, archaeal OTUs are more likely to co-occur with other OTUs in same community. Importantly, we also found that the topological features for archaeal OTUs were different between the northern and southern regions, but were not significant for bacterial and fungal OTUs. These results suggest that the variation of topological features was primarily associated with archaea rather than bacteria and fungi.
We performed an MRM-based analysis to identify environmental factors that explain topological variations across the climatic regions. Soil physicochemical properties were significantly associated with topological features for sub-networks. The overlap explanation ratio between geographic distance and regional climatic factors may be indicative of a common tendency or, alternatively, a common response of the soil microbiota to geographic distance and regional climatic factors. Soil physicochemical properties, however, can separately explain a part of the variation in topological features. We identified soil organic matter and iron as the major soil properties affecting the topological features of co-occurrence networks. One key role of organic matter and iron in soils is that they act as electron shuttles for bioreduction processes in soils (Chacon et al., 2006; Kang and Choi, 2008). Given that co-occurrence relationships reflect the interactions in a community, soil organic matter and iron are expected to explain the topological features of microbial co-occurrence networks as they can influence bacterial interactions.
Our topology-based system approach has also suggested candidate keystone microbial species in co-occurrence networks. Keystone species in co-occurrence networks exert large effects on other community components. Most keystone bacterial nodes in our study belonged to the phyla Actinobacteria and Proteobacteria, which were also the most abundant phyla. Within the Proteobacteria, keystone species belonged to orders Rhizobiales, known for their nitrogen-fixing abilities (Brown et al., 2012), and Gaiellales, which was recently identified and remains poorly understood (Albuquerque et al., 2011). The occurrence of keystone species of the order Rhizobiales may be indicative of the influence of root activities on microbial co-occurrence relationships in soil. Pezizomycotina sp., a fungus contributing to plant organic matter degradation (Ertz and Tehler, 2011), was represented by six OTUs among fungal keystone species. Future work focusing on uncultured keystone species is crucial to better understand the role of these organisms in co-occurrence networks.
This study focused on the spatial trend of topological features in co-occurrence networks. Despite the usefulness of network analysis, one must be cautious when inferring interactions from these co-occurrence networks as they only represent associations between two variables and do not prove a direct interaction association. Although the co-occurrence relationship in the present study was optimized using a deconvolution protocol (Feizi et al., 2013) to remove indirect associations, the output association network is a statistical correlation and does not directly prove microbial interactions. Therefore, future co-occurrence network investigations should focus on more reasonable inferring methods that are validated by literature or microscopy-based experiments, as have been shown for marine samples (Lima-Mendez, 2015).
This study has contributed to microbial ecology research in the same way as network analysis advanced genomics, by appreciating the complex interactions among microbes and the impact of these interactions on community dynamics. However, further investigations identifying specific sets of microbial species responsible for system-level patterns, characterizing the implications of various topological variations, and linking this variation to changes in species composition and functional potential are essential to better understand the interactions in soil microbial communities. Yet, this network approach provides a complementary viewpoint to microbial biogeography by exploring geographic patterns for co-occurrence and interaction relationships and through identification of keystone species for further validation.
Acknowledgments
This research was financially supported by the National Natural Science Foundation of China (41520104001, 41130532), the 111 Project (B06014) and the Fundamental Research Funds for the Central Universities. This work was supported in part by the US Dept. of Energy under Contract DE-AC02-06CH11357.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adamic LA, Huberman BA. (2000). Power-law distribution of the World Wide Web. Science 287: 2115–2115. [Google Scholar]
- Albuquerque L, França L, Rainey FA, Schumann P, Nobre MF, da Costa MS. (2011). Gaiellaocculta gen. nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov.and Gaiellales ord. nov. Syst Appl Microbiol 34: 595–599. [DOI] [PubMed] [Google Scholar]
- Barabási AL. (2009). Scale-Free Networks: A Decade and Beyond. Science 325: 412–413. [DOI] [PubMed] [Google Scholar]
- Barabási AL, Jeong H, Neda Z, Ravasz E, Schubert A, Vicsek T. (2002). Evolution of the social network of scientific collaborations. Phys Stat Mech Its Appl 311: 590–614. [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. (2011). Examining the global distribution of dominant archaeal populations in soil. ISME J 5: 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D. (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93: 491–507. [Google Scholar]
- Bergman A, Siegal ML. (2003). Evolutionary capacitance as a general feature of complex gene networks. Nature 424: 549–552. [DOI] [PubMed] [Google Scholar]
- Berry D, Widder S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 5: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X et al. (2012). Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA 109: 1697–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon N, Silver WL, Dubinsky EA, Cusack DF. (2006). Iron reduction and soil phosphorus solubilization in humid tropical forests soils: the roles of labile carbon pools and an electron shuttle compound. Biogeochemistry 78: 67–84. [Google Scholar]
- Chaffron S, Rehrauer H, Pernthaler J, Mering C von. (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res 20: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro JM, SheflinAM, Manter DK, Vivanco JM. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. BiolFertil Soils 48: 489–499. [Google Scholar]
- Chow C-ET, Kim D, Sachdeva R, Caron D, Fuhrman J. (2013). Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J 8: 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. (2006). The igraph software package for complex network research. InterJournal Complex Systems. (1695).
- Ellouze W, Hamel C, Vujanovic V, Gan Y, Bouzid S, St-Arnaud M. (2013). Chickpea genotypes shape the soil microbiome and affect the establishment of the subsequent durum wheat crop in the semiarid North American Great Plains. Soil Biol Biochem 63: 129–141. [Google Scholar]
- Ertz D, Tehler A. (2011). The phylogeny of Arthoniales (Pezizomycotina) inferred from nucLSU and RPB2 sequences. Fungal Divers 49: 47–71. [Google Scholar]
- Faust K, Raes J. (2012). Microbial interactions: from networks to models. Nat Rev Microbiol 10: 538–550. [DOI] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti J, Izard J, Segata N, Gevers D, Raes J et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8: e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizi S, Marbach D, Médard M, Kellis M. (2013). Network deconvolution as a general method to distinguish direct dependencies in networks. Nat Biotechnol 31: 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS et al. (2013). Reconstructing the microbial diversity and function of pre-agricultural Tallgrass Prairie soils in the United States. Science 342: 621–624. [DOI] [PubMed] [Google Scholar]
- Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA 109: 21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. (2009). Microbial community structure and its functional implications. Nature 459: 193–199. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA. (2008). Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol 53: 69–81. [Google Scholar]
- Greenblum S, Turnbaugh PJ, Borenstein E. (2012). Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA 109: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemstra PH, Pebesma EJ, Twenhöfel CJ, Heuvelink GB. (2009). Real-time automatic interpolation of ambient gamma dose rates from the Dutch radioactivity monitoring network. Comput Geosci 35: 1711–1721. [Google Scholar]
- Hultman J, Waldrop MP, Mackelprang R, David MM, McFarland J, Blazewicz SJ et al. (2015). Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 521: 208–212. [DOI] [PubMed] [Google Scholar]
- Kang S-H, Choi W. (2008). Oxidative degradation of organic compounds using zero-valent iron in the presence of natural organic matter serving as an electron shuttle. Environ Sci Technol 43: 878–883. [DOI] [PubMed] [Google Scholar]
- Kara EL, Hanson PC, Hu YH, Winslow L, McMahon KD. (2013). A decade of seasonal dynamics and co-occurrences within freshwater bacterioplankton communities from eutrophic Lake Mendota, WI, USA. ISME J 7: 680–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. (2012). Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw 46: i11. [PMC free article] [PubMed] [Google Scholar]
- Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F et al. (2015). Determinants of community structure in the global plankton interactome. Science 348: 1262073. [DOI] [PubMed] [Google Scholar]
- Luo F, Zhong J, Yang Y, Scheuermann RH, Zhou J. (2006). Application of random matrix theory to biological networks. Phys Lett A 357: 420–423. [Google Scholar]
- Lupatini M, Suleiman AKA, Jacques RJS, Antoniolli ZI, de Siqueira Ferreira A, Kuramae EE et al. (2014). Network topology reveals high connectance levels and few key microbial genera within soils. Front Environ Sci 2: 10. [Google Scholar]
- Miltner A, Bombach P, Schmidt-Brücken B, Kästner M. (2012). SOM genesis: microbial biomass as a significant source. Biogeochemistry 111: 41–55. [Google Scholar]
- Newman M. (2003). The structure and function of complex networks. SIAM Rev 45: 167–256. [Google Scholar]
- Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J. (2015). Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J 9: 980–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F. (2006). Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22: 2532–2538. [DOI] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5: 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wolcott RD, Dowd SE. (2011). Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. Methods Mol Biol 733: 129–141. [DOI] [PubMed] [Google Scholar]
- Xu X, Thornton PE, Post WM. (2013). A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob Ecol Biogeog 22: 737–749. [Google Scholar]
- Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X et al. (2014). Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J 8: 818–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y-M, Cao P, Fu B, Hughes JM, He J-Z. (2013). Ecological drivers of biogeographic patterns of soil archaeal community. PLoS One 8: e63375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.